Abstract
The effects of plant–microbe interactions between the hyperaccumulator Arabidopsis halleri and eight bacterial strains, isolated from the rhizosphere of A. halleri plants grown in a cadmium- and zinc-contaminated site, were analysed for shoot metal accumulation, shoot proteome, and the transcription of genes involved in plant metal homeostasis and hyperaccumulation. Cadmium and zinc concentrations were lower in the shoots of plants cultivated in the presence of these metals plus the selected bacterial strains compared with plants grown solely with these metals or, as previously reported, with plants grown with these metals plus the autochthonous rhizosphere-derived microorganisms. The shoot proteome of plants cultivated in the presence of these selected bacterial strains plus metals, showed an increased abundance of photosynthesis- and abiotic stress-related proteins (e.g. subunits of the photosynthetic complexes, Rubisco, superoxide dismutase, and malate dehydrogenase) counteracted by a decreased amount of plant defence-related proteins (e.g. endochitinases, vegetative storage proteins, and β-glucosidase). The transcription of several homeostasis genes was modulated by the microbial communities and by Cd and Zn content in the shoot. Altogether these results highlight the importance of plant-microbe interactions in plant protein expression and metal accumulation and emphasize the possibility of exploiting microbial consortia for increasing or decreasing shoot metal content.
Keywords: Arabidopsis halleri, cadmium; heavy metals; plant–microbe interactions; proteomic analysis
Introduction
Cadmium (Cd) is one of the most phytotoxic non-essential heavy metals released into the environment by urban traffic, refuse dumping, metal-working industries and the intense use of phosphate fertilizers (Sanità di Toppi and Gabbrielli, 1999; Polle and Schützendübel, 2003). Some species (c. 450) accumulate remarkably high concentrations of Cd and other metals such as Zn, Ni, Mn, Cu, and Co in their above-ground biomass without showing signs of toxicity (Reeves and Baker, 2000; Milner and Kochian, 2008; Krämer, 2010). Typically, these hyperaccumulators show in their tissues, concentrations of up to four orders of magnitude higher than in other adjacent plants (Reeves and Baker, 2000). This unusually high concentration occurs even when the metal is present at a low external concentration, suggesting that plants may benefit from such metal accumulation rather than being injured by it (Sors et al., 2005). The evolution of these plants is under investigation to determine what benefits have served as a selective pressure for the development of the hyperaccumulation trait although, to date, none of the proposed hypotheses has been fully accredited (Whiting et al., 2003; Noret et al., 2005).
Above-ground metal hyperaccumulation appears to be the ideal property for phytoextraction of heavy metals from contaminated soils (phytoremediation), or for the concentration and extraction of valuable metals (phytomining). Moreover, the identification of genes responsible for metal tolerance and hyperaccumulation in these species, and their expression in transgenic trees with far-reaching roots and adapted to growing in particular soil and climate conditions, render these phytotechnologies very attractive (Pilon-Smits and Pilon, 2002; Krämer, 2010). Investigations are still needed to identify mechanisms of gene regulation that allow hyperaccumulator plants to extract, transport, and sequestrate heavy metals. Within this context Arabidopsis halleri is a species suitable for molecular studies of metal tolerance and hyperaccumulation (Becher et al., 2004; Weber et al., 2004; Hanikenne et al., 2008). It naturally accumulates and tolerates leaf concentrations as high as 2.2% Zn and 0.028% Cd in dry biomass (Dahmani-Muller et al., 2000). Zn and Cd accumulations have been explored in a large number of European populations growing in metal-contaminated and non-contaminated soils, and the results suggested that they are probably constitutive traits in A. halleri (Bert et al., 2002). As an extremophile sister species of A. thaliana, with which it shares about 94% DNA sequence within the coding regions (Becher et al., 2004), it is an excellent model for molecular studies on both metal tolerance and accumulation (Mitchell-Olds, 2001) and on plant adaptation to extreme metalliferous environments (Becher et al., 2004; Craciun et al., 2006). Molecular mechanisms of metal hyperaccumulation have recently been reviewed (Verbruggen et al., 2009). Current knowledge suggests that these species have probably evolved in metal-rich soils through the adaptation of metal homeostasis processes. In addition, although the function of many genes up-regulated in hyperaccumulators is still unknown, molecular studies suggest that genes potentially involved in hypertolerance and hyperaccumulation are differentially regulated and expressed, in these species, rather than species-specific (Verbruggen et al., 2009).
Heavy metal hyperaccumulation depends not only on the plant, but also on the interaction between plant roots and rhizosphere microorganisms which must necessarily be metal resistant. Indeed, root proliferation and effective root uptake mechanisms are among the key processes in the rhizosphere that distinguish metal hyperaccumulators from normal plants (Abou-Shanab et al., 2003). The high bacterial density in the rhizosphere is due to the high level of nutrients such as amino acids, sugars and organic acids, exuded from the plant roots and capable of supporting microbial growth. On the other hand, rhizosphere bacteria can promote plant growth by providing compounds or by facilitating nutrients uptake from the environment (Glick, 2003). They can also improve plant competitiveness, response to external stress factors, and promote plant health by stimulating root growth and enhancing water uptake (Mantelin and Touraine, 2004; Pilon-Smits, 2005). In this complex and dynamic microenvironment, bacteria may increase metal mobility and availability to the plant or else decrease metal solubility due to precipitation.
In a previous study, these interactions were considered in an ecotype of A. halleri harvested in Auby (Northern France) in heavy metal-contaminated soils characterized by a pH of 6.8, total metal contents of c. 400 μg g−1 Cd and 41750 μg g−1 Zn (Farinati et al., 2009), and extractable amounts (determined as exchangeable and electrostatically weakly bound phase) up to 150 μg g−1 in Cd and 850 μg g−1 in Zn (S Farinati et al., unpublished data). Shoot proteomes of plants grown with or without microbes derived from the autochthon rhizosphere were compared (Farinati et al., 2009). Meanwhile, bacterial strains were selected in vitro, from the autochthon population of the rhizosphere, for their ability to grow in the presence of high Cd (2 mM) or high Zn (20 mM) concentration. Further selection was performed by growing the resulting strains, resistant to Cd or Zn in vitro, in the presence of both heavy metals (1 mM CdSO4 and 10 mM ZnSO4). In this way, eight bacterial strains were predominantly represented, and therefore isolated and identified. The objectives of the study presented here were (i) to characterize these bacteria phylogenetically; (ii) to conduct a comparative shoot proteomic analysis between A. halleri plants grown in the greenhouse and cultivated with or without Cd and Zn and with the addition of the eight isolated bacterial strains; (iii) to test the effect of these bacterial strains on Cd and Zn above-ground accumulation when added to the nutrient solution together or individually; and (iv) to analyse the effect of culturable rhizosphere-derived microorganisms and the eight selected bacteria strains on the expression of genes involved in plant metal homeostasis and hyperaccumulation.
Materials and methods
Plant material and growth conditions
Seeds of A. halleri were collected from plants growing on a heavy metal contaminated site in Auby (Nord Pas-du-Calais, France, latitude 50°25'0'' N, longitude 3°3'0'' E, elevation 21 m) (Farinati et al., 2009). After surface-sterilization in 70% ethanol for 1 min and 1.5% v/v sodium hypochlorite for 20 min, seeds were germinated on MS medium (Murashige and Skoog, 1962) containing 0.7% agar and grown in a growth chamber under 16/8 h light/dark regime (light intensity around 50 μE m−2 s−1) at 22/18 °C. After 30 d, plants were transplanted to perlite-filled 1.0 liter polyethylene pots wetted with half-strength Hoagland solution (Hoagland and Arnon, 1938) to substrate saturation and were moved to the greenhouse for an additional month under 16/8 h light/dark conditions (light intensity ranging from 130 μE m−2 s−1 to 300 μE m−2 s−1). Plants were inoculated with the bacteria as described below and cultured in the same conditions for a further month. The experiment was conducted from September to November.
Isolation and identification of Cd- and Zn-resistant bacterial strains
Two grams of A. halleri rhizospheric soil, collected from the contaminated site, were suspended in a 150 ml Erlenmeyer flask containing 18 ml of a 0.9% w/v NaCl solution. The flask was incubated at 28 °C in the dark for 2 h on a rotary shaker (250 rpm). Serial dilutions of the suspensions were then prepared. Afterwards, Petri dishes containing Nutrient Broth and agar (Oxoid, Garbagnate Milanese, Italy) were streaked with 0.1 ml per plate of differently diluted suspensions and aerobically incubated for 5 d at 28 °C in order to elicit mainly bacterial growth. In addition, plates with solidified Nutrient Broth with CdSO4 (up to 2 mM) or ZnSO4 (up to 20 mM) were used to select for resistant bacterial strains among those growing on simply solidified medium. Strains resistant to either Cd or Zn were then cultivated on medium containing both metals at final concentrations of 1 mM and 10 mM respectively, to mimic the actual levels of Cd and Zn in the rhizospheric soil. Eight different isolates were identified on the basis of morphological and growth features. Genomic DNA was extracted from each strain by using the NucleoSpin Tissue kit (Clontech, Palo Alto, CA) and a PCR amplification of the 16S rDNA gene was carried out with primers F8 and R11 (Weisburg et al., 1991). Amplification products were directly sequenced and subjected to the BLASTN analysis (Altshul et al., 1997).
