Abstract
Cold storage of tubers of potato (Solanum tuberosum L.) compromises tuber quality in many cultivars by the accumulation of hexose sugars in a process called cold-induced sweetening. This is caused by the breakdown of starch to sucrose, which is cleaved to glucose and fructose by vacuolar acid invertase. During processing of affected tubers, the high temperatures involved in baking and frying cause the Maillard reaction between reducing sugars and free amino acids, resulting in the accumulation of acrylamide. cDNA clones with deduced proteins homologous to known invertase inhibitors were isolated and the two most abundant forms, termed INH1 and INH2, were shown to possess apoplastic and vacuolar localization, respectively. The INH2 gene showed developmentally regulated alternative splicing, so, in addition to the INH2α transcript encoding the full-length protein, two hybrid mRNAs (INH2β*A and INH2β*B) that encoded deduced vacuolar invertase inhibitors with divergent C-termini were detected, the result of mRNA splicing of an upstream region of INH2 to a downstream region of INH1. Hybrid RNAs are common in animals, where they may add to the diversity of the proteome, but are rarely described in plants. During cold storage, INH2α and the hybrid INH2β mRNAs accumulated to higher abundance in cultivars resistant to cold-induced sweetening than in susceptible cultivars. Increased amounts of invertase inhibitor may contribute to the suppression of acid invertase activity and prevent cleavage of sucrose. Evidence for increased RNA splicing activity was detected in several resistant lines, a mechanism that in some circumstances may generate a range of proteins with additional functional capacity to aid adaptability.
Keywords: Cold-induced sweetening, cold stress, hybrid RNA, invertase inhibitor, RNA splicing, Solanum tuberosum
Introduction
Due to their sessile nature, plants must adapt their metabolism to cope with abiotic stresses such as drought, high salinity, and low temperature. These three environmental stresses are related to some extent, and there are genes expressed in response to all three challenges as well as those specific to a particular stress (Kreps et al., 2002; Seki et al., 2002; Rabbani et al., 2003). Some of these stress-responsive genes encode regulatory proteins, whereas others protect cells by causing the accumulation of metabolic proteins and cellular protectants including sugars (Yamaguchi-Shinozaki and Shinozaki, 2006). These stress-induced responses enable the plant to adapt its physiology and survive.
Potato is now the third most important food crop in the world, and the most important non-grain food crop (FAOSTAT, 2007). Metabolic stability of potato tubers during long-term cold storage is one of the prime trait targets for breeding programmes worldwide. Potatoes need to be cool-stored throughout the year to maintain uninterrupted supply to industry, and storage at low temperature (<8 °C) is beneficial because it reduces bacterial soft rots, decreases water and dry matter loss, and prevents sprouting without the need to add sprout inhibitor chemicals (Sowokinos, 2001). However, in many cultivars, low temperature storage causes glucose and fructose to accumulate at substantial levels (Sowokinos, 2001; Blenkinsop et al., 2004). This is termed cold-induced sweetening, and is one of the most serious problems facing the processing industry. When affected potatoes are processed into crisps or chips, the high temperature associated with frying or baking causes these reducing sugars to react with free amino acids by the Maillard reaction, resulting in an unacceptable blackening and unpalatability of the product (Kumar et al., 2004) and accumulation of the potent neurotoxin and probable carcinogen acrylamide (Mottram et al., 2002; Stadler et al., 2002). Variability in resistance to cold-induced sweetening between cultivars is presumably due to a composite of differences in mRNA expression patterns and post-transcriptional events.
Cold-induced sweetening occurs due to an imbalance between the rate of breakdown of starch and metabolism of the resulting sucrose, some of which enters the vacuole where it can be irreversibly cleaved by acid invertase to the hexose sugars glucose and fructose (Sowokinos, 2001; Blenkinsop et al., 2004). The pathway from starch breakdown to hexose sugars is complex (Malone et al., 2006), and several enzymes are potentially involved. Cold storage has been found to cause increases in mRNA abundance or enzyme activity of forms of β-amylase, UDP-glucose pyrophosphorylase, sucrose phosphate synthase, and acid invertase (Zrenner et al., 1993; Hill et al., 1996; Nielsen et al., 1997; Reimholz et al., 1997; McKenzie et al., 2005). Transgenic manipulations have also suggested roles in cold-induced hexose accumulation for UDP-glucose pyrophosphorylase (Borovkov et al., 1996), sucrose phosphate synthase (Krause et al., 1998), sucrose phosphatase (Chen et al., 2008), glucan-water dikinase, and phosphorylase-L (Rommens et al., 2006). However, the activity of acid invertase is the most significant factor in determining the accumulation of hexose sugars, and correlations between acid invertase activity and the hexose:sucrose ratio have been made in cultivars exhibiting varying resistance to cold-induced sweetening (Richardson et al., 1990; Zrenner et al., 1996; Matsuura-Endo et al., 2004; McKenzie et al., 2005). Indeed, transgenic suppression of acid invertase prevents hexose accumulation and cold-induced sweetening of stored tubers (Zrenner et al., 1996; Bhaskar et al., 2010).
Invertase activity may be controlled post-translationally in planta by invertase inhibitors, which have long been known to be present in potato tubers (Schwimmer et al., 1961; Pressey, 1966, 1967; Pressey and Shaw, 1966). Storage of susceptible potato tubers results in a cold-induced increase in acid invertase activity over ∼3 weeks, followed by a decline that appears to be due to accumulation of the invertase inhibitor protein (Pressey and Shaw, 1966). Overexpression of a tobacco (Nicotiana tabacum L.) vacuolar invertase inhibitor in transgenic potato tubers strongly reduced acid invertase activity and the formation of reducing sugars (Greiner et al., 1999). These observations suggest that endogenous potato invertase inhibitors, if present in particular cultivars, could potentially be important in determining the extent of cold-induced sweetening.
Invertase inhibitors are small (∼17 kDa) proteins that bind to invertases, forming an inactive complex (Pressey, 1967; Bracho and Whitaker, 1990a, b; Rausch and Greiner, 2004). Invertase inhibitors are closely related to pectin methylesterase inhibitor proteins (Hothorn et al., 2004a, b) and are of two types, found in the apoplast and vacuole, respectively. The sequences of invertase inhibitors have been determined from several species including tobacco, Arabidopsis, soybean, sweetpotato, maize, rice, and tomato (Greiner et al., 1998, 1999; Bate et al., 2004; Link et al., 2004; Rausch and Greiner, 2004; Reca et al., 2008; Jin et al., 2009), but not so far from potato. The aim of this work was to characterize the invertase inhibitor genes of potato, and to examine the expression of the members of this gene family both in a range of tissues and in the mature tuber during cold stress. Invertase inhibitor mRNA accumulation was assessed in a range of lines to determine if invertase inhibitor gene expression correlates with resistance to cold-induced sweetening.
Materials and methods
Plant material
Flowers were collected from greenhouse-grown plants and separated into sepal, petal, stamen, and gynoecium. Leaves and stems were collected from young plants. Tubers were harvested from immature developing plants and staged by size, then peeled and central tissues collected. All tissues were immediately frozen in liquid nitrogen and stored at –80 °C until required. For cold storage studies, mature tubers of breeding lines 1021/1 (resistant to cold-induced sweetening), 937/3 (sensitive to cold-induced sweetening), and selected lines from a mapping population were harvested from field-grown plants after the above-ground vegetative organs had died, and stored in the dark at ambient temperature to equilibrate for 1 week. After this time (T0), tubers were randomly sorted into groups and stored for 2 or 4 weeks at either 4 °C to develop cold-induced sweetening or at 10 °C as a control. At the end of the storage period, tubers were peeled and central regions chopped into cubes of ∼5 mm then snap-frozen in liquid nitrogen and stored at –80 °C until required.
cDNA cloning and sequence analysis
Total RNA was prepared using the hot borate method (Wan and Wilkins, 1994) from tubers of 937/3 and 1021/1 that had been stored for 2 weeks at 4 °C as above. mRNA was selected using a Dynabeads mRNA purification kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. A cDNA library for each cultivar was constructed using 5 μg of poly(A)+ RNA and a ZAP-cDNA synthesis kit (Stratagene, La Jolla, CA, USA) following the manufacturer's instructions. To generate a template for probe synthesis, degenerate primers INHdF and INHdR (Supplementary Table S1 available at JXB online) were designed to conserved amino acid domains in tomato and tobacco invertase inhibitors, and used to PCR-amplify a band of ∼300 bp from potato tuber cDNA, which was ligated into pBluescript and some clones sequenced. A clone with homology to known invertase inhibitors was used as a template for preparing a labelled probe by the random prime method (Feinberg and Vogelstein, 1983). The labelled, purified probe was used to screen both cDNA libraries at moderate temperature (57 °C) according to the manufacturer's instructions. Positive plaques were subjected to a second round of purification followed by in vivo excision and sequencing. Sequences were aligned and compared with existing invertase inhibitors using CLUSTALW2 (Larkin et al., 2007), and the major forms designated INH1 and INH2. The unusual C-terminus of the deduced protein of INH2 did not show conservation with a similar sequence from tobacco (GenBank accession no. Y12806), so the downstream region of the cDNA was further analysed by 3′-RACE (rapid amplification of cDNA ends) using nested primers INH2-O and INH2-I (Supplementary Table S1), oligo d(T)17 primer, and potato tuber cDNA as a template. This confirmed the sequence originally found (termed INH2β) and led to the discovery of an additional form, termed INH2α. Full-length clones of INH2α and INH2β were amplified from cDNA or genomic DNA as below. Signal peptide prediction was carried out using SignalP 3.0 (http://www.cbs.dtu.dk./services/SignalP/).
