Abstract
The DREB genes code for important plant transcription factors involved in the abiotic stress response and signal transduction. Characterization of DREB genes and development of functional markers for effective alleles is important for marker-assisted selection in foxtail millet. Here the characterization of a cDNA (SiDREB2) encoding a putative dehydration-responsive element-binding protein 2 from foxtail millet and the development of an allele-specific marker (ASM) for dehydration tolerance is reported. A cDNA clone (GenBank accession no. GT090998) coding for a putative DREB2 protein was isolated as a differentially expressed gene from a 6 h dehydration stress SSH library. A 5' RACE (rapid amplification of cDNA ends) was carried out to obtain the full-length cDNA, and sequence analysis showed that SiDREB2 encoded a polypeptide of 234 amino acids with a predicted mol. wt of 25.72 kDa and a theoretical pI of 5.14. A theoretical model of the tertiary structure shows that it has a highly conserved GCC-box-binding N-terminal domain, and an acidic C-terminus that acts as an activation domain for transcription. Based on its similarity to AP2 domains, SiDREB2 was classified into the A-2 subgroup of the DREB subfamily. Quantitative real-time PCR analysis showed significant up-regulation of SiDREB2 by dehydration (polyethylene glycol) and salinity (NaCl), while its expression was less affected by other stresses. A synonymous single nucleotide polymorphism (SNP) associated with dehydration tolerance was detected at the 558th base pair (an A/G transition) in the SiDREB2 gene in a core set of 45 foxtail millet accessions used. Based on the identified SNP, three primers were designed to develop an ASM for dehydration tolerance. The ASM produced a 261 bp fragment in all the tolerant accessions and produced no amplification in the sensitive accessions. The use of this ASM might be faster, cheaper, and more reproducible than other SNP genotyping methods, and thus will enable marker-aided breeding of foxtail millet for dehydration tolerance.
Keywords: Allele-specific marker, dehydration-responsive element-binding protein 2, dehydration stress, gene expression, Setaria italica, single nucleotide polymorphism
Introduction
Foxtail millet (Setaria italica L.), an elite drought-tolerant crop, is an important food and fodder grain crop in arid and semi-arid regions of Asia and Africa. Its genome is being sequenced by the US Department of Energy Joint Genomic Institute and BGI (formerly the Beijing Genomics Institute), China. Foxtail millet together with proso millet (Panicum miliaceum) ranks second in the total world production of millets (FAO STAT data 2005; http://faostat.fao.org/). It is a diploid (2n=2x=18), self-pollinating, C4 panicoid crop, with a small genome of ∼490 Mb having a low repetitive DNA content (30%) and a highly conserved genome structure relative to the ancestral grass lineage, making it a suitable model species for genetic and molecular studies (Devos et al., 1998; Jayaraman et al., 2008). Further, it is a close relative of several important biofuel crops such as, switchgrass, napiergrass, and pearl millet, and hence has been suggested to represent an appropriate model for this class of crop species (Doust et al., 2009). Water use efficiency of foxtail millet has been shown to be higher than that of maize, wheat, and sorghum (Gu et al., 1987). The adaptation of foxtail millet to low water conditions has been ascribed to its relatively small leaf area, the cell arrangement in its epidermis, its thick cell walls, and its ability to form a dense root system (Li, 1987). However, the molecular mechanism of its drought adaptation is still not clear. To gain a better understanding of the molecular responses of this crop to dehydration stress, two SSH forward libraries were constructed (Lata et al., 2010). A differentially expressed expressed sequence tag (EST) from a 6 h dehydration stress library was found to encode a dehydration-responsive element-binding protein 2 (DREB2).
The DREBs are important transcription factors that induce a set of abiotic stress-related genes and impart stress tolerance to plants. They could broadly be grouped as DREB1 and DREB2, involved in two separate signal transduction pathways under low temperature and dehydration, respectively (Agarwal et al., 2006). They belong to the ERF (ethylene-responsive element-binding factor) family, a subfamily of AP2/EREBP (APETALA2/ethylene-responsive element-binding protein) plant-specific transcription factors (Riechmann et al., 2000). These proteins share a conserved DNA-binding domain of ∼58–59 amino acids that bind to two cis-elements, namely a GCC box and a CRT/DRE (C-repeat/dehydration-responsive element) motif playing an important role in regulating gene expression in response to dehydration, high salinity, and cold in plants (Yamaguchi-Shinozaki and Shinozaki, 2005; Agarwal et al., 2006). The 14th and 19th amino acids of the AP2/ERF domain are highly distinctive as they distinguish the DREB (valine and glutamic acid, respectively) from the ERF (alanine and aspartic acid, respectively) gene classes (Sakuma et al., 2002). Abscisic acid (ABA)-independent stress-responsive gene expression is generally regulated by DREBs that bind to DRE cis-elements (5′-TACCGACAT-3′). The first cDNA clone for a DREB, CBF1 (CRT-binding factor 1) was isolated in a yeast one-hybrid screening (Stockinger et al., 1997). Since then several DREBs have been identified from Arabidopsis, rice, maize, and other plants that specifically interact with the DRE sequence (Liu et al., 1998; Dubouzet et al., 2003; Chen et al., 2007; Qin et al., 2007). Overexpression of Arabidopsis DREB2A results in significant drought stress tolerance, water stress, and heat stress tolerance, but only slight freezing tolerance in transgenic plants (Sakuma et al., 2006a, b). GmDREB2 also conferred drought and high-salinity tolerance to transgenic Arabidopsis and tobacco plants (Chen et al., 2007). However, to the best of our knowledge, no studies on AP2/EREBP transcription factors in foxtail millet have been reported.
Single nucleotide polymorphisms (SNPs) are the source of DNA variation in most of the plant and animal genomes (Garcés-Claver et al., 2007). SNPs are single base changes or indels at a specific nucleotide position and have recently been developed into genetic markers. SNPs can serve as a powerful tool for marker-assisted selection (MAS) and map-based cloning since they are considered as highly stable markers and often contribute directly to a phenotype (Andersen and Luebberstedt, 2003; Kim et al., 2005). Among the many SNP genotyping methods, allele-specific PCR largely satisfies the requirements for MAS since it is simple, user friendly, cost-effective, and reproducible.
In the present study, the cloning and characterization of a novel DREB2 homologous gene, SiDREB2, isolated from foxtail millet and classified into the A-2 subgroup in the DREB subfamily, are reported. An SNP in the SiDREB2 gene was also identified and an allele-specific PCR-based marker associated with dehydration tolerance was developed and validated in a core set of 45 foxtail millet accessions.
Materials and methods
Plant materials and growth conditions
Seeds of a core set of 45 foxtail millet [S. italica (L.)] accessions originating from different eco-geographic regions were obtained from the National Bureau of Plant Genetic Resources, Hyderabad and the University of Agricultural Sciences, GKVK, Bangalore, India (Table 1). Seeds were surface sterilized in 3% sodium hypochlorite for 20 min and rinsed 10–12 times (1 min each time) in distilled water. Five seeds of each accession were germinated, and sown in square black pots (9×9×9.8 cm) containing composite soil (peat compost to vermiculite, 3:1) in triplicate. The seedlings were grown in a plant growth chamber containing two cabinets (PGC-6L; Percival Scientific Inc., USA) for 10 d after germination at 28±1 °C day/23±1 °C night/70±5% relative humidity with a photoperiod of 14 h and a photosynthetic photon flux density of 500 μmol m−2 s−1. The plants were watered daily with one-third strength Hoagland's solution. Ten-day-old seedlings were used for dehydration treatment. They were pre-cultured for 24 h in one-third strength Hoagland's solution. Dehydration stress was applied by transferring seedlings into the same solution containing 20% polyethylene glycol (PEG-6000). The control plants were cultured in the same way as the dehydration treatments but without the addition of PEG (Zhang et al., 2007). Whole seedlings were collected and used as such. All experimental data are the means of at least three independent experiments and, for each experiment, ∼100 mg seedling samples were collected by random sampling.