Experimental design
Ten plants of A. halleri per pot were cultivated on a perlite bed wetted to saturation with half-strength Hoagland solution and subjected to the following treatments, each performed in three pots obtaining three independent biological replicates, for one month. (i) Control: untreated plants grown solely in the nutrient solution (Ctr); (ii) only metals: 1 mM CdSO4 and 10 mM ZnSO4 were added to the nutrient solution (Mt); (iii) metals plus the eight selected bacterial strains: 1 mM CdSO4 and 10 mM ZnSO4 together with the eight bacterial strains (each at 1×107 CFU g−1 perlite) isolated from the native rhizosphere of A. halleri plants (Mt+8S); (iv) metals plus each selected bacterial strain considered singly: 1 mM CdSO4 and 10 mM ZnSO4 together with each strain (1×107 CFU g−1 perlite) added singly (named from A to H); (v) metals plus native rhizosphere microbial coenosis: 1 mM CdSO4 and 10 mM ZnSO4 together with the microbial community (1×107 CFU g−1 perlite) associated with the native rhizosphere of A. halleri plants (Mt+Mc).
In order to obtain bacterial inocula (treatments iii and iv), axenic cultures were prepared by growing each strain separately in 250 ml Erlenmeyer flasks containing 100 ml Nutrient broth and then incubated on an orbital shaker at 180 rpm at 28 °C for 42 h to reach the stationary phase. The final concentration of 1×107 CFU in the substrate for plant cultivation was achieved by collecting a proper aliquot of each bacterial culture and centrifuging them at 4000 g for 10 min at 4 °C. Pellets were then resuspended in 100 ml half-strength Hoagland's solution. Finally, microbial suspensions were uniformly distributed throughout the perlite bed. Preparation of the inoculum with the rhizospheric microbial population (treatment v) was as previously described by Farinati et al. (2009). The inoculation was performed once. Pots were periodically checked and distilled water was added in case of dehydration of the perlite beds. At the end of the experiment, for each treatment, shoots from the three pots were harvested and weighed separately. Shoot samples were then frozen in liquid nitrogen, ground to powder, and kept at –80 °C until use. Roots were also collected and samples were frozen in liquid nitrogen and stored at –80 °C until use. Perlite attached to the roots was washed in physiological solution (0.9% NaCl), slowly stirred for 1 h at RT, and then aliquots were plated onto nutrient medium solidified with agar and incubated at 28 °C for 5 d to elicit bacterial growth. Genomic DNA was extracted from phenotipically different colonies and PCR amplification of the 16S rDNA gene was carried out (with subsequent sequencing of the amplicons) to confirm that the inoculated bacterial strains were effectively still present and growing.
Proteome isolation and protein pattern differential analysis
Total proteins were extracted from the shoots of two biological replicates corresponding to different pots (for treatments i, ii, and iii) and separated by 2-DE; MS/MS analysis and protein identification were performed as described in Farinati et al. (2009). The image analysis of the 2-DE gels (five replicates were performed for each biological replicate and treatment considered) was carried out using PDQuest software (BioRad) version 7.3. Each gel was analysed for spot detection, background subtraction, and protein spot OD intensity quantification. The gel image showing the highest number of spots and the best protein pattern was chosen as a reference template. All spot comparisons and analysis were co-ordinated through the reference template and its spots were then matched across all gels. Gels were divided into three separate groups (control-Ctr, only metals-Mt, metals plus the eight bacterial strains-Mt+8S) and, for each protein spot, the quantity was normalized in each gel by dividing the raw quantity of each spot by the total quantity, in that gel, of all the spots found in the reference template (Arora et al., 2005). For each protein spot, the average spot quantity value and its variance coefficient in each group were determined. Statistical analysis (Student's t test) was performed to identify proteins that were significantly (P <0.05) increased or decreased in the three sets of samples. The comparisons were as follows: (i) Mt+8S versus Ctr; and (ii) Mt+8S versus Mt.
Western blot analysis
Total protein samples for Western blots were collected as one aliquot of shoot sample per pot (three independent biological replicates) and three technical replicates were performed (i.e. three independent SDS-PAGE gels). Tissues were homogenized in solubilization buffer (100 mM TRIS pH 8, 50 mM EDTA pH 8, 0.25 M NaCl, 1 mM DTT, and 0.7% SDS). The homogenate was heated at 65 °C for 10 min and centrifuged at 16 000 g for 10 min at RT to remove cellular debris. Prior to the electrophoresis fractionations, proteins were precipitated with ice-cold acetone following standard protocols and resuspended in SDS-loading buffer (6 M urea, 50 mM TRIS-Cl pH 6.8, 100 mM dithiothreitol, 2% SDS, 10% glycerol, and 0.1% bromophenol blue). Eight μg of total protein were loaded on 12% acrylamide SDS-PAGE-gels and transferred to PVDF membranes (Millipore, Bedford, MA, USA) according to standard protocols. Specific antibodies against monodehydroascorbate reductase (MDAR), chaperonin 20 (CPN20), two photosystem II proteins (PSBP1 and PSBO2), and the β-subunit of plastidic ATPase were used for hybridization. Detection was achieved using the ECL detection system (Amersham, Buckinghamshire, UK). The intensity of the chemiluminescence response was measured by scanning the films and processing the image with the Quantity One software Version 4.4 (Bio-Rad, Hercules, CA, USA).
Measurement of Cd and Zn contents
Cd and Zn contents in shoots were determined in three sample-aliquots taken from each of the three shoot samples corresponding to the different experimental conditions. Samples were oven-dried at 85 °C for 3 d, weighed, and homogenized using a Wiley mill. Cd and Zn analyses were performed after microwave-assisted acid digestion by means of inductively coupled plasma MS analysis (EPA 3051A 2007 and EPA 6010C 2007). Metal concentrations were also determined for the eight microbial strains grown axenically in 100 ml Nutrient Broth with the addition of 100 μM CdSO4 (c. 11.3 ppm Cd) or 100 μM ZnSO4 (c. 6.5 ppm Zn) and incubated on a orbital shaker (180 rpm) at 28 °C for 48 h. Microbial cells were collected by centrifugation (6000 g, 15 min, 4 °C) and rinsed twice in 50 ml of 0.9% w/v NaCl. Cd and Zn contents were determined in supernatants and rinsing solutions together, after filtration through 0.22 μm membranes. Each analysis was performed in triplicate.
Determination of chlorophyll content
To measure chlorophyll content, three technical replicates (about 0.1 g each) were made from each of the three shoot samples for each experimental condition described. Pigments were extracted with 80% acetone. Absorbance was measured at 646.6 nm and 663.6 nm. Chlorophyll content was calculated according to classical equations as previously described by Porra (2002).
RNA isolation, cDNA synthesis, and quantification of transcripts by real-time PCR
Total RNA was isolated from roots and shoots of A. halleri plants subjected to all treatments as one RNA-sample for each of the three pots as explained in the ‘Experimental design’ section, using the Trizol Reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized using the ImProm-IITM Reverse Transcriptase (Promega, Madison, WI). Real-time PCR was performed using the ABI Prism sequence detection system (Applied Biosystems, Foster City, CA,) with the Platinum SYBR Green qPCR SuperMix UDG (Invitrogen). For each condition tested, real-time PCR analysis was carried out in triplicate on three different cDNA samples. Data were organized according to the 2–ΔΔCT method (Livak and Schmittgen, 2001). Real-time PCRs were performed to analyse the transcription level of phytochelatin synthetase (AhPCS1), heavy metal ATPase 4 (AhHMA4), Zn and Fe-regulated transporter protein 6 (AhZIP6), metal transporter protein (AhMTP1), a vacuolar transporter that exchanges protons with Mg, Zn, and Fe ions (AhMHX), and nicotinamine synthase 3 (AhNAS3). Transcripts were normalized by amplifying a fragment of the A. halleri actin cDNA. For each primer pair, primer design was optimized by analysing the run of serial 10-fold dilutions of the cDNA [corresponding to the Ctr, treatment (i)] and generating a standard curve by plotting the dilution factor of the template, against the CT value obtained during amplification of each dilution. Only primer pairs that showed an R2 value >0.98 were considered. Primers used are reported in Table 1. The cycling programme was the following: 95 °C for 5 min, [94 °C for 15 s, 57 °C for 30 s, 72 °C for 30 s]×40. After the run, the melting curve analysis was performed which resulted in single product at specific temperatures. The absence of primer-dimers was confirmed by running the products of the 40 cycles onto high resolution gel electrophoresis.
Table 1.