To examine the diversity of INH2β forms present in cDNA, primers were designed to the non-coding flanking regions of the cDNA for INH2β (sense primer INH2F and antisense primer INH2R2; Supplementary Table S1). cDNA was synthesized from tuber RNA of both 937/3 and 1021/1 using SuperScript reverse transcriptase (Invitrogen) and oligo d(T)17 according to the manufacturer's instructions. PCR amplification using the primer pair, cDNA template, and PCR Extender proofreading Taq DNA polymerase (5 Prime Co., Gaithersburg, MD, USA) resulted in a band of ∼800 bp, which was ligated into pBluescript and six clones sequenced for each cultivar.
Genomic characterization
Genomic DNA was prepared from young leaves of cultivars 937/3 and 1021/1 using a urea method. For investigation of the allelic diversity of INH1 and INH2, primers were designed to the non-coding flanking regions of the cDNA for INH1 (sense primer INH1F2 and antisense primer INH1R4) and for INH2 (sense primer INH2F and antisense primer INH2R; Supplementary Table S1). Genomic DNA was PCR-amplified using the above primer pairs and Pwo (Roche, Auckland, New Zealand), TripleMaster (Eppendorff, Hamburg, Germany), or HiFidelity (Qiagen, Valencia, CA, USA) proofreading DNA polymerase. Two clones (for INH1) or at least 12 clones per genotype (for INH2) were sequenced on both strands.
For DNA gel blot analysis, aliquots of genomic DNA of the tetraploid cultivar 1021/1 and the diploid cultivar 3T were exhaustively digested with the indicated restriction enzymes, and either 20 μg (1021/1) or 12 μg (3T) of DNA was loaded per lane onto a 1% agarose gel. After separation of the DNA fragments by electrophoresis, the gel was blotted to Hybond-XL membrane (Amersham, Auckland, New Zealand) overnight. Short probe templates (∼150 bp) were prepared from flanking non-coding or divergent coding regions that had previously been amplified by PCR from plasmids (for INH1, sense primer INH1F2 and antisense primer INH1R2; for INH2, sense primer INH2F4 and antisense primer INH2R4). These templates were labelled by PCR as previously described (Brummell et al., 1999), using the antisense primer, [α-32P]dATP, unlabelled nucleotides (dCTP, dGTP, dTTP), and Taq polymerase with 10 cycles of 94 °C for 1 min, 50 °C for 1 min, and 72 °C for 1 min. The denatured probes were hybridized with the gel blot in Church and Gilbert (1984) solution at 65 °C overnight, then washed several times in 0.5× SSC/0.1% SDS at 65 °C and exposed to Kodak (Rochester, NY, USA) Biomax-MS film.
Subcellular protein localization using green fluorescent protein (GFP) fusions
For INH1, primers INH1GFPF and INH1GFPR (Supplementary Table S1) were used to PCR-amplify a fragment encoding the N-terminal 32 amino acids, and for INH2 primers INH2GFPF and INH2GFPR were used to amplify a fragment encoding the N-terminal 52 amino acids. In each case, an EcoRI site upstream of the translation start codon and a downstream BamHI site were used to ligate the fragment into the CaMV35S-GFP expression vector. The in-frame BamHI site added a glycine and a serine residue to the predicted protein between the INH fragment and the GFP. Plasmid DNA (1 μg) of the 35S:INH1Nterm:GFP:ocs and 35S:INH2Nterm:GFP:ocs constructs or 35S:GFP:ocs alone (as a control) was mixed with 1 μm gold particles and bombarded into the adaxial epidermis of bulb of onion (Allium cepa) or white petals of lisianthus (Eustoma grandiflorum) as described (Shang et al., 2007). Tissues were incubated on hormone-free Murashige and Skoog medium at 25 °C in the dark for 36 h, and transformed cells were examined under a Leica laser scanning confocal fluorescence microscope or an Olympus fluorescence microscope.
mRNA analysis
Total RNA prepared as above (10 μg per lane) was separated by electophoresis in 1.2% agarose denaturing formaldehyde gels, then blotted to Hybond-XL membrane (Amersham) overnight. DNA templates and labelled probes were produced as for genomic DNA gel blot analysis, and hybridized with the RNA gel blots in Church and Gilbert (1984) solution at 65 °C overnight. Blots were washed in 0.5× SSC/0.1% SDS at 65 °C and exposed to Kodak Biomax-MS film.
For quantitative real-time PCR analysis, DNA was eliminated from RNA samples by treatment with DNase I (Invitrogen). Aliquots of RNA (1 μg) were converted to cDNA using oligo d(T)20, dNTPs, and SuperScript III reverse transcriptase (Invitrogen). Products were diluted 25-fold in water prior to use and aliquots of 2.5 μl used per reaction (10 μl final volume) in the presence of 0.5 μM clone-specific primers (Supplementary Table S1) and Light Cycler 480 SYBR Green I master mix (Roche). Primer pairs used were: INH1, INH1RTF+INH1RTR; INH2α, INH2RTF+INH2αRTR; INH2β, INH2RTF+INH2βRTR; and actin, ActRTF+ActRTR (Supplementary Table S1). Real-time PCR was performed in triplicate on a Rotor-Gene RG-3000 (Corbett Research, Sydney, Australia) following the manufacturer's instructions. Experiments were repeated using a minimum of two independent RNA preparations. Data acquisition and analysis were carried out using Rotor-Gene Series 6000 software version 1.7 (Corbett Research). Actin was used as the reference gene for calculation of relative transcript abundance, using the Relative Quantitation feature of the software. Melt analysis and visualization of the final products on agarose gels were used to ensure that products were a single band.
Sugar analysis
Frozen potato tissue was powdered in liquid nitrogen and sugars were extracted with 62.5% (v/v) methanol at 55 °C for 1 h. Sucrose, fructose, and glucose concentrations in the clarified extract were estimated against standards using a Sucrose, D-Fructose and D-Glucose assay kit (Megazyme, Bray, Ireland) according to the manufacturer's instructions.
Recombinant protein expression
Synthetic genes encoding INH2α*A1, INH2β*A1, and INH2β*B1, in each case harbouring an N-terminal tobacco etch virus (TEV) protease cleavage site and flanked by Gateway recombination sequences, were synthesized by GenScript (Piscataway, NJ, USA). For all three proteins, the encoded sequence began with the tyrosine residue at amino acid 27, since this appeared to be the mature protein based on structural studies (Hothorn et al., 2004a). Genes were ligated into the pDEST566 vector (Addgene plasmid 11517, www.addgene.org) by Gateway recombination, to add an N-terminal His6-maltose-binding protein (MBP) domain. Plasmids were transformed into Escherichia coli Rosetta Gami 2 cells (Novagen, San Diego, CA, USA) and grown at 37 °C in Terrific Broth until they reached a density of A600=0.6, then induced by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG), and grown at 20 °C for 18 h. Harvested cells were resuspended in buffer containing 20 mM TRIS-HCl, pH 8.0, 300 mM NaCl, and antiprotease cocktail (Roche), and lysed using a C5 emulsifier (Avestin, Mannheim, Germany). The supernatant after centrifugation at 20 000 g for 30 min was loaded on a His-Trap HP column (GE Healthcare, Piscataway, NJ, USA), washed with 20 mM TRIS, pH 8.0, 300 mM NaCl, and fusion proteins were eluted with buffer containing 20 mM TRIS, pH 8.0, 150 mM NaCl, 300 mM imidazole. Fractions containing the fusion protein were pooled, and dialysed against 20 mM TRIS, pH 8.0, 50 mM NaCl at 4 °C for 18 h. The fusion proteins were then further purified by anion exchange on a HiTrap Q HP column (GE Healthcare), using a 50–500 mM NaCl linear elution gradient, and size exclusion chromatography on a Superdex 75pg HiLoad 16/60 column (GE Healthcare) using 20 mM TRIS, pH 8.0, 150 mM NaCl as eluant. In all cases protein was collected as soluble aggregates in the void volume of the column. The control was produced by TEV cleavage of a His-MBP-α/β hydrolase (from Arabidopsis) fusion protein and purification of the His-MBP domain as above. Protein concentrations were calculated from absorption at 280 nm, using extinction coefficients calculated from their respective amino acid sequences. For SDS–PAGE analysis, samples were denatured for 30 min at 37 °C with either SDS loading buffer or SDS loading buffer containing 6 M urea.
