Abstract
Arabidopsis cyclic nucleotide-gated ion channels (AtCNGCs) form a large family consisting of 20 members. These channels have so far been reported to be involved in a diverse range of physiological phenomena. For example, AtCNGC18 was reported to play an important role in pollen tube growth, while AtCNGC2, 4, 11, and 12 were implicated in mediating pathogen defence. To identify additional functions for AtCNGC11 and 12, various physiological aspects were analysed using both AtCNGC11 and 12 single knockout mutants as well as a double mutant. Although AtCNGC11 and 12 can function as K+ and Ca2+ channels in yeast, it was found that the loss of AtCNGC11 and 12 in Arabidopsis caused increased sensitivity to Ca2+ but not K+, indicating a specific function for these genes in Ca2+ signalling in planta. However, they did not show an alteration in Ca2+ accumulation, suggesting that AtCNGC11 and 12 are not involved in general Ca2+ homeostasis but rather in the endogenous movement of Ca2+ and/or Ca2+ signalling. Furthermore, these channels synergistically contribute to the generation of a Ca2+ signal that leads to gravitropic bending. Finally, AtCNGC11 and 12 gene expression was induced during dark-induced senescence and AtCNGC11 and 12 knockout mutants displayed enhanced chlorophyll loss, which was even more pronounced in the double mutant, also indicating synergistic roles in senescence. The findings indicate that (i) some CNGC family members have multiple physiological functions and (ii) some plant CNGCs share the same biological function and work in a synergistic manner.
Keywords: Calcium, CNGC, cyclic nucleotide-gated ion channel, gravitropism, senescence
Introduction
Plants respond to a myriad of external stimuli throughout their life cycle in order to cope with an ever-changing environment. After perception of these stimuli, secondary messengers are required to amplify the signal leading to a physical response. One such secondary messenger is Ca2+. Spatial and temporal changes of cytosolic free Ca2+ concentrations ([Ca2+]cyt) are frequently observed as an immediate response to many stimuli (Rudd and Franklin-Tong, 1999; Dodd et al., 2010). In the case of gravitropism, Ca2+ fluxes have been shown to occur in the roots seconds after their angle of gravity was changed (Plieth and Trewavas, 2002). Additionally, it has been reported that Ca2+ chelators reduce the responsiveness of roots to gravity stimuli (Hasenstein and Evans, 1986), clearly indicating the importance of Ca2+ in gravitropism.
Perturbations of Ca2+ homeostasis can also affect developmental processes such as senescence. For example, treatment with excess amounts of Ca2+ has been shown to suppress dark-induced senescence in a number of plant species (Poovaiah and Leopold, 1973; Ferguson et al., 1983; Cheour et al., 1992). However, the identities of the channels which facilitate movement of Ca2+ upon each stimulus are not known. Possible candidates are cyclic nucleotide-gated ion channels (CNGCs; Kaplan et al., 2007; Dietrich et al., 2010; Dodd et al., 2010). These channels were first identified in animals, where they play key roles in light and olfactory signalling (Zagotta and Siegelbaum, 1996; Craven and Zagotta, 2006). In Arabidopsis thaliana, there is a large CNGC family consisting of 20 members (Mäser et al., 2001). All Arabidopsis CNGCs (AtCNGCs) tested so far have been shown to transport K+, and a further subset, which includes AtCNGC1, 2, 11, 12, and 18, have been shown to translocate Ca2+ (Leng et al., 1999; Ali et al., 2005; Frietsch et al., 2007; Urquhart et al., 2007). Furthermore, it has been reported that these five AtCNGCs localize to the plasma membrane, suggesting that they transport cations from the apoplast into the cell (Mercier et al., 2004; Ma et al., 2006; Frietsch et al., 2007; Urquhart et al., 2007). These data indicate a biological role for AtCNGCs in K+ or Ca2+ uptake and homeostasis.
To date, a number of studies have implicated AtCNGCs in Ca2+ signalling and homeostasis during development using knockout (KO) mutants. For example, root growth of seedlings of the AtCNGC1 KO mutant plant, cngc1, displayed insensitivity to Ca2+ and reduced endogenous Ca2+ levels, indicating that AtCNGC1 plays a role in maintaining proper Ca2+ homeostasis during development (Ma et al., 2006). In addition, AtCNGC1 as well as AtCNGC10 have been shown to play a role in root gravitropism, as cngc1 and AtCNGC10 antisense lines exhibited reduced responsiveness to gravity (Ma et al., 2006; Borsics et al., 2007). On the other hand, the AtCNGC2 KO mutants cngc2-1 and cngc2-2 showed increased sensitivity to Ca2+. Furthermore, gene expression profiling of cngc2-2 showed strong similarity to the expression pattern of wild-type plants grown at elevated Ca2+ levels, indicating abnormal Ca2+ sensing in cngc2 (Chan et al., 2008). Adult cngc2 mutants displayed decreased size and fertility when grown on Ca2+-supplemented medium (Chan et al., 2003; Chaiwongsar et al., 2009). Pollen tube growth is another developmental process that is driven by Ca2+ fluxes (Michard et al., 2009), and it has been shown that the loss of AtCNGC18 causes abnormalities in pollen tube growth (Frietsch et al., 2007; Chang et al., 2008).
In addition to developmental aspects, plant CNGCs have been shown to be involved in Ca2+ signalling during pathogen defence. Loss-of-function AtCNGC2 and 4 mutants (dnd1 and dnd2/hlm1, respectively) exhibited increased resistance to pathogen infection but fail to induce a hypersensitive response (HR) after infection with some avirulent pathogens (Clough et al., 2000; Balague et al., 2003; Jurkowski et al., 2004). HR is the localized programmed cell death at and around the site of pathogen infection and is a Ca2+-dependent process (Xu and Heath, 1998). AtCNGC11 and 12 KO mutant plants exhibited decreased resistance to an avirulent isolate of the oomycete pathogen Hyaloperonospora arabidopsidis as well as avirulent strains of the bacterial pathogen Pseudomonas syringae, but they did not show any alteration in susceptibility to virulent isolates, indicating a specific role for these two channels in pathogen defence (Yoshioka et al., 2006; Moeder et al., 2011). Interestingly, the cpr22 mutant, which has a deletion between AtCNGC11 and AtCNGC12 resulting in the creation of a novel, but functional chimeric CNGC comprised of the front half of AtCNGC11 and the second half of AtCNGC12, exhibits increased resistance to pathogen infection similar to that of dnd1 and dnd2/hlm1 (Yoshioka et al., 2001, 2006; Moeder et al., 2011). Since AtCNGC2 has roles in both defence and development, as mentioned above (Chan et al., 2003; Chaiwongsar et al., 2009), it is possible that AtCNGC11 and 12 may have other biological functions in addition to defence.
In this study, the expression profiles of AtCNGC11 and 12 are investigated and the roles of AtCNGC11 and 12 beyond their involvement in defence signalling are explored. As outlined above, cpr22 shares many phenotypes with the Ca2+-sensitive mutant dnd1. Furthermore, it has been shown that both AtCNGC11 and 12 transport Ca2+ (Urquhart et al., 2007). Thus, this prompted the testing of the involvement of AtCNGC11 and 12 in a broad range of Ca2+-dependent responses beyond that of pathogen defence.
Materials and methods
Plant materials and their growth conditions
The A. thaliana insertion mutant lines of AtCNGC11 and 12, salk_026568 and salk_092657, were obtained from the SALK Institute (Alonso et al., 2003) and were named cngc11-1 and cngc12-1, respectively. Information for T-DNA insertion positions, morphological phenotypes, and gene expression analysis were previously reported (Yoshioka et al., 2006). Plants grown in Petri dishes were plated on 0.5× Murashige and Skoog (MS) medium (Sigma, http://www.sigmaaldrich.com/sigma-aldrich/home.html) containing 0.8% agar (BioShop Canada, http://www.bioshopcanada.com) buffered with MES to a pH of 5.7. The growth conditions were as follows: 24 h light at an intensity of 70 μE s−1 m−2 at 24 °C. For those plants grown on soil, Pro-Mix soil (Premier Horticulture Inc., http://www.premierhort.com/) was used and the plants were grown at normal relative humidity (∼60%) and 22 °C under a 9 h light/15 h dark cycle at ∼140 μE m−2 s−1. All seeds were stratified for 2 d, except for seeds for the germination assay which were stratified for 4 d.