Forward and reverse primers used in real-time PCR analysis
| Primer name | Primer sequence (5′–3′) |
| AhPCS1-F | GTCACTTCCATCAAACTTCAAC |
| AhPCS1-R | CTTGACTTCTCCTCTGCGCT |
| AhHMA4-F | TCAGCTTGCGTCGCCATTGT |
| AhHMA4-R | ACCACAGGGACATCCACTGA |
| AhNAS3-F | AACACATGGCTCCTGGTGCT |
| AhNAS3-R | CCTCGAACCCCTGAAGATCA |
| AhZIP6-F | ACGATGATCAGAACCCGAATG |
| AhZIP6-R | CAATGCAATTAGATCAACCAAG |
| AhMHX-F | AGACACTCCACCCGATTCTG |
| AhMHX-R | TTCCAACGTTATTGCATCCAC |
| AhMTP1-F | ACATAGCCATAGCCATGGGG |
| AhMTP1-R | TGCTGGTCCTCTCCATGACT |
Results
Phylogenetic affiliation of Cd- and Zn-resistant bacterial strains
Cd- and Zn-resistant bacterial strains were isolated, as axenic cultures, from the rhizosphere of A. halleri plants grown in Cd and Zn (and Pb) polluted soil (van Rossum et al., 2004), by selection on CdSO4 (up to 2 mM) or ZnSO4 (up to 20 mM). About 40 bacterial strains were selected on Cd-supplemented medium, whereas approximately 30 were found to be resistant on Zn-supplemented medium. All these strains were further streaked on medium supplemented with both metals (1 mM CdSO4 and 10 mM ZnSO4). Eight morphologically different colonies were predominantly represented and therefore analysed further. For phylogenetic affiliation, 16S rDNA gene was amplified from these eight selected strains. Fragments of about 500 bp were sequenced and compared with NCBI database entries. All sequences showed at least 97% similarity to known sequences. Notably, matches to the eight identified bacterial strains could mostly be attributed to isolates from environmental matrices with related pollution characteristics (Table 2).
Table 2.
Phylogenetic affiliation of bacteria strains isolated from the rhizosphere of A. halleri plants grown in heavy metal polluted site
| Strain | Closest NCBI match | (Accession no.) | % Homologya | Identification experimentb | Phylogenetic groupc (class) |
| A | Pseudomonas putida | (FN600411) | 100% | Fenamiphos and oxamyl degrading bacteria | Gammaproteobacteria |
| Pseudomonas plecoglossicida fA2 | (FJ947052) | 99% | Isolated from sewage of waste water treatment | Gammaproteobacteria | |
| Uncultured bacterium clone ncd999a04c1 | (HM335248) | 99% | Isolated from human clinical samples | unclassified | |
| B | Chryseobacterium joostei LMG18208 | (AY468479) | 98% | Isolated from diseased aquatic animals | Flavobacteria |
| Chryseobacterium sp. M547 | (AB461832) | 97% | Isolated from stems of field-grown soybeans | Flavobacteria | |
| Chryseobacterium joostei LMG 18212 | (NR025387) | 97% | Isolated from dairy environment | Flavobacteria | |
| C | Chryseobacterium sp. BW2 | (DQ985274) | 100% | Isolated from wetland soil | Flavobacteria |
| Chryseobacterium sp. CI06 | (DQ530068) | 99% | Rhizosphere bacteria | Flavobacteria | |
| Bacterium PSB-1-20 | (AY822563) | 99% | Bacteria associated with sand dune plants | Unclassified | |
| D | Chryseobacterium sp. RI39 | (DQ530125) | 98% | Rhizosphere bacteria | Flavobacteria |
| Chryseobacterium sp. HX31 | (EF601823) | 98% | Soil bacteria from alpine grassland | Flavobacteria | |
| Uncultured bacterium clone 50p8_2116 | (FJ935614) | 98% | Maple sap bacteria | Unclassified | |
| E | Tsukamurella strandjordae | (AM284997) | 99% | Arsenic-resistant soil bacteria | Actinobacteria |
| Tsukamurella tyrosinosolvens | (AB478957) | 99% | Isolated from clinical samples | Actinobacteria | |
| Tsukamurella paurometabola | (CP001966) | 99% | Isolated from soil and sludge | Actinobacteria | |
| F | Escherichia coli O55:H7 str. CB9615 | (CP001846) | 99% | Isolated from clinical samples | Gammaproteobacteria |
| Escherichia coli A34 | (GU594295) | 99% | Isolated from clinical samples | Gammaproteobacteria | |
| Uncultured bacterium clone RP_2aaa03b11 | (EU778474) | 99% | Mammals gut bacteria | Unclassified | |
| G | Sphingomonas sp. BIR2-rlima | (EF153191) | 99% | R-limonene-degrading bacteria | Alphaproteobacteria |
| Novosphingobium sp. FND-3 | (DQ831000) | 99% | Carbofuran-degrading bacteria | Alphaproteobacteria | |
| Uncultured bacterium clone SSmCB08-81 | (AB176242) | 99% | Isolated from sub-seafloor hydrothermal field | Alphaproteobacteria | |
| H | Curtobacterium pusillum I3 | (DQ086779) | 99% | Isolated from potato tubers | Actinobacteria |
| Curtobacterium sp. 2340 | (AY688358) | 99% | Isolated from human clinical samples | Actinobacteria | |
| Uncultured bacterium clone ncd523a03c1 | (HM276345) | 98% | Isolated from human clinical samples | Unclassified |
Tentative identification and percent similarity values were determined by BLAST and are based on approximately 500 bp of the 16S rDNA gene sequence for each clone (NCBI nucleotide database).
Environmental matrices where the isolates are mostly attributed.
Classification according to the Bergey's Manual of systematic bacteriology (2nd edn).
Plant growth and metal accumulation
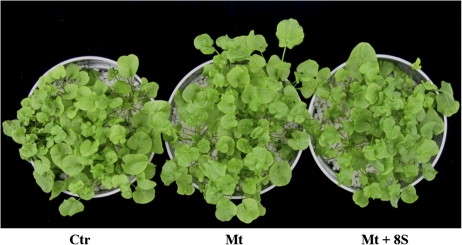
In the present study, the effect on plant status, plant metal accumulation, and shoot protein expression modulation, of eight bacterial strains, isolated from the autochthonous rhizosphere of A. halleri plants collected in a heavy metal contaminated site, and selected in vitro for their ability to tolerate high Zn and Cd concentrations was addressed. Plants underwent different treatments for a month to allow the establishment of plant–microbe interactions. At the end of the experiment, plants treated with Cd and Zn and plants treated with these metals plus bacterial strains displayed the same phenotype as untreated control plants maintained in the solely nutrient solution (Fig. 1). No differences were found in chlorophyll content between control plants and plants co-cultivated with the eight bacterial strains plus metals (Table 3). Conversely, chlorophyll content was significantly higher in plants cultivated solely with Cd and Zn and in plants cultivated with these heavy metals plus the all rhizosphere-derived microorganisms (Table 3; Farinati et al., 2009). Measurement of shoot fresh weight indicated that neither metals nor bacteria added to the nutrient solution had an effect on biomass production (Table 3).
Fig. 1.
A. halleri plants grown for a month in the greenhouse subjected to different treatments. In Hoagland nutrient solution (Ctr); with the addition of 1 mM CdSO4 and 10 mM ZnSO4 (Mt); and with the addition of 1 mM CdSO4 and 10 mM ZnSO4 plus eight selected bacterial strains (Mt+8S). (This figure is available in colour at JXB online.)
Table 3.
Shoot fresh weight and leaf chlorophyll content of plants submitted to different treatments
| Treatment | Shoot Fresh Weight (g) | Chlorophyll (mg Chl a+b g−1) |
| Ctr | 1.6±0.2 | 0.983±0.065 a* |
| Mt | 2.1±0.2 | 1.247±0.023 b* |
| Mt+8S | 1.8±0.2 | 1.022±.076 a |
| Mt+Mc | 1.9±0.2 | 1.456±0.114 b* |
Untreated control plants (Ctr); plants treated with 1 mM CdSO4 and 10 mM ZnSO4 (Mt); plants treated with 1 mM CdSO4 and 10 mM ZnSO4 and the addition of the eight selected bacterial strains (Mt+8S); plants treated with 1 mM CdSO4 and 10 mM ZnSO4 plus autochthonous rhizosphere-derived microorganisms (Mt+Mc). Values are the average of 10 plants for three replicates ±SE. Chlorophyll content, different letters correspond to significant differences (P <0.05).
Results also reported in Farinati et al. (2009).
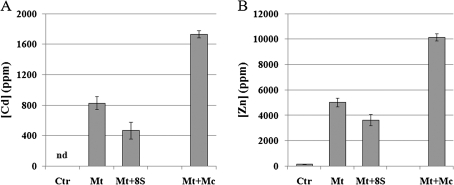
After harvesting, Cd and Zn concentrations were measured in dried shoots. A similar trend for Cd and Zn accumulation was noted. As expected, metal concentrations increased in plants treated with metals compared with control plants grown solely in Hoagland solution. Plants treated with the eight bacterial strains plus metals showed much lower Cd and Zn contents in the shoot than plants treated solely with these metals (Fig. 2). The presence of these strains therefore hinders metal accumulation in the shoots. On the other hand, the highest shoot metal content was observed when plants were cultivated in the presence of the culturable rhizosphere-derived microbial population plus metals (Fig. 2).
Fig. 2.
Cd and Zn content in shoots of plants submitted to different treatments. Untreated control plants (Ctr); plants treated with 1 mM CdSO4 and 10 mM ZnSO4 (Mt); plants treated with 1 mM CdSO4 and 10 mM ZnSO4 and the addition of the eight selected bacterial strains (Mt+8S); and plants treated with 1 mM CdSO4 and 10 mM ZnSO4 plus autochthonous rhizosphere-derived microorganisms (Mt+Mc). nd: not determined. With the exception of Mt+8S, results are also reported in Farinati et al. (2009). Values shown represent the average of ten plants for three biological replicates (all values are highly statistically significant, P <0.001). Bar, SE.