Table 1.
Description of 45 foxtail millet (Setaria italica) accessions used in the present study along with the level of lipid peroxidation and relative water content in the control and after 24 h of dehydration stress
| Sl no. | Accession no.a | Origin | Lipid peroxidationb | 24 h ±SD | Relative water contentc | 24 h ±SD | Allele | Overall graded |
| Control ±SD | Control ±SD | |||||||
| 1 | GS-464 | USA | 8.22±1.1 | 4.11±0.5 | 91.61±2.01 | 36.40±5.26 | A | HT |
| 2 | IC-403579 | India | 24.5±0.34 | 15.2±0.99 | 96.91±1.11 | 42.07±2.65 | A | HT |
| 3 | Prasad | India | 39.1±0.86 | 24.6±1.28 | 91.78±2.06 | 40.47±3.86 | A | HT |
| 4 | IC-403476 | India | 25.1±0.8 | 16.6±1.56 | 96.19±1.26 | 38.40±3.87 | A | HT |
| 5 | IC-436928 | India | 37.5±1.29 | 24.7±2.37 | 95.31±1.45 | 34.34±2.07 | A | T |
| 6 | IC-328716 | India | 29.0±1.96 | 20.8±1.38 | 96.51±1.18 | 35.34±0.73 | A | T |
| 7 | GS-1646 | Russia | 15.06±0.54 | 11.62±0.74 | 94 90±1.98 | 36.40±3.26 | A | T |
| 8 | IC-426735 | India | 29.2±0.87 | 25±2.41 | 97.60±1.49 | 36.30±1.43 | A | T |
| 9 | IC-436863 | India | 32.4±0.48 | 27.8±2.39 | 96.60 ±1.08 | 34.73±1.08 | A | T |
| 10 | IC-438725 | India | 32.4±1.57 | 27.9±2.36 | 87.74±2.03 | 33.18±2.43 | A | T |
| 11 | IC-519642 | India | 47.4±0.26 | 41.7±2.37 | 94.77±1.94 | 32.97±1.27 | A | T |
| 12 | IC-438725-1 | India | 33.5±1.26 | 29.6±0.77 | 94.36±0.85 | 34.50±2.42 | A | T |
| 13 | GS-1636 | Pakistan | 16.19±4.63 | 5.55±0.18 | 95.04±1.20 | 34.61±3.62 | A | T |
| 14 | GS-455 | USA/Africa | 15.06±5.75 | 24.94±0.35 | 93.34±1.98 | 32.54±1.98 | A | T |
| 15 | GS-496 | Kenya | 10.67±0.35 | 10.79±1.44 | 94.57±2.46 | 32.27±2.86 | A | T |
| 16 | GS-493 | Turkey | 7.79±0.49 | 8.01±0.32 | 93.29±1.65 | 28.06±2.52 | G | S |
| 17 | GS-457 | USA/Africa | 11.97±1.08 | 12.57±0.74 | 95.1±3.13 | 28.42±2.87 | G | S |
| 18 | GS-459 | USA | 9.60±0.35 | 10.91±0.41 | 93.85±1.43 | 27.34±2.31 | G | S |
| 19 | GS-494 | Kenya | 12.45±0.00 | 14.23±0.62 | 94.38±2.14 | 26.89±2.01 | G | S |
| 20 | GS-1928 | Bangladesh | 5.77±0.96 | 6.79±0.11 | 90.25±2.04 | 25.83±1.92 | G | S |
| 21 | IC-404103 | India | 38.7±0.44 | 58.44±1.39 | 96.31±1.79 | 26.89±1.83 | G | S |
| 22 | GS-1926 | Bangladesh | 5.23±0.18 | 8.12±0.18 | 95.04±2.01 | 27.01±3.22 | G | S |
| 23 | IC-479989 | India | 38.6±1.27 | 60.3±2.36 | 96.7±2.31 | 27.06±3.38 | G | S |
| 24 | IC-403871-A | India | 44.1±1.56 | 78.8±0.69 | 89.16±4.31 | 23.93±3.00 | G | S |
| 25 | IC-479481 | India | 41.1±1.32 | 75.3±1.38 | 94.57±2.46 | 27.28±2.86 | G | S |
| 26 | IC-345014 | India | 38.2±3.28 | 70.3±3.68 | 93.34±1.98 | 27.54±1.98 | G | S |
| 27 | IC-480185 | India | 38.1±1.35 | 76.2±1.06 | 85.16±3.54 | 24.6±2.16 | G | S |
| 28 | IC-403834 | India | 48.8±3.16 | 101.8±0.98 | 88.34±3.05 | 23.53±2.50 | G | S |
| 29 | IC-404133 | India | 37.3±1.28 | 77.9±2.37 | 93.6±1.91 | 25.89±2.17 | G | S |
| 30 | IC-479442 | India | 35.8±3.65 | 81±1.68 | 94.12±2.17 | 27.67±1.87 | G | S |
| 31 | IC-404144 | India | 26.0±0.39 | 59.39±2.64 | 91.5±1.85 | 25.8±2.37 | G | S |
| 32 | IC-479985 | India | 28.6±0.42 | 65.6±1.36 | 92.17±2.14 | 25.21±1.70 | G | S |
| 33 | IC-479479 | India | 34.3±2.4 | 78.9±2.39 | 92.08±1.81 | 23.80±1.53 | G | S |
| 34 | IC-403832 | India | 37.8±2.28 | 89±1.27 | 89±1.74 | 24.86±1.94 | G | S |
| 35 | IC-403739 | India | 37.0±1.49 | 87.4±1.24 | 89.7±3.67 | 25.33±2.86 | G | S |
| 36 | IC-480104 | India | 36.6±1.23 | 87±1.39 | 93.10±2.05 | 25.20±1.70 | G | S |
| 37 | IC-403522-A | India | 38.6 ±1.28 | 93.9±1.69 | 92.2±2.75 | 24.63±2.51 | G | S |
| 38 | IC-403522 | India | 40.2±1.59 | 97.9±1.68 | 91.54±2.54 | 23.73±2.67 | G | S |
| 39 | IC-403717 | India | 42.1±1.45 | 108.3±2.37 | 91.90±1.69 | 24.73±3.63 | G | S |
| 40 | IC-479586 | India | 30.5±0.89 | 79.5±1.6 | 94.8±1.31 | 25.23±1.63 | G | S |
| 41 | IC-479731 | India | 33.7±0.76 | 91.9±1.38 | 91.08±4.00 | 26.46±2.59 | G | S |
| 42 | IC-403521 | India | 31.1±2.56 | 85.2±1.24 | 89.93±3.61 | 21.81±1.53 | G | HS |
| 43 | IC-404178 | India | 31.3±0.76 | 89.09±1.38 | 91.36±2.54 | 21.65±2.35 | G | HS |
| 44 | Lepakshi | India | 31.5±1.26 | 91.5±1.07 | 88.45±1.66 | 17.97±3.01 | G | HS |
| 45 | IC-480117 | India | 29.4±0.92 | 87.4±1.02 | 89.94±4.28 | 18.4±2.74 | G | HS |
Standard deviations (±SD) were calculated from three independent experiments and were statistically significant at P <0.05.
Accession no. refers to the serial number of the accession preserved in NBPGR(IC), Hyderabad and UAS (GS), Bangalore, India.
The values of lipid peroxidation (LP) obtained in the control and after 24 h dehydration stress are expressed as μmol g−1 FW MDA concentration.
The values of the relative water content (RWC) obtained in the control and after 24 h dehydration stress are expressed as the percentage in 20% polyethylene glycol (PEG-6000).
HT, highly tolerant (>30% decline in LP, 0–60% decline in RWC); T, tolerant (<30% decline in LP, 61–66% decline in RWC); S, sensitive (<170 % increase in LP, 70–75% decline in RWC); HS, highly sensitive (>170% increase in LP, >75% decline in RWC).
