Abstract
Ascorbate (AsA) is a redox buffer and enzyme cofactor with various proposed functions in stress responses and growth. The aim was to identify genes whose transcript levels respond to changes in leaf AsA. The AsA-deficient Arabidopsis mutant vtc2-1 was incubated with the AsA precursor L-galactono-1,4-lactone (L-GalL) to increase leaf AsA concentration. Differentially expressed genes screened by DNA microarray were further characterized for AsA responsiveness in wild-type plants. The analysis of 14 candidates by real-time PCR identified an aspartyl protease gene (ASP, At1g66180) and a C3HC4-type RING zinc finger gene (AtATL15, At1g22500) whose transcripts were rapidly responsive to increases in AsA pool size caused by L-GalL and AsA supplementation and light. Transgenic Arabidopsis plants expressing an AtATL15 promoter::luciferase reporter confirmed that the promoter is L-GalL, AsA, and light responsive. The expression patterns of ASP and AtATL15 suggest they have roles in growth regulation. The promoter of AtATL15 is responsive to AsA status and will provide a tool to investigate the functions of AsA in plants further.
Keywords: Arabidopsis thaliana, ascorbate, L-galactonolactone, gene expression, vtc2-1 mutant
Introduction
Ascorbic acid (AsA) has diverse physiological roles in plants. AsA scavenges reactive oxygen species (ROS; Noctor and Foyer, 1998), both non-enymatically and enzymatically via AsA peroxidase (APX; Ishikawa and Shigeoka, 2008). It also acts as a cofactor for violaxanthin de-epoxidase, an enzyme involved in the photoprotective xanthophyll cycle (Smirnoff, 2000). It is a cofactor for 2-oxoglutarate-dependent dioxygenases including prolyl hydroxylase (Arrigoni et al., 1977; Smirnoff, 2000) and the enzymes participating in the biosynthesis of plant hormones, such as abscisic acid (ABA), gibberellic acid (GA), and ethylene (Arrigoni and De Tullio, 2002; Mirica and Klinman, 2008) as well as a range of enzymes involved in hydroxyproline, flavonoid, and glucosinolate biosynthesis (Davey et al., 2000; Kliebenstein et al., 2001; Turnbull et al., 2004). This wide range of functions is reflected in the necessity of AsA for post-germination growth of Arabidopsis seedlings (Dowdle et al., 2007). The precise role for AsA in growth, beyond its role as an enzyme cofactor, has not been established but there are suggestions that it could affect cell division, cell expansion, and ROS-mediated stomatal closure by influencing the cellular redox state (Fry, 1998; Horemans et al., 2000; Barth et al., 2004; Chen and Gallie, 2004; Potters et al., 2004; Foyer and Noctor, 2005; Tokunaga et al., 2005). Many of the proposed roles for AsA have been confirmed by the investigation of AsA-deficient A. thaliana mutants, which are susceptible to ozone, UV-B, and photooxidative stress and are smaller and altered in development and pathogen resistance compared with WT plants (Conklin et al., 1996; Barth et al., 2004; Mukherjee et al., 2010).
Considering the various effects of AsA, the question of how changes in AsA status affect gene expression arises. Previous investigations have shown that genes encoding PR proteins are more highly expressed in vtc1 and vtc2 (Barth et al., 2004; Colville and Smirnoff, 2008). Transcriptome analysis of vtc1 showed changes in expression of a number of genes and also in response to AsA supplementation (Kiddle et al., 2003; Pastori et al., 2003). For instance, gene families related to PR proteins and a gene encoding 9-cis-epoxicarotenoid dioxygenase, a key enzyme for ABA biosynthesis located in plastids, were up-regulated in vtc1 mutant (Pastori et al., 2003). Changed gene expression in the vtc mutants could result from secondary effects of long-term deficiency, so to investigate the more immediate effect of AsA status, screening for candidate AsA-responsive genes was done in the AsA-deficient vtc2-1 mutant (Conklin et al., 1996, 2000) supplemented with the AsA precursor L-galactonolactone (L-GalL) by using microarray analysis. Several candidate L-GalL-responsive genes were validated as AsA-responsive by real-time PCR and expression of a promoter::luciferase construct for one gene (AtATL15) showed that it was both light- and AsA-responsive. The results show that gene expression can respond rapidly to AsA status and provide tools to investigate this response in more detail.
Materials and methods
Plant materials and growth conditions
Seeds from Arabidopsis wild-type Columbia-0 (Col-0) were purchased from the Nottingham Arabidopsis Stock Centre. Seeds of Arabidopsis AsA-deficient mutant vtc2-1 were kindly supplied by PL Conklin (Conklin et al., 2000). All seeds were sterilized with 10% bleach and 10% bleach/75% ethanol separately, for 4 min, washed three times with sterilized H2O, and then spread onto 0.8% (w/v) agar plates containing Murashige and Skoog (MS, Sigma) medium and 2.5% sucrose. The seeds were stratified by placing them in darkness for 2 d at 4 °C, and then incubating in growth chambers under 14 h of 80 μmol photons m−2 s−1 light and 10 h of darkness at 21 °C. Furthermore, sterilized seeds were grown on Jiffy soil (Jiffy Products International AS, Norway), stratified for 2 d in darkness at 4 °C and then transferred to a controlled environment room at 21 °C with a 14 h light (80 μmol photons m−2 s−1) and a 10 h dark cycle.
L-galactonolactone (L-GalL), D-galactonolactone (D-GalL), AsA, and D-glucose (D-Glu feeding to detached leaves
Six-week-old wild type and the vtc2-1 mutant (Conklin et al., 2000) were used. A total of 20 leaves were harvested from five independent rosettes of each genotype. The cut leaves were then placed in a Petri dish, containing 20 ml of 5 mM D-Glu, L-GalL, D-GalL, and AsA, respectively. AsA was dissolved in 10 mM 3-(N-morpholino)-propanesulphonic acid (MOPS) buffer, pH 6.0, as described by Pastori et al. (2003). Leaves were also incubated in MOPS buffer separately as a control. Light intensity was 80 μmol photons m−2 s−1. After incubation, leaves were washed three times with Milli-Q water, gently dried, and then frozen with liquid nitrogen and stored at –80 °C. Leaves incubated in the dark were frozen while still in the dark.