Proteomic analysis of differentially expressed proteins and functional classification
Although roots are the site of interaction with bacterial strains and the first organ that perceives the presence of heavy metals, the proteomic analysis was carried out on the above-ground tissues to identify proteins differentially expressed in the shoot and potentially involved in shoot metal accumulation or exclusion. This could contribute to the identification of genes responsible for metal tolerance and above-ground hyperaccumulation, considering their potential biotechnological application eventually for phytoremediation purposes. Our interest was centred on the differentially expressed proteins in the total protein extract without protein fractionation and subcellular localization of the gene product. To focus attention on the effects of diverse treatments, gels were performed in five replicates and differences in spot intensity were confirmed by statistical analysis. In general, the spot distribution pattern allowed a comparison of the protein profile. Similarity in protein distribution and positioning was observed between the three experimental groups and the protein spots mainly ranged from pI 4 to 9. Spots showing intensity variation across all the 2-DE replica maps were considered (with a total of 47 spots regarding the Mt+8S versus Ctr comparison and 16 in the Mt+8S versus Mt match), excised from the gel and subjected to MS analysis. A total of 35 spots could be identified and assigned to 24 proteins classified as up- or down-regulated (P <0.05) among the different treatments (Table 4).
Table 4.
Shoot proteomic analysis of A. halleri plants subjected to different treatments
| No | SSP | Protein name | AC number (gi NCBI) | MW (kDa)/pI theoretical | Peptide | Mascot | Fold variationsa | ||||
| and reference organism | count | score | Mt+8S versus Ctr | Mt+8S versus Mt | |||||||
| Photosynthesis, light reactions | |||||||||||
| 1 | 1204 | PSBP1b | At1g06680 | 28078/6.90 | 11 | 445 | ↑4.45 | ||||
| Arabidopsis thaliana | |||||||||||
| 2 | 2204b | PSBP1b | At1g06680 | 20078/6.90 | 10 | 369 | ↑2.67 | ||||
| Arabidopsis thaliana | |||||||||||
| 3 | 5101 | PSBP2 | At2g30790 | 27704/9.08 | 6 | 204 | ↓2.43 | ||||
| Arabidopsis thaliana | |||||||||||
| 4 | 1302 | PSBO2b | At3g50820 | 34998/5.92 | 9 | 388 | ↑2.43 | ||||
| Arabidopsis thaliana | |||||||||||
| 5 | 1303 | PSBO2b | At3g50820 | 34998/5.92 | 15 | 775 | ↑2.58 | ||||
| Arabidopsis thaliana | |||||||||||
| 6 | 1311 | PSBO1-like | At4g37230 | 35114/5.68 | 17 | 672 | n.i.c | ||||
| Arabidopsis thaliana | |||||||||||
| 7 | 2407 | HCF136 | At5g23120 | 44076/6.79 | 9 | 417 | ↑3.27 | ||||
| Arabidopsis thaliana | |||||||||||
| 8 | 2103 | δ-subunit ATPase | At4g09650 | 25653/9.04 | 7 | 346 | ↑2.78 | ||||
| Arabidopsis thaliana | |||||||||||
| 9 | 4004 | β-subunit ATPaseb | AtCg00480 | 52625/5.28 | 5 | 194 | ↑3.02 | ||||
| Arabidopsis thaliana | |||||||||||
| 10 | 2601a | β-subunit ATPaseb | AtCg00480 | 52905/5.10 | 14 | 749 | ↑3.0 | ||||
| Arabidopsis thaliana | |||||||||||
| Photosynthesis, dark reactions | |||||||||||
| 11 | 1106 | Rubisco: large subunit | AtCg00490 | 52922/5.88 | 5 | 148 | ↑6.03 | ||||
| Arabidopsis thaliana | |||||||||||
| 12 | 4611 | Rubisco: large subunit | ArhiCp028 | 52933/5.96 | 15 | 614 | ↑2.93 | ||||
| Arabis hirsuta | |||||||||||
| 13 | 5704 | Rubisco: large subunit | ArhiCp028 | 52933/5.96 | 19 | 761 | ↑2.44 | ||||
| Arabis hirsuta | |||||||||||
| 14 | 3008 | Rubisco: small subunit | At5g38430 | 20273/7.59 | 4 | 160 | ↑7.20 | ||||
| Arabidopsis thaliana | |||||||||||
| 15 | 4001 | Rubisco: small subunit | gi|406727 | 20178/8.24 | 4 | 95 | ↑3.48 | ||||
| Brassica napus | |||||||||||
| 16 | 8001 | Rubisco: small subunit | At1g67090 | 20448/7.59 | 6 | 253 | ↑2.41 | ||||
| Arabidopsis thaliana | |||||||||||
| 17 | 6007 | Rubisco: small subunit | At5g38430 | 20273/7.59 | 5 | 223 | ↑2.04 | ||||
| Arabidopsis thaliana | |||||||||||
| 18 | 1505 | Phosphoribulokinase | At1g32060 | 44436/5.71 | 13 | 475 | ↑2.16 | ↑2.13 | |||
| Arabidopsis thaliana | |||||||||||
| 19 | 2503 | Rubisco activase | At2g39730 | 52006/5.69 | 10 | 446 | ↑2.4 | ||||
| Arabidopsis thaliana | |||||||||||
| Stress | |||||||||||
| 20 | 7507 | AtMDAR1b | At3g52880 | 46458/6.41 | 9 | 439 | ↓2.5 | ||||
| Arabidopsis thaliana | |||||||||||
| 21 | 3504 | AtMDAR2b | At5g03630 | 47450/5.24 | 16 | 496 | ↓2 | ||||
| Arabidopsis thaliana | |||||||||||
| 22 | 4203 | Fe-Superoxide | At4g25100 | 25409/6.30 | 3 | 165 | ↑2.16 | ||||
| dismutase, FSD1 | Arabidopsis thaliana | ||||||||||
| 23 | 4101 | Allene oxide | At3g25770 | 21199/5.65 | 6 | 247 | ↓4.54 | ↓4.05 | |||
| cyclase | Arabidopsis thaliana | ||||||||||
| 24 | 7307 | Endochitinase | gi|7798632 | 32004/5.93 | 5 | 156 | ↓5 | ||||
| (Class I) | Arabis holboellii | ||||||||||
| 25 | 8005 | Endochitinase | gi|7798632 | 32004/5.93 | 3 | 133 | ↓3.85 | ||||
| (Class I) | Arabis holboellii | ||||||||||
| 26 | 5003 | Malate dehydrogenase | At1g04410 | 35548/6.11 | 3 | 97 | ↑5.13 | ||||
| Arabidopsis thaliana | |||||||||||
| 27 | 6101 | Germin-like protein | At5g20630 | 21849/6.81 | 2 | 72 | ↑2.44 | ↑2 | |||
| AtGER3 | Arabidopsis thaliana | ||||||||||
| 28 | 5102 | Lipocalin, TIL1 | At5g58070 | 21421/5.97 | 2 | 84 | ↑2.65 | ||||
| Arabidopsis thaliana | |||||||||||
| 29 | 2601b | Enolase | At2g36530 | 47689/5.54 | 9 | 433 | ↑3.0 | ||||
| LOS2 | Arabidopsis thaliana | ||||||||||
| Cellular metabolism and organization | |||||||||||
| 30 | 2204a | Plastidic chaperonin, | At5g20720 | 26785/8.86 | 11 | 572 | ↑2.67 | ||||
| AtCPN20b | Arabidopsis thaliana | ||||||||||
| 31 | 5209 | Vegetative storage | At5g24770 | 29806/6.17 | 6 | 239 | ↓4.16 | ||||
| protein, VSP2 | Arabidopsis thaliana | ||||||||||
| 32 | 5710 | α-subunit ATPase | AtMg01190 | 55011/6.23 | 20 | 1046 | ↓2.27 | ||||
| mitochondrion | Arabidopsis thaliana | ||||||||||
| 33 | 6712 | β-glucosidase 1 | At1g52400 | 60462/6.89 | 11 | 383 | ↓6.25 | ↓2.48 | |||
| Arabidopsis thaliana | |||||||||||
| 34 | 6713 | β-glucosidase 1 | At1g52400 | 60462/6.89 | 13 | 434 | ↓<10 | ||||
| Arabidopsis thaliana | |||||||||||
| 35 | 7702 | β-glucosidase 1 | At1g52400 | 60462/6.89 | 7 | 228 | ↓3.45 | ||||
| Arabidopsis thaliana |
Untreated control plants (Ctr); plants treated with 1 mM CdSO4 and 10 mM ZnSO4 (Mt); plants treated with 1 mM CdSO4 and 10 mM ZnSO4 and the addition of the eight selected bacterial strains (Mt+8S).
Fold variations in spot abundance between the different treatments and the corresponding control. No value in the table indicate that, for the corresponding spot, was not possible to identify a change that exceeds both a fold of variation >2 and a P <0.05.
Proteins whose expression pattern have been confirmed by Western analysis.
Newly induced.