Estimation of relative water content (RWC)
Leaf RWC was measured in control and stressed seedlings as reported earlier (Lata et al., 2010). Fully expanded leaves were excised and fresh weight (FW) was recorded immediately; then leaves were soaked for 4 h in distilled water at room temperature under a constant light, and the turgid weight (TW) was recorded. Total dry weight (DW) was recorded after drying for 24 h at 80 °C in a hot air oven. RWC was calculated according to the formula given by Barrs and Weatherley (1962): RWC (%)=[(FW–DW)/(TW–DW)]×100.
Estimation of lipid peroxidation (LP)
The LP level in plant tissues was determined by measuring the malondialdehyde (MDA) content via the 2-thiobarbituric acid (TBA) reaction (Hodgson and Raison, 1991). Seedlings (100 mg) were homogenized in 1 ml of 10 mM sodium phosphate buffer (pH 7.4) and centrifuged at 4000 g for 5 min at room temperature. A 200 μl aliquot of the supernatant was added to a reaction mixture containing 100 μl of 8.1% (w/v) SDS, 750 μl of 20% (w/v) acetic acid (pH 3.5), 750 μl of 0.8% (w/v) aqueous TBA, and 200 μl of Milli-Q water. An identical reaction mixture in which 200 μl of supernatant was substituted by an equal volume of buffer was simultaneously set up as a blank. Both reaction mixtures were then incubated at 98 °C for 1 h. After cooling to room temperature the mixtures were centrifuged for 5 min. Absorbance at 535 nm was measured and corrected for non-specific absorbance at 600 nm. The level of LP was expressed as μmol of MDA formed derived from the difference in absorbance at 535 nm and 600 nm using an extinction coefficient of 156 mM−1 cm−1.
Statistical analysis
All experimental data are the means of at least three independent experiments and the results are presented as the mean values ±SD. The significance of differences between mean values of control and each dehydration-stressed samples was statistically determined using one-way analysis of variance (ANOVA) and comparison among means was carried out using the Tukey–Kramer multiple comparisons test. The differences in the effects of PEG-induced dehydration stress on various parameters in 45 foxtail millet accessions were considered statistically significant at P <0.05, P <0.01, and P <0.001.
Stress treatments and total RNA extraction
For time course analysis of SiDREB2 expression, 21-day-old seedlings (Prasad, IC-403579, Lepakshi, IC-480117) were exposed for 0, 0.5,1, 3, 6, 12, 24, or 48 h to the following treatments: dehydration, solutions containing 20% PEG 6000; salinity, saline solutions containing 250 mM NaCl; cold, chilling at 4 °C in a growth chamber; ABA, solutions containing 100 μM ABA; SA, solutions containing 100 μM salicylic acid; methyl jasmonate, solutions containing 100 μM MeJA; and ethylene, solutions containing 100 μM ethephone or water. The seedlings were then transferred to tubes containing fresh, distilled water (Dubouzet et al., 2003; Lata et al., 2010). Total RNA was extracted by the modified hot phenol method using lithium chloride (Longeman et al., 1987). DNA contamination was removed from the RNA samples using RNase-free DNase I (50 U μl−1, Fermentas).
Rapid amplification of 5' cDNA ends (5' RACE)
An EST sequence, FIII E5 (GenBank accession no. GT090998), was chosen from a 6 h PEG-induced forward SSH library, and was used to design three gene-specific reverse PCR primers using Primer 3.0. 5′ RACE was performed using the 5′ RACE kit version 2.0 (Invitrogen) following the manufacturer's instructions. First-strand cDNA was synthesized using GSP (5′-GTGCCGTTACAGAGAAATAACT-3′). The adaptor-ligated first-strand cDNA was used as template for PCR amplification with GSP1 (5′-GAAGCGTTTCCTAACAGCTAGAAC-3′) and NGSP1 (5′- CTAATATGCAAAAAGACTAAATC-3′). A 5 μl aliquot of the RACE reaction product was separated by electrophoresis in a 1.2% (w/v) agarose gel. The band was then excised from the gel using a Qiagen gel extraction kit, cloned into the pGEM-T Easy vector (Promega), and sequenced in both directions.
Cloning of the SiDREB2 gene
The SiDREB2 gene was amplified using the primer pair SiDREB2 ORF_F (5′-ATGAGGAGGAAAAGCACTGG-3′) and NGSP1. PCR thermal cycles were performed as follows: initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 1 min, and 72 °C for 1 min, and then an extension at 72 °C for 10 min. A PCR product of 705 bp was then purified from the gel, cloned into the pGEM-T Easy vector, and sequenced. Similarly, the primer pair SiDREB2 FL_F (5′-CAACTCAGGAAGAAGAGGAT-3′) and SiDREB2 FL_R (5′-ATCTGGCACAAAAGGTAAGA-3′) was used to amplify the entire full-length cDNA of the SiDREB2 gene. A PCR product of 1103 bp was then precipitated by sodium acetate and sequenced.
Sequence analysis and structure prediction
Sequencing of the recombinant plasmids and the PCR amplification products was performed according to Lata et al. (2010). Alignments were carried out by ClustalW with default parameters (Thompson et al., 1994). The phylogenetic tree for the SiDREB2 gene was built using the software program MEGA 4.0 based on protein sequences. The phylogenetic tree was set up with the distance matrix using the Neighbor–Joining (NJ) method with 1000 bootstrap replications. Secondary structure prediction of the SiDREB2 protein was performed using the program PSIPRED (Jones, 1999). The ab intio structure prediction of the protein was done with the help of I-TASSER (Zhang, 2008). Automated homology model building of the DNA-binding domain was performed using the protein structure modelling program MODELLER which models protein tertiary structure by satisfaction of spatial restraints. The input for MODELLER consisted of the aligned sequences of 1gcc and the SiDREB2, a steering file that gives all the necessary commands to the MODELLER to produce a homology model of the target on the basis of its alignment with the template. Energy minimization was performed by the steepest descent followed by the conjugate gradient method using a 20 Å non-bonded cut-off and a constant dielectric of 1.0. Evaluation of the predicted model involved analyses of the geometry and the stereochemistry of the model. The reliability of the model structure was tested using the ENERGY commands of MODELLER (Sali and Blundell, 1993). The modelled structures were also validated using the program PROSA (Wiederstein and Sippl, 2007).
Southern blot analysis
Genomic DNA of foxtail millet was extracted from leaves using the cetyltrimethylammonium bromide (CTAB) method (Saghai-Maroof et al., 1984), digested with PvuII and HindIII (New England Biolabs), fractioned in a 1.0% agarose gel, and blotted on a Hybond N+ membrane (Amersham). The blots were hybridized to a 705 bp SiDREB2 probe radioactively labelled with [α-32P] dCTP using a High Prime DNA labeling kit (Roche, USA). Hybridization was carried out in 0.5 M sodium phosphate (pH 7.2), 7% SDS, and 1 mM EDTA.
Subcellular localization of the SiDREB2 protein
The SiDREB2 gene was fused to the 5' end of the green fluorescent protein (GFP) reporter gene using the pCAMBIA 1302 plant expression vector without a stop codon between the NcoI and SpeI sites. Recombinant DNA constructs encoding the SiDREB2–GFP fusion protein downstream of the cauliflower mosaic virus (CaMV) 35S promoter were introduced into onion epidermal cells by gold particle bombardment using the PDS-1000 system (Bio-Rad) at 1100 psi helium pressure. Onion cells were also transiently transformed with the pCAMBIA 1302-GFP vector as a control. Transformed cells were placed on MS solid medium at 22 °C and incubated for ∼48 h before being examined. The subcellular localization of GFP fusion proteins was visualized with a confocal microscope (TCS_SP2; Leica).