Invertase activity
Potato samples frozen in liquid nitrogen were powdered using a mortar and pestle in 3 vols of buffer containing 50 mM HEPES-KOH, pH 7.5, 15 mM MgCl2, 2 mM EDTA, 2 mM dithiothreitol, 10% (v/v) glycerol, and 2% (w/v) polyvinylpolypyrrolidone. Crude extracts were centrifuged at 14 000 g and the supernatants desalted using Sephadex G-25 columns according to the manufacturer's instructions. The extracts were shaken for 90 min rapidly enough to generate foaming, to minimize the activity of endogenous invertase inhibitor proteins (Pressey, 1966). Enzyme assays contained 60 μl of protein extract, 60 mM phosphate-citrate buffer, pH 5.0, and 40 mM sucrose. Reactions were carried out at 30 °C for 30 min, and were stopped by heating to 94 °C for 1 min. Control reactions for each sample were performed by heating the reaction aliquots of protein extract to 94 °C for 1 min before addition of substrate and buffer. Glucose formed was determined against a standard curve using Sumner's reagent (Sumner, 1921). Protein was determined using a protein assay kit (Bio-Rad, Hercules, CA, USA) with bovine serum albumin as a standard. Activity was expressed as nmol of glucose formed h−1 mg−1 protein.
In experiments where the activity of invertase inhibitors was examined, aliquots of crude protein extract from cold-stored tubers of cultivar Summer Delight prepared as above were incubated with dilutions of purified recombinant invertase inhibitor fusion proteins at 37 °C for 30 min to allow binding, prior to assay for invertase activity.
Results
Characterization of INH1 and INH2 genes and mRNAs
Library screening and PCR analysis identified several cDNAs with deduced proteins showing homology to known invertase inhibitors (Table 1). All deduced proteins possessed four conserved cysteine residues believed to be important in invertase inhibitor protein three-dimensional structure, but were of two distinct types. The first type had deduced proteins with a predicted signal peptide of 19 amino acids and a putative apoplastic localization (Fig. 1A). Two closely related cDNAs were isolated, probably representing allelic forms of the same gene. This gene was termed INH1, and the allelic forms INH1*A and INH1*B. INH1*A and INH1*B deduced proteins differed in only six amino acids, and both were highly homologous to invertase inhibitors from Nicotiana tabacum (NtINH1, GenBank accession no. Y12805) and Solanum lycopersicum (SlINH, AJ010943) (Fig. 1A). Variants of SlINH have been termed SolyCIF (Reca et al., 2008) and SlINHINV1 (Jin et al., 2009). Apoplastic invertase inhibitors do not appear to be glycosylated (Weil et al., 1994; Reca et al., 2008), and INH1*A and B lacked sites for N-glycosylation. Sequencing of two potato genomic clones found that both were INH1*B, and that the gene possessed an intron of 1242 bp located after nucleotide +297 (GenBank accession no. GU980592). The location of the intron was almost identical to that in the orthologous tomato gene SolyCIF (Reca et al., 2008).
Table 1.
INH forms isolated from potato genotypes 1021/1 and 937/3 using a combination of cDNA library screening and PCR amplification from genomic DNA and cDNA.
| Name | Introns | Deduced amino acids | Sites for N-glycosylation | Putative localization | Accession no. |
| INH1*A | 1 | 171 | 0 | Apoplastic | AY864819 |
| INH1*B | 1 | 171 | 0 | Apoplastic | AY864820 |
| INH2α*A1 | 0 | 181 | 1 | Vacuolar | FJ810205 |
| INH2α*A2 | 0 | 181 | 1 | Vacuolar | FJ810206 |
| INH2α*B | 0 | 180 | 1 | Vacuolar | FJ810207 |
| INH2α*C | 0 | 178 | 1 | Vacuolar | FJ810208 |
| INH2α*D | 0 | 178 | 1 | Vacuolar | FJ810209 |
| INH2β*A1 | Cryptic? | 153 | 0 | Vacuolar | AY864821 |
| INH2β*A2 | Cryptic? | 153 | 0 | Vacuolar | GU980593 |
| INH2β*B1 | Cryptic? | 158 | 1 | Vacuolar | GU980594 |
| INH2β*B2 | Cryptic? | 158 | 1 | Vacuolar | GU980595 |
INH2 produced multiple transcripts from the same gene, with INH2α representing allelic versions of mRNAs encoding the full-length protein, and INH2β*A and B representing spliced, hybrid mRNAs encoding deduced proteins that are truncated and altered at the C-terminus.
Fig. 1.
Deduced amino acid sequences of StINH1 and StINH2α. (A) Two allelic forms of StINH1 aligned with NtINH1 from tobacco (GenBank accession no. Y12805) and SlINH from tomato (AJ010943). Predicted signal sequences are shown in lower case. (B) Four allelic forms of StINH2α aligned with NtINH-h from tobacco (Y12806). Potential sites for N-glycosylation are underlined. Four cysteine residues believed to be important in the formation of two disulphide bridges (conserved in both apoplastic and vacuolar invertase inhibitors) are marked with crosses. Amino acid identity is represented by asterisks, highly conservative changes by double dots, and conservative changes by dots.
The second type of invertase inhibitor lacked a predicted signal peptide, and existed in several forms differing at the C-terminus of the deduced proteins and in the 3'-untranslated regions (UTRs). These were termed INH2α (encoding the full-length protein) and INH2β (encoding truncated and altered variants of the INH2 protein). Five alleles of INH2α were identified from sequencing of cDNA and genomic clones amplified from both potato cultivars (937/3 and 1021/1), although not all alleles were present in each cultivar. The alleles possessed four different deduced amino acid sequences, which were designated INH2α*A–D, all of which showed homology throughout their length to a putative vacuolar invertase inhibitor from tobacco (Fig. 1B). INH2α*A–D differed from each other predominantly in the number of amino acids present in a run of consecutive asparagine residues near the N-terminus of the protein. INH2α*C and D also differed from each other by three amino acid residues elsewhere in the protein, as well as extensive differences in the 3′-UTR (data not shown). Two versions of INH2α*A were detected, differing only in the length of a stretch of adenine nucleotides upstream of the translation start codon. All of the potato INH2α forms had a single site for potential N-glycosylation, located near the C-terminus of the protein (Fig. 1B). No introns were detected in any INH2α genomic clones.
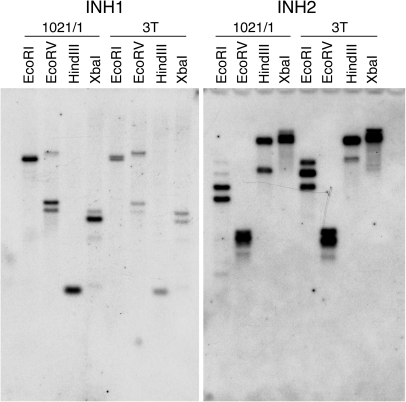
DNA gel blot analysis revealed that INH1 and INH2 are both likely to be encoded by very small gene families, probably two genes per haploid genome for INH1 and with the possibility of a small number of closely related members for INH2 (Fig. 2). Preliminary data released by the Potato Genome Sequencing Consortium (2009) have so far identified a single gene for each of these two families, both located on chromosome 12 with a tandem orientation (Fig. 3).
Fig. 2.
DNA gel blot analysis. Genomic DNA of the potato tetraploid variety 1021/1 and the diploid variety 3T was digested with the indicated restriction enzyme and separated by gel electrophoresis. A blot of the gel was hybridized with labelled probes specific for INH1 or INH2 as shown, and washed at high stringency.
Fig. 3.
Tandem location of INH2 and INH1 genes on potato chromosome 12. Numbers show nucleotide positions on the StBGI01_000038 Scaffold of version 1 of the potato (S. tuberosum RH) assembly (Potato Genome Sequencing Consortium, 2009). (This figure is available in colour at JXB online.)
Subcellular localization of INH1 and INH2 proteins
Initial experiments using DNA constructs that fused the entire coding regions of INH1 and INH2 to GFP did not result in visible fluorescence upon transient transformation into onion bulb epidermis or flower petals. Consequently, constructs were produced that fused the N-terminal quarter of each INH protein, including any putative signal sequence, to GFP. As a control, GFP alone was transiently expressed in onion bulb epidermis cells by particle bombardment, and was detected as relatively weak GFP fluorescence present only in the cytoplasm (Fig. 4A–C). In contrast, the INH1Nterm–GFP fusion protein was detected strongly in the cytoplasm, but also in the cell wall of epidermal cells of both onion bulb (Fig. 4D–F) and lisianthus petal (Fig. 4J). Plasmolysis of lisianthus cells caused the cytoplasm to withdraw from the apoplast, revealing clear GFP fluorescence in the cell wall (Fig. 4K). INH2Nterm–GFP fusion protein was detected transiently in onion bulb epidermal cells, but degraded immediately upon exposure to light (data not shown). Expression of INH2Nterm–GFP fusion protein in lisianthus petals, where vacuolar proteins are more stable, showed that the majority of the fusion protein was in the cytoplasm, but that the protein also accumulated in the vacuole (Fig. 4G–I). These data show that although the bulk of INH1Nterm–GFP and INH2Nterm–GFP proteins was present in the cytoplasm, where proteins are synthesized, the signal sequences present on these fusion proteins caused a proportion to enter the cell wall or the vacuole, respectively.
Fig. 4.