Germination assay
Protocols from the Arabidopsis Gantlet Project Conditional Phenotype Analysis website (http://www.gantlet.org/) were used as a basis for the analysis. In triplicate, 30 seeds for each line were plated on 0.5× MS agar medium (basal Ca2+ levels in MS medium <5 mM, basal K+ levels in MS medium <15 mM) augmented with 0, 50, 70, or 100 mM CaCl2 or 100, 150, or 180 mM KCl. The seeds were stratified for 5 d and then moved to light. The emergence of the radicle after 3 d was scored as germination.
Root length assay
Protocols from the Arabidopsis Gantlet Project Conditional Phenotype Analysis website (http://www.gantlet.org/) were used as a basis for the analysis. Seeds were grown vertically on 0.5× MS agar medium for 7 d. In triplicate, eight seedlings were transferred to new plates containing 0.5× MS agar medium with or without added CaCl2 (30, 50, or 70 mM) or KCl (40, 80, or 120 mM). The seedlings were then grown vertically for another 7 d, after which the new primary root growth was measured using the program, ImageJ (http://rsbweb.nih.gov/ij/).
Promoter:GUS (β-glucuronidase) reporter transgenic lines
Promoter regions of AtCNGC11 (At2g46440) and 12 (At2g46450) were amplified from Arabidopsis thaliana ecotype Columbia (Col) genomic DNA using the primer combinations: PromoCNGC11F, 5'-CTCCTAGGCCAGTAAAGAGCTTTATGTG-3'; and PromoCNGC11R, 5'-CTCCTAGGGTTTTTATCTGTCAATCTTC-3'; or PromoCNGC12F, 5'-CTCCTAGGTGTTGCCTCAGAAACCAGCC-3'; and PromoCNGC12R, 5'-CTCCTAGGTGTTGCCTCAGAAACCAGCC-3', respectively. The amplified areas are the 800 bp upstream sequence for AtCNGC11 and the 1100 bp upstream sequence for AtCNGC12 that included the 5'-untranslated regions (UTRs) of the gene of interest and a portion of the 3'-UTRs of the upstream gene. These fragments were then cloned into the pGEM T-easy vector (Promega Corporation, Madison, WI, USA). After the sequence was confirmed, the promoter regions were subcloned into pBI101.2 (Clontech, Mountain View, CA, USA). The vectors were then transformed into Columbia wild-type plants by vacuum infiltration (Bechtold and Pelletier, 1998).
AtCNGC11 and 12 expression analysis during development and senescence
Single insertion, homozygous AtCNGC11 and 12 promoter:GUS reporter transgenic lines of the T3 generation were grown on 0.5× MS agar medium for up to 3 weeks. Seedlings were harvested at 5, 7, 14, and 21 d after germination. Harvested seedlings were placed in GUS buffer composed of 100 mM sodium phosphate buffer, 10 mM NaEDTA (pH 7.5), 0.5 mM K ferrocyanide, 0.5 mM K ferricyanide, and 2 mM 5-bromo-4-chloro-3-indolyl glucuronide. The buffer was vacuum-infiltrated into the samples and incubated at 37 °C for 90 min. After incubation, the tissues were fixed and cleared with a 3:1 solution of ethanol and acetic acid. Lastly, the samples were washed with 70% ethanol before mounting. For dark-induced senescence, the process was the same except the plants were grown for 6 weeks on soil and the tips of the leaf samples were cut just before mounting to be able to mount whole leaves on slides.
Ion content measurement by inductively coupled plasma optical emission spectrometry (ICP-OES)
Seedlings were grown vertically on 0.5× MS agar medium for 2 weeks. The seedlings were transferred to fresh 0.5× MS agar medium with or without 50 mM CaCl2. Samples were taken in triplicate at 0 h, 3 h, 1 d, and 5 d after transfer. Each sample contained 10 seedlings at a weight of ∼20 mg. The samples were dissolved in 500 μl of 68% (v/v) nitric acid at 95 °C and then diluted with water to a final volume of 5 ml. The samples were analysed using a Perkin Elmer Optima 7300 DV ICP-OES and Win32 for ICP V 4.0 control software (Perkin Elmer, Waltham, MA, USA).
Gravitropic responsiveness
The experiment was conducted following the procedure from Ma et al. (2006) with some modifications. Seedlings were grown on 0.5× MS agar medium for 2 weeks. Six seedlings per plate were transferred to fresh 0.5× MS agar medium with or without added CaCl2 (50 mM or 100 mM) and four plates per treatment were tested. After transfer, the plates were rotated 90 ° and the angle of root curvature was measured after 6, 12, and 24 h.
Senescence assay
Plants were grown for 6 weeks on soil, and leaf 7 from each plant was collected (Weaver et al., 1998; Riefler et al., 2006) and fresh weights were measured. Sets of three leaves were floated in water and placed in the dark for 0, 3, 4, or 5 d. Chlorophyll was extracted by homogenizing the individual leaves in a solution of 80% (v/v) acetone and 25 mM HEPES at pH 7.5. The absorbance of the supernatant was measured using a Beckman Coulter DU730 spectrophotometer (Beckman Coulter, Brea, CA, USA) and the equation 17.76 (A646)+7.34(A663) was used to determine the total chlorophyll content (Porra et al., 1989).
Quantitative reverse transcriptase PCR (RT-PCR)
The plants were grown for 6 weeks on soil, and leaf 7 was collected from three plants and floated on water in the dark for 0, 3, and 5 d. The leaves from each time point were pooled, followed by RNA extraction and cDNA generation as described previously (Urquhart et al., 2007). The primers used to analyse the expression of SAG12 (At5g45890) were: forward primer SAG12F, 5'-AGGCACATCGAGTGGATGAC-3'; and reverse primer SAG12R, 5'-TCAATGCGTTCGACGTTGTT-3' (Swartzberg et al., 2008). This primer set amplifies a fragment of ∼100 bp. The quantitative RT-PCR was conducted using an Applied Biosystems Power SYBR Green PCR Master Mix according to the manufacturer's specifications (http://www3.appliedbiosystems.com/AB_Home/index.htm). Amplification was detected using a Bio-Rad Chromo4 real-time PCR detector and analysed using Opticon Monitor version 3.1 (Bio-Rad). Samples were normalized using an elongation factor-1α (At5g60390) primer set: forward EF-1αF, 5'-TGAGCACGCTCTTCTTGCTTTCA-3'; and reverse EF-1αR, 5'-GGTGGTGGCATCCATCTTGTTACA-3' (Czechowski et al., 2005).
Sequence alignment
Sequence alignment was conduced by ALIGN Query (http://xylian.igh.cnrs.fr/bin/align-guess.cgi).