Quantitative real-time PCR (qRT-PCR) analysis
About 2 μg of total RNA was used to synthesize first-strand cDNA primed with oligo(dT) in a 20 μl reaction mix using Protoscript M-MuLV reverse transcriptase (New England Biolabs, USA) following the manufacturer's instructions. qRT-PCR was performed using Power SYBER-Green dye (Applied Biosystems) on a step one real-time PCR system of Applied Biosysytems in triplicate. The constitutive actin gene (CAA33874.1) was used as the endogenous control (Actin RT_F, 5′-CTGACGCCGAGGATATCCA-3′; and Actin RT_R, 5′-GCCTTGACCATACCAGTTCCA-3′). The amount of transcript accumulated for the SiDREB2 gene (SiDREB2 RT_F, 5′-GCCTTGTAGTCATTTGTGGTTT-3′; and SiDREB2 RT_R, 5′-CCTCACAACTCCTTTTCTCAAGCT-3′) normalized to the internal control actin was analysed using the 2-ΔΔCt method (Livak and Schmittgen, 2001). The primers used for real-time PCR analysis were designed by using Primer Express Version 3.0. The PCR cycling conditions were: initial denaturation at 95 °C for 10 min, 95 °C for 15 s, 60 °C for 1 min for 40 cycles, 95 °C for 15 s, and 60 °C for 1 min and 95 °C for 15 s.
SNP assay
A core set of 45 foxtail millet accessions, listed in Table 1, were analysed for SNP discovery by re-sequencing of the SiDREB2 gene. Genomic DNA was extracted as mentioned earlier. The SiDREB2 FL_F/SiDREB2 FL_R primer pair was used to amplify full-length SiDREB2 in all 45 accessions. PCR was performed using Sigma Taq DNA polymerase (1 U) in a 20 μl reaction volume containing ∼50 ng of gemonic DNA, 1× PCR buffer (1.5 mM MgCl2), 10 μM of each dNTP, and 10 pmol of each primer. PCR cycling was done as mentioned earlier. Amplified PCR products were precipitated and sequenced in both directions. Sequences were aligned by ClustalW.
Development of a marker for an allele of SiDREB2
To obtain primers specific to the identified SNP, the segment of the SiDREB2 sequence containing the SNP site was entered into the Primer 3.0 program. Three primers designed corresponding to the SNP, namely DREB.AP2_F, SNP.TS_R, and SNP.Tol_R, were tested using standard PCR amplification as mentioned above. Amplified products were separated on a 1% agarose gel to estimate each allele in the SNP site as the presence or absence of a band.
Results and Discussion
Screening of foxtail millet accessions for dehydration tolerance
In this study, a core set of 45 foxtail millet (S. italica L.) accessions were studied for their dehydration tolerance on the basis of LP and RWC. The total amount of TBA-reacting substances reflects the level of LP resulting from oxidative stress (Guo et al., 2006). The increased accumulation of lipid peroxides upon oxidative stress indicates enhanced production of toxic reactive oxygen species (ROS). ROS are responsible for stress-induced peroxidation of membrane lipids and are often used as an indicator of increased oxidative damage (Smirnoff, 1995). The level of MDA increased significantly in the sensitive accessions during 24 h of dehydration stress with respect to the control (sensitive, <100% increase; highly sensitive, >100% increase; P <0.05), whereas the level of LP in the tolerant accessions declined significantly in comparison with the control (highly tolerant, >30% decline; tolerant, <30% decline; P <0.05) (Table 1). Sreenivasulu et al. (1999) also observed an increase in MDA content in the sensitive foxtail millet cv. Lepakshi as compared with the tolerant cv. Prasad in response to 150 mM salinity stress in 5-day-old seedlings. LP was demonstrated as a function of membrane integrity in chickpea plants exposed to dehydration stress for 7 d and, together with electrolytic leakage, has been shown to be a direct indicator of dehydration stress tolerance (Bhushan et al., 2007). Further, enhanced H2O2 scavenging ability and low levels of LP provide tolerance to various crop plants including pigeonpea, sesame, and alfalfa (Shigeoka et al., 2002; Fazeli et al., 2007; Kumutha et al., 2009).
The choice of RWC as the best representation of plant water status for assessing genetic differences in dehydration tolerance is supported by genetic association between RWC and plant production under dehydration (Blum, 1996; Bhushan et al., 2007). Although having the same water potential, genotypes may vary in their RWC due to the respective difference in osmotic adjustment. Leaf RWC was determined in 45 foxtail millet accessions exposed to 24 h of PEG-induced dehydration stress. Stress treatments significantly decreased the RWC in all accessions (Table 1). The tolerant accessions showed the capacity to maintain a relatively high RWC in comparison with the sensitive accessions during stress (0–60% decline in highly tolerant, 61–66% decline in tolerant; 70–75% decline in sensitive, >75% decline in highly sensitive; P <0.05). The above observations are in accordance with previous reports on several other water-stressed crop plants such as Cenchrus sp., chickpea, groundnut, wheat, sorghum, and maize (Nagy et al., 1995; El Hafid et al., 1998; Madhusudan et al., 2006; Bhushan et al., 2007; Chandra and Dubey, 2010).
Cloning and sequence analysis of the SiDREB2 gene
An EST (GenBank accession no. GT090998) coding for putative DREB2 was isolated from a 6 h SSH library as a differentially expressed sequence in response to PEG-induced dehydration stress. The original 529 bp truncated cDNA clone was complete at its 3' end and aligned to a maize gene encoding a DREB2. Hence, a 5' RACE was carried out to obtain the 5' end of the cDNA. Finally the open reading frame (ORF) of this gene was amplified by RT-PCR and cloned. This gene, designated as SiDREB2 (GenBank ID HQ132744), had a full-length cDNA of 1119 bp, with an ORF of 705 nucleotides. The deduced protein contained 234 amino acids with a predicted molecular mass of 25.72 kDa and a theoretical pI of 5.14. The SiDREB2 protein has a highly conserved AP2/ERF DNA-binding domain of 58 amino acids. It also had two conserved functional amino acids (valine and glutamic acid) at the 14th and 19th residues, respectively, in the DNA-binding domain, thought to be crucial sites for the binding of DREBs and DRE core sequences (Liu et al., 1998). An alkaline N-terminal amino acid region (RRK), which might act as a nuclear localization signal, and a conserved Ser/Thr-rich region adjacent to the AP2/ERF DNA-binding domain were also present (Fig. 1A). The protein contained an acidic C-terminal region which might be functional in trans-activation activity (Stockinger et al., 1997). Most of the positively charged residues are found to be conserved in the N-terminal domain of SiDREB2.
Fig. 1.