AsA and dehydroascorbate (DHA) measurement
Frozen leaves were ground to a powder within a mortar and using liquid nitrogen. The frozen leaf powder was homogenized in 0.1 M HCl and 1 mM EDTA. The homogenate was centrifuged at 12 000 g for 5 min. The supernatant was transferred into a new tube and the total AsA (reduced and oxidized) concentration was assayed by the method of Kampfenkel et al. (1995). In this method AsA reduces Fe3+ under acidic conditions and Fe2+ is detected by the red-coloured complex it forms with bipyridyl. Although other compounds (particularly phenols with ortho-hydroxyl groups) can reduce Fe3+, they are not sufficiently abundant in A. thaliana to interfere with this assay.
RNA isolation and cDNA preparation
RNA was isolated from frozen tissue (around 100 mg for each sample), after homogenizing the sample with liquid nitrogen, 1 ml RNAiso (Takara, Japan) was added and mixed well, 200 μl chloroform was then added and the samples were shaken vigorously for 1 min, left for 5 min and then centrifuged at 13 000 rpm for 15 min. The supernatant was transferred to new tubes, the same volume of isopropanol was added and mixed well. After 10 min the samples were centrifuged at 13 000 rpm for 10 min. The supernatant was decanted and the RNA pellets dried under vacuum. The crude RNA was treated with 10 units of DNase I (Takara, Japan) and further purified with an RNeasy Plant Mini kit (Qiagen) according to the manufacturer's instructions. The concentration of total RNA was determined with an Eppendorf BioPhotometer. cDNA was prepared from 500 ng total purified RNA template with Perfect Real Time (Takara, Kyoto, Japan) according to the manufacturer's instructions.
Microarray analysis
Total RNA samples were isolated from the vtc2-1 leaves fed with 5 mM L-GalL and H2O, respectively, under light at 80 μmol photons m−2 s−1 for 16 h, using RNAiso (Takara) following the manufacturer's instructions. The RNA samples were further purified according to an RNAeasy Mini kit (Qiagen) and qualities were checked using an Agilent 2100 bioanalyser (Agilent) and Nano Drop (Thermo Scientific) before labelled cRNA was synthesized. Biotinylated cRNA samples were synthesized from 1 μg total RNA using Affymetrix One Cycle Synthesis kits (Affymetrix) as described by the manufacturer's protocol. cRNA was hybridized to the ATH1 GeneChip (Affymetrix), which contains around 22 500 oligonucleotide probes. The signal intensities from each GeneChip were normalized to the 50th percentile using GeneSpring (Agilent Technologies) software. The fold change in expression between the control and L-GalL-treated samples was calculated for each gene and genes flagged as marginal or absent in the L-GalL treatment were ignored.
Real-time PCR
For the real-time PCR, cDNA (50 ng) was mixed with 10 μl SYBR Premix Ex Tag and 1 μl of 10 μM mixed primer (forward and reverse), H2O was added to up to 25 μl and the reaction was performed with Thermal Cycler (Dice Real Time System TP800, Takara, Japan). Transcript abundance was calculated using the relative expression software tool (Dice Real Time V2.10B), which takes into account the volume of different templates and calculates the statistical significance of the observed differences. To validate reference genes under experimental conditions, expression levels of three reference genes, ACTIN-2, UBQ10, and EF1α were compared. The expression level of ACTIN-2 was the most stable in its transcript level across the samples than the other two reference genes (see Supplementary Fig. S6 at JXB online). Therefore, ACTIN-2 was used as an internal control to normalize each sample for variations in the amount of initial RNA. The gene-specific primers were: ACTIN-2 (At3g18780-F): GGCAAGTCATCACGATTGG; R: CAGCTTCCATTCCCACAAAC; EF1α (At1g07940-F): ACCACGAGTCTCTTCTTGAGGCAC; R: TGGCAGGGTCATCCTTGGAG; UBQ-10 (At4g05320-F): AACTTTGGTGGTTTGTGTTTTGG; R: TCGACTTGTCATTAGAAAGAAAGAGATAA; At1g80240-F: GGCTTGTGTTCAAGGCAGTG; R: GGAGTTTCCCAACCGCTACC; At5g25460-F: CTTTACCTGGATGGATGGTG; R: CCAATGACAGTCCGAACAC; At5g12940-F: TGGTCACCTTGACGTGAGC; R: AATTCCCAAGAGGCTTTCCAC; At4g13420-F: TCAGTCATCTCGCAATCTCTACG; R: GATGCATGCAAGCATGAGC; At1g73120-F: GAGAAAGTGATGCAAGATGTGCC; R: CGAGTCTTCGACGAGATCGAG; At1g66180-F: TGATCCCACGGTTGATAGGAG; R: GCTCCAAGCATACTCGACCG; At1g77760-F: CAAAGGAGCGTCAGCTTGAGA; R: TGATCCATGCAGAGTCAGCAG; At4g16890-F: ATCCTCGATCTCAGTGGTTGC; R: CATCATAAGTACAGTGAGCCTCGTG; At1g22500-F: AGAACCGGAAGCCTAGGGAC; R: AATCTCATCCGGAGACTGTATCG; At1g52190-F: CACAAGTGAAGCCATCGCC; R: CCAAATGCTAAGGAACATGGTCTG; At3g52720-F: CAAATGCATTGGCACACTCC; R: CCTTCATCTGAGAGAGGAAAGGC; At4g08040-F: CAGGTTTGTTCTGTTGGGTTGAC; R: AACCAGGTTCATCGCAATGAC; At1g06490-F: GGACTCTCTTGGGCTATCTTGTTG; R: AAGAGAAGTGCCTTGAGAATCCTG; At5g17700-F: ATGGGTGCAGCATGTGTACG; R: ATATCACACCGATCACCGCTG.