Subcellular localization of INH1 and INH2 N-terminal peptides fused with GFP. GFP (control) and INH1Nterm–GFP and INH2Nterm–GFP fusion proteins were expressed transiently in adaxial epidermal cells of onion bulb or lisianthus flower petals and visualized using confocal laser scanning microscopy (A–I) or fluorescence microscopy (J and K) 36 h after particle bombardment with a gene gun. (A, D, G) Fluorescence image; (B, E, H) differential interference contrast image; (C, F, I) merged image. (A–C) Accumulation of GFP in the cytoplasm of an onion epidermal cell (0.2 μm optical section). (D–F) Accumulation of INH1Nterm–GFP fusion protein in the cytoplasm and cell wall of an onion epidermal cell (0.2 μm optical section). (G–I) Accumulation of INH2Nterm–GFP fusion protein in the cytoplasm and vacuole of a lisianthus petal cell (0.5 μm optical section). (J) Accumulation of INH1Nterm–GFP fusion protein in the cytoplasm and cell wall of a lisianthus petal cell. (K) A similar cell to that shown in J plasmolysed in 1 M sucrose for 10 min, showing fluorescence remaining in the wall after the protoplast has withdrawn. Inset boxed: the plasmolysed region of the cell with contrast enhanced to show GFP clearly in the cell wall.
Characterization of INH2β mRNAs
INH2β (subsequently termed INH2β*A1) was originally isolated from a cDNA library screen. An additional transcript termed INH2β*A2 was isolated from cDNA of cultivar 1021/1 by PCR amplification, and differed from INH2β*A1 by 10 nucleotides and four deduced amino acids. The deduced amino acid sequences of INH2β*A were highly similar to that of INH2α (almost identical to the INH2α*C allele) throughout most of the length, but diverged sharply near the C-terminus (Fig. 5A). At this point the DNA sequence differed from INH2α completely, and, surprisingly, the sequence downstream of this junction was identical to INH1 (Fig. 5B) with a splice junction in the first exon (Fig. 5C). This apparent fusion of the mRNA sequences of INH2 and INH1 provided the hybrid INH2β*A mRNA with an alternative and longer 3′-UTR to that of INH2α, but had the effect of adding just 11 amino acid residues to the predicted protein before a translation stop occurred, so that INH2β*A was 25 amino acid residues shorter than INH2α*C (Fig. 5A, C).
Fig. 5.
INH2β is a hybrid RNA homologous to INH2 upstream and to INH1 downstream of the junction. (A) Alignment of the deduced amino acid sequence of four allelic forms of INH2β with that of INH2α*C. Potential sites for N-glycosylation are underlined, and four conserved cysteine residues are marked with crosses. Amino acid identity is represented by asterisks, highly conservative changes by double dots, and conservative changes by dots. (B) Part of the cDNA sequence of INH2β*A and B aligned with that of INH2α and INH1. The region shown is the junction where sequence conserved with that of INH2α (shown in green) is spliced to sequence conserved with that of INH1 (shown in blue). Two different splice junctions are evident, INH2β*A possessing more upstream sequence derived from INH1 than does INH2β*B. Putative short homologous sequences present in both INH1 and INH2α, located at the apparent splice sites, are underlined in red. Translation stop codons are boxed. Nucleotide identity is indicated by asterisks, and gaps introduced into the alignment by dashes. (C) Schematic diagram showing the regions of INH2α (green) and INH1 (blue) mRNA that make up the spliced INH2β*A transcript. Whether the intron derived from INH1 appears in the pre-mRNA of INH2β*A and is then processed out, or splicing occurs after formation of the mRNA is unclear. Exons are shown as boxes, and introns and untranslated regions as lines. The exons derived from INH1 are out-of-frame in INH2β, resulting in a shorter coding region. (D) Schematic diagram showing the regions of INH2α (green) and INH1 (blue) mRNA that make up the spliced INH2β*B transcript. The exon derived from INH1 is out-of-frame in INH2β*B, resulting in a shorter coding region. Potential sites for N-linked glycosylation in the corresponding deduced proteins are represented by Y-shaped symbols.
Subsequent isolation of further INH2β cDNA clones by PCR amplification found that INH2β*A1 was the predominant form, but that a variant, termed INH2β*B, also existed in both genotypes (Fig. 5A, B). In INH2β*B, the splice junction occurred further downstream, at the intron splice boundary at the beginning of exon 2 of INH1 (Fig. 5D). A comparison of INH2β*B sequences isolated from genotypes 1021/1 and 937/3 found four single nucleotide differences, resulting in two amino acid residue changes, all of these differences being in the region of the DNA conserved with INH2α. These forms were termed INH2β*B1 and INH2β*B2, respectively (Fig. 5A).
Although INH2β*B possessed the entire exon 2 from INH1, the exon was in a different reading frame so that translation caused the addition of only 16 amino acid residues before a translation stop codon occurred (Fig. 5A). This frameshift meant that there was no conservation in amino acid sequence between the C-terminal regions of INH2β*B and either INH2β*A or INH2α, although, like INH2α, the INH2β*B form had a site for potential N-glycosylation of the protein (Fig. 5A, D).
It was not possible to carry out genomic DNA gel blot analysis for INH2β, since this does not possess any unique DNA. Attempts to isolate genomic versions of INH2β using an array of nested primers (12 different primer pair combinations) spanning the apparent INH2/INH1 junction were unsuccessful, although all primer pairs successfully amplified a product from tuber cDNA (data not shown). Data released by the Potato Genome Sequencing Consortium (2009) have also found no evidence for the existence of such hybrid sequences in the genome. These data suggest that INH2β may exist as a transcript but not as a gene.
Presence of alleles of INH2α and INH2β in cultivars 1021/1 and 937/3
Although several different allelic forms of the various genes are described above, it should be noted that not all alleles were found in each cultivar. This was particularly clear for INH2α, where cultivar 1021/1 had alleles A1, A2, and D, while cultivar 937/3 had alleles A1, B, and C (data not shown). Since the number of clones sequenced was limited and because only particular primer pairs were used, it is of course possible that other alleles of INH2 (or related genes) are present in these genotypes. Both cultivars had the INH2β*A1 mRNA, and both cultivars had an INH2β*B mRNA, although these were INH2β*B1 and INH2β*B2, respectively.
Expression of invertase inhibitors in potato plants
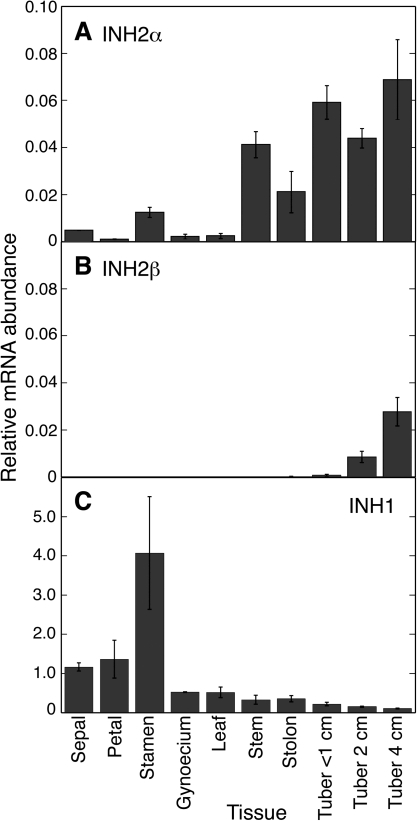
Examination of various tissues of potato plants, including flower parts, vegetative tissues, and developing tubers, found that INH2α mRNA accumulated in all tissues, though to varying extents (Fig. 6A). INH2α mRNA was present at very low abundance in some flower parts (sepal, petal, and gynoecium) and leaf, and at higher abundance in stamen, stem, stolon, and developing tubers. In contrast, INH2β mRNA was undetectable in flower parts, leaf, stem, and stolon, and accumulated at low but increasing abundance in tubers as they developed (Fig. 6B). When present, the abundance of INH2β mRNA was several fold less than the accumulation of the INH2α transcript. These data suggest that INH2β transcript accumulation may be specific to tubers. INH1 mRNA was present at very high abundance in flower parts, and at levels exceeding those of INH2α in all other tissues (Fig. 6C).
Fig. 6.
mRNA accumulation of INH1, INH2α, and INH2β in various tissues of potato plants determined using quantitative real-time PCR. (A) Relative INH2α mRNA abundance. (B) Relative INH2β mRNA abundance. (C) Relative INH1 mRNA abundance (note scale difference from A and B). Data shown are for cultivar 1021/1 (cultivar 937/3 was similar; data not shown). Bars denote SDs from a minimum of two replicate RNA preparations.
Abundance of invertase inhibitor mRNAs during potato tuber storage
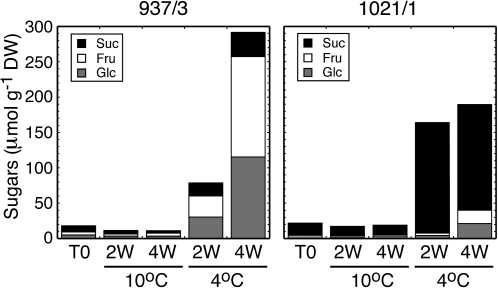
In order to produce cold-induced sweetening, tubers of the susceptible cultivar 937/3 and the resistant cultivar 1021/1 were stored at 4 °C for 2 or 4 weeks and compared with tubers stored at 10 °C. In 937/3, soluble sugars accumulated drastically at 4 °C, increasing by 3-fold between 2 and 4 weeks (Fig. 7). The majority of this sugar was the reducing sugars glucose and fructose, with only a minor proportion being retained as sucrose. In contrast, in 1021/1, soluble sugars increased during cold storage to a lesser extent than in 937/3, and the majority of the soluble sugar was sucrose, with hexoses providing only one-fifth of the total sugar on a molar basis. In both genotypes, soluble sugar showed little change in amount during storage at 10 °C.