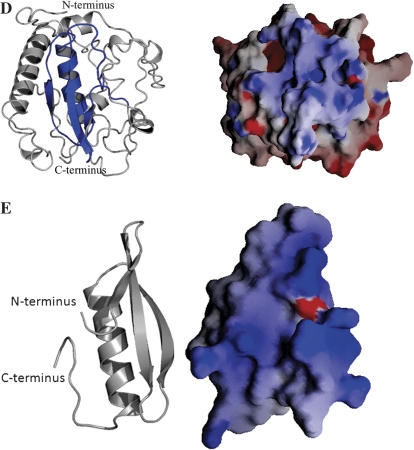
Homologue analysis of the deduced amino acid sequence of the SiDREB2 protein. (A) Sequence analysis of the SiDREB2 gene. The nuclear localization signal (RRK) in the N-terminus is underlined in bold. The serine/threonine-rich region has dashed underlining. Two conserved amino acid residues, valine (V) and glutamic acid (E), in the AP2 domain of the SiDREB2 protein are double underlined. The AP2/ERF conserved domain of SiDREB2 is single underlined. The start (ATG) and stop (TAG) codons are boxed. Full-length primer-binding sites are marked with arrows. (B) Comparison of the deduced amino acid sequence of the AP2/ERF domain of the SiDREB2 protein with other DREB proteins using CLC free workbench version 3.0. The amino acid residues with 100% homology are light in colour while the partially conserved residues are in different colours. The consensus sequence is shown in bold in black at the bottom of the alignment. (C) Phylogenetic analysis of AP2/ERF domains of published AP2/EREBP proteins in the NCBI database. The phylogenetic tree was generated by MEGA 4.0 software. Branch lengths indicate the distance. A-1 to A-6 indicate the groups proposed by Sakuma et al. (2002). SiDREB2 is indicated by a circle. The appended proteins are as follows: Arabidopsis thaliana DREB1 (BAA33434), DREB1B (BAA33435), DREB1C (BAA33436), RAP2.1 (NP564496), RAP2.10 (NP195408), RAP2.3 (NP188299), RAP2.4 (NP177931), RAP2.6 (AAC36019), CBF1 (NP567721), DREB2A (AAU93685), DREB2B (BAA36706), DREB2C (Q8LFR2), ABI4 (AF085279), RAV1 (NP172784), RAV2 (AB013887), ERF1 (AB008103), ERF3 (AB008105), TINY (AAC29139); Oryza sativa OsDREB1A (AAN02486), OsDREB1B (AAX28958), OsDREB2 (AAN02487), OsDREB2B (Q5W6R4), OsDREB2C (Q84ZA1), OsERF3 (NP_001064506), OsDBF1 (AAP56252); Zea mays ZmDREB2A (ACG47772), ZmDBF1 (AAM80486), ZmDBF3 (NP 001105651), Glossy15 (NP_001105890); Triticum aestivum TaDBF (AAL37944); Sorghum bicolor SbDREB2 (ACA79910); Hordeum vulgare HvDBF2 (AAM13419), HvCBF1 (AAX23686), BCBF1 (AAK01088), BCBF3 (AAK01089), HvDRF1.1 (AAO38209), HvDRF1.3 (AAO38211); Festuca arundinacea FeDREB1 (CAG30550); Phyllostachys edulis PeDREB2 (ABY19376); Solanum lycopersicum (formerly Lycopersicon esculentum) LeCBF1 (AAS77820); Nicotiana tabacum NtDREB2 (ACE73694), Tsl1 (AAC14323); Gossypium hirsutum GhDREB1A (AAP83936); Glycine max GmTINY (ACP40513); Catharanthus roseus ORCA1 (CAB93939); Brassica juncea BjDREB1B (ABX00639), Brassica napus BnCBF7 (AAM18959); Thellungiella halophila ThDREB2B (ABV08790). (D) 3D structure of the full-length SiDREB2 protein. The DNA-binding domain which spans from 56 to 113 amino acid residues in the peptide chain is coloured blue. The electrostatic surface potential is shown in a colour gradient from positive (blue) to negative (red). The DNA-binding domain is highly positively charged which complements the negatively charged DNA double helix. (E) The N-terminal DNA-binding domain of SiDREB2 is shown on the left as a cartoon, and on the right the electrostatic surface potential is shown in a colour gradient from positive (blue) to negative (red).
Multiple sequence alignment revealed that SiDREB2 has high similarity to DREB proteins from other plants (Supplementary Fig. S1 available at JXB online). High sequence similarity in the AP2/ERF domain indicated that DREB genes were well conserved among gramineous crops (Fig. 1B). To determine further the relatedness of SiDREB2 to the other AP2/EREBP proteins, a systematic phylogenetic analysis was carried out based on the similarities of AP2 domains in the proteins isolated from Arabidopsis, rice, maize, wheat, sorghum, barley, Festuca, Phyllostachys, tomato, tobacco, cotton, Catharanthus, and Brassica species using MEGA 4.0 by the NJ method (Fig. 1C). SiDREB2 was classified into the A-2 subgroup of the DREB subfamily on the basis of the phylogenetic tree constructed.
Structure of the full-length protein
Knowledge of the three-dimensional (3D) structure of a protein is a necessary prerequisite for understanding its molecular function. To create a 3D structure of SiDREB2, a BLAST search was performed in protein databases for proteins with similar sequence and known 3D structure, but no significant homologous template was found to model the complete sequence. Thus, ab initio modelling of SiDREB2 was done with the help of the I-TASSER server. In brief, this is a hierarchical protein structure modelling approach based on the secondary structure-enhanced Profile–Profile threading alignment (Zhang, 2008). The model structure was validated with PROSA (z-score of –5.58) which was in the range of native conformations of other experimentally determined protein structures of the same size (Wiederstein and Sippl, 2007). The overall structure has an α–β fold and the DNA-binding domain contained antiparallel β-sheets and an α-helix packed approximately parallel to the β-sheet (Fig. 1D). This domain, which is highly positively charged, binds the negatively charged DNA double helix. The tertiary structure is stabilized by 224 intramolecular hydrogen bonds as determined by the program HBPLUS (McDonald and Thornton, 1994). The majority of these hydrogen bonds (44%) are formed by atoms where both the acceptor and donor are from the main chain of the amino acid residues. Only in 25% of the cases are both the acceptor and donor atoms the side chain atoms. The rest (31%) are observed where both main chain and side chain atoms are participating.
Structure of the DNA-binding domain
The structure of the AP2 domain of AtERF1 from Arabidopsis thaliana (PDB code 1gcc) was taken as a template for the comparative modelling of the DNA-binding domain of SiDREB2 (Fig. 1E) (Allen et al., 1998). The sequence alignment between the target and the template is shown in Supplementary Fig. S1 at JXB online. In addition, the secondary structure of this domain as predicted by several secondary structure prediction servers (PHDsec: www.predictprotein.org/; PSIPRED: http://bioinf.cs.ucl.ac.uk/psipred/) is quite similar to the secondary structure of 1gcc (Supplementary Fig. S1, and Fig. 1E). This finding supports the results obtained from several fold recognition servers (FUGUE: www-cryst.bioc.cam.ac.uk/∼fugue/; 3DPSSM: www.sbg.bio.ic.ac.uk/), suggesting that SiDREB2 has the fold of the DNA-binding domain of the DREB superfamily. Based on the above information, the ‘A’ subunit of 1gcc was used as a template to predict the 3D structure of SiDREB2 by applying the comparative modelling strategy as described in the Materials and methods. The overall rmsd (root mean square deviation) between the template and the target structure is 0.2 Å. The calculated angle between the β-sheet and α-helices is ∼172 °. The amino acid residues Arg6, Arg8, Trp10, Glu16, Arg18, Arg25, and Trp27 that are known to make direct contact with the DNA double helix for deciphering the DNA-binding activity are also fully conserved.
Genomic organization of SiDREB2
The copy number of SiDREB2 in the foxtail millet genome was determined by DNA gel blot (Southern analysis). Genomic DNA was digested with PvuII and HindIII restriction enzymes, and transferred to the membrane. After digestion with HindIII, which did not cut this gene, a single hybridization band was detected, whereas with PvuII, which cuts the gene once, two major hybridization bands of high intensity with a few minor bands were detected (Supplementary Fig. S2 at JXB online). These results indicate that this gene has a low copy number in the foxtail millet genome; however, the minor bands indicate the presence of a low number of homologous copies of SiDREB2. Such an observation has also been confirmed in wheat for Wdreb2 (Egawa et al., 2006). In addition, a comparison of the genomic clone with the cDNA clone showed that SiDREB2 is an intron-less gene (Supplementary Fig. S3).
Nuclear localization of SiDREB2 protein
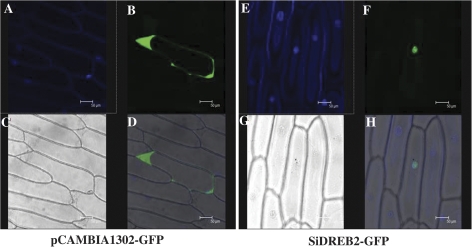
To investigate the subcellular localization of the SiDREB2 protein, the coding region of the SiDREB2 gene was fused in-frame to the GFP gene, and the resulting construct was introduced into onion epidermal cells by particle bombardment. Confocal microscopic observation demonstrated that GFP fluorescence was dispersed throughout the entire cell when bombarded with the control plasmid 35S–GFP. In contrast, the SiDREB2–GFP fusion protein was localized exclusively to the nucleus, indicating that SiDREB2 is a nuclear-localized protein (Fig. 2A–H). The result obtained confirms all previous reports that suggest DREBs to be nuclear localized (Mizuno et al., 2006; Liu et al., 2008; Zhao et al., 2010).