Genomic DNA isolation
Arabidopsis wild-type leaves (around 20 mg) were homogenized with 0.5 ml buffer containing 0.6 M NaCl, 0.1 M TRIS-HCl (pH 7.5), 40 mM EDTA (pH 8.0), and 1% SDS. The same volume of chloroform/phenol mixture (1:1 v/v) was then added and mixed after which the sample was centrifuged at 15 000 rpm for 5 min. The supernatant was transferred to a new Eppendorf tube and 100% ethanol was added to precipitate genomic DNA followed by centrifugation at 15 000 rpm for 5 min. The DNA pellet was washed with 1 ml 70% ethanol and dried under vacuum for 5 min. Finally, the DNA pellet was dissolved in 20 μl TE buffer (10 mM TRIS, 1 mM EDTA, pH 8.0) and stored at –20°C.
Promoter reporter system construct and Arabidopsis transformation
The promoters (about 2 kb upstream of the start codon) of the genes At1g66180 and At1g22500 were amplified from Arabidopsis wild-type genomic DNA with Blend Taq polymerase (Toyobo). The gene-specific primers used were: At1g66180 F: AGAGAAACCGCGTTCCATATTTCAC; R: GAAGAGTGGTTTTGGAATGGGAGa; At1g22500 F: AGAGCTAGCGTGCTGCCATAAGATGA; R: CATCGATGGTTGCAAATGGTTTGA. Promoters were inserted into the entry vector pDONR201, and then cloned into the Gateway binary vector pGWB 3435 (containing the LUC reporter gene) by Gateway LR reaction using LR clonase (Invitrogen). The construct was transferred into Agrobacterium tumefaciens strain C58 and transformed into wild-type plants by the floral dip method (Clough and Bent, 1998). The transformed plants were selected on 0.8% (w/v) agar MS medium plates containing 2.5% sucrose and 30 μg ml−1 kanamycin as the selection agent. The seeds were stratified for 2–3 d and then moved to the culture chamber (22°C, 80 μmol photons m−2 s−1). Three weeks later, healthy seedlings were transferred into Jiffy-7 soil and grown in a controlled environment room (23°C with a 14/10 h light/dark cycle).
Detection of luciferase activity
Transgenic Arabidopsis seedlings were sprayed with 0.4 mM D-luciferin (potassium salt, Wako) and luciferase activity was detected with an EM-CCD digital camera (imagEM, Hamamatsu Photonics, Japan). The raw luciferase activity data were analysed with HAKAWO imaging software (version 2.1, Hamamatsu Photonics, Japan).
Data analysis
Significance of differences between data sets was evaluated by Student's t test. Calculations were carried out with Microsoft Excel software.
Results
Identification of L-GalL-responsive genes by microarray screening
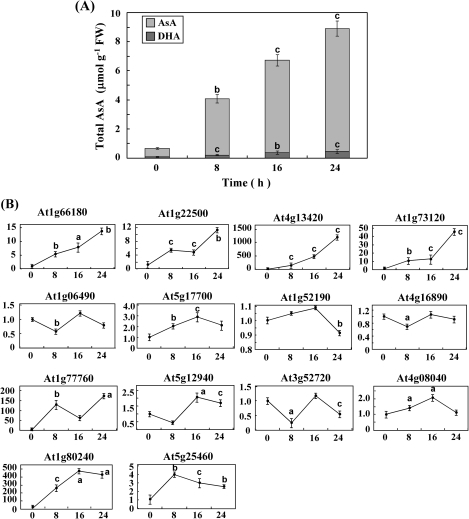
The strategy to identify putative AsA-responsive genes was to determine which genes respond to supply of the AsA precursor L-GalL. L-GalL is the immediate precursor of AsA and effectively increases AsA pool size (Wheeler et al., 1998). The AsA-deficient mutant vtc2-1 was used (Conklin et al., 2000), because it exhibited the lowest level of leaf AsA among all the established vtc mutant lines. This mutant is affected in the AsA biosynthesis enzyme GDP-L-galactose phosphorylase and its AsA concentration can be rescued by L-galactose feeding (Dowdle et al., 2007). Leaves from 6-week-old mutant plants were cut off and incubated with 5 mM L-galactose or water (as a control) under continuous illumination at 80 μmol photons m−2 s−1. The leaves were sampled at 4, 8, 12, and 16 h, respectively. The AsA concentration (a sum of AsA and DHA) rose constantly over the incubation time and increased 3.7-fold after 16 h incubation (see Supplementary Fig. S1 at JXB online).
To identify the genes which respond to L-GalL feeding, total RNA was extracted from the 16 h L-GalL-incubated and control samples, and transcript levels were determined by hybridization to an Affymetrix ATH1 GeneChip. 337 transcripts showed greater than 2-fold response to L-GalL feeding (see Supplementary Table S1 at JXB online). Among these genes, 14 transcripts were up-regulated by 4-fold or more, and were shown to be the most sensitive to feeding of L-GalL and the consequently self-synthesized AsA (see Supplementary Table S2 at JXB online). By contrast, 518 transcripts were decreased by 2-fold or more. Amongst these, 36 transcripts were reduced by 10-fold or more (see Supplementary Table S1 at JXB online). The results identified a substantial number of candidate genes whose expression is influenced by supplying vtc2-1 mutant leaves with the AsA precursor L-GalL. Thus the microarray was used as a screening tool that helped in the identification of the above 14 genes, which possibly included the hypothetical AsA-responsive genes. These 14 candidate genes were analysed further.
Evaluation of candidate L-GalL-responsive genes in vtc2-1 by real-time PCR
The effect of L-GalL feeding on transcript levels of 14 candidate genes identified by DNA microarray analysis (see Supplementary Table S2 at JXB online) was investigated further using real-time PCR. Changes in transcript level were followed over 24 h after feeding 5 mM L-GalL to vtc2-1 leaves (Fig. 1A). Four transcripts, At1g66180, At1g22500, At4g13420, and At1g73120, increased significantly at the end of L-GalL feeding (Fig. 1B). The fold changes of the expression of these four genes were all higher than that of the AsA content. Other candidate genes did not show clear trends in expression with increasing AsA level. It is interesting that the transcript level of At4g13420 increased by almost 1200-fold of control after 24 h incubation in L-GalL.