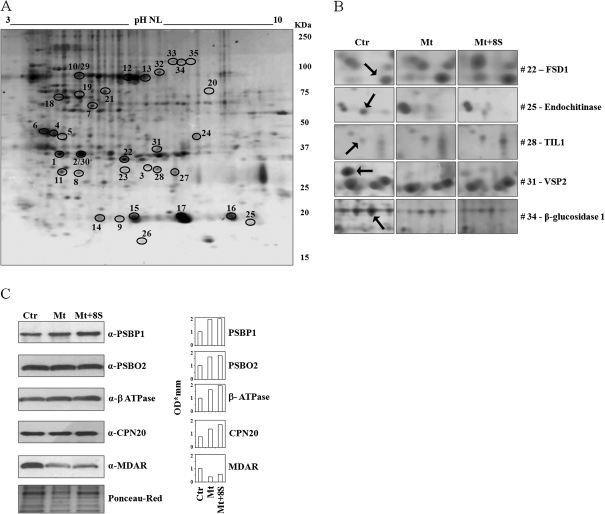
A 2-DE gel representative of the Mt+8S sample showing protein spots with significant changes in intensity among tested conditions, is reported in Fig. 3A. Selected differentially accumulated protein spots are highlighted in Fig. 3B. Several proteins were identified in more than one spot. This may be explained by different splicing variants, or post-translational modified or cleaved isoforms of the same protein (Weiss and Görg, 2007). Of the 24 differentially expressed proteins, 11 were grouped as proteins involved in the photosynthetic processes, another nine were classified as involved in stress response and four were clustered as taking part in cell metabolism and organization. To confirm the shoot proteomic pattern, several candidate proteins chosen to represent all functional categories, were selected for immunoblot analysis (Fig. 3C). In all cases, Western blots confirmed the trend in protein expression modulation observed in proteomic analysis.
Fig. 3.
Shoot proteome analysis. (A) Standard 2-DE map of proteins extracted from A. halleri shoots corresponding to the Mt+8S sample, with an average of 346±49 spots (Ctr and Mt samples showed an average of 562±21 and 377±11 spots, respectively). The 35 protein spots showing statistically significant differences between the conditions tested are marked by an open circle and numbered according to Table 4. (B) Close-up view of spots (indicated by arrows) representative of differentially expressed proteins (P <0.05) between control and treatments. (C, left) Validation of five differentially expressed proteins by Western blot (a representative gel of three repetition is reported). Ponceau-Red staining is shown for control of protein loading. (C, right) Densitometric analysis of immunoblot signals.
Among the proteins involved in the photosynthetic process, in shoots of plants treated with bacterial strains plus metals compared with control plants, up-regulation was observed for the PSBP1 subunit of photosystem II, responsible for electron transport, for the PSBO2 subunit of the oxygen-evolving complex, and for HCF136, a necessary protein for the assembly of the photosystem II reaction centre. A newly induced protein, PSBO1-like, was also found in the shoots of plants treated with bacterial strains plus metals, whereas a reduced expression was only detected for PSBP2. Similarly, an overproduction of the Rubisco large subunit was detected in three protein spots and the Rubisco small subunit was found to be up-regulated in another four spots. The Rubisco over-expression was mainly detected in plants co-cultivated with bacterial strains plus metals compared with untreated plants. This Rubisco overproduction was accompanied by the up-regulation of phosphoribulokinase and Rubisco activase. An increased expression of phosphoribulokinase was also seen as an effect of the bacteria strains. According to these data on photosynthesis-related proteins, an increased expression of the δ- and β-subunits of the plastidic ATPase was also detected.
Taking into account stress-related proteins, shoot proteome analysis showed a down-regulation of monodehydroascorbate reductase (MDAR1 and MDAR2), an enzyme required for the regeneration of reduced ascorbate, in plants treated with bacteria and metals compared with untreated plants. By contrast, an increment was detected for Fe-Superoxide Dismutase (FSD1), a key enzyme of the cellular antioxidative stress defence mechanism. A consistent decreased expression was noted for an Allene Oxide Cyclase (AOC), a crucial enzyme of the jasmonic acid biosynthesis, and for an endochitinase, potentially involved in plant defence mechanisms. In shoots of plants treated with heavy metals and bacterial strains, malate dehydrogenase, a germin-like protein (GLP) and a lipocalin showed a significantly elevated expression compared with control plants. Furthermore, an enolase (LOS2) and the above-mentioned GLP were shown to be specifically induced by the plant-bacteria interactions (Table 4, last column).
Among the proteins included in the functional category of cellular metabolism and organization, a plastidic chaperonin, that could be involved in folding and assembly of the Rubisco, was found to be up-regulated in plants treated with metals plus bacteria compared witho control plants. A down-regulation induced by bacterial strains plus metals was detected for a Vegetative Storage protein (VSP). A similar decline in protein expression was seen for the mitochondrial α-subunit of the ATPase in shoots of plants co-cultivated with heavy metals plus the selected bacterial strains. Finally, a strongly reduced expression was observed for a β-glucosidase in plants grown in the presence of bacteria and metals compared with untreated plants. It is worth noting that the AOC and β-glucosidase were specifically down-regulated in plants maintained with both heavy metals plus bacterial strains compared with plants grown only in the presence of metals (Table 4, last column).
Transcription analysis of genes involved in metal homeostasis and hyperaccumulation
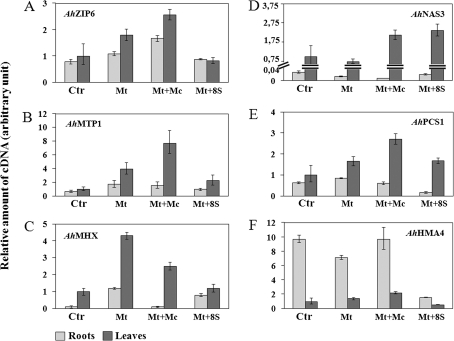
Since the shoot proteome was performed on the total protein extract, proteins involved in metal homeostasis and hyperaccumulation, but only expressed at a low level, were undetected. For this reason, the transcription level of several genes in the shoots and roots of untreated plants was compared by real-time PCR with those of plants treated with (i) only Cd and Zn, (ii) with these metals plus the autochthonous rhizosphere-derived microorganisms, and (iii) with Cd and Zn plus the eight selected bacterial strains (Fig. 4). A generally increased expression, compared with untreated control plants, was observed for AhZIP6, AhMTP1, and AhMHX (Fig. 4A, B, C) in leaves of plants treated with Cd and Zn or with these metals plus the autochthonous rhizosphere-derived microorganisms. When the heavy metals were added to the culture medium with the eight bacterial strains, the expression of these genes in the leaves was similar to that observed in control plants. A slightly increased expression of AhZIP6 and AhMTP1 was also seen in the roots of plants grown solely with metals or with metals plus rhizosphere-derived microorganisms. An analysis of the AhMHX mRNA level in roots showed a higher expression when plants were treated with metals or with metals plus the eight bacterial strains compared with control plants. Furthermore, an increased transcription for AhPCS1 and AhNAS3 was observed in leaves when plants were maintained with Cd and Zn plus rhizosphere-derived microorganisms or plus the eight microbial strains. For both genes, the expression in the roots was only slightly modulated by the different treatments (Fig. 4D, E). The mRNA level of AhHMA4 was constitutively higher in the roots compared with the leaves and a drastic down-regulation in the roots was caused by the eight bacterial strains plus metals, while expression in the leaves was lightly modulated by the different treatments (Fig. 4F).
Fig. 4.
Expression analysis of metal homeostasis and hyperaccumulator genes. AhZIP6, AhMTP1, AhMHX, AhNAS3, AhPCS1, and AhHMA4 transcript levels in the shoot and root of A. halleri plants subjected to different treatments, by real-time PCR. The experiment was carried out in triplicate (±SE).
Effects of each bacterial strain on shoot metal accumulation
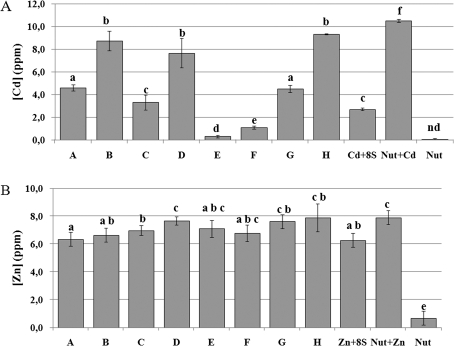
Before checking for the effects on shoot metal accumulation due to each bacterial isolate, the eight selected bacteria were individually grown to the stationary phase in liquid Nutrient Broth with 100 μM CdSO4 (c. 11.3 ppm Cd) or 100 μM ZnSO4 (c. 6.5 ppm Zn) and Cd and Zn contents in the respective supernatants and rinsing solutions were analysed to quantify the metal removed by the different strains (Fig. 5). This analysis showed that Cd concentrations in solution from strains E and F were very low, suggesting that these bacteria are able to accumulate or adsorb and/or precipitate Cd causing this metal to be no longer available. Instead, Cd measurements relative to strains B, D, and H indicated that almost all the Cd added to the culture was still present in the solution, whereas strains A, C, and G, as well as the eight strains cultured together, left Cd only partially available in the solution (Fig. 5A). Different behaviours were found for the same bacterial strains grown in 100 μM ZnSO4. With the exception of strain A, which behaves as the eight strains grown together, the others left higher amounts of Zn in the suspension culture (Fig. 5B).