Fig. 2.
Subcellular localization of the SiDREB2 protein in onion epidermal cells. Onion epidermal cells were transiently transformed with constructs containing either control pCAMBIA1302–GFP or SiDREB2–GFP under the control of the CaMV 35S promoter by the particle bombardment method. The subcellular localization of the SiDREB2–GFP fusion protein or control was viewed with a fluorescent confocal microscope ∼48 h after bombardment. DAPI (4',6-diamidino-2-phenylindole) images (A and E), fluorescence images (B and F), bright-field images (C and G), and corresponding overlaid images (D and H) of representative cells expressing pCAMBIA1302-GFP or SiDREB2–GFP fusion protein are shown.
Expression characteristics of SiDREB2 in response to various stresses and hormones
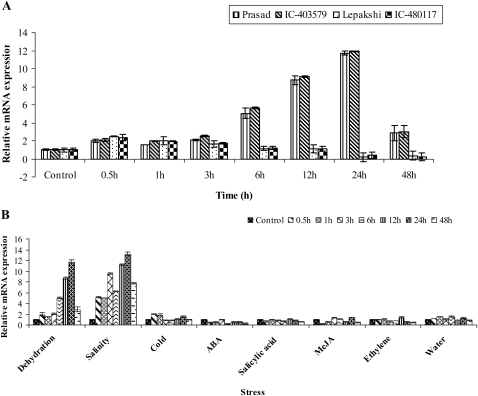
The transcript of SiDREB2 was significantly induced in response to dehydration stress in highly tolerant accessions, namely Prasad and IC-403579, while its expression declined slowly after initial induction in both highly sensitive accessions, namely Lepakshi and IC-480117 (Fig. 3A). The up-regulation of this gene (up to 12-fold) in tolerant accessions suggests that it might impart drought tolerance capacity to the tolerant accessions in comparison with the sensitive ones. These observations were supported by a recent study which suggested that the susceptibility of IR64 was probably attributable to the significant down-regulation of regulatory components including DREBs that confer drought tolerance in rice (Lenka et al., 2010).
Fig. 3.
qRT-PCR analysis of the SiDREB2 gene. (A) Time course expression analysis in two dehydration-tolerant (Prasad and IC-403579) and two sensitive accessions (Lepakshi and IC-480117) of foxtail millet in response to dehydration stress. (B) Time course expression analysis of the SiDREB2 gene under various stresses: 20% PEG 6000; 250 mM NaCl; cold at 4 °C; 100 μM ABA; 100 μM SA; 100 μM MeJA; 100 μM ethephone; and water for 0.5, 1, 3, 6, 12, 24, and 48 h. Bars indicate the standard error (±SE) calculated from three independent experiments.
In response to salinity, a strong gradually increasing induction pattern was observed from 0.5 h to 24 h with a slight decline at 6 h and at 48 h post-stress. However, its expression in other stresses was much less in comparison with the control (Fig. 3B). The strong responsiveness of the SiDREB2 gene to dehydration and salinity implies that its role in the transduction of abiotic stress signals is very similar to those of Arabidopsis and rice, and occurs more or less through an ABA-independent pathway (Liu et al., 1998; Sakuma et al., 2002; Dubouzet et al., 2003).
Tissue-specific expression of SiDREB2
The organ-specific expression of SiDREB2 in foxtail millet was detected by qRT-PCR. SiDREB2 expression can be detected in leaves, roots, and young and mature spikes under normal conditions (Fig. 4). Its transcripts were mainly expressed in leaves followed by roots. This expression pattern indicates that SiDREB2 may function in the normal programme of plant growth and development in this grass species. In Arabidopsis, DREB2 transcript accumulation could also be detected in roots, leaves, and stems, while DREB1 was not expressed under normal growth conditions (Liu et al., 1998). This also indicated that SiDREB2 was more similar to DREB2. The transcript accumulation in young and mature spikes indicated that SiDREB2 may also play an important role in development and procreation. Such an observation has also been reported by Liu et al. (2008).
Fig. 4.
Expression patterns of SiDREB2 in various intact tissues of foxtail millet. Total RNAs were extracted from whole seedling, leaves, roots, young spikelets, and mature spikelets. The results are relative to expression in whole seedlings. Actin was used as endogenous control. Bars indicate the standard error (±SE) calculated from three independent experiments.
Identification and validation of an allele-specific marker (ASM) for the DREB2 locus
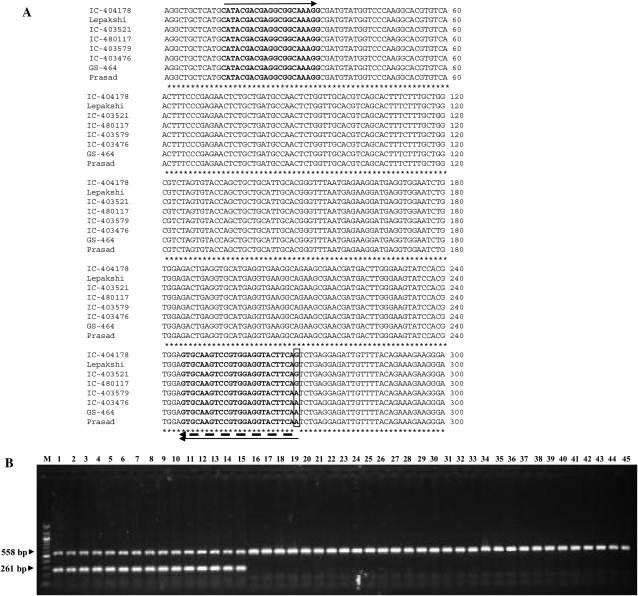
A core set of 45 foxtail millet accessions were screened for their dehydration tolerance on the basis of LP and RWC (Table 1). All the accessions were subjected to 24 h of dehydration stress as compared with the respective controls. The amplified genomic DNA sequence (SiDREB2 FL_F and SiDREB2 FL_R primers) analysis of SiDREB2 from four highly dehydration-tolerant (GS-464, IC-403579, Prasad, and IC-403476) and four highly sensitive (IC-403521, IC-404178, Lepakshi, and IC-480117) accessions revealed the synonymous SNP at base pair 558 (A/G transition) (Fig. 5A). This A/G transition was further validated by sequencing of the SiDREB2 gene with the same primers in an additional 11 and 26 dehydration-tolerant and sensitive accessions, respectively (Table 1). Based on this sequence analysis, locus-specific primers (LSPs) and allele-specific primers (ASPs) were developed (Table 2). PCR products of LSPs (SiDREB2 FL_F and SNP.TS_R) and ASPs (DREB.AP2_F and SNP.Tol_R) in all 45 accessions were generated at the same time and checked by resolving them on a 1% agarose gel independently. Later, the PCR products were mixed and resolved on an agarose gel for both locus-specific markers and ASMs (Fig. 5B). LSPs gave a 558 bp fragment in all the accessions, while ASPs gave a 261 bp fragment in 15 dehydration-tolerant accessions only. Therefore, the dehydration-tolerant accessions were proven to be linked to the ‘A’-specific allele (Table1).
Fig. 5.
SNP (A/G) transition of a single SNP in four dehydration-tolerant and sensitive accessions. (A) ClustalW alignment between partial sequence of SiDREB2 containing the SNP in four dehydration-sensitive (IC-404178, Lepakshi, IC-403521, and IC-480117) and four tolerant (IC-403579, IC-403476, GS-464, and Prasad) foxtail millet accessions. Asterisks denote similar sequences, the box represents the SNP, solid arrows represent allele-specific primers (DREB.AP2_F and SNP.Tol_R), and the dashed arrow represents the locus-specific reverse primer (SNP.TS_R). The locus-specific forward primer is shown as a dashed arrow in Fig. 1A. (B) Agarose gel image of the allele-specific and locus-specific SNP-derived allele-specific marker (ASM) associated with dehydration tolerance. The ASM can distinguish the A allele (tolerant), which generates a 261 bp fragment, from the G allele (sensitive), which generates no PCR fragment. The 558 bp fragment is common to both alleles and it is used as a positive control. The order of samples is the same as in Table 1.