Fig. 1.
Quantitative real-time PCR measurement of the transcript levels of 14 genes previously identified as increased over 4-fold in vtc2-1 leaves fed with L-GalL by microarray analysis. Increase in total AsA content of L-GalL fed vtc2-1 leaves over a 24 h time-course following feeding with 5 mM L-GalL (A). Transcript levels of 14 genes over a 24 h time-course following feeding with 5 mM L-GalL (B). The gene transcript levels were normalized against actin-2. Data are the mean values ±SD of three independent experiments. Values that are significantly different from the control according to Student's t test are indicated; a, P <0.05; b, P <0.01; c, P <0.001.
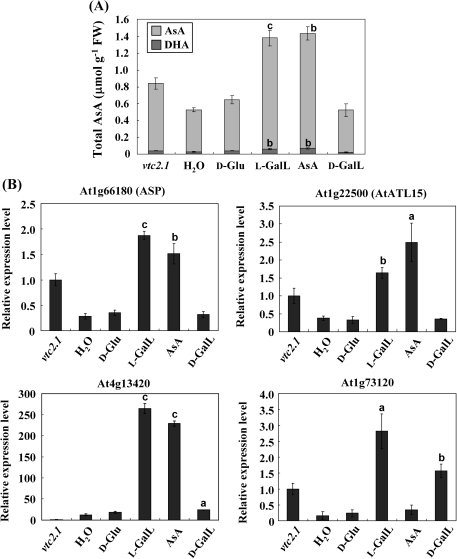
While L-GalL is a precursor for AsA biosynthesis, it is necessary to demonstrate that the effect on gene expression is actually AsA-dependent and not caused by its action as a possible carbon source. Therefore the effect of D-Glu, L-GalL, and AsA feeding was compared in the dark. D-galactonolactone (D-GalL), an optical isomer of L-GalL, was also selected. L-GalL and AsA both increased the total AsA concentration in the leaves while D-Glu and D-GalL had no effect (Fig. 2A). It should be noted that the AsA concentrations reached in the dark are substantially lower than those in the light (Fig. 1A). The transcript levels of four of the L-GalL-responsive genes were investigated by real-time PCR. Three of these (At1g66180, At1g22500, and At4g13420) responded to both L-GalL and AsA, and no significant increase was caused by feeding with D-Glu and D-GalL, while At1g73120 responded to L-GalL and its optical isomer only (Fig. 2B). None of the genes responded to D-Glu. The expression level of At4g13420 increased by more than 200-fold by feeding with L-GalL in the dark (Fig. 2B). In addition, both wild type and vtc2.1 were fed with 5 mM DHA for 3 h and 6 h. As a result, both DHA and total AsA contents, and the expression level of At1g66180 and At1g22500 in both plants were not affected by DHA feeding within at least 6 h (see Supplementary Figs S2 and S3 at JXB online), indicating that these genes were highly responsive to AsA and L-GalL, not DHA.
Fig. 2.
Effect of D-Glu, L-GalL, D-GalL, and AsA feeding in the dark on AsA concentration and transcript levels of four L-GalL-responsive genes in vtc2-1. Total AsA concentration in leaves after feeding 5 mM D-Glu, L-GalL, D-GalL, and AsA to detached leaves in the dark for 16 h (A). The left hand bar (‘untreated’) indicates the AsA concentration in intact plants. Level was detected of each sample including the leaves of vtc2-1 without incubation. Relative transcript levels of four genes following feeding 5 mM D-Glu, L-GalL, D-GalL, and AsA to detached leaves in the dark (B). The left hand bar (‘untreated’) indicates the expression level in intact plants. Total RNA was extracted from each above samples and reverse transcribed to cDNA that is used as templates for real-time PCR analysis with specific primers of the four up-regulated genes selected out from the previous real-time PCR results. The transcript levels were normalized against gene actin-2. Data are the mean values ±SD of three independent experiments (n=3). Values that are significantly different from control according to Student's t test are indicated; a, P <0.05; b, P <0.01; c, P <0.001.
AsA and light responsiveness of At1g66180, At1g22500, and At4g13420 in wild-type Arabidopsis
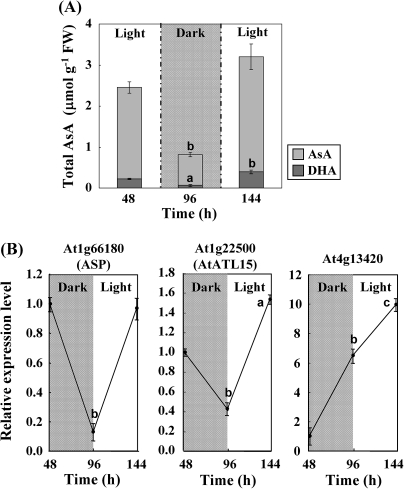
The previous analysis identified three AsA-responsive candidate genes (At1g66180, At1g22500, and At4g13420) in the vtc2-1 mutant background, so further experiments were carried out to determine if they are also AsA-responsive in wild-type plants. Since light is a key factor in generating the AsA pool size (Müller-Moulé et al., 2004; Dowdle et al., 2007; Yabuta et al., 2007), the response in light and dark was compared. Arabidopsis wild-type leaves were exposed to light (80 μmol photons m−2 s−1) for 48 h, and then moved to the dark for another 48 h, and finally transferred to light for 48 h at the same light intensity. At the end of each treatment, the leaves were collected. The leaves of the wild-type plants illuminated continuously for 48 h contained more than 5-fold the total AsA of 48 h dark-treated plants (Fig. 3A). Results of real-time PCR showed that the transcript levels of At1g66180 (aspartyl protease family protein, ASP) and At1g22500 (C3HC4-type RING finger protein, AtATL15) were higher after periods in the light, while the K+ transporter gene At4g13420 (HAK5 potassium transporter) steadily increased over the experimental period (Fig. 3B). Therefore, the AsA responsiveness of At1g66180 and At1g22500 in wild-type plants was investigated further.
Fig. 3.