Results
AtCNGC11 and 12 are tandemly repeated on chromosome II and show similarities in their promoter sequences
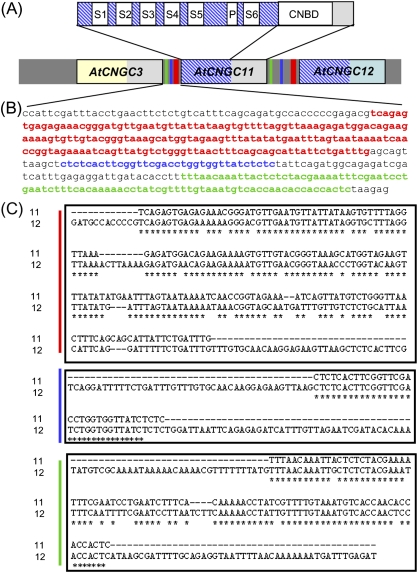
In Arabidopsis, the CNGC family is comprised of 20 members and has two tandemly repeated gene clusters; AtCNGC3, 11, and 12 are tandemly repeated at the south end of chromosome II (Fig. 1A; Yoshioka et al., 2006), while AtCNGC19 and 20 are tandemly repeated on chromosome III (Kugler et al., 2009). The structure of plant CNGCs is similar to that of the voltage-gated outward rectifying (Shaker) K+-selective ion channels, including a cytoplasmic N-terminus, six membrane-spanning regions (S1–S6), a pore domain located between S5 and S6, and a cytoplasmic C-terminus that contains important regulatory domains such as a cyclic nucleotide-binding domain (CNBD; Chin et al., 2009; Fig. 1A). Sequence alignment revealed that AtCNGC11 and 12 share 82% overall sequence identity in their coding sequence (CDS), whereas AtCNGC3 shares only 68% and 62.1% identity with AtCNGC11 and 12, respectively. Interestingly, the initial 1276 bp and 1572 bp of the CDS of AtCNGC11 and AtCNGC12, respectively, share extremely high identity (97.6%), whereas the rest of the CDS exhibits only 55.2% identity. The highly similar region ranges from the start of the gene to just before the CNBD (Fig. 1A). This is reflected in the amino acid sequence, which shows 95% identity in the area before the CNBD and only 47% identity in the C-terminal half. In contrast, AtCNGC3 shares very low identity with AtCNGC11 in the first 1572 bp, but extremely high identity in the C-terminal area starting from the CNBD. This is also reflected in their amino acid sequence; 52% identity in the area before the CNBD and 94% identity starting from the CNBD. Thus, in terms of CDS, AtCNGC11 consists of two parts: the N-terminal half, which is similar to AtCNGC12, and the C-terminal half, which is similar to AtCNGC3 (Fig. 1A).
Fig. 1.
Similarity of the coding sequence and promoter sequence of AtCNGC3, 11, and 12. (A) Domain structure of AtCNGC genes and similarity of the coding sequence and promoter sequence of AtCNGC3, 11, and 12. S1–S6, transmembrane domains; P, pore region; CNBD, cyclic nucleotide-binding domain; grey coloured box, highly similar area between AtCNGC3 and AtCNGC11 containing the CNBD; blue hatched box, highly similar area between AtCNGC11 and AtCNGC12 containing the pore region. Black framed boxes indicate coding regions. Green, blue, and red bars in the intergenic sequence correspond to the sequences with the same colour in B. (B) The 424 bp intergenic upstream sequence of AtCNGC11. Three segments that are colour coded by red, blue, and green indicate the areas that show high similarity to the intergenic upstream sequence of AtCNGC12. (C) Alignments of three areas in the 424 bp intergenic upstream sequence of AtCNGC11 with that of AtCNGC12. The colour coding is the same as in B. Asterisks indicate identical nucleotides.
The intergenic upstream sequence of AtCNGC11, which probably contains the promoter region, is only 424 bp in length (Fig. 1B), whereas that of AtCNGC12 spans 834 bp (TAIR sequence viewer, http://www.arabidopsis.org/). Simple alignment of these two regions results in only 42% identity. However, the intergenic upstream sequence of AtCNGC11 contains three segments that share extremely high similarity with that of AtCNGC12. From position –55 to –238, the upstream sequence of AtCNGC11 (red in Fig. 1B) shares 83% identity; from position –251 to –284 (blue in Fig. 1B) it shares 97% identity; and from position –331 to –418 (green in Fig. 1B) it shares 90% identity (Fig. 1C). Interestingly, no high similarity segments were observed in the upstream sequence of AtCNGC3 (data not shown), suggesting that the expression pattern of AtCNGC3 may not be similar to that of the other two genes although these three genes are tandemly repeated. Based on the analysis with Athena (http://www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/home.pl), there are some known motifs, such as a MYB4-binding motif in both upstream regions; however, the three high similarity segments do not contain any shared known cis-acting elements (data not shown).
AtCNGC11 and 12 expression profiles
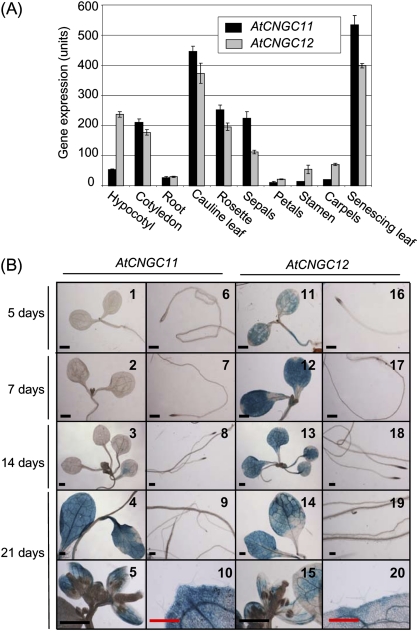
The physiological function of a gene can, in some cases, be associated with its tissue-specific expression pattern. For this reason, expression analyses of AtCNGC11 and AtCNGC12 were carried out using publicly available microarray expression data and promoter:GUS reporter gene constructs. Based on data obtained from the Bio-Array Resource for Plant Functional Genomics (BAR; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Schmid et al., 2005; Winter et al., 2007), the expression of AtCNGC11 and 12 has similar patterns in mature plants but shows differences in seedlings (Fig. 2A). AtCNGC11 is expressed at a level of 210.25±12.02 units in the cotyledons but has a considerably lower expression in the hypocotyls (53.43±2.18 units) and in the roots (27.23±3.56 units). AtCNGC12 has similar expression in the roots (29.91±2.14 units) and cotyledons (177±9.37 units), but significantly higher expression in the hypocotyls (237.14±8.6 units) compared with AtCNGC11. In mature plants, AtCNGC11 and 12 are expressed throughout the shoot, with particularly high expression in cauline leaves and senescing leaves. Neither AtCNGC gene is highly expressed in the flower, but there is some expression in the sepals. However, it should be noted that the probe sets used to obtain the above microarray data are unable to discern between AtCNGC11 and AtCNGC3; thus, the expression level annotated as AtCNGC11 represents the expression of both AtCNGC11 and 3. To separate the expression of AtCNGC11 from that of AtCNGC3 and to confirm further the expression profile of AtCNGC11 and 12, expression analysis was conducted using promoter:GUS reporter constructs. The intergenic upstream sequence of AtCNGC11 or 12 together with their UTRs was cloned from genomic DNA of Arabidopsis ecotype Columbia (Col) wild-type plants and fused to the GUS reporter gene. Stable homozygous transgenic lines were created and the data presented here are representative of two independent transformant lines. In 0.5× MS agar medium-grown seedlings, AtCNGC11 expression could not be detected until 14 d after germination (Fig. 2B, 1–3), whereas AtCNGC12 showed strong expression in both cotyledons and hypocotyls from 5 d after germination (Fig. 2B, 11–13). For neither AtCNGC11 nor 12 could any expression be detected in the root (Fig. 2B, 6–9, 16–19. (Note: the dark areas in the root tips are not GUS staining. These are due to the high density of cells.) In adult plants, the expression patterns of AtCNGC11 and 12 were similar. The expression of both AtCNGC11 and 12 was detected throughout the shoot tissue, with very strong expression in the rosette and cauline leaves (Fig. 2B, 4 and 14). Expression was, however, not detected in the roots, trichomes, and most of the flower organs except for the sepals (Fig. 2B, 9, 10, 19, 20, 5, and 15). This expression pattern was confirmed using soil-grown plants. Although the expression was weaker, the overall expression patterns for both AtCNGC11 and AtCNGC12 agree well with the results from plate-grown plants (Supplementary Fig. S1 available at JXB online). The only exception was that very weak expression was detected in the root. This was also confirmed by semi-quantitative RT-PCR (data not shown). Thus, these data indicate that both genes are expressed in roots, but at a level too low to be detected by GUS staining, which is in agreement with Fig. 2A.
Fig. 2.