Fig. 5.
Measurement of Cd and Zn content in bacteria supernatants and rinsing solutions. (A) Cd content of singly grown bacterial strains (A–H), as indicated in Table 2 and of the eight selected bacterial strains grown together (Cd+8S). (B) Zn content of singly grown bacterial strains (A–H), as indicated in Table 2 and of the eight selected bacterial strains grown together (Zn+8S). Nut: nutrient solution. Cd+nut and Zn+nut were added as positive control. Data are the mean ±SD (n=3). Differences between means were scored for significance according to the Duncan test. Values sharing a common letter are not significantly different at P <0.05.
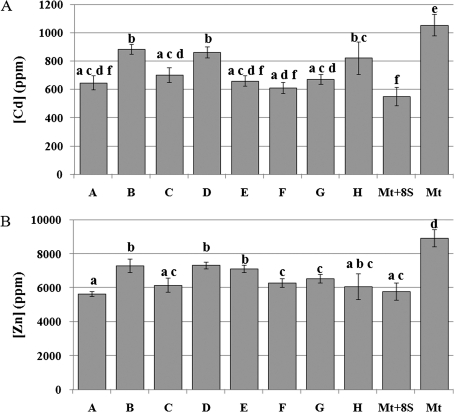
Cd and Zn concentrations were then measured in the shoots of plants cultivated with these metals plus each selected rhizosphere bacterium (experimental design, treatment iv) for one month to test the effect of each bacterial strain on shoot metal accumulation (Fig. 6). These analyses confirmed the significant difference (P <0.05) on shoot metal accumulation in plants cultivated with Cd and Zn plus bacteria either added together or singly, compared with plants grown solely with these metals (Fig. 6A, B). The different values measured in this analysis, for the plants grown with the eight bacterial strains compared with the previous one (see Fig. 2) are explained by the variation in greenhouse conditions at different times of year. With regard to the effect of each single strain on shoot Cd content, the main significant differences (P <0.05) were observed when plants were cultivated with strains B, D, and H (Fig. 6A) whereas the concentration of Zn was significantly different when plants were grown with strains B, D, and E (Fig. 6B).
Fig. 6.
Measurement of Cd and Zn content in shoots of A. halleri plants. (A) Cd content (B) Zn content in shoots of plants grown with 1 mM CdSO4 and 10 mM ZnSO4 plus the eight bacterial strains (added singly: A–H); added together (Mt+8S); without bacteria (Mt). Data are the mean ±SD (n=3), subjected to analysis of variance. Differences between means were scored for significance according to the Duncan test. Values sharing a common letter are not significantly different at P <0.05.
Discussion
The majority of bacteria in the environment are unable to grow on in vitro culture. Nevertheless, culturable rhizosphere bacteria are required for studying plant–microbe interactions and may be used in bioaugmentation applications (Pilon-Smits, 2005).
In this study, eight bacterial strains were selected in vitro from the rhizosphere-derived microorganisms of A. halleri plants grown in heavy metal contaminated soil (van Rossum et al., 2004). They were all added together, plus 1 mM CdSO4 and 10 mM ZnSO4, to the plant nutrient solution and tested for their effects on Cd and Zn accumulation, on shoot biomass, chlorophyll content, and shoot protein expression. In addition, each strain was added individually plus the metals to the nutrient solution to check the influence of each one on shoot metal accumulation. In these experiments, the bacterial population selected for co-cultivation with A. halleri plants has probably greatly changed during the experimental period due to greenhouse conditions and competition among different strains. Nevertheless, as shown in Fig. 2, a clear effect of the eight selected bacterial strains, compared with the effects of rhizosphere-derived microorganisms or of the metals alone, was evident on shoot metal concentration. The addition of the eight selected bacterial strains clearly impaired Cd and Zn accumulation in the shoot. Moreover, when these bacterial strains were added individually to the nutrient solution, some of them affected the metal content in the shoot more than others. It is known that certain bacteria can solubilize metals and adsorb them to their biomass and/or precipitate them with a consequent decrease in metal bioavailability (Gadd, 2000). The results shown in Figs 5A and 6A, point to a correlation between the metal accumulated in plant shoots and the metal remaining available in the bacterial nutrient solution, and therefore not accumulated in the bacteria or precipitated by them. Several bacterial strains (A, C, E, F, and G) contributed to decrease Cd content in the nutrient solution and, when co-cultivated with plants, they determined a lower Cd accumulation in the shoots. On the other hand, the presence of the indigenous rhizosphere microflora may increase the plant metal accumulation capacity (de Souza et al., 1999; Farinati et al., 2009). These results emphasize the important role of microbial processes in shoot metal accumulation and point out the possibility of to a certain extent controlling metal accumulation in the shoots by selecting the appropriate bacteria consortium. The influence of these selected strains was also indirectly observed on chlorophyll content. It was previously experimented that the increased shoot metal concentration in A. halleri was accompanied by an increased chlorophyll content (Farinati et al., 2009). It was found here that a reduced metal concentration in the shoot, caused by the addition of selected bacteria, comes with a reduced chlorophyll content.
In plants susceptible to heavy metals, leaf chlorosis, as a result of damage to the photosynthetic apparatus (Smeets et al., 2005; Craciun et al., 2006), is one of the first symptoms of stress. In this study, the treatment of A. halleri plants with heavy metals plus eight selected rhizosphere-derived bacteria had the effect, as previously observed with the autochthonous rhizosphere-derived microorganisms (Farinati et al., 2009), of enhancing synthesis of the proteins belonging to the photosynthetic machinery, such as components of the oxygen-evolving complex, of the PSII and ATPase. Such a general increase could be explained, considering that, being an energy-demanding process, heavy metal detoxification requires enhanced production of ATP. Few enzymes of the Calvin-Benson cycle have been detected as being up-regulated by treatment with heavy metals plus selected bacterial strains: Rubisco (both the large and the small subunits are up-regulated), responsible for CO2 fixation, and phosphoribulokinase, which generates ribulose 1,5-bisphosphate, the substrate for Rubisco. Due to its intrinsic oxygenase activity, the higher Rubisco content could caused an elevated rate of photorespiration. The malate dehydrogenase enzyme plays a role in this pathway, which takes place in the peroxisome, and its expression is indeed enhanced in response to the treatment with metals plus the eight selected bacterial strains. PSBP2 is an exception to this massive up-regulation of the photosynthetic process, its abundance decreases in plants treated with selected bacterial strains and heavy metals compared with control plants. This could be explained as a ‘redundancy effect’ considering that PSBP2 is homologous to PSBP1, which is in fact up-regulated, and that both proteins participate in PSII activity and stability. In contrast to what was detected in the previous work (Farinati et al., 2009) increased expression of light-harvesting chlorophyll a/b binding proteins was not detected. Indeed, the treatment with the eight selected bacterial strains plus metals did not affect chlorophyll content in the leaves.
The expression of MDAR1 and MDAR2 is down-regulated by the treatment with metals plus the eight selected bacterial strains. These enzymes are required for the regeneration of reduced ascorbate, correlated with the activity of the ascorbate peroxidase. Since the latter was found to be significantly decreased in the presence of high Cd concentration (Metwally et al., 2005), it is rational that less MDAR is also required. A general down-regulation of VSP, β-glucosidase, endochitinase, and AOC was observed in plants treated with Cd and Zn plus the bacterial strains. These proteins are implicated in different biological processes but they are all also involved in deterring herbivorous insects and pathogen attack (Mattiacci et al., 1995; Kasprzewska, 2003; Morant et al., 2008). For instance, VSPs can accumulate in various plant organs, act as storage of amino acids, and their biosynthesis and degradation are regulated accordingly to storage needs (Liu et al., 2005). For this reason, VSP2 was included in the functional category of cellular metabolism in this study, although a role of VSP in defence against herbivorous insects is also well documented (Liu et al., 2005; de Vos et al., 2006). Likewise, β-glucosidases are involved in phytohormone activation, lignin synthesis, and cell wall degradation, although their crucial role as elicitor of the plant defence response has been proved (Mattiacci et al., 1995; Morant et al., 2008). AOC is a key enzyme of jasmonate biosynthesis whereas chitinases are implicated in plant defence mechanisms but also involved in growth processes (Kasprzewska, 2003). That metal hyperaccumulator plants benefit from protection against herbivores is supported by experimental data (Jhee et al., 1999, 2005; Jiang et al., 2005; Freeman et al., 2007). The results presented here point to a switch of gene expression: in the absence of heavy metals the defence mechanisms are switched on whereas, in hyperaccumulating shoots, Cd and Zn are efficient as protectors and defence genes are down-regulated. This suggests that a defence system is costly and that elemental accumulation allows plants to forgo these costs when defence is provided by other means. Furthermore, shoot metal accumulation necessitates energy as manifested by the up-regulation of photosynthesis-related genes, but energy can be saved if genes in charge of defence against biotic stresses are provisionally down-regulated. This points to the advantage of employing a network of signals to satisfy the necessity for plants to cope with ever-changing environmental conditions and co-evolving root-associated microbial communities and of converting these signals into well-defined responses. It is noteworthy that a specific effect of the eight bacterial strains plus Cd and Zn, that was not observed when plants were treated with the whole rhizosphere-derived microorganisms plus metals (Farinati et al., 2009), was the down-regulation of AOC. This enzyme contributes to jasmonate biosynthesis, which, in turn, by regulating growth and defence, optimizes plant fitness in rapidly changing environments (Chung et al., 2008). Therefore, the eight selected bacterial strains plus metals, by inhibiting the AOC protein expression may regulate a trade-off between plant defense and fitness.