Fig. 7.
Hexose sugars accumulate in cold-stored tubers of the cold-induced sweetening-sensitive cultivar 937/3, whereas sucrose accumulates in the resistant cultivar 1021/1. Tubers of two genotypes of potato with differing sensitivity to cold-induced sweetening were stored at 10 °C or 4 °C for 2 or 4 weeks (W). Contents of sucrose and the reducing sugars glucose and fructose were determined in extracts and are expressed on a molar basis.
For examination of invertase inhibitor mRNA abundances (Fig. 8), RNA was prepared from the same samples of stored tubers as shown in Fig. 7. Since it was not possible to design a probe for RNA gel blot analysis that would be specific for INH2β and not show cross-hybridization with either INH2α or INH1, quantitative real-time PCR was used to discriminate between INH2α and INH2β mRNAs (Fig. 8B). INH2α and INH2β mRNAs accumulated at much higher levels in the resistant cultivar 1021/1 than they did in 937/3, especially after storage at 4 °C. In 1021/1, INH2α mRNA abundance increased several fold during treatment at 10 °C, and at 4 °C accumulated slightly later but to even higher levels. INH2β mRNA remained at very low abundance in 937/3 after all treatments, and in 1021/1 did not accumulate at 10 °C but increased during treatment at 4 °C.
Fig. 8.
mRNAs of INH2α and β accumulate during cold storage of tubers in the cold-induced sweetening-resistant cultivar 1021/1 but not in the sensitive cultivar 937/3. (A) RNA gel blot hybridized with a probe that will detect both INH2α and β mRNAs. (B) Relative abundance of mRNAs of INH2α and β determined using quantitative real-time PCR. (C) Relative abundance of mRNA of INH1 determined using quantitative real-time PCR (note scale difference from B). Tubers were stored at 10 °C or 4 °C for 2 or 4 weeks (W). Bars denote SDs from three replicate RNA preparations.
The estimations of mRNA abundances by real-time PCR (Fig. 8B) were confirmed using RNA gel blot analysis (Fig. 8A). Hybridization of an RNA gel blot with a probe that would detect the sum of both INH2α and INH2β transcripts found that INH2α+β mRNA was at low abundance in cultivar 937/3 in all treatments, but accumulated substantially in cultivar 1021/1 (Fig. 8A). INH2α+β mRNA abundance increased during storage at 10 °C, and increased more slowly at 4 °C but to a greater extent by 4 weeks of treatment. The mRNA abundances determined by RNA gel blot analysis were in good agreement with those estimated by quantitative real-time PCR. These data show that accumulation of INH2α and INH2β mRNAs is independently developmentally regulated, and suggest that in cultivars resistant to cold-induced sweetening accumulation of the INH2α mRNA is promoted by both chilling (10 °C) and cold (4 °C), whereas INH2β mRNA accumulation is promoted by cold treatment. INH1 mRNA was induced strongly after 2 weeks at 4 °C, and accumulated at much higher levels than INH2α or INH2β (Fig. 8C). However, the increase in mRNA accumulation was transient and occurred in both susceptible and resistant genotypes.
To investigate the wider applicability of these observations, lines with known cold-induced sweetening properties were selected from a mapping population, and fresh tubers were stored at 4 °C for 4 weeks. In the cold-induced sweetening-susceptible lines Karaka, Summer Delight, K056, S343, S190, and S130, cold storage resulted in a high accumulation of soluble sugars of which the majority was glucose and fructose (Fig. 9A). In the cold-induced sweetening-resistant lines K184, S028, S341, S217, S166, 1021/1, and K068, soluble sugars accumulated to low levels after storage and hexoses formed a minor proportion of the total. In general, the cold-induced sweetening-susceptible lines exhibited high acid invertase activity after storage, whereas the resistant lines had low activity (Fig. 9B). An exception was line S130, which had a high hexose sugar content but low acid invertase activity. Examination of a time course of treatment would be necessary to determine if acid invertase activity had been higher at earlier points during storage in this line. In the majority of lines, low acid invertase activity was correlated with moderate or high accumulation of the sum of INH2α plus INH2β mRNAs (Fig. 9C), suggesting that invertase inhibitors may contribute to lower levels of acid invertase activity in certain cultivars. The correlation was not complete, however, with line S217 having low INH2α+β abundance and low acid invertase activity, and lines S130 and K184 having similar INH2α+β mRNA abundance and acid invertase activity but different sugar accumulation. It is interesting that most of the cold-induced sweetening-resistant lines had mRNAs of both INH2α and INH2β, whereas most of the susceptible lines had either INH2α or INH2β mRNA but not both (Fig. 9C). The significance of this, if any, remains to be established. The abundance of INH1 mRNA did not correlate with acid invertase activity, sugar accumulation, or cold-induced sweetening status (Fig. 9D).
Fig. 9.
Sugar content, acid invertase activity, and abundance of INH mRNAs in cold-stored tubers of lines selected from a mapping population. (A) Content and composition of soluble sugars. The least significant difference (indicated by a bar) at P=0.05 was 11.0 μmol g−1 DW. (B) Soluble acid invertase activity. Data are means of triplicate determinations from at least two independent protein extractions ±SE. (C) Relative abundance of mRNAs of INH2α and β determined using quantitative real-time PCR. (D) Relative abundance of mRNA of INH1 determined using quantitative real-time PCR. Tubers were stored at 4 °C for 4 weeks. In C and D, bars denote SDs of triplicate measurements from two independent cDNA preparations.
Activity of invertase inhibitor variants
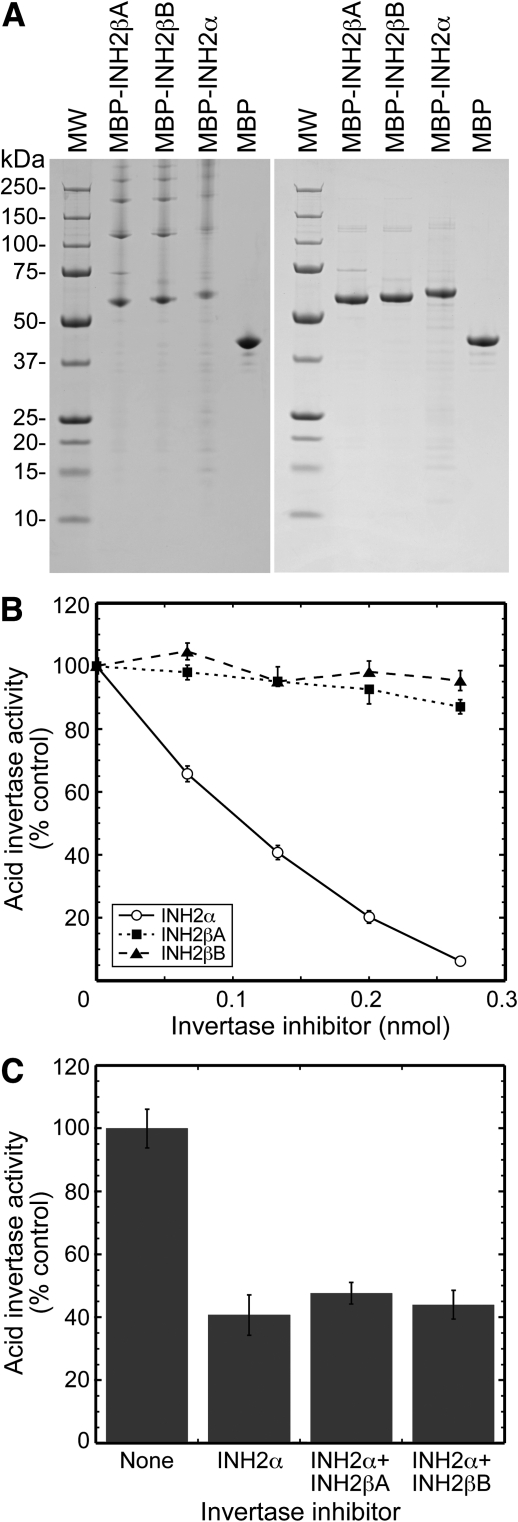
Attempts to express INH proteins in E. coli or Pichia pastoris did not result in soluble or active protein and, although soluble protein could be obtained using a wheat germ cell-free system, no activity could be detected (data not shown). However, expression as fusion proteins with MBP in E. coli did result in soluble protein, although the fusion proteins were not susceptible to cleavage by TEV protease. Consequently, after purification, the fusion proteins were tested for inhibitory activity against acid invertase. All three of the recombinant His-MBP–INH fusion protein preparations were found to form high-ordered complexes, eluting in the void volume of the size-exclusion column. When examined by SDS–PAGE, they all showed the presence of a band of the expected size plus substantial amounts of higher molecular weight complexes (Fig. 10A). These complexes disappeared when samples were denatured in SDS buffer containing 6 M urea prior to SDS–PAGE analysis, showing that the preparations were reasonably pure with no major contaminants (Fig. 10A). The His-MBP control protein did not form high molecular mass complexes, suggesting that the presence of the INH domain was responsible for the aggregation of the proteins. The presence of mild detergents or 50 mM arginine and 50 mM glutamic acid reduced, but did not prevent, protein aggregation (data not shown), and recombinant fusion proteins were therefore tested for activity as high-ordered complexes.