Expression pattern of AtCNGC11 and 12. (A) Tissue-specific expression profile of AtCNGC11 (black bar) and AtCNGC12 (grey bar) based on the publicly available AtGenExpress data set at the Botany Array Resource (BAR). The expression values are measured in expression units, with the average of all genes expressed being 100 units whereas the expression background is 20 expression units (Schmid et al., 2005; Winter et al., 2007). (B) Tissue-specific expression analysis of AtCNGC11 and 12 using transgenic Arabidopsis plants carrying promoter:GUS reporter constructs. The tissue was stained with X-Gluc solution for 1.5 h and then cleared with ethanol/acetic acid. AtCNGC11 expression is undetectable in both shoot and roots in 5- and 7-day-old seedlings (1, 2, 6, and 7). AtCNGC11 expression is relatively low in 14-day-old seedling leaves (3) and is absent in the roots (8). AtCNGC12 expression is ubiquitous throughout the leaves at all developmental stages (11–14) and is absent in the roots (16–19). In 21-day-old plants, AtCNGC11 and 12 are similarly expressed in all leaves (4 and 14) and sepals (5 and 15), but are absent from trichomes (10 and 20) and roots (9 and 19). Black and red scale bars represent 1 mm and 0.5 mm, respectively. Note: the dark areas in the root tips are not GUS staining. They are due to the high density of cells.
Fig. 3.
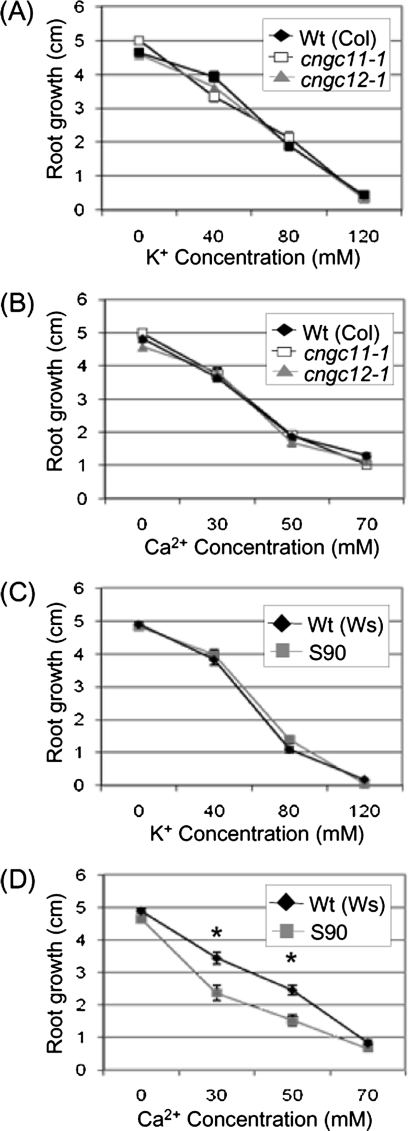
Root growth during K+ or Ca2+ stress. cngc11-1, cngc12-1, S90, and their respective wild-type seeds were grown on 0.5× MS agar medium for 7 d before being transferred to 0.5× MS agar medium with various concentrations of Ca2+or K+. Root lengths were measured 7 d after transferring the seedlings to plates with elevated levels of K+ or Ca2+. Data represent the mean of three replicate plates (n=8) ± SE. Asterisks indicate statistically significant differences (Student's t-test, P <0.05). All experiments were repeated at least twice with similar results. (A and B) cngc11-1, open square; cngc12-1, grey triangle; wild type (Col), filled diamond. (C and D), S90, grey square; wild type (Ws), filled diamond.
Fig. 6.
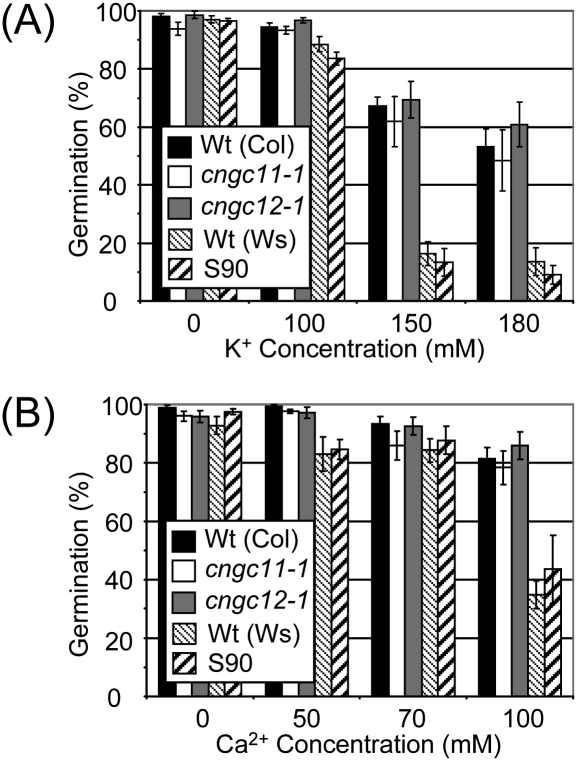
Effect of elevated Ca2+ on germination. cngc11-1, cngc12-1, S90, and their respective wild-type seeds were germinated on 0.5× MS agar medium with various concentrations of Ca2+. Data represent the mean of three independent experiments ±SE. (A) The effect of K+ on germination. (B) The effect of Ca2+ on germination.
Fig. 4.
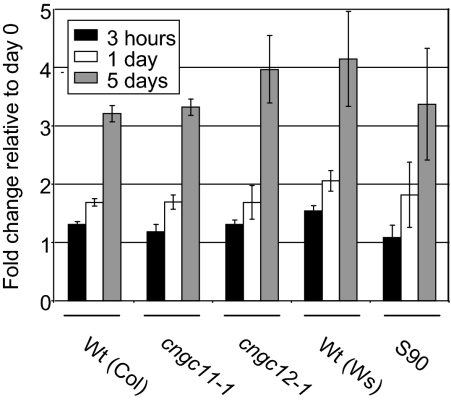
Endogenous Ca2+ levels in cngc11-1, cngc12-1, and S90 mutant plants. Seedlings were grown on 0.5× MS agar medium for 2 weeks before being transferred to 50 mM Ca2+ plates and samples were collected 0 h, 3 h, 1 d, and 5 d after transfer. Ca2+ content was determined by ICP-OES. The data are shown as fold change compared with the 0 h value. The presented data are the mean ±SE. The experiment was repeated three times and data from one representative experiment are shown.
Overall, the promoter:GUS analysis is consistent with publicly available microarray data except for the expression of AtCNGC11 in cotyledons. The expression of AtCNGC11 in cotyledons in the microarray data is likely to be due to the expression of AtCNGC3, which agrees well with the AtCNGC3 promoter:GUS analysis previously reported (Gobert et al., 2006). Interestingly, Gobert et al. (2006) reported that the expression of AtCNGC3 in cotyledons occurs in small clusters of cells that tend to concentrate near the leaf vasculature and cotyledon periphery. This was not the case for AtCNGC11 or AtCNGC12. Such a difference in the expression patterns supports the aforementioned observation that there is no similarity in the promoter region of AtCNGC3 to those of AtCNGC11 and AtCNGC12.
Root growth of AtCNGC11 and 12 KO mutants on media with elevated K+ or Ca2+
Both AtCNGC11 and 12 have been shown to conduct K+ and Ca2+ in heterologous expression systems using yeast mutants and, thus, may contribute to cation transport or signalling in Arabidopsis (Yoshioka et al., 2006; Urquhart et al., 2007). To test the involvement of AtCNGC11 and 12 in cation stress responses, root growth was analysed on media with various concentrations of K+ and Ca2+ using KO mutant lines for AtCNGC11 or 12, named cngc11-1 and cngc12-1 (homozygous T-DNA insertion lines, salk_026568 and salk_092657, respectively; Yoshioka et al., 2006).