Effect of light and dark on AsA concentration and transcript levels of three L-GalL-responsive genes in wild-type Arabidopsis. Plants previously grown under a 14 h light period at 80 μmol photons m−2 s−1 were transferred to a cycle of 48 h light, 48 h dark, and 48 h light at the same light intensity. Total AsA level concentration in leaves (A). Relative transcript levels of three L-GalL-responsive genes (B). The transcript levels were normalized against actin-2. Data are the mean values ±SD of three independent experiments. Values that are significantly different from control according to Student's t test are indicated; a, P <0.05; b, P <0.01; c, P <0.001.
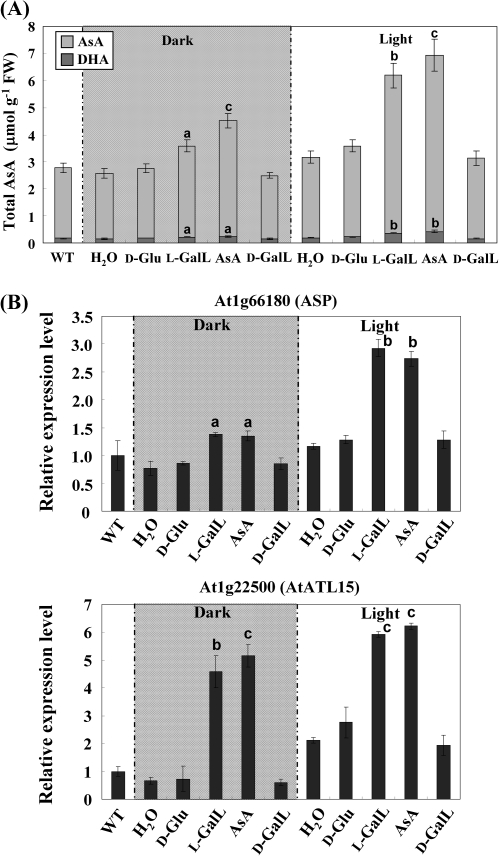
Arabidopsis wild-type leaves were incubated with 5 mM D-Glu, L-GalL, AsA, D-GalL, and H2O under dark and light (80 μmol photons m−2 s−1) conditions for 16 h. AsA concentration increased after 16 h dark incubation with L-GalL and AsA: the contents were 3.6 and 4.5 μmol g−1 FW, respectively, they were a little higher than the control, and no increase was observed after D-Glu and D-GalL feeding. Samples supplied with L-GalL and AsA in the light showed an increase in AsA concentration of over 2-fold compared with the control and D-Glu (Fig. 4A). There was no significant difference in the redox status of AsA after the feeding. The relative transcript levels of both genes were higher in the light and both were substantially induced by L-GalL and AsA feeding but not D-Glc and D-GalL (Fig. 4B). The results suggested that both At1g66180 and At1g22500 transcript levels were responsive to the AsA status of the leaf and to light in both the wild type and the AsA-deficient vtc2-1 mutant.
Fig. 4.
Effect of D-Glu, L-GalL, D-GalL, and AsA feeding on AsA concentration and transcript levels of two L-GalL-responsive genes in Arabidopsis wild type. Excised leaves from wild-type plants were fed with 5 mM D-Glu, L-GalL, D-GalL, and AsA in the light (80 μmol photons m−2 s−1) or in the dark for 16 h. Total AsA concentration in leaves (A). The left hand bar (‘untreated’) indicates the AsA concentration in intact plants. Relative transcript levels of genes At1g66180 and At1g22500 (B). The left hand bar (‘untreated’) indicates the expression level in intact plants. The transcript levels were normalized against actin-2. Data are the mean values ±SD of three independent experiments. Values that are significantly different from control according to Student's t test are indicated; a, P <0.05; b, P <0.01; c, P <0.001.
AsA-responsive expression of C4HC4-type RING finger (At1g22500) promoter::luciferase constructs in transgenic Arabidopsis
Next, promoter/reporter analysis was carried out to evaluate the AsA-responsiveness of the aspartyl protease (ASP, At1g66180) and C3HC4-type RING finger (AtATL15, At1g22500) genes further. The promoter region of each gene (∼2 kb upstream) was fused with luciferase and transformed into wild-type Arabidopsis. Homozygous lines carrying ASP promoter::LUC and AtATL15 promoter::LUC were produced. The AtATL15 promoter::LUC lines showed luminescence but ASP promoter::LUC transformants were not luminescent.
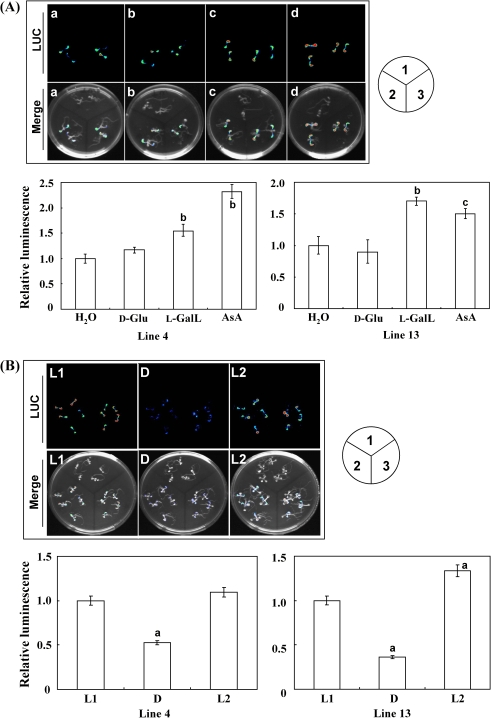
One-week-old transgenic seedlings of AtATL15 promoter::LUC, grown on MS medium, were supplemented with 5 mM of D-Glu, L-GalL, and AsA under light for 16 h, respectively. The luciferase activities of both homozygous lines 4 and 13 fed with L-GalL and AsA were significantly higher than the control and D-Glc-fed seedlings (Fig. 5A). Compared with the seedlings fed with H2O, the luciferase activity of line 4 was increased by 54% and 132% by feeding with L-GalL and AsA, and only by 17% with D-Glu. Line 13 was increased by 70% and 50% after incubation with L-GalL and AsA, and decreased by 11% with D-Glu. The AtATL15 promoter::LUC construct was also light-responsive. Seedlings kept in the light for 24 h had higher luciferase activity than those kept in the dark for 24 h (Fig. 5B). These results were consistent with the previous real-time PCR results.