Table 2.
Sequences of primer pairs specific to the A/G single nucleotide polymorphism of the SiDREB2 gene used in PCR and sizes and phenotypes of the PCR products generated by each primer pair
| Allele | Primer sequence (5′–3′) | PCR product size | Phenotype |
| Locus specific | SiDREB2 FL_F: CAACTCAGGAAGAAGAGGAT | 558 bp | Monomorphic |
| SNP.TS_R: TGAAGTACCTCCACGGACTTGCACT | |||
| DREB.AP2_F: ATGCATACGACGAGGCGGCAAAGG | 261 bp | Polymorphic | |
| Allele specific | SNP.Tol_R: TTGAAGTACCTCCACGGACTTGCAC |
The importance of allele-specific PCR systems for reliable SNP typing has been demonstrated several times in previous studies (Wu et al., 1989; Wei et al., 2006). Introduction of additional mismatch bases has improved the specificity of this technique (Drenkard et al., 2000). Its application is especially important for an accurate discrimination of different alleles in MAS. In the present study, only one pair of ASPs was used to amplify the specific products from the dehydration-tolerant and sensitive accessions. Therefore, the ASM can be effectively used to type SNPs as well as to avoid unambiguous false scoring. The functional differences in trait performance mainly caused by SNPs has also been reported in previous studies on several cloned genes (Peng et al., 1999; Takahashi et al., 2001; Liu et al., 2002; Jin et al., 2003; Toshiyuki et al., 2003; Kim et al., 2005; Garcés-Claver et al., 2007; Fan et al., 2009). Molecular marker-assisted breeding technology is a rapid and accurate method for any candidate gene, providing a very effective tool for backcross breeding (Collard and Mackill, 2008). MAS efficiency is influenced by several complex factors such as recombination between the marker and the candidate gene, a low level of polymorphism between the parents with contrasting traits, and lower resolution of quantitative trait loci (QTLs) due to environmental interactions. In the present study, the ASM is part of the candidate gene, thus eliminating the main disadvantage of MAS. However, the ASM developed in this study is a dominant marker, and therefore it cannot distinguish the heterozygotes. The marker has high reliability and efficiency, and the desired PCR product can be identified easily on a simple agarose gel. With the help of this marker, dehydration-tolerant accessions can be selected at a variety of life stages. This is particularly true when the target is the dehydration-tolerant allele in backcross breeding and thus the foxtail millet breeding process can be accelerated. Further, the ASM identified would facilitate allele mining of foxtail millet germplasm resources, thereby leading to identification and utilization of newer alleles in crop improvement.
Conclusions
In summary, the findings from the current study suggest that SiDREB2 is a novel DREB2-like transcription factor of the AP2/EREBP family since the sequence transition of A–G in this gene was associated with the differences in the dehydration tolerance levels among the foxtail millet accessions. It mediates various stress responses and developmental processes in foxtail millet. The structure determination of the SiDREB2 gene and the DNA-binding domain may provide the framework for understanding the functions of DREBs at the molecular level. The higher expression level of SiDREB2 in tolerant accessions in comparison with sensitive accessions could probably be due to some deficiency of a stress-responsive regulatory cis-element in the promoter regions of the sensitive accessions or due to post-transcriptional modification of its gene. However, this remains to be demonstrated before any conclusions can be reached. The ASM developed in this study requires validation before it can be implemented routinely to assist selection for dehydration tolerance in foxtail millet breeding. To the best of our knowledge, this is the first report of molecular cloning, characterization of a DREB2-like gene SiDREB2 in foxtail millet, and development of an ASM for dehydration tolerance. We are also currently exploring the biological functions of SiDREB2 in stress and developmental responses of foxtail millet using overexpression, gene silencing, and promoter analysis.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Sequence alignment of full-length SiDREB2 protein with other DRE-binding transcription factors: Sorghum SbDREB2 (ACA79916), maize ZmDREB2A (ACG47772), Buchloe BdDREB (ABP52086), Cynodon CdDREB1 (AAS46284), Zoysia ZjDREB1 (ACV42429), Festuca FaDREB2A (CAG30547), Poa PpDREB (AAS59530), oat AsDREB2 (ABS11171), barley HvDREB1 (AAY25517), wheat DREB6 (AAX13289), rice OsDREB2A (A2WL19.2), OsDREB2 (NP_001042107), Phyllostachys PeDREB2 (ABY19375), and tomato SlDREB (AAN77051). Identical amino acid residues in the DNA-binding domain are coloured red, strongly similar in green, and weakly similar in blue. Predicted secondary structures, helix (H), sheet (S), and coil (C) are also shown.
Figure S2. Southern gel blot analysis of SiDREB2 cDNA. A 10 μg aliquot of foxtail millet genomic DNA was digested with PvuII and HindIII and fractioned in a 1% agarose gel. The SiDREB2 ORF (705 bp) was used as a probe.
Figure S3. Genome organization of SiDREB2. The 1103 bp genomic clone as well as the cDNA clone was PCR amplified using the primer pair SiDREB2 FL_F and SiDREB2 FL_R, and electrophoresed on a 0.8% agarose gel.
Supplementary Material
Acknowledgments
This study was supported by the Department of Biotechnology (grant no. BT/PR9851/AGR/02/521/2007), the Government of India, New Delhi, and a core grant from the National Institute of Plant Genome Research. CL acknowledges the award of a SRF from the UGC, New Delhi. We are grateful to NBPGR, Hyderabad and UAS, GKVK, Bangalore, India for providing the seed materials, and to Dr N. K. Singh, IARI, and Dr. D. Chattopadhyay, NIPGR, New Delhi for helpful discussions. We would like to thank the reviewers for their constructive comments.