cngc11-1 and cngc12-1 were grown on 0.5× MS agar medium for 7 d and then transferred to plates containing elevated concentrations of Ca2+ or K+. The mutant seedlings were then grown for another 7 d before the new primary root growth was measured. As shown in Fig. 3A and B, root growth of single mutants on medium with elevated levels of either K+ or Ca2+ was not significantly different from that of wild-type plants (P <0.05), indicating that there is no significant alteration in sensitivity to either cation in the single KOs (Fig. 3A, B; Supplementary Fig. S2 at JXB online). Next, experiments were carried out to test whether a double KO mutant line would show an alteration of this phenotype. For this test, a cpr22 suppressor mutant, S90, was used. As mentioned, AtCNGC11 and 12 are tandemly repeated and <1 kb apart; therefore, it is extremely difficult to obtain double KOs by cross-pollination. S90 was identified though a suppressor screen of the cpr22 mutant (AtCNGC11/12; Baxter et al., 2008) and it has a mutation in the fused AtCNGC11/12 gene causing a premature stop codon situated before the pore region (Supplementary Fig. S3). Therefore, S90 has lost both tandemly repeated genes, AtCNGC11 and12, through the creation of the fused AtCNGC11/12 in cpr22 and has further lost the function of this fused gene due to the premature stop codon by single nucleotide substitution. Thus, S90 is equivalent to an AtCGNC11 and 12 double KO line. Strikingly, while S90, like the single KOs, did not show increased sensitivity to elevated K+ (Fig. 3C; Supplementary Fig. S2), it did show a greater decrease in root growth on medium with 30 mM and 50 mM Ca2+ than its corresponding wild type (P <0.05) (ecotype Wassilewskija; Ws), indicating an increased sensitivity to Ca2+ (Fig. 3D; Supplementary Fig. S2). The difference observed in Ca2+ stress conditions is not due to osmotic stress since none of the mutants showed a difference in root growth on plates with a comparable molarity of K+. Thus, these data indicate that there is functional redundancy between AtCNGC11 and 12 and that both genes are involved in Ca2+ but not K+ uptake or signalling in planta.
AtCNGC11 and 12 KO mutants do not show alterations in endogenous levels of Ca2+
The root growth assays indicated that AtCNGC11 and 12 may have a role in the uptake of Ca2+. To test this, the endogenous levels of Ca2+ content in mutants were measured using ICP-OES. cngc11-1 and cngc12-1 as well as S90 and their respective wild-type seedlings were grown on regular 0.5× MS medium and then transferred to medium with or without 50 mM Ca2+ after 2 weeks of growth. The seedlings were then harvested at 0 h, 3 h, 1 d, and 5 d after transfer in order to test short-term uptake and long-term accumulation. At 0 h, no significant difference was observed between the plant lines tested here (data not shown). As shown in Fig. 4, no statistically significant difference in the rate of Ca2+ uptake 3 h after they were transferred to Ca2+-enriched medium was observed (based on Student's t-test). Furthermore, no statistically significant difference in accumulation of Ca2+ was observed at later time points (5 d in Fig. 4). Taken together, these data suggest that AtCNGC11 and 12 are not involved in general Ca2+ homeostasis.
Gravitropic responsiveness in AtCNGC11 and 12 KO mutants
Although cngc11-1 and cngc12-1 mutants showed no difference in Ca2+ content and thus may not be involved in general Ca2+ homeostasis, it is possible that they play a role in Ca2+ signalling. Indeed, cngc1 and AtCNGC10 antisense lines have been shown to have altered Ca2+-dependent gravitropic responsiveness of roots (Ma et al., 2006; Borsics et al., 2007). Therefore, the gravitropic response of cngc11-1 and cngc12-1 as well as S90 was tested. All lines were grown vertically on 0.5× MS agar medium for 2 weeks and then transferred to 0.5× MS agar medium with or without 50 mM or 100 mM Ca2+. At the time of transfer, the 0.5× MS medium plates were rotated 90 °. After 6, 12, and 24 h, the angle of root bending was measured (Ma et al., 2006). At 6 h post-rotation, the bending of cngc11-1 and cngc12-1 was not significantly different from that of the wild type (Table 1). Gravitropic responsiveness was greatly inhibited for seedlings on medium containing 100 mM Ca2+; however, no significant difference was observed between single KO mutants and their corresponding wild type (Col). The wild type for S90, Ws, showed a similar trend to that of the Col wild type, with a decreasing responsiveness to gravity as Ca2+ levels increased. Interestingly, S90 had a decreased level of curvature in response to rotation in medium without added Ca2+, with a bending angle of only 33.1±2.0 ° (Ws wild type: 40.1±2.7). However, in the presence of excess amounts of Ca2+, S90 responded more strongly (bending angle of 52.6±5.9 ° on 100 mM Ca2+) than Ws wild type (13.9±2.7 °). Thus, S90 showed a deficiency to adapt to changes in gravity but exhibited a stronger response in the presence of elevated external Ca2+ than wild-type plants. This phenomenon was confirmed in another premature stop codon mutant of cpr22, S98, as well as S73, which has lost its channel activity due to a point mutation (Baxter et al., 2008; Supplementary Table S1 at JXB online). These differences observed at the 6 h time point were still seen at 12 h but were less pronounced. However, all plants showed the same bending angle (∼90 °; data not shown) at 24 h. Taken together, AtCNGC11 and 12 seem to contribute to the generation of a Ca2+ signal that leads to gravitropic bending synergistically.
Table 1.
Gravitropic bending in cngc11-1, cngc12-1, and S90a
| Lines | Ca2+ concentration (mM) |
||
| 0 | 50 | 100 | |
| Col (Wt) | 44.9±5.8 | 35.6±7.5 | 12.3±3.3 |
| cngc11-1 | 47.5±3.0 | 53.4±5.6 | 9.6±3.5 |
| cngc12-1 | 41.1±5.9 | 50.7±6.6 | 11.1±2.2 |
| Ws (Wt) | 40.1±2.7 | 46.5±4.5 | 13.9±2.7 |
| S90 | 33.1±2.0* | 65.6±5.4* | 52.6±5.9** |
Degree of root curvature ±SE at 6 h post-rotation. The experiment was repeated >3 times with similar results.
Values marked with asterisks are statistically different from their wild-type (Student's t-test, *P <0.05; **P <0.001.)
The effect of dark-induced senescence
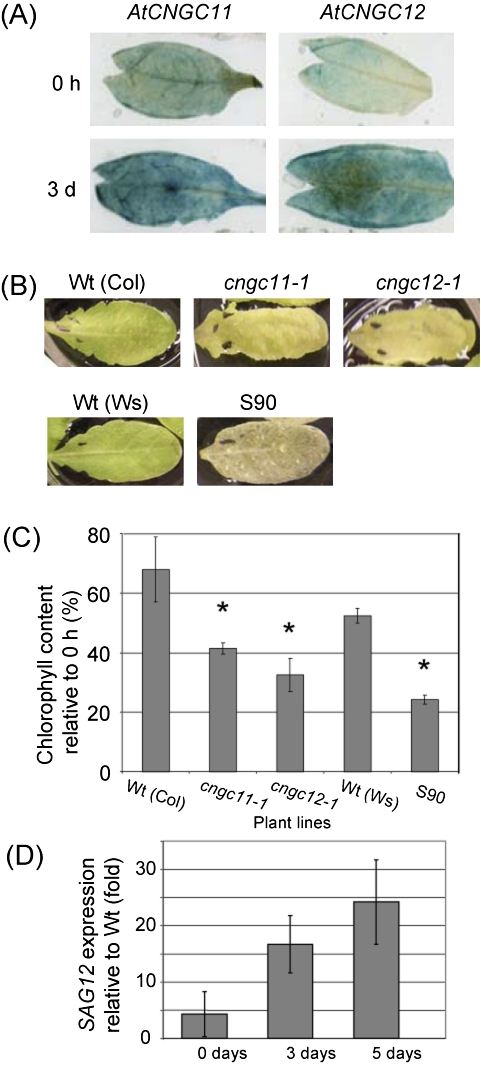
As shown in Fig. 2B, the expression level of AtCNGC11 and 12 is much higher in above-ground tissue than in the roots, and thus these channels may have important functions in the shoot. Ca2+ has been suggested to be involved in dark-induced senescence (Ferguson et al., 1983; Poovaiah and Leopold, 1973; Fujiki et al., 2005), and AtCNGC1 and 2 have been shown to be up-regulated during senescence (Köhler et al., 2001; Ma et al., 2006). Thus, it is possible that AtCNGC11 and 12 are also involved in senescence signalling. To investigate the possible involvement of AtCNGC11 and 12 in senescence, their expression patterns were investigated using promoter:GUS transgenic lines. Leaves from these promoter:GUS transgenic lines were cut and floated on water in the dark for 3 d (Riefler et al., 2006). The expression levels of AtCNGC11 and 12 were strongly induced after 3 d in the dark (Fig. 5A). This finding was consistent with microarray data from the BAR that also show the expression of AtCNGC11 and 12 being up-regulated during senescence (Fig. 2A). Chlorophyll loss is associated with leaf senescence, and yellowing of the leaf progresses as chlorophyll is being degraded (Ougham et al., 2008). Interestingly, after 4 d in the dark the leaves of cngc11-1, cngc12-1, and S90 plants showed an elevated degree of yellowing compared with their wild types (Fig. 5B). This increased rate of chlorophyll loss was also quantified by measuring the remaining chlorophyll after 5 d. As shown in Fig. 5C, cngc11-1 and cngc12-1 as well as S90 have increased rates of chlorophyll loss, with cngc11-1 and cngc12-1 retaining only 41% and 32% of their chlorophyll content, respectively, whereas Col wild type maintained 68%. Likewise, S90 retained only 24% of its chlorophyll content while the Ws wild type maintained 52% after 5 d in the dark (Fig. 5C). To confirm accelerated senescence at the molecular level, the expression level of SAG12, a senescence marker gene, was determined (Weaver et al., 1998). Relative to Ws wild type, S90 leaves started with a similar SAG12 expression level. However, a much stronger accumulation of SAG12 was observed in S90 leaves than in the Ws wild type, with SAG12 levels being nearly 25 times higher in the former after 5 d in darkness (Fig. 5D).