Proteins that participate in oxidative stress tolerance such as a GLP, and a lipocalin were found to be specifically up-regulated by the treatment with selected bacterial strains plus metals. GLPs have been found to be up-regulated, by biotic and abiotic stress, in a wide variety of plant tissues, although their function in plants is still unclear (Doll et al., 2003). In wheat, for example, the expression of a germin, with oxalate oxidase activity, was stimulated by some heavy metals, particularly by Cd (Berna and Bernier, 1999) whereas in Medicago truncatula a specific GLP was induced by arbuscular mycorrhiza (Doll et al., 2003). In this work, GLP expression modulation was detected specifically when plants were co-cultivated with the selected bacterial strains and metals suggesting an explicit effect of these bacteria (Table 4) that was not detected when plants were maintained with the entire population of rhizosphere-derived microorganisms plus Cd and Zn (Farinati et al., 2009). Likewise, specific up-regulation induced by the interaction between plants and selected bacteria was observed for the enolase LOS2. This enzyme is important for chilling resistance (Lee et al., 2002), but our data suggest that it is critical for an optimal cellular response to other environmental cues.
Comparative transcriptomic analyses of related hyperaccumulators and non-hyperaccumulators species confirmed a substantial constitutive over-expression of metal homeostasis genes even in the absence of excess of metallic ions (Weber et al., 2004, 2006; Becher et al., 2004; Talke et al., 2006). It was reported that high expression of AhMHX in leaves was found to be constitutive and not significantly affected by the Cd, Zn and Mg status of the plants (Elbaz et al., 2006). Similarly, the transcript level of AhMTP1 was reported as being constitutively high in leaves and almost unaffected by different Zn concentrations, whereas in roots it was found to be slightly modulated by Zn after a 4 d treatment (Dräger et al., 2004). Of the ZIP family gene transcripts analysed, AhZIP6 expression was predominant in the shoot and was not influenced by the Zn exposure (Becher et al., 2004). In this study a modulation in expression of these genes caused by metals and by metals plus microbial communities was observed. In particular, the high Cd and Zn accumulation in leaves of plants treated solely with metals or with metals plus the rhizosphere-derived microorganisms is accompanied by the up-regulation of AhMHX, AhMTP1, and AhZIP6, whereas the lower Cd and Zn accumulation in shoots of plants treated with these metals and the selected bacterial strains is coupled with the lower expression of these genes. Although it cannot be excluded that long-term metal treatments have induced other mechanisms such as iron deficiency, which in turn contribute to the up-regulation of metal transporters (Thomine et al., 2000), these results clearly showed the different effects of the total rhizosphere-derived microbial community and selected bacterial strains on the expression of some metal homeostasis genes. The influence of these bacterial strains is even more marked for AhHMA4, a gene mainly expressed in the root, which plays a role in root-to-shoot metal translocation and is crucial for metal hyperaccumulation (Verret et al. 2004; Hanikenne et al., 2008; Verbruggen et al., 2009). Its expression is down-regulated by the interaction between plants and selected bacterial strains. Several NAS genes, particularly NAS3, showed higher transcript level in A. halleri compared with A. thaliana suggesting a role for nicotianamine in metal hyperaccumulation (Becher et al., 2004; Talke et al., 2006). In agreement with these studies, a higher NAS3 transcription was observed in the shoot than in the root independently of the presence of metals, but a clear effect of plant–microbe interaction on NAS3 transcription level was also noticed. A constitutive expression has been reported for PCS in A. thaliana (Vatamaniuk et al., 2004), its transcription is not influenced by the presence of Cd (Ha et al., 1999) but its activity is very much dependent on the presence of a heavy metal (Vatamaniuk et al., 2004). A perturbation of AhPCS1 transcription was observed, especially in the shoots of strongly metal accumulating plants.
These results call attention to the role played by the microbial community directly on Cd and Zn accumulation in the shoot and indirectly on the expression of several genes. Modulation of plant metal uptake may be due to a stimulatory effect of microorganisms on root growth or to the microbial effect on metal bioavailability (De Souza et al., 1999) and, as seen in this study, to the expression modulation of metal transporters. In addition, decreased metal bioavailability could be due to microorganisms causing metal precipitation (Gadd, 2000). The eight bacterial strains used in this study were isolated from the autochthonous rhizosphere population based on their ability to tolerate high levels of Cd and Zn and, at least for some of them, very likely involved in microbial processes that decrease Cd and Zn bioavailability. Therefore, the identification of microbial consortia able to increase or decrease the plant's potential for Cd and Zn accumulation may become important in phytoremediation programmes or for decreasing metal translocation and accumulation in shoots of crop species.
Acknowledgments
We thank Professor P Saumitou-Laprade (University of Lille, France) for his help in collecting A. halleri plants, seeds and contaminated soil in Bois des Asturies (Auby, France). We are grateful to Professor D Leister (University of München, Germany) for providing the antibodies against PSBP1, PSBO2, and the β-subunit-ATPase, to Dr S Sano (Kyoto Prefectural University, Japan) for donating the anti-cucumber MDAR antibody, to Professor A Azem (University of Tel Aviv, Israel) for the antibody against chaperonin 20. We kindly acknowledge Dr G Zapparoli (University of Verona, Italy) for statistical analysis, and Dr D Cecconi and Professor G Vallini (University of Verona) for many valuable suggestions and discussions.
References
- Abou-Shanab RA, Angle JS, Delorme TA, Chaney RL, van Berkum P, Moawad H, Ghanemand K, Ghozlan A. Rhizobacterial effects on nickel extraction from soil and uptake by. Alyssum murale. New Phytologist. 2003;158:219–224. [Google Scholar]
- Altshul SF, Madden TL, Shaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora PS, Yamagiwa H, Srivastava A, Bolander ME, Sarkar G. Comparative evaluation of two two-dimensional gel electrophoresis image analysis software applications using synovial fluids from patients with joint disease. Journal of Orthopaedic Science. 2005;10:160–166. doi: 10.1007/s00776-004-0878-0. [DOI] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Kramer U. Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator. Arabidopsis halleri. The Plant Journal. 2004;37:251–268. doi: 10.1046/j.1365-313x.2003.01959.x. [DOI] [PubMed] [Google Scholar]
- Berna A, Bernier F. Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Molecular Biology. 1999;39:539–549. doi: 10.1023/a:1006123432157. [DOI] [PubMed] [Google Scholar]
- Bert V, Bonnin I, Saumitou-Laprade P, de Laguérie P, Petit D. Do Arabidopsis halleri from nonmetallicolous populations accumulate zinc and cadmium more effectively than those from metallicolous populations? New Phytologist. 2002;155:47–57. doi: 10.1046/j.1469-8137.2002.00432.x. [DOI] [PubMed] [Google Scholar]
- Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivore. Plant Physiology. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciun AR, Courbot M, Bourgis F, Salis P, Saumitou-Laprade P, Verbruggen N. Comparative cDNA-AFLP analysis of Cd-tolerant and -sensitive genotypes derived from crosses between the Cd hyperaccumulator Arabidopsis halleri and Arabidopsis lyrata ssp. Petraea. Journal of Experimental Botany. 2006;57:2967–2983. doi: 10.1093/jxb/erl062. [DOI] [PubMed] [Google Scholar]
- Dahmani-Muller H, van Oort F, Gelie B, Balabane M. Strategies of heavy metal uptake by three plant species growing near a metal smelter. Environmental Pollution. 2000;109:231–238. doi: 10.1016/s0269-7491(99)00262-6. [DOI] [PubMed] [Google Scholar]
- De Souza MP, Chu D, Zhao M, Zayed AM, Ruzin SE, Schichnes D. Rhizosphere bacteria enhance selenium accumulation and volatilization by indian mustard. Plant Physiology. 1999;119:565–573. doi: 10.1104/pp.119.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Zaanen W, Koornneef A, Korzelius JP, Dicke M, Van Loon LC, Pieterse CMJ. Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiology. 2006;142:352–363. doi: 10.1104/pp.106.083907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll J, Hause B, Demchenko K, Pawlowski K, Krajinski F. A member of the germin-like protein family is a highly conserved mycorrhiza-specific induced gene. Plant and Cell Physiology. 2003;44:1208–1214. doi: 10.1093/pcp/pcg153. [DOI] [PubMed] [Google Scholar]