Fig. 5.
Analysis of AtATL15 promoter activity with LUC reporter. The promoter of gene AtATL15 was characterized with reporter gene LUC within two homozygous T3 transgenic lines. One-week-old T3 transgenic homozygous lines were incubated with H2O, 5 mM D-Glu, 5 mM L-GalL, and 5 mM AsA under light for 16 h. AtATL15 gene promoter activity was observed through the detection of LUC bioluminescence with a CCD camera (A). Two-week-old T3 transgenic homozygous lines were treated over a time-course, the first 24 h light (L1), 24 h dark (D), and then 24 h light (L2) (B). Light intensity was 80 μmol photons m−2 s−1. LUC bioluminescence was observed at the end of each course. The relative luminescence intensity compared with the control was calculated for each case. Merge, mixed picture; 1, wild type; 2, line 4; 3, line 13. Data are the mean values ±SD of three independent experiments. Values that are significantly different from control according to Student's t test are indicated; a, P <0.05; b, P <0.01; c, P <0.001.
Discussion
The aim of this work was to identify genes whose expression is responsive to leaf AsA status. An initial screen using microarray analysis of the AsA deficient vtc2-1 mutant supplied with the AsA precursor L-GalL provided a number of candidate genes. A subset of these genes were investigated further and two (ASP and AtATL15) were confirmed to be highly responsive to leaf AsA concentration and light.
Identification of L-GalL and AsA-responsive genes
In the initial screen for potentially AsA-responsive genes the AsA-deficient mutant vtc2-1 was supplemented with the AsA precursor L-GalL. This mutant is blocked in AsA synthesis at the step involving GDP-L-galactose phosphorylase (Dowdle et al., 2007). This is upstream of L-GalL dehydrogenase, the mitochondrial enzyme that oxidizes L-GalL to AsA (Østergaard et al., 1997; Siendones et al., 1999; Bartoli et al., 2000). Supplying L-GalL to vtc2-1 was therefore effective in increasing foliar AsA. The extent of AsA accumulation from L-GalL was increased by light, as has been noted previously (Bartoli et al., 2006). It is interesting that feeding AsA itself also resulted in greater AsA accumulation in the light than in the dark, suggesting that AsA uptake or turnover are light responsive. Screening for L-GalL-responsive genes in vtc2-1 using the Affymetrix ATH1 GeneChip identified 337 genes whose transcripts increased more than 2-fold and 518 transcripts that decreased by more than 2-fold. Since the transcriptome analysis was unreplicated, a detailed analysis of the classes of genes affected by L-GalL feeding and a comparison with previously published analysis of the vtc1 transcriptome (Pastori et al., 2003) would be speculative. However, a high proportion of genes identified by this screen proved to be L-GalL-responsive after real-time PCR. Gene Ontology (GO) analysis does not reveal over-representation of any specific functional categories or pathways (see Supplementary Fig S4 at JXB online). Further analysis of the expression of four L-GalL up-regulated genes by real-time PCR in vtc2-1 confirmed that three of them (ASP; AtATL15, and At4g13420/HAK5, a potassium transporter) were also up-regulated by AsA feeding but not D-Glc and D-GalL, while At1g73120, encoding a protein of unknown function was responsive to L-GalL and D-GalL but not to AsA (Fig. 2B), indicating that the optical isomer can also regulate the gene expression. Because the three genes respond to both L-GalL and AsA and L-GalL increases the AsA pool size (Fig. 2B), it seems likely that the L-GalL response is mediated by AsA. Furthermore, the transcripts of these genes increased by 8 h after L-GalL feeding suggesting a relatively rapid response to AsA status. It is worth noting that the expression level of At4g13420 (K+ transporter) was strongly up-regulated by exogenous L-GalL and AsA (Fig. 2). From the eFP database (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), the expression level of gene At4g13420 was highest in mature pollen, but moderate in leaves at all growth stages. Interestingly, the results from our time-course experiment showed that the expression of this gene was light-independent and did not seem to respond to the endogenous AsA level (Fig. 3B). The mechanism and physiological function is still largely unknown, but it might be highly responsive to broad cellular redox status.
Further investigation of At1g66180 and At1g22500 confirmed that their transcript levels increased in WT plants in response to both L-GalL and AsA supplementation. Furthermore, these genes were also light-responsive, showing greater expression in the light than the dark. It is not clear if their expression is affected by the endogenous AsA pool, which was higher in the light under the experimental conditions used, or if they respond to light and AsA independently. To provide a tool to follow the expression of the two genes, promoter::luciferase fusions were made. Luminescence could be detected for the AtATL15 but not the ASP construct. The 2 kb region upstream of the AtATL15 gene directed luciferase expression in an AsA-, L-GalL-, and light-dependent manner and will provide a tool to identify the cis elements responsible for AsA and light-dependent expression. The approach used in this study to identify genes whose expression responds fairly rapidly to the altered AsA status has yielded several genes, but it is likely that further investigation of the candidate genes identified by microarray analysis will yield more.