Fig. 5.
cngc11-1, cngc12-1, and S90 show altered senescence phenotypes. All experiments were repeated >3 times with similar results. (A) Expression analysis of AtCNGC11 and 12 using detached leaves from promoter:GUS transgenic plants. The leaves were floated on water in the dark for 0 d or 3 d. (B) cngc11-1 and cngc12-1, as well as S90, showed a more rapid loss of chlorophyll when detached leaves were placed in darkness for 4 d. (C) Quantitative analysis of chlorophyll degradation after 5 d in darkness. Asterisks indicate statistically significant differences (Student's t-test, P <0.05) (D) SAG12 expression levels were measured by quantitative RT-PCR in detached S90 leaves after 0, 3, and 5 d in darkness and graphed relative to SAG12 expression in Ws wild-type controls. The data shown here are the mean of three technical repeats using one cDNA sample. The same experiment was conducted three times using different cDNAs (biological repeats). The data shown here are one representative.
Sensitivity to Ca2+ was not observed in the germination phenotype in AtCNGC11 and 12 KO mutants
Gobert et al. (2006) reported that KO mutants for AtCNGC3, which locates next to AtCNGC11 (Fig. 1A), exhibit reduced germination on media containing high levels of Na+, but not K+. Since enhanced sensitivity to Ca2+ was observed in S90 in the root growth assay (Fig. 3), it was hypothesized that KO mutants for AtCNGC11, AtCNGC12, or S90 may also exhibit enhanced sensitivity to Ca2+ during germination. Thus, seeds of cngc11-1, cngc12-1, and S90 were germinated on 0.5× MS agar medium containing increasing concentrations of Ca2+ or K+. However as shown in Fig. 6, no statistically significant difference was observed, not only for cngc11-1 and cngc12-1, but also for S90. These data indicate that AtCNGC11 and AtCNGC12 play a role in Ca2+ signalling in specific aspects of plant physiology, while they seem not to play a role in seed germination.
Discussion
Plants have to respond continuously to an ever-changing environment. Ca2+ is often employed as a secondary messenger that transforms the physical stimulus into a signal. For example, it has been reported that a change in the angle of gravity induces an increase in cytosolic Ca2+ within specific root cells from 30 nM to 225 nM (Gehring et al., 1990). This Ca2+ increase is not only spatially specific, occurring only in the lower cells, but it is also temporal, lasting for only ∼25 s (Plieth and Trewavas, 2002). The channels that are responsible for this Ca2+ influx, however, have not been identified yet. Possible candidates are members of the CNGC gene family which have already been shown in animal systems to modulate ion fluxes induced by light and olfactory stimuli (Zagotta and Siegelbaum, 1996; Craven and Zagotta, 2006). To understand the biological roles of plant CNGCs, the focus of this study was on the closely related CNGCs, AtCNGC11 and AtCNGC12. They belong to group I of the CNGC family, which also contains AtCNGC1, 3, 10, and 13. AtCNGC3, 11, and 12 are tandemly repeated (Fig. 1) and are located at the south end of chromosome II (Mäser et al., 2001). Using heterologous expression systems, it has been shown that AtCNGC3 can conduct K+ and Na+, whereas AtCNGC11 and 12 can conduct both K+ and Ca2+ and are plasma membrane localized (Gobert et al., 2006; Yoshioka et al., 2006; Urquhart et al., 2007). In terms of biological function, AtCNGC11 and 12 are involved in pathogen defence signalling, whereas AtCNGC3 is not (Gobert et al. 2006; Yoshioka et al., 2006; Moeder et al., 2011). In addition, sequence analysis in this study revealed a high degree of similarity in the promoter region of AtCNGC11 and 12, whereas this similarity is not shared with AtCNGC3, further indicating common functions of AtCNGC11 and 12 and a distinct biological role for AtCNGC3.
In this study, it was demonstrated that AtCNGC11 and 12 have a much broader physiological role and contribute to a range of Ca2+-dependent signalling pathways. Furthermore, their role may be redundant. Based on AtCNGC11 or 12 promoter:GUS reporter gene analysis, although the expression of AtCNGC11 starts later during development compared with that of AtCNGC12, the overall expression profile of the two genes is quite similar (Fig. 2). Furthermore, AtCNGC11 and 12 are tandemly repeated genes which share 97% identity in the front half that includes the pore region, which is responsible for ion selectivity. To test for possible redundancy, a cpr22 suppressor mutant, S90, which has a premature stop codon resulting in the deletion of both AtCNGC11 and 12 (Supplementary Fig. S1 at JXB online), was tested. The results obtained from these assays suggest two things: the root growth and root gravitropic assays both indicate that there is functional redundancy between AtCNGC11 and 12 since neither single KO showed an altered phenotype, whereas S90 was significantly different from its wild type, Ws. However, in tissue other than the root, AtCNGC11 and 12 appear to share biological functions, and their loss in S90 synergistically enhanced their phenotype. Thus, this is the first report that demonstrates that specific AtCNGC genes show redundancy in various biological functions and that they may work in a synergistic manner in specific tissues.