Fig. 10.
Activity of recombinant His-MBP–INH fusion proteins. (A) SDS–PAGE of the three putative vacuolar His-MBP–INH fusion proteins and the His-MBP control. Samples were denatured in SDS loading buffer (left hand panel) or SDS–urea loading buffer (right hand panel) for 30 min at 37 °C prior to loading aliquots of 1.3 μg of protein per lane. Expected molecular masses were: His-MBP–INH2β*A, 59.4 kDa; His-MBP–INH2β*B, 60.2 kDa; His-MBP–INH2α, 62.5 kDa; His-MBP, 43.9 kDa. (B) Inhibition of acid invertase activity by recombinant His-MBP–INH fusion proteins. Dilutions of fusion protein were incubated at 37 °C for 30 min with crude protein extracts (∼12.5 μg of protein) from potato tuber, prior to assay for acid invertase activity. Activity is shown as a percentage of that with no addition of recombinant protein. His-MBP was not inhibitory to acid invertase activity, and has been omitted for clarity. Means are calculated from three experiments carried out in triplicate ±SE. (C) Effect on acid invertase activity of the presence of 0.133 nmol of His-MBP–INH2α alone, or with an equivalent amount of either His-MBP–INH2β*A or His-MBP–INH2β*B. Means are calculated from one experiment carried out in triplicate ±SD.
The His-MBP–INH2α fusion protein was strongly inhibitory to acid invertase activity, with 94% inhibition being observed at the highest concentration of fusion protein tested (Fig. 10B). The His-MBP protein alone was not active (data not shown), and evidently the presence of the His-MBP domain in the fusion protein did not prevent interaction of the INH2α protein domain with acid invertase. However, the His-MBP–INH2β*A and His-MBP–INH2β*B fusion proteins were substantially less active. The His-MBP–INH2β*A protein inhibited invertase by only ∼13% at the highest concentration tested, and so possessed only approximately one-seventh of the activity of His-MBP–INH2α. His-MBP–INH2β*B did not possess significant activity in the concentration range examined.
Since the presence of both INH2α and INH2β transcripts was detected in several of the lines most resistant to cold-induced sweetening, the effects of mixing His-MBP–INH2α with either His-MBP–INH2β*A or His-MBP–INH2β*B were examined (Fig. 10C). However, the presence of either His-MBP–INH2β*A or His-MBP–INH2β*B did not result in an inhibition of acid invertase activity greater than that due to His-MBP–INH2α alone.
Discussion
The deduced proteins of invertase inhibitors isolated from potato were highly similar to deduced proteins from tobacco (Fig. 1), which is also a solanaceous species. Indeed, INH1 and INH2 were more similar to their orthologues from tobacco (86% and 71% for mature proteins, respectively) than they were to each other (43%). INH1 had a predicted signal peptide, which would target the mature proteins to the endoplasmic reticulum for eventual secretion to the apoplast, and the protein was shown to accumulate in the cell wall (Fig. 4). In tobacco, direct protein sequencing of NtINH1 determined that a signal peptide of 19 amino acids was removed to produce the mature protein (Greiner et al., 1998). NtINH1 was localized in the apoplast and acted as an inhibitor of cell wall invertase (Weil et al., 1994; Greiner et al., 1998), as has also been shown for the tomato orthologue SolyCIF/SlINVINH1 (Reca et al., 2008; Jin et al., 2009).
The INH2 gene showed developmentally regulated alternative splicing, with the unspliced INH2α mRNA encoding the full-length protein and the two hybrid INH2β mRNAs encoding altered INH2 proteins. Analysis of the deduced protein sequences of the INH2α and β transcripts with SignalP 3.0 did not unambiguously determine the intracellular target. The homologous protein NtINH-h from tobacco (Fig. 1B) has been assigned a putative vacuolar localization, since overexpression of NtINH-h in potato strongly inhibited vacuolar invertase but not cell wall invertase (Greiner et al., 1999). A fusion of the N-terminus of potato INH2 to GFP was detected in the vacuole (Fig. 4), and the high homology between the INH2α and β forms suggests that all are probably localized to the vacuole.
INH1 mRNA accumulated to the highest levels in potato flower parts and was low in roots (Fig. 6C), similar to NtINH1 mRNA in tobacco (Greiner et al., 1998) and SolyCIF in tomato (Reca et al., 2008). INH1 mRNA could, however, accumulate to high abundance in cold-stored tubers (Fig. 8C). The mRNA accumulation was substantial, but occurred transiently in both lines 937/3 and 1021/1 about 2 weeks into cold storage and had declined by 4 weeks (Fig. 8C). The reason for this transient increase is unclear, but may possibly be a part of the initial mechanism of coping with chilling stress. In two of the cold-induced sweetening-susceptible lines, S343 and S190, the abundance of INH1 mRNA was very high after 4 weeks of cold storage (Fig. 9D), higher than in the other lines examined, although it is possible that the transient accumulation of INH1 mRNA was occurring later in these two lines.
Resistance to cold-induced sweetening correlated, in general, with low acid invertase activity, several fold lower than in susceptible lines (Fig. 9B). The susceptible line 937/3 (Fig. 7) is also known to have high acid invertase activity, up to 7-fold greater than that of the resistant line 1021/1 (McKenzie et al., 2005). These observations are consistent with analysis of other populations that also found a correlation between low acid invertase activity and either low sugar accumulation or a low hexose:sucrose ratio (Zrenner et al., 1996; Matsuura-Endo et al., 2004; McKenzie et al., 2005), and indicate that the level of acid invertase activity is a major component of resistance to cold-induced sweetening. However, other factors may be involved since total invertase activity does not always correlate with hexose accumulation (line S130 of Fig. 9, and Richardson et al., 1990), and quantitative trait loci (QTLs) for several genes other than acid invertase have been implicated (Menéndez et al., 2002). The abundance of vacuolar invertase inhibitor proteins may be one such factor. INH2α mRNA was most abundant in potato tubers (Fig. 6A), particularly after cold stress (Fig. 8B). Lines resistant to cold-induced sweetening in general had greater accumulation of the sum of INH2α plus β mRNAs (Fig. 9C), presumably resulting in greater abundance of invertase inhibitor proteins. Attempts to destroy endogenous invertase inhibitors through foaming (Pressey, 1966) resulted in highly irreproducible data, although some increase in acid invertase activity was always seen (data not shown).
Cold stress can result in altered expression of genes, but also in altered processing of transcripts. In potato plants, exposure to cold stress caused exon skipping in an invertase mRNA, and presumably led to the formation of a version of the protein with altered properties (Bournay et al., 1996). In stored tubers of some potato lines, cold stress caused the formation of the spliced INH2β mRNAs. There are two explanations for how a hybrid INH2β mRNA could arise. INH2 and INH1 are both located on chromosome 12, and have a tandem orientation, with INH2 being upstream (Fig. 3). The genes are separated by ∼5.5 kb with no obvious intervening genes. Although the two genes exhibit distinct developmental expression patterns, and the very different transcript abundances in different tissues indicate that they are probably independently transcribed (Figs 6, 8), one possibility is that a single transcript could be derived by read-through transcription from the INH2 promoter into the downstream INH1 gene. This large pre-mRNA transcript could then be processed in cis using cryptic splice sites into the INH2β*A and INH2β*B mRNAs observed. The tissue-specific accumulation of INH2β is related to that of INH2α (Fig. 6), which could be considered consistent with this possibility. Alternative splicing of exons in cis is a common feature of plant mRNAs (Wang and Brendel, 2006; Reddy, 2007). However, the INH2 gene does not possess introns, and the splice junction found in the major INH2 transcript (INH2β*A) did not correspond to an intron splice site in INH1 (Fig. 5C). RNA folding into extensive secondary structures is exacerbated at low temperatures (Zhu et al., 2007) which, combined with increased RNA splicing in the cold (Lee et al., 2006), could result in multiple splicing products in cis using cryptic splice sites.
mRNAs can be also be spliced via trans mechanisms. Trans splicing occurs when two (or more) independently transcribed pre-mRNAs are trans-spliced to form a single functional mRNA. It has been best described in animals (Dorn et al., 2001; Fischer et al., 2008), but also occurs in organelles and seeds of plants (Chapdelaine and Bonen, 1991; Kawasaki et al., 1999; Schmitz-Linneweber et al., 2006). However, trans-splicing uses canonical splice sites to join together exons from different pre-mRNAs (Horiuchi and Aigaki, 2006), which is not the case for INH2β since INH2 does not have an intron. The splicing of two separate mRNAs without the involvement of canonical splice sites produces what have been termed chimeric RNAs and, while rarely described in plants, 45 000 examples have been described in yeast, fruit fly, mouse, and human (Li et al., 2009). In these organisms, about half of the chimeric RNAs had an non-conserved short homologous sequence of at least four nucleotides present at the junction site in both donor mRNAs. This was observed for both INH2β*A and INH2β*B, where a six nucleotide domain (AAACTT) and a five nucleotide domain (TAATT) were common to both INH2 and INH1 at the splice site, respectively (Fig. 5B). This short homologous sequence has been shown to be important for production of the spliced RNA (Li et al., 2009).