- Dräger DB, Desbrosses-Fonrouge AG, Krach C, Chardonnens AN, Meyer RC, Saumitou-Laprade P, Krämer U. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. The Plant Journal. 2004;39:425–439. doi: 10.1111/j.1365-313X.2004.02143.x. [DOI] [PubMed] [Google Scholar]
- Elbaz B, Shoshani-Knaani N, David-Assael O, Mizrachy-Dagri T, Mizrahi K, Saul H, Brook E, Berezin I, Shaul O. High expression in leaves of the zinc hyperaccumulator Arabidopsis halleri of AhMHX, a homolog of an Arabidopsis thaliana vacuolar metal/proton exchanger. Plant, Cell and Environment. 2006;29:1179–1190. doi: 10.1111/j.1365-3040.2006.01500.x. [DOI] [PubMed] [Google Scholar]
- Farinati S, DalCorso G, Bona E, Corbella M, Lampis S, Cecconi D, Polati R, Berta G, Vallini G, Furini A. Proteomic analysis of Arabidopsis halleri shoots in response to the heavy metals cadmium and zinc and rhizosphere microorganisms. Proteomics. 2009;9:4837–4850. doi: 10.1002/pmic.200900036. [DOI] [PubMed] [Google Scholar]
- Freeman JL, Lindblom SD, Quinn CF, Fakra S, Marcus MA, Pilon-Smits EAH. Selenium accumulation protects plants from herbivory by Orthoptera via toxicity and deterrence. New Phytologist. 2007;175:490–500. doi: 10.1111/j.1469-8137.2007.02119.x. [DOI] [PubMed] [Google Scholar]
- Gadd GM. Bioremedial potential of microbial mechanisms of metal mobilization and immobilization. Current Opinion in Biotechnology. 2000;11:271–279. doi: 10.1016/s0958-1669(00)00095-1. [DOI] [PubMed] [Google Scholar]
- Glick BR. Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechology Advances. 2003;21:383–393. doi: 10.1016/s0734-9750(03)00055-7. [DOI] [PubMed] [Google Scholar]
- Ha SB, Smith P, Howden R, Dietrich WM, Bugg S, O'Connell MJ, Goldsbrough PB, Cobbett CS. Phytochelatin synthase genes from Arabidopsis and the yeast. Schizosaccharomyces pombe. The Plant Cell. 1999;11:1153–1163. doi: 10.1105/tpc.11.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of. HMA4. Nature. 2008;453:391–396. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. The water culture method for growing plants without soil. 1938. University of California, Agricultural Experimental Station, 347–353. [Google Scholar]
- Jhee EM, Boyd RS, Eubanks MD. Nickel hyperaccumulation as an elemental defense of Streptanthus polygaloides (Brassicaceae): influence of herbivore feeding mode. New Phytologist. 2005;168:331–344. doi: 10.1111/j.1469-8137.2005.01504.x. [DOI] [PubMed] [Google Scholar]
- Jhee EM, Dandridge KL, Christy AM, Jr, Pollard AJ. Selective herbivory on low-zinc phenotypes of the hyperaccumulator. Thlaspi caerulescens (Brassicaceae). Chemoecology. 1999;9:93–95. [Google Scholar]
- Jiang RF, Ma DY, Zhao FJ, McGrath SP. Cadmium hyperaccumulation protects Thlaspi caerulescens from leaf feeding damage by thrips (Frankliniella occidentalis) New Phytologist. 2005;167:805–814. doi: 10.1111/j.1469-8137.2005.01452.x. [DOI] [PubMed] [Google Scholar]
- Kasprzewska A. Plant chitinases: regulation and function. Cellular and Molecular Biology Letters. 2003;8:809–824. [PubMed] [Google Scholar]
- Krämer U. Metal hyperaccumulation in plants. Annual Review of Plant Biology. 2010;61:517–534. doi: 10.1146/annurev-arplant-042809-112156. [DOI] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO Journal. 2002;21:2692–2702. doi: 10.1093/emboj/21.11.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ahn JE, Datta S, Salzman RA, Moon J, Huyghues-Despointes B, Pittendrigh B, Murdock LL, Koiwa H, Zhu-Salzman K. Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiology. 2005;139:1545–1556. doi: 10.1104/pp.105.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantelin S, Touraine B. Plant growth-promoting bacteria and nitrate availability: impacts on root development and nitrate uptake. Journal of Experimental Botany. 2004;55:27–34. doi: 10.1093/jxb/erh010. [DOI] [PubMed] [Google Scholar]
- Mattiacci L, Dicke M, Posthumus MA. β-Glucosidase: an elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proceedings of the National Academy of Sciences, USA. 1995;92:2036–2040. doi: 10.1073/pnas.92.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metwally A, Safronova VI, Belimov AA, Dietz KJ. Genotypic variation of the response to cadmium toxicity in Pisum sativum L. Journal of Experimental Botany. 2005;56:167–178. doi: 10.1093/jxb/eri017. [DOI] [PubMed] [Google Scholar]
- Milner MJ, Kochian LV. Investigating heavy-metal hyperaccumulation using Thlaspi caerulescens as a model system. Annals of Botany. 2008;102:3–13. doi: 10.1093/aob/mcn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends in Ecology and Evolution. 2001;16:693–700. [Google Scholar]
- Morant AV, Jørgensen K, Jørgensen C, Paquette SM, Sánchez-Pérez R, Møller BL, Bak S. β-Glucosidases as detonators of plant chemical defence. Phytochemistry. 2008;69:1795–1813. doi: 10.1016/j.phytochem.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Noret N, Meerts P, Tolrà RP, Poschenrieder C, Barcelò D, Escarré J. Palatability of Thlaspi caerulescens for snails: influence of Zn and glucosinolates. New Phytologist. 2005;165:763–772. doi: 10.1111/j.1469-8137.2004.01286.x. [DOI] [PubMed] [Google Scholar]
- Pilon-Smits E. Phytoremediation. Annual Review of Plant Biology. 2005;56:15–39. doi: 10.1146/annurev.arplant.56.032604.144214. [DOI] [PubMed] [Google Scholar]
- Pilon Smits E, Pilon M. Phytoremediation of metals using transgenic plants. Critical Reviews in Plant Sciences. 2002;21:439–456. [Google Scholar]
- Polle A, Schützendübel A. Heavy metal signalling in plants: linking cellular and organismic responses. In: Hirt H, Shinozaki K, editors. Plant responses to abiotic Stress. Berlin, Heidelberg: Springer-Verlag; 2003. pp. 187–215. [Google Scholar]
- Porra RJ. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and. b. Photosynthesis Research. 2002;73:149–156. doi: 10.1023/A:1020470224740. [DOI] [PubMed] [Google Scholar]
- Reeves RD, Baker AJM. Metal-accumulating plants. In: Raskin I, Ensley BD, editors. Phytoremediation of toxic metals: using plants to clean-up the environment. New York: John Wiley; 2000. pp. 193–229. [Google Scholar]
- Sanità di Toppi L, Gabbrielli R. Response to cadmium in higher plants. Environmental and Experimental Botany. 1999;41:105–130. [Google Scholar]
- Smeets K, Cuypers A, Lambrechts A, Brahim S, Hoet P, Van Laere A, Vangronsveld J. Induction of oxidative stress and antioxidative mechanisms in Phaseolus vulgaris after Cd application. Plant Physiology and Biochemistry. 2005;43:437–444. doi: 10.1016/j.plaphy.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Sors TG, Ellis DR, Salt DE. Selenium uptake, translocation, assimilation and metabolic fate in plants. Photosynthesis Research. 2005;86:373–389. doi: 10.1007/s11120-005-5222-9. [DOI] [PubMed] [Google Scholar]
- Talke IN, Hanikenne M, Krämer U. Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator. Arabidopsis halleri. Plant Physiology. 2006;142:148–167. doi: 10.1104/pp.105.076232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NG, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopisis with homology to Nramp genes. Proceedings of the National Academy of Sciences, USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossum F, Bonnin I, Fénart S, Pauwels M, Petit D, Saumitou-Laprade P. Spatial genetic structure within a metallicolous population of Arabidopsis halleri, a clonal, self-incompatible and heavy-metal-tolerant species. Molecular Ecology. 2004;13:2959–2967. doi: 10.1111/j.1365-294X.2004.02314.x. [DOI] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lang A, Chalasani S, Demkiv LO, Rea PA. Phytochelatin synthase, a dipeptidyltransferase that undergoes multisite acylation with γ-glutamylcysteine during catalysis. Journal of Biological Chemistry. 2004;279:22449–22460. doi: 10.1074/jbc.M313142200. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. Molecular mechanisms of metal hyperaccumulation in plants. New Phytologist. 2009;181:759–776. doi: 10.1111/j.1469-8137.2008.02748.x. [DOI] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Letters. 2004;576:306–312. doi: 10.1016/j.febslet.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Weber M, Harada E, Vess C, Roepenack-Lahaye E, Clemens S. Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. The Plant Journal. 2004;37:269–281. doi: 10.1046/j.1365-313x.2003.01960.x. [DOI] [PubMed] [Google Scholar]
- Weber M, Trampczynska A, Clemens S. Comparative transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd2+-hypertolerant facultative metallophyte. Arabidopsis halleri. Plant, Cell and Environment. 2006;29:950–963. doi: 10.1111/j.1365-3040.2005.01479.x. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss W, Görg A. Two-dimensional electrophoresis for plant proteomics. Methods in Molecular Biology. 2007;355:121–143. doi: 10.1385/1-59745-227-0:121. [DOI] [PubMed] [Google Scholar]
- Whiting SN, Neumann PM, Baker AJM. Nickel and zinc hyperaccumulation by Alyssum murale and Thlaspi caerulescens (Brassicaceae) do not enhance survival and whole-plant growth under drought stress. Plant, Cell and Environment. 2003;26:351–360. [Google Scholar]