Expression and functional analysis of At1g66180/aspartyl protease and At1g22500/C3HC4-type RING finger protein
What are the possible functions of AsA and L-GalL-induced genes? At1g22500 is an C3HC4-type RING finger protein (AtATL15), which has been demonstrated to have E3 ubiquitin ligase activity and is therefore very likely to be involved in proteasome-mediated protein degradation (Stone et al., 2005). Furthermore, it is reported that mitotic activity in the nucellar projection, tissues that are responsible for nutrient transfer, endosperm/embryo nutrition, and grain development in barley grain, is indicated by the high amount of mRNA of different histones and the preferred expression of C3HC4-type RING finger ubiquitin ligases (Thiel et al., 2008). C3HC4-type RING finger proteins were found to play a key role in cell cycle regulation of Arabidopsis (Fleury et al., 2007; Liu et al., 2007). Because the cellular AsA content can modulate the plant cell cycle (Potters et al., 2004), regulate elongation growth (Fry, 1998; Tokunaga et al., 2005), and control cell division (Horemans et al., 2000), it is hypothesized that AsA may perform the above functions through this C3HC4-type RING finger gene (At1g22500). At1g66180 is a predicted A1 family aspartyl protease (ASP) which has 59 other members in Arabidopsis (Beers et al., 2004). It has a weakly predicted transmembrane domain at the N-terminus and is strongly predicted to be located in the secretory pathway and weakly-predicted to be chloroplast-targeted (http://aramemnon.botanik.uni-koeln.de). Protease activity of At1g66180 has not been verified experimentally. A comparison of the response of both genes to stimuli using Genevestigator (Zimmermann et al., 2004) showed that they are both down-regulated by cold, glucose, low nitrogen, drought, syringolin A, and induced programmed cell death (PCD) (see Supplementary Fig. S5 at JXB online). At1g66180 is induced after exposure of the flu mutant of Arabidopsis, to high light, suggesting that it responds to singlet oxygen (op den Camp et al., 2003). Its transcript levels are also increased 25-fold in the srk2d/e/i triple mutants which have enhanced insensitivity to abscisic acid and reduced drought tolerance (Nakashima et al., 2009). Extending the comparison to the eight genes induced by L-GalL, all are down-regulated by low nitrogen, glucose, syringolin A and seven out of eight are down-regulated by heat shock-induced PCD. Looking further at the top 50 L-GalL-induced genes from the Affymetrix array, 68% are down-regulated by low N, 62% by syringolin, and 52% by induced PCD. There are no stimuli that consistently up-regulate any of the gene sets (see Supplementary Table S1 at JXB online). This analysis suggests that conditions that tend to limit growth down-regulate the genes that are up-regulated by L-GalL/AsA. The preponderance of genes that are down-regulated by syringolin A is interesting because this non-ribosomal peptide, synthesized by Pseudomonas syringae, is a proteasome inhibitor (Groll et al., 2008). Further investigation is needed to determine the functions of these AsA-responsive genes. However, it is interesting to note that AsA has possible roles in plant growth because vtc mutants are small (Conklin et al., 1996), have altered flowering time (Barth et al., 2006; Kotchoni et al., 2009), and are more resistant to pathogens (Barth et al., 2004; Colville and Smirnoff, 2008). Seedling growth is arrested in vtc2/vtc5 double mutants that are unable to synthesise AsA (Dowdle et al., 2007). Since many hormone signalling pathways involve proteasome-mediated protein degradation (Santner and Estelle, 2010), the induction of an E3 ubiquitin ligase and an aspartyl protease may reflect activation of certain growth processes by AsA supplementation.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. List of transcripts either induced or repressed in leaves of the Arabidopsis vtc2-1 mutant by feeding with AsA precursor L-GalL.
Supplementary Table S2. L-GalL responsive genes in the vtc2-1 mutant identified with Affymetrix ATH1 GeneChips.
Supplementary Fig. S1. Effect of L-GalL supplementation on total AsA concentration in the AsA-deficient Arabidopsis mutant vtc2-1.
Supplementary Fig. S2. Effect of DHA feeding on AsA concentration and transcript levels of four L-GalL-responsive genes in the vtc2-1 mutant.
Supplementary Fig. S3. Effect of DHA feeding on AsA concentration and transcript levels of four L-GalL-responsive genes in the Arabidopsis thaliana wild type.
Supplementary Fig. S4. GO analysis of 337 genes whose transcript levels were increased 2-fold or more by L-GalL.
Supplementary Fig. S5. Expression pattern over a range of experimental treatments of L-GalL-responsive genes.
Supplementary Fig. S6. Transcripts levels of reference genes in each group of samples.
Acknowledgments
This work was partly supported by Grants-in-aid for Scientific Research (A) (SS: 22248042) and (B) (TI: 21380207) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by the Japan Society for the Promotion of Science, and the Royal Society under the Japan-UK Research Cooperative Program (TI).
References
- Arrigoni O, Arrigoni-Liso R, Calabrese G. Ascorbic acid requirement for biosynthesis of hydroxyproline-containing proteins in plants. FEBS Letters. 1977;82:135–138. doi: 10.1016/0014-5793(77)80903-4. [DOI] [PubMed] [Google Scholar]
- Arrigoni O, De Tullio MC. Ascorbic acid: much more than just an antioxidant. Biochimica et Biophysica Acta. 2002;1569:1–9. doi: 10.1016/s0304-4165(01)00235-5. [DOI] [PubMed] [Google Scholar]
- Barth C, De Tullio M, Conklin PL. The role of ascorbic acid in the control of flowering time and the onset of senescence. Journal of Experimental Botany. 2006;57:1657–1665. doi: 10.1093/jxb/erj198. [DOI] [PubMed] [Google Scholar]
- Barth C, Moeder W, Klessig DF, Conklin PL. The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1. Plant Physiology. 2004;134:1784–1792. doi: 10.1104/pp.103.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Pastori GM, Foyer CH. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiology. 2000;123:335–344. doi: 10.1104/pp.123.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Yu J, Gómez F, Fernández L, McIntosh L, Foyer CH. Interrelationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. Journal of Experimental Botany. 2006;57:1621–1631. doi: 10.1093/jxb/erl005. [DOI] [PubMed] [Google Scholar]
- Beers EP, Jones AM, Dickerman AW. The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry. 2004;65:43–58. doi: 10.1016/j.phytochem.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Chen Z, Gallie DR. The ascorbic acid redox state controls guard cell signaling and stomatal movement. The Plant Cell. 2004;16:1143–1162. doi: 10.1105/tpc.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colville L, Smirnoff N. Antioxidant status, peroxidase activity, and PR protein transcript levels in ascorbate-deficient Arabidopsis thaliana vtc mutants. Journal of Experimental Botany. 2008;59:3857–3868. doi: 10.1093/jxb/ern229. [DOI] [PubMed] [Google Scholar]