References
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reports. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO Journal. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JR, Lübberstedt T. Functional markers in plants. Trends in Plant Science. 2003;8:554–560. doi: 10.1016/j.tplants.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Barrs HD, Weatherley PE. A re-examination of the relative turgidity technique for estimating water deficit in leaves. Australian Journal of Biological Sciences. 1962;15:413–428. [Google Scholar]
- Bhushan D, Pandey A, Choudhary MK, Datta A, Chakraborty S, Chakraborty N. Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Molecular and Cellular Proteomics. 2007;6:1868–1884. doi: 10.1074/mcp.M700015-MCP200. [DOI] [PubMed] [Google Scholar]
- Blum A. Developing drought and low N-tolerant maize. In: Edmeades GO, Banziger M, Mickelson HR, Pena-Valdivia CB, editors. Proceedings of a Symposium at the International Maize and Wheat Improvement Cente. 1996. (CIMMYT), El-Batan, March 25–29, 1996. El-Batan, Mexico: CIMMYT, 131–135. [Google Scholar]
- Chandra A, Dubey A. Effect of ploidy levels on the activities of Δ-pyrroline-5-carboxylate synthetase, superoxide dismutase and peroxidase in Cenchrus species grown under water stress. Plant Physiology and Biochemistry. 2010;48:27–34. doi: 10.1016/j.plaphy.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Chen M, Wang QY, Cheng XG, Xu ZS, Li LC, Ye XG, Xia LQ, Ma YZ. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochemical and Biophysical Research Communications. 2007;353:299–305. doi: 10.1016/j.bbrc.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Collard BC, Mackill DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Wang Z, Beales CJ, Sasaki T, Gale MD. Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa) Theoretical and Applied Genetics. 1998;96:63–68. [Google Scholar]
- Doust AN, Kellogg EA, Devos KM, Bennetzen JL. Foxtail millet: a sequence-driven grass model system. Plant Physiology. 2009;149:137–141. doi: 10.1104/pp.108.129627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E, Richter BG, Rozen S, et al. A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiology. 2000;124:1483–1492. doi: 10.1104/pp.124.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Egawa C, Kobayashi F, Ishibashi M, Nakamura T, Nakamura C, Shiego T. Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes and Genetic Systems. 2006;81:77–91. doi: 10.1266/ggs.81.77. [DOI] [PubMed] [Google Scholar]
- El Hafid R, Smith DH, Karrou M, Samir K. Physiological attributes associated with early-season drought resistance in spring durum wheat cultivars. Canadian Journal of Plant Science. 1998;78:227–237. [Google Scholar]
- Fan C, Yu S, Wang C, Xing Y. A causal C–A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theoretical and Applied Genetics. 2009;118:465–472. doi: 10.1007/s00122-008-0913-1. [DOI] [PubMed] [Google Scholar]
- Fazeli F, Ghorbanli M, Niknam V. Effect of drought on biomass, protein content, lipid peroxidation and antioxidant enzymes in two sesame cultivars. Biologia Plantarum. 2007;51:98–107. [Google Scholar]
- Garcés-Claver A, Fellman SM, Gil-Ortega R, Jahn M, Arnedo-Andrés MS. Identification, validation and survey of a single nucleotide polymorphism (SNP) associated with pungency in Capsicum spp. Theoretical and Applied Genetics. 2007;115:907–916. doi: 10.1007/s00122-007-0617-y. [DOI] [PubMed] [Google Scholar]
- Gu SL, Liu J, Ren HR, et al. Foxtail millet cultivation in China. (In Chinese) Bejing: China Agricultural Press; 1987. The relationship between foxtail millet and its environment. In: Shanxi Academy of Agricultural Sciences, ed; pp. 64–65. [Google Scholar]
- Guo Z, Ou W, Lu S, Zhong Q. Differential responses of antioxidative system to chilling and drought in four rice cultivars differing in sensitivity. Plant Physiology and Biochemistry. 2006;44:828–836. doi: 10.1016/j.plaphy.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Hodgson RAJ, Raison JK. Lipid peroxidation and superoxide dismutase activity in relation to photoinhibition induced by chilling in moderate light. Planta. 1991;185:215–219. doi: 10.1007/BF00194063. [DOI] [PubMed] [Google Scholar]
- Jayaraman A, Puranik S, Rai NK, Vidapu S, Sahu PP, Lata C, Prasad M. cDNA-AFLP analysis reveals differential gene expression in response to salt stress in foxtail millet (Setaria italica L.) Molecular Biotechnology. 2008;40:241–251. doi: 10.1007/s12033-008-9081-4. [DOI] [PubMed] [Google Scholar]
- Jin Q, Waters D, Cordeiro GM, Henry RJ, Reinke RF. A single nucleotide polymorphism (SNP) marker linked to the fragrance gene in rice (Oryza sativa L.) Plant Science. 2003;165:359–364. [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kim MY, Van K, Lestari P, Moon JK, Lee SH. SNP identification and SNAP marker development for a GmNARK gene controlling supernodulation in soybean. Theoretical and Applied Genetics. 2005;110:1003–1010. doi: 10.1007/s00122-004-1887-2. [DOI] [PubMed] [Google Scholar]
- Kumutha D, Ezhilmathi K, Sairam RK, Srivastava GC, Deshmukh PS, Meena RC. Waterlogging induced oxidative stress and antioxidant activity in pigeonpea genotypes. Biologia Plantarum. 2009;53:75–84. [Google Scholar]
- Lata C, Sahu PP, Prasad M. Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochemical and Biophysical Research Communications. 2010;393:720–727. doi: 10.1016/j.bbrc.2010.02.068. [DOI] [PubMed] [Google Scholar]
- Lenka SK, Katiyar A, Chinnusamy V, Bansal KC. Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnology Journal. 2010 doi: 10.1111/j.1467-7652.2010.00560.x. (in press) [DOI] [PubMed] [Google Scholar]
- Li YM. Drought-resistant mechanism and genetic expression of foxtail millet. In: Li YM, editor. Foxtail millet breeding. Beijing: China Agricultural Press; 1987. pp. 433–434. [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proceedings of the National Academy of Sciences, USA. 2002;99:13302–13306. doi: 10.1073/pnas.162485999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhu K, Yang Y, Wu J, Chen F, Yu D. Molecular cloning, expression profiling and trans-activation property studies of a DREB2-like gene from chrysanthemum (Dendranthema vestitum) Journal of Plant Research. 2008;121:215–226. doi: 10.1007/s10265-007-0140-x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Longeman J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Analytical Biochemistry. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Madhusudan KV, Giridarakumar S, Ranganayakulu GS, Reddy PC, Sudhakar C. Effect of water stress on some physiological responses in two groundnut (Arachis hypogea L.) cultivars with contrasting drought tolerance. Journal of Plant Biology. 2002;29:199–202. [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. Journal of Molecular Biology. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Mizuno S, Hirasawa Y, Sonoda M, Nakagawa H, Sato T. Isolation and characterization of three DREB/ERF-type transcription factors from melon (Cucumis melo) Plant Science. 2006;170:1156–1163. [Google Scholar]
- Nagy Z, Tuba Z, Zsoldos F, Erdei L. CO2 exchange and water relation responses of sorghum and maize during water stress. Journal of Plant Physiology. 1995;145:539–544. [Google Scholar]
- Peng J, Richards DE, Hartley NM, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Qin F, Kakimoto M, Sakuma Y, et al. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. The Plant Journal. 2007;50:54–69. doi: 10.1111/j.1365-313X.2007.03034.x. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factor: genome wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Biyaschev RM, Yang GP, Zhang Q, Allard RW. Extraordinary polymorphism microsatellite DNA in barley: species diversity, chromosomal location and population dynamics. Proceedings of the National Academy of Sciences, USA. 1984;91:5466–5470. doi: 10.1073/pnas.91.12.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding-specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A involved in drought-responsive gene expression. The Plant Cell. 2006a;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress responsive and heat-stress-responsive gene expression. Proceedings of the National Academy of Sciences, USA. 2006b;103:18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell T. Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, et al. Regulation and function of ascorbate peroxidase isoenzymes. Journal of Experimental Botany. 2002;53:1305–1319. [PubMed] [Google Scholar]
- Smirnoff N. Antioxidant systems and plant response to the environment. In: Smirnoff N, editor. Environment and plant metabolism. Oxford: BIOS Scientific Publishers; 1995. pp. 217–243. [Google Scholar]
- Sreenivasulu N, Ramanjulu S, Ramachandra-Kini K, et al. Total peroxidase activity and peroxidase isoforms as modified by salt stress in two cultivars of foxtail millet with differential salt tolerance. Plant Science. 1999;141:1–9. [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proceedings of the National Academy of Sciences, USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Shomura A, Sasaki T, Yano M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the subunit of protein kinase CK2. Proceedings of the National Academy of Sciences, USA. 2001;98:7922–7927. doi: 10.1073/pnas.111136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshiyuki K, Shizo O, Nobuhiko M, et al. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.) The Plant Journal. 2003;37:315–325. doi: 10.1046/j.1365-313x.2003.01961.x. [DOI] [PubMed] [Google Scholar]
- Wei B, Jing RL, Wang CS, Chang XP. Assaying single nucleotide polymorphism in wheat (Triticum aestivum L.) with allele-specific PCR. Scientia Agricola Sinica. 2006;39:1313–1320. [Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research. 2007;35:407–410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Ugozzoli L, Pal BK, Wallace RB. Allele-specific enzymatic amplification of β- globin genomic DNA for diagnosis of sickle cell anemia. Proceedings of the National Academy of Sciences, USA. 1989;86:2757–2760. doi: 10.1073/pnas.86.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends in Plant Science. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu T, Fu J, Zhu Y, et al. Construction and application of EST library from Setaria italica in response to dehydration stress. Genomics. 2007;90:121–131. doi: 10.1016/j.ygeno.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;23:9–40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Hu Y, Chong K, Wang T. ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice. Annals of Botany. 2010;105:401–409. doi: 10.1093/aob/mcp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.