It remains unclear whether the INH2β mRNA arises though splicing in cis or trans, since the presence at the splice junctions of weak cryptic splice sites (data not shown) and short homologous sequences (Fig. 5B) is consistent with either mechanism. The detection of INH2β mRNA only in tubers implies a tissue-specific regulation of this splicing event, which produces at least two variants. In Arabidopsis, alternative splicing of pre-mRNAs of serine/arginine-rich genes was also regulated in a tissue-specific manner (Palusa et al., 2007), and was greatly increased by abiotic (particularly temperature) stress (Reddy, 2007). Indeed, increased RNA splicing under cold stress conditions appears to be part of the acclimation mechanism of plants (Lee et al., 2006).
The deduced proteins of INH2α and both forms of INH2β possessed the four cysteine residues believed to be important in the formation of two disulphide bridges that stabilize the protein three-dimensional structure (Scognamiglio et al., 2003; Hothorn et al., 2004a). INH2β shared sequence identity with INH2α in the upstream region of the cDNA sequence, encoding most of the length of the deduced proteins. The sequence diverged within the codon for the fourth conserved cysteine, but in both INH2β*A and B the fusion with the mRNA of INH1 provided the nucleotides necessary to complete the codon as a cysteine. However, when assayed as His-MBP–INH fusion proteins, only INH2α had strong inhibitory activity against potato acid invertase (Fig. 10B). INH2β*A exhibited weak inhibitory activity against acid invertase, while activity could not be detected for INH2β*B. Examination of the likely three-dimensional structure of invertase inhibitors (Hothorn et al., 2004a) suggests that both INH2β*A and INH2β*B are likely either to be partially lacking or to possess a substantially altered fourth α-helix.
Low temperature stress causes large changes in the metabolome and transcriptome (Cook et al., 2004; Lee et al., 2005; Vogel et al., 2005; Kaplan et al., 2007), resulting in the accumulation of small molecules such as sucrose and other sugars that may be part of the mechanism for stabilizing proteins and membranes and maintaining cell osmotic pressure (Yamaguchi-Shinozaki and Shinozaki, 2006). Comparisons of different species of potato (S. tuberosum and S. phureja) have shown wide differences in their ability to acclimate to cold stress, which was correlated with differences in global gene expression including carbohydrate metabolism (Oufir et al., 2008). In potato tuber many cold-responsive genes involved in carbohydrate metabolism showed a biphasic pattern of mRNA accumulation, with a rapid increase over 3–5 d being followed by a decrease and, in many cases, a subsequent sustained increase over the next 20 d (Bagnaresi et al., 2008). Acid invertase mRNA showed this pattern (Bagnaresi et al., 2008), and in a potato cultivar resistant to cold-induced sweetening (1021/1), INH2α mRNA also accumulated to high levels during the 28 d of cold storage (Fig. 8). Overall, the data are consistent with low acid invertase activity being the primary cause of resistance to cold-induced sweetening. Whether this is due to reduced expression of acid invertase or the expression of alleles with low activity will vary between genotypes, but may be assisted in some cultivars by a high mRNA accumulation of vacuolar invertase inhibitors. These proteins therefore play an important role in the resistance of certain potato cultivars to cold-induced sweetening by suppressing acid invertase activity.
Plants have several ways other than gene expression in which to modify the transcriptome. Three mechanisms used to produce more than one protein product from a single gene are alternative transcription initiation, alternative splicing of exons, and alternative translation initiation (Reddy, 2007; Wamboldt et al., 2009; Tanaka et al., 2010). Alternative splicing is regulated in a specific manner, since it can vary between tissues, during development, and in response to stresses (Reddy, 2007). Increased RNA splicing activity was detected in the tubers of many potato lines resistant to cold-induced sweetening, and the existence of such a mechanism under cold stress conditions may be a way for the plant to expand the variety of proteins beyond the one-gene one-protein model, and generate a range of proteins with the potential for additional and more diverse functional capacity to aid adaptability.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used for PCR amplification and quantitative real-time PCR.
Acknowledgments
We thank Russell Genet, John Anderson, and Jeanne Jacobs for supplying tubers with known cold-induced sweetening properties, the Potato Genome Sequencing Consortium (www.potatogenome.net) for use of the draft potato genome sequence prior to publication, Mark Fiers and Susan Thomson for ascertaining gene location from these data, Jianyu Chen for confocal microscopy at the Manawatu Microscopy and Imaging Centre of Massey University, Tony Conner for providing diploid 3T tissue and useful discussions, Samantha Baldwin for sequencing, and Kevin Davies and Roger Hellens for comments on the manuscript. We are grateful to Tony Conner, Ian Ferguson, and Ross Lill for their support of this project. This work was funded by the New Zealand Foundation for Research, Science and Technology (FRST), and by Vital Vegetables, a Trans-Tasman research project jointly funded and supported by Horticulture Australia Ltd, the New Zealand Institute for Plant & Food Research Limited, FRST, the Victorian Department of Primary Industries, Horticulture New Zealand, and the Australian Vegetable and Potato Growers Federation Inc.
References
- Bagnaresi P, Moschella A, Beretta O, Vitulli F, Ranalli P, Perata P. Heterologous microarray experiments allow the identification of the early events associated with potato tuber cold sweetening. BMC Genomics. 2008;9:176. doi: 10.1186/1471-2164-9-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate NJ, Niu XP, Wang YW, Reimann KS, Helantjaris TG. An invertase inhibitor from maize localizes to the embryo surrounding region during early kernel development. Plant Physiology. 2004;134:246–254. doi: 10.1104/pp.103.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PB, Wu L, Busse JS, Whitty BR, Hamernik AJ, Jansky SH, Buell CR, Bethke PC, Jiang J. Suppression of the vacuolar invertase gene prevents cold-induced sweetening in potato. Plant Physiology. 2010;154:939–948. doi: 10.1104/pp.110.162545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkinsop RW, Yada RY, Marangoni AG. Metabolic control of low-temperature sweetening in potato tubers during postharvest storage. Horticultural Reviews. 2004;30:317–354. [Google Scholar]
- Borovkov AY, McClean PE, Sowokinos JR, Ruud SH, Secor GA. Effect of expression of UDP-glucose pyrophosphorylase ribozyme and antisense RNAs on the enzyme activity and carbohydrate composition of field-grown transgenic potato plants. Journal of Plant Physiology. 1996;147:644–652. [Google Scholar]
- Bournay AS, Hedley PE, Maddison A, Waugh R, Machray GC. Exon skipping induced by cold stress in a potato invertase gene transcript. Nucleic Acids Research. 1996;24:2347–2351. doi: 10.1093/nar/24.12.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracho GE, Whitaker JR. Characteristics of the inhibition of potato (Solanum tuberosum) invertase by an endogenous proteinaceous inhibitor in potatoes. Plant Physiology. 1990a;92:381–385. doi: 10.1104/pp.92.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracho GE, Whitaker JR. Purification and partial characterization of potato (Solanum tuberosum) invertase and its endogenous proteinaceous inhibitor. Plant Physiology. 1990b;92:386–394. doi: 10.1104/pp.92.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Dunsmuir P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Molecular Biology. 1999;39:161–169. doi: 10.1023/a:1006130018931. [DOI] [PubMed] [Google Scholar]
- Chapdelaine Y, Bonen L. The wheat mitochondrial gene for subunit I of the NADH dehydrogenase complex—a transsplicing model for this gene-in-pieces. Cell. 1991;65:465–472. doi: 10.1016/0092-8674(91)90464-a. [DOI] [PubMed] [Google Scholar]
- Chen S, Hajirezaei MR, Zanor MI, Hornyik C, Debast S, Lacomme C, Fernie AR, Sonnewald U, Bornke F. RNA interference-mediated repression of sucrose-phosphatase in transgenic potato tubers (Solanum tuberosum) strongly affects the hexose-to-sucrose ratio upon cold storage with only minor effects on total soluble carbohydrate accumulation. Plant, Cell and Environment. 2008;31:165–176. doi: 10.1111/j.1365-3040.2007.01747.x. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proceedings of the National Academy of Sciences, USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of. Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R, Reuter G, Loewendorf A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in. Drosophila. Proceedings of the National Academy of Sciences, USA. 2001;98:9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT. 2007. Food and Agriculture Organization of the United Nations Statistical Database. http://faostat.fao.org. [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Analytical Biochemistry. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fischer SEJ, Butler MD, Pan Q, Ruvkun G. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature. 2008;455:491–496. doi: 10.1038/nature07274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Krausgrill S, Rausch T. Cloning of a tobacco apoplasmic invertase inhibitor—proof of function of the recombinant protein and expression analysis during plant development. Plant Physiology. 1998;116:733–742. doi: 10.1104/pp.116.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Rausch T, Sonnewald U, Herbers K. Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nature Biotechnology. 1999;17:708–711. doi: 10.1038/10924. [DOI] [PubMed] [Google Scholar]