- Conklin PL, Saracco SA, Norris SR, Last RL. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics. 2000;154:847–856. doi: 10.1093/genetics/154.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Williams EH, Last RL. Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proceedings of the National Academy of Sciences, USA. 1996;93:9970–9974. doi: 10.1073/pnas.93.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MW, Van Montagu M, Inzé D, Sanmartin M, Kanellis A, Smirnoff N, Benzie IJJ, Strain JJ, Favell D, Fletcher J. Plant L-ascorbic acid: chemistry, function, metabolism, bioavailability and effects of processing. Journal of the Science of Food and Agriculture. 2000;80:825–860. [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. The Plant Journal. 2007;52:673–689. doi: 10.1111/j.1365-313X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. The Plant Cell. 2007;19:417–432. doi: 10.1105/tpc.106.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. The Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. Oxidative scission of plant cell wall polysaccharides by ascorbate-induced hydroxyl radicals. Biochemistry Journal. 1998;332:507–515. doi: 10.1042/bj3320507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Schellenberg B, Bachmann AS, Archer CR, Huber R, Powell TK, Lindow S, Kaiser M, Dudler R. A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism. Nature. 2008;452:755–758. doi: 10.1038/nature06782. [DOI] [PubMed] [Google Scholar]
- Horemans N, Foyer CH, Asard H. Transport and action of ascorbate at the plant plasma membrane. Trends in Plant Science. 2000;5:263–267. doi: 10.1016/s1360-1385(00)01649-6. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Shigeoka S. Recent advances in ascorbate biosynthesis and the physiological significance of ascorbate peroxidase in photosynthesizing organisms. Bioscience, Biotechnology and Biochemistry. 2008;72:1143–1154. doi: 10.1271/bbb.80062. [DOI] [PubMed] [Google Scholar]
- Kampfenkel K, Van Montagu M, Inzé D. Extraction and determination of ascorbate and dehydroascorbate from plant-tissue. Analytical Biochemistry. 1995;225:165–167. doi: 10.1006/abio.1995.1127. [DOI] [PubMed] [Google Scholar]
- Kiddle G, Pastori GM, Bernard S, Pignocchi C, Antoniw J, Verrier PJ, Foyer CH. Effects of leaf ascorbate content on defense and photosynthesis gene expression in Arabidopsis thaliana. Antioxidants and Redox Signaling. 2003;5:23–32. doi: 10.1089/152308603321223513. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate–dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. The Plant Cell. 2001;13:681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchoni SO, Larrimore KE, Mukherjee M, Kempinski CF, Barth C. Alterations in the endogenous ascorbic acid content affect flowering time in Arabidopsis. Plant Physiology. 2009;149:803–815. doi: 10.1104/pp.108.132324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Koornneef M, Soppe WJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. The Plant Cell. 2007;19:433–444. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirica LM, Klinman JP. The nature of O2 activation by the ethylene-forming enzyme 1-aminocyclopropane-1-carboxylic acid oxidase. Proceedings of the National Academy of Sciences, USA. 2008;105:1814–1819. doi: 10.1073/pnas.0711626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Larrimore KE, Ahmed NJ, Bedick TS, Barghouthi NT, Traw MB, Barth C. Ascorbic acid deficiency in Arabidopsis induces constitutive priming that is dependent on hydrogen peroxide, salicylic acid, and the NPR1 gene. Molecular Plant–Microbe Interactions. 2010;23:340–351. doi: 10.1094/MPMI-23-3-0340. [DOI] [PubMed] [Google Scholar]
- Müller-Moulé P, Golan T, Niyogi KK. Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiology. 2004;134:1163–1172. doi: 10.1104/pp.103.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1, and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant and Cell Physiology. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. ASCORBATE AND GLUTATHIONE: keeping active oxygen under control. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. The Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard J, Persiau G, Davey MW, Bauw G, Van Montagu M. Isolation of a cDNA coding for L-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. Journal of Biological Chemistry. 1997;272:30009–30016. doi: 10.1074/jbc.272.48.30009. [DOI] [PubMed] [Google Scholar]
- Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, Foyer CH. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. The Plant Cell. 2003;15:939–951. doi: 10.1105/tpc.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters G, Horemans N, Bellone S, Caubergs RJ, Trost P, Guisez Y, Asard H. Dehydroascorbate influences the plant cell cycle through a glutathione-independent reduction mechanism. Plant Physiology. 2004;134:1479–1487. doi: 10.1104/pp.103.033548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. The Plant Journal. 2010;61:1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siendones E, Gonzalez-Reyes JA, Santos-Ocana C, Navas P, Cordoba F. Biosynthesis of ascorbic acid in kidney bean: L-galactono-γ-lactone dehydrogenase is an intrinsic protein located at the mitochondrial inner membrane. Plant Physiology. 1999;120:907–912. doi: 10.1104/pp.120.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N. Ascorbate biosynthesis and function in photoprotection. Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences. 2000;355:1455–1464. doi: 10.1098/rstb.2000.0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiology. 2005;137:113–130. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel J, Weier D, Sreenivasulu N, Strickert M, Weichert N, Melzer M, Czauderna T, Wobus U, Weber H, Weschke W. Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells: a microdissection-based transcriptome study of young barley grains. Plant Physiology. 2008;148:1436–1452. doi: 10.1104/pp.108.127001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga T, Miyahara K, Tabata K, Esaka M. Generation and properties of L-ascorbic acid overproducing transgenic tobacco cells expressing sense RNA for L-galactono-1,4-lactone dehydrogenase. Planta. 2005;220:854–863. doi: 10.1007/s00425-004-1406-3. [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Nakajima J, Welford RWD, Yamazaki M, Saito K, Schofield CJ. Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis. Journal of Biological Chemistry. 2004;279:1206–1216. doi: 10.1074/jbc.M309228200. [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Mieda T, Rapolu M, Nakamura A, Motoki T, Maruta T, Yoshimura K, Ishikawa T, Shigeoka S. Light regulation of ascorbate biosynthesis is dependent on the photosynthetic electron transport chain but independent of sugars in Arabidopsis. Journal of Experimental Botany. 2007;58:2661–2671. doi: 10.1093/jxb/erm124. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.