In terms of ion sensitivity, it is reasonable to expect that the loss of an AtCNGC that is involved in inward conductance of a specific ion will result in a KO plant that is less sensitive than the wild type to media containing excess amounts of ions. This is, however, not the case for S90, which displayed increased sensitivity to Ca2+ in root growth assays. There was, however, no significant difference between S90 and the wild type when K+ was tested, indicating that the ion sensitivity of the mutant line was specific to Ca2+ and not a broadly increased sensitivity to all cations. There are a number of possibilities for the increased Ca2+ sensitivity of S90. One possible role for AtCNGC11 and 12 is that they translocate Ca2+ from one area of the plant to another during Ca2+ stress or that they are involved in the generation of specific Ca2+ signals. A second possibility is that the two channels control the uptake of Ca2+ into the plant and that their loss has disrupted Ca2+ homeostasis. The former function has been suggested for a number of other AtCNGCs. For example, cngc3 seeds have increased sensitivity to Na+ during germination when compared with the wild type, and it is suggested that AtCNGC3 shuttles Na+ from areas of high sensitivity to areas more tolerant to Na+ (Gobert et al., 2006). AtCNGC19 and 20 may also contribute to the internal movement of Na+ during salt stress (Kugler et al., 2009). On the other hand, a role for maintaining ion homeostasis has been suggested for AtCNGC10, which, when heterologously expressed in human kidney (HEK293) cells, channels K+ both inwardly and outwardly (Christopher et al., 2007). AtCNGC3 has also been suggested to regulate the uptake of both K+ and Na+ in growing seedlings, demonstrated by the finding that cngc3 has decreased accumulation and uptake of these two ions (Gobert et al., 2006). Lastly, cngc1 has decreased internal Ca2+ levels, implicating AtCNGC1 in a role of Ca2+ uptake/homeostasis. For AtCNGC11 and 12, a role in Ca2+ distribution and/or signalling is more likely than in homeostasis since the Ca2+ content was not altered in mutant seedlings. This indicates that the endogenous level of Ca2+ was not significantly different from that of the wild type and that AtCNGC11 and 12 may affect the tissue-specific distribution of Ca2+ within the plant rather than being involved in ion uptake. Tissue-specific distribution of Ca2+ has been shown to occur (Leigh and Storey, 1993). For example, in barley, Ca2+ is preferentially stored in epidermal cells while K+ is accumulated in the mesophyll. It has been speculated that this compartmentalization of ions prevents the undesirable formation of insoluble CaHPO4 (Leigh and Storey, 1993). Ion compartmentalization is also present in dicots. For example, in citrus leaves, Ca2+ accumulates in the mesophyll cells and, at a tissue level, Ca2+ is located at a higher concentration in the centre of coriander leaves than at the outer regions (Storey and Leigh, 2004; Kerton et al., 2009). Thus, it is possible that in the absence of AtCNGC11 and 12, Ca2+ distribution is abnormal. This would result in detrimental Ca2+ concentrations occurring at a tissue or cellular level even though the overall Ca2+ concentration remains normal at the whole-plant level. These detrimental Ca2+ concentrations may result in Ca2+ toxicity or alteration in signalling.
The process of gravitropism is suggested to be Ca2+ dependent and it has been shown that an uneven distribution of Ca2+ precedes root bending in response to changes in gravity (Chandra et al., 1982; Chen et al., 1999). For example, gravity-sensing cells have a higher level of internal Ca2+, and an apoplastic Ca2+ gradient is generated across the root cap shortly after rotation (Chandra et al., 1982; Chen et al., 1999). Additionally, in maize coleoptiles, a 180 ° rotation induces a near 10-fold increase of the Ca2+ concentration specifically on the lower side (Gehring et al., 1990). Prior to this study, two AtCNGC genes had been shown to be involved in gravitropic sensitivity. The loss of AtCNGC1 or 10 decreased the responsiveness of roots to a change in the angle of gravity (Ma et al., 2006; Borsics et al., 2007). Here it was demonstrated that the loss of AtCNGC11 and 12 also reduces the rate of gravitropic responsiveness. A reduction of Ca2+ in the growth medium has been shown to reduce wild-type root curvature in response to gravity (Ma et al., 2006), and it is therefore possible that S90 is not generating an adequate Ca2+ signal to induce root bending. This possibility is further supported by the finding that the addition of Ca2+ corrected the reduced bending of S90 to a rate similar to that of the wild type on normal medium. Although the GUS reporter analysis presented here could not detect expression of AtCNGC11 and 12 in the roots of plate-grown plants, microarray data and the present RT-PCR analysis indicated expression in the root, although at a low level. Furthermore, the BAR data showed low expression of AtCNGC11 and 12 in particular types of tissues in the root (i.e. the cortex and the procambium). Thus, it is possible that AtCNGC11 and 12 may, in part, contribute to the creation of Ca2+ differentials across specific root areas, leading to gravitropic bending.
Unlike the root, the leaf has very high levels of AtCNGC11 and 12 expression, indicating that both genes may have roles in leaf-specific responses. It has been shown that AtCNGC1 and 2 both play a role in senescence (Köhler et al., 2001; Ma et al., 2006, 2010). Here it is demonstrated that AtCNGC11 and 12 also have increased expression in response to senescence. Interestingly, the loss of AtCNGC11 and 12 enhances dark-induced senescence in a synergistic manner. It has been suggested that Ca2+ signals defer the onset of dark-induced senescence (Poovaiah and Leopold, 1973; Ferguson et al., 1983; Cheour et al., 1992); therefore, it is possible that AtCNGC11 and 12 negatively regulate dark-induced senescence by modulating Ca2+ movement within the leaf.
Taken together, the findings of this study indicate that the role of AtCNGC11 and 12 is probably not to maintain general Ca2+ homeostasis but rather to contribute to internal Ca2+ translocation and/or signalling during a variety of developmental processes in addition to pathogen defence. Furthermore, AtCNGC11 and 12 could share the same biological roles, and some functions of the two channels can be redundant depending on the tissue type and stimulus. This study clearly expands the biological role of AtCNGC11 and 12 to include functions other than pathogen defence and sets a foundation from which the biological relationships of AtCNGC genes, in the context of Ca2+ signalling, can be further examined.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Tissue-specific expression analysis of AtCNGC11 and 12 using soil-grown transgenic Arabidopsis plants carrying promoter:GUS reporter constructs. The tissue was stained with X-Gluc solution for 1.5 h and then cleared with ethanol/acetic acid. AtCNGC11 expression is undetectable in both shoots and roots in 5-day-old seedlings (1, 4, and 7). AtCNGC11 expression is very low in 14-day-old seedling leaves (2) and is absent in the roots (8), but became stronger as plants aged (3). AtCNGC12 expression is ubiquitous throughout the leaves at all developmental stages (9, 10, and 11), but it became stronger when the plants aged (11). In the roots, expression of both genes was not detectable in 5- and 14-day-old plants (7, 8, 15, and 16), but very weak expression was observed in 30-day-old plants (6 and 14). Bar=0.1 cm
Figure S2. Root growth during K+ or Ca2+ stress. cngc11-1, cngc12-1, S90, and their respective wild-type seeds were grown on 0.5× MS agar medium for 7 d before being transferred to 0.5× MS agar medium with the indicated concentrations of CaCl2 or KCl. Root lengths were measured 7 d after transferring the seedlings to plates with elevated levels of K+ or Ca2+. Red lines indicate the position of root tips when they were transferred.
Figure S3. Configuration of AtCNGC3, AtCNGC11, and AtCNGC12 in the wild type, cpr22, and its suppressor mutants, S73, S90, and S98. cpr22 has an ∼3 kb deletion between AtCNGC11 and AtCNGC12, creating an in-frame chimeric AtCNGC11/12 gene (Yoshioka et al., 2006). S73 has a single amino acid substitution in the cyclic nucleotide-binding domain, resulting in loss of channel function (Baxter et al., 2008). S90 and S98 both have premature stop codons before the pore region. The filled triangle shows the mutation position in S73. Hatched boxes show the area after the premature stop codons.
Table S1. Gravitropic bending in the allelic premature stop codon mutant of cpr22, S98, and the loss-of-function mutant, S73.
Acknowledgments
We would like to thank Dr S. Lumba and Ms C. To (Life Technologies) for their technical assistance. This research was supported by the Natural Science and Engineering Research Council of Canada (NSERC), Canadian Foundation for Innovation (CFI), the Ontario Research Fund, and the Ontario Ministry of Research and Innovation to KY, and through an NSERC graduate scholarship to WU and Ontario Graduate Scholarship (OGS) to KC.