- Hill LM, Reintholz R, Schroder R, Nielsen TH, Stitt M. The onset of sucrose accumulation in cold stored potato tubers is caused by an increased rate of sucrose synthesis and coincides with low levels of hexose phosphates, an activation of sucrose phosphate synthase and the appearance of a new form of amylase. Plant, Cell and Environment. 1996;19:1223–1237. [Google Scholar]
- Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biology of the Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- Hothorn M, D'Angelo I, Marquez JA, Greiner S, Scheffzek K. The invertase inhibitor Nt-CIF from tobacco: a highly thermostable four-helix bundle with an unusual N-terminal extension. Journal of Molecular Biology. 2004a;335:987–995. doi: 10.1016/j.jmb.2003.10.066. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K. Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. The Plant Cell. 2004b;16:3437–3447. doi: 10.1105/tpc.104.025684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ni DA, Ruan YL. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose levels. The Plant Cell. 2009;21:2072–2089. doi: 10.1105/tpc.108.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. The Plant Journal. 2007;50:967–981. doi: 10.1111/j.1365-313X.2007.03100.x. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Okumura S, Kishimoto N, Shimada H, Higo K, Ichikawa N. RNA maturation of the rice SPK gene may involve trans-splicing. The Plant Journal. 1999;18:625–632. doi: 10.1046/j.1365-313x.1999.00493.x. [DOI] [PubMed] [Google Scholar]
- Krause KP, Hill L, Reimholz R, Nielsen TH, Sonnewald U, Stitt M. Sucrose metabolism in cold-stored potato tubers with decreased expression of sucrose phosphate synthase. Plant, Cell and Environment. 1998;21:285–299. [Google Scholar]
- Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Singh BP, Kumar P. An overview of the factors affecting sugar content of potatoes. Annals of Applied Biology. 2004;145:247–256. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. The Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Kapoor A, Zhu JH, Zhu JK. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover. and stress tolerance in Arabidopsis. The Plant Cell. 2006;18:1736–1749. doi: 10.1105/tpc.106.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao L, Jiang HF, Wang W. Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. Journal of Molecular Evolution. 2009;68:56–65. doi: 10.1007/s00239-008-9187-0. [DOI] [PubMed] [Google Scholar]
- Link M, Rausch T, Greiner S. In Arabidopsis thaliana, the invertase inhibitors AtC/VIF1 and 2 exhibit distinct target enzyme specificities and expression profiles. FEBS Letters. 2004;573:105–109. doi: 10.1016/j.febslet.2004.07.062. [DOI] [PubMed] [Google Scholar]
- Malone JG, Mittova V, Ratcliffe RG, Kruger NJ. The response of carbohydrate metabolism in potato tubers to low temperature. Plant and Cell Physiology. 2006;47:1309–1322. doi: 10.1093/pcp/pcj101. [DOI] [PubMed] [Google Scholar]
- Matsuura-Endo C, Kobayashi A, Noda T, Takigawa S, Yamauchi H, Mori M. Changes in sugar content and activity of vacuolar acid invertase during low-temperature storage of potato tubers from six Japanese cultivars. Journal of Plant Research. 2004;117:131–137. doi: 10.1007/s10265-003-0137-z. [DOI] [PubMed] [Google Scholar]
- McKenzie MJ, Sowokinos JR, Shea IM, Gupta SK, Lindlauf RR, Anderson JAD. Investigations on the role of acid invertase and UDP-glucose pyrophosphorylase in potato clones with varying resistance to cold-induced sweetening. American Journal of Potato Research. 2005;82:231–239. [Google Scholar]
- Menéndez CM, Ritter E, Schäfer-Pregl R, Walkemeier B, Kalde A, Salamini F, Gebhardt C. Cold sweetening in diploid potato: mapping quantitative trait loci and candidate genes. Genetics. 2002;162:1423–1434. doi: 10.1093/genetics/162.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram DS, Wedzicha BL, Dodson AT. Acrylamide is formed in the Maillard reaction. Nature. 2002;419:448–449. doi: 10.1038/419448a. [DOI] [PubMed] [Google Scholar]
- Nielsen TH, Deiting U, Stitt M. A β-amylase in potato tubers is induced by storage at low temperature. Plant Physiology. 1997;113:503–510. doi: 10.1104/pp.113.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oufir M, Legay S, Nicot N, Van Moer K, Hoffman L, Renaut J, Hausman JF, Evers D. Gene expression in potato during cold exposure: changes in carbohydrate and polyamine metabolisms. Plant Science. 2008;175:839–852. [Google Scholar]
- Palusa SG, Ali GS, Reddy ASN. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. The Plant Journal. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium. 2009. Public data release, version 1. www.potatogenome.net. [Google Scholar]
- Pressey R. Separation and properties of potato invertase and invertase inhibitor. Archives of Biochemistry and Biophysics. 1966;113:667–674. doi: 10.1016/0003-9861(66)90246-3. [DOI] [PubMed] [Google Scholar]
- Pressey R. Invertase inhibitor from potatoes: purification, characterization, and reactivity with plant invertases. Plant Physiology. 1967;42:1780–1786. doi: 10.1104/pp.42.12.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey R, Shaw R. Effect of temperature on invertase, invertase inhibitor, and sugars in potato tubers. Plant Physiology. 1966;41:1657–1661. doi: 10.1104/pp.41.10.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analysis. Plant Physiology. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Greiner S. Plant protein inhibitors of invertases. Biochimica et Biophysica Acta. 2004;1696:253–261. doi: 10.1016/j.bbapap.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Reca IB, Brutus A, D'Avino R, Villard C, Bellincampi D, Giardina T. Molecular cloning, expression and characterization of a novel invertase inhibitor from tomato (Solanum lycopersicum) and its use to purify a vacuolar invertase. Biochimie. 2008;90:1611–1623. doi: 10.1016/j.biochi.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Reddy ASN. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annual Review of Plant Biology. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- Reimholz R, Geiger M, Haake V, Deiting U, Krause KP, Sonnewald U, Stitt M. Potato plants contain multiple forms of sucrose phosphate synthase, which differ in their tissue distributions, their levels during development, and their responses to low temperature. Plant, Cell and Environment. 1997;20:291–305. [Google Scholar]
- Richardson DL, Davies HV, Ross HA, MacKay GR. Invertase activity and its relation to hexose accumulation in potato tubers. Journal of Experimental Botany. 1990;41:95–99. [Google Scholar]
- Rommens CM, Ye JS, Richael C, Swords K. Improving potato storage and processing characteristics through all-native DNA transformation. Journal of Agricultural and Food Chemistry. 2006;54:9882–9887. doi: 10.1021/jf062477l. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. The Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer S, Makower RU, Rorem ES. Invertase & invertase inhibitor in potato. Plant Physiology. 1961;36:313–316. doi: 10.1104/pp.36.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scognamiglio MA, Ciardiello MA, Tamburrini M, Carratore V, Rausch T, Camardella L. The plant invertase inhibitor shares structural properties and disulfide bridges arrangement with the pectin methylesterase inhibitor. Journal of Protein Chemistry. 2003;22:363–369. doi: 10.1023/a:1025342207831. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shang Y, Schwinn KE, Bennett MJ, Hunter DA, Waugh TL, Pathirana NN, Brummell DA, Jameson PE, Davies KM. Methods for transient assay of gene function in floral tissues. Plant Methods. 2007;3:1. doi: 10.1186/1746-4811-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowokinos JR. Biochemical and molecular control of cold-induced sweetening in potatoes. American Journal of Potato Research. 2001;78:221–236. [Google Scholar]
- Stadler RH, Blank I, Varga N, Robert F, Hau J, Guy PA, Robert MC, Riediker S. Acrylamide from Maillard reaction products. Nature. 2002;419:449–450. doi: 10.1038/419449a. [DOI] [PubMed] [Google Scholar]
- Sumner JB. Dinitrosalicylic acid: a reagent for the estimation of sugar in normal and diabetic urine. Journal of Biological Chemistry. 1921;47:5–9. [Google Scholar]
- Tanaka T, Koyanagi KO, Itoh T. Highly diversified molecular evolution of downstream transcription start sites in rice and Arabidopsis. Plant Physiology. 2010;149:1316–1324. doi: 10.1104/pp.108.131656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. The Plant Journal. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Wamboldt Y, Mohammed S, Elowsky C, Wittgren C, de Paula WBM, Mackenzie SA. Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. The Plant Cell. 2009;21:157–167. doi: 10.1105/tpc.108.063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Analytical Biochemistry. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proceedings of the National Academy of Sciences, USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil M, Krausgrill S, Schuster A, Rausch T. A 17 kDa Nicotiana tabacum cell-wall peptide acts as an in-vitro inhibitor of the cell-wall isoform of acid invertase. Planta. 1994;193:438–445. doi: 10.1007/BF00201824. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Zhu J, Dong CH, Zhu JK. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology. 2007;10:290–295. doi: 10.1016/j.pbi.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Schuler K, Sonnewald U. Soluble acid invertase determines the hexose-to-sucrose ratio in cold-stored potato tubers. Planta. 1996;198:246–252. doi: 10.1007/BF00206250. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Willmitzer L, Sonnewald U. Analysis of the expression of potato uridinediphosphate-glucose pyrophosphorylase and its inhibition by antisense RNA. Planta. 1993;190:247–252. doi: 10.1007/BF00196618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.