References
- Ali R, Zielinski RE, Berkowitz GA. Expression of plant cyclic nucleotide-gated cation channels in yeast. Journal of Experimental Botany. 2005;57:125–138. doi: 10.1093/jxb/erj012. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Balague C, Lin BQ, Alcon C, Flottes G, Malmstrom S, Köhler C, Neuhaus G, Pelletier G, Gaymard F, Roby D. HLM1, an essential signalling component in the hypersensitive response, is a member of the cyclic nucleotide-gated ion channel family. The Plant Cell. 2003;15:365–379. doi: 10.1105/tpc.006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J, Moeder W, Urquhart W, Shahinas D, Chin K, Christendat D, Kang HG, Angelova M, Kato N, Yoshioka K. Identification of a functionally essential amino acid for Arabidopsis cyclic nucleotide gated ion channels using the chimeric AtCNGC11/12 gene. The Plant Journal. 2008;56:457–469. doi: 10.1111/j.1365-313X.2008.03619.x. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods in Molecular Biology. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Borsics T, Webb D, Andeme-Ondzighi C, Staehelin LA, Christopher DA. The cyclic nucleotide-gated calmodulin-binding channel AtCNGC10 localizes to the plasma membrane and influences numerous growth responses and starch accumulation in Arabidopsis thaliana. Planta. 2007;225:563–573. doi: 10.1007/s00425-006-0372-3. [DOI] [PubMed] [Google Scholar]
- Chaiwongsar S, Strohm AK, Roe JR, Godiwalla RY, Chan CWM. A cyclic nucleotide-gated channel is necessary for optimum fertility in high-calcium environments. New Phytologist. 2009;183:76–87. doi: 10.1111/j.1469-8137.2009.02833.x. [DOI] [PubMed] [Google Scholar]
- Chan CWM, Scorrak LM, Smith RK, Bent AF, Sussman MR. A cyclic nucleotide-gated ion channel, CNGC2, is crucial for plant development and adaptation to calcium stress. Plant Physiology. 2003;132:728–731. doi: 10.1104/pp.102.019216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CWM, Wohlbach DJ, Rodesch MJ, Sussman MR. Transcriptional changes in response to growth of Arabidopsis in high external calcium. FEBS Letters. 2008;582:967–976. doi: 10.1016/j.febslet.2008.02.043. [DOI] [PubMed] [Google Scholar]
- Chandra S, Chabot JF, Morrison GH, Leopold AC. Localization of Ca2+ in amyloplasts of root cap cells using ion microscopy. Science. 1982;216:1221–1223. doi: 10.1126/science.216.4551.1221. [DOI] [PubMed] [Google Scholar]
- Chang F, Yan A, Zhao L-N, Wu W-H, Yang Z. A putative calcium-permeable cyclic nucleotide-gated channel, CNGC18, regulates polarized pollen tube growth. Journal of Integrative Plant Biology. 2008;49:1261–1270. [Google Scholar]
- Chen R, Rosen E, Masson PH. Gravitropism in higher plants. Plant Physiology. 1999;120:343–350. doi: 10.1104/pp.120.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheour F, Arult J, Makhlouf J, Willemont C. Delay of membrane lipid degradation by calcium treatment during cabbage leaf senescence. Plant Physiology. 1992;100:1656–1660. doi: 10.1104/pp.100.4.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, Moeder W, Yoshioka K. Biological roles of cyclic-nucleotide-gated ion channels in plants: what we know and don't know about this 20 member ion channel family. Botany. 2009;87:668–677. [Google Scholar]
- Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK, Jr., Bent AF. The Arabidopsis dnd1 ‘defense, no death’ gene encodes a mutated cyclic nucleotide-gated ion channel. Proceedings of the National Academy of Sciences, USA. 2000;97:9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher DA, Borsics T, Yuen CY, Ullmer W, Andeme-Ondzighi C, Anders MA, Kang BH, Staehelin LA. The cyclic nucleotide gated cation channel AtCNGC10 traffics from the ER via Golgi vesicles to the plasma membrane of Arabidopsis root and leaf cells. BMC Plant Biology. 2007;7:48. doi: 10.1186/1471-2229-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven KB, Zagotta WN. CNG and HCN channels: two peas, one pod. Annual Review of Physiology. 2006;68:375–401. doi: 10.1146/annurev.physiol.68.040104.134728. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich P, Anschütz U, Kugler A, Becker D. Physiology and biophysics of plant ligand-gated ion channels. Plant Biology. 2010;12:80–93. doi: 10.1111/j.1438-8677.2010.00362.x. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. The language of calcium signalling. Annual Review of Plant Biology. 2010;61:4. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- Ferguson IB, Watkins CB, Harman JE. Inhibition by calcium of senescence of detached cucumber cotyledons. Plant Physiology. 1983;71:182–186. doi: 10.1104/pp.71.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietsch S, Wang YF, Sladek C, Poulsen LR, Romanowsky SM, Schroeder JL, Harper JF. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proceedings of the National Academy of Sciences, USA. 2007;104:14531–14536. doi: 10.1073/pnas.0701781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Nakagawa Y, Furumoto T, Yoshida S, Biswal B, Ito M, Watanabe A, Nishida I. Response to darkness of late-responsive dark-inducible genes is positively regulated by leaf age and negatively regulated by calmodulin-antagonist-sensitive signalling in Arabidopsis thaliana. Plant and Cell Physiology. 2005;46:1741–1746. doi: 10.1093/pcp/pci174. [DOI] [PubMed] [Google Scholar]
- Gehring CA, Williams DA, Cody SH, Parish RW. Phototropism and geotropism in maize coleoptiles are spatially correlated with increases in cytosolic free calcium. Nature. 1990;345:528–530. doi: 10.1038/345528a0. [DOI] [PubMed] [Google Scholar]
- Gobert A, Park G, Amtmann A, Sanders D, Maathuis FJM. Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. Journal of Experimental Botany. 2006;57:791–800. doi: 10.1093/jxb/erj064. [DOI] [PubMed] [Google Scholar]
- Hasenstein K, Evans ML. Calcium dependence of rapid auxin action in maize roots. Plant Physiology. 1986;81:439–443. doi: 10.1104/pp.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski GI, Smith RK, Yu IC, Ham JH, Sharma SB, Klessig DF, Fengler KA, Bent AF. Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the ‘defense, no death’ phenotype. Molecular Plant-Microbe Interactions. 2004;17:511–520. doi: 10.1094/MPMI.2004.17.5.511. [DOI] [PubMed] [Google Scholar]
- Kaplan B, Sherman T, Fromm H. Cyclic nucleotide-gated channels in plants. FEBS Letters. 2007;581:2237–2246. doi: 10.1016/j.febslet.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Kerton M, Newbury HJ, Hand D, Pritchard J. Accumulation of calcium in the centre of leaves of coriander (Coriandrum sativum L.) is due to an uncoupling of water and ion transport. Journal of Experimental Botany. 2009;60:227–235. doi: 10.1093/jxb/ern279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Merkle T, Roby D, Neuhaus G. Developmentally regulated expression of a cyclic nucleotide-gated ion channel from Arabidopsis indicates its involvement in programmed cell death. Planta. 2001;213:327–332. doi: 10.1007/s004250000510. [DOI] [PubMed] [Google Scholar]
- Kugler A, Köhler B, Palme K, Wolff P, Dietrich P. Salt-dependent regulation of a CNG channel subfamiliy in Arabidopsis. BMC Plant Biology. 2009;9:140. doi: 10.1186/1471-2229-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh RA, Storey R. Intercellular compartmentation of ions in barley leaves in relation to potassium nutrition and salinity. Journal of Experimental Biology. 1993;44:755–762. [Google Scholar]
- Leng Q, Mercier RW, Yao W, Berkowitz GA. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiology. 1999;121:753–761. doi: 10.1104/pp.121.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Ali R, Berkowitz GA. Characterization of plant phenotypes associated with loss-of-function of AtCNGC1, a plant cyclic nucleotide gated cation channel. Plant Physiology and Biochemistry. 2006;44:494–505. doi: 10.1016/j.plaphy.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Ma W, Smigel A, Walker RK, Moeder W, Yoshioka K, Berkowitz GA. Leaf senescence signaling the Ca2+-conducting Arabidopsis cyclic nucleotide gated channel2 acts through nitric oxide to repress senescence programming. Plant Physiology. 2010;154:733–743. doi: 10.1104/pp.110.161356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiology. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier RW, Rabinowitz NM, Gaxiola RA, Ali R, Berkowitz GA. Use of hygromycin hypersensitivity of a K+ uptake yeast mutant as a functional assay of plant cyclic nucleotide gated cation channels. Plant Physiology Biochemistry. 2004;42:529–536. doi: 10.1016/j.plaphy.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Michard E, Alves F, Feijo JA. The role of ion fluxes in polarized cell growth and morphogenesis: the pollen tube as an experimental paradigm. International Journal of Developmental Biology. 2009;53:1609–1622. doi: 10.1387/ijdb.072296em. [DOI] [PubMed] [Google Scholar]
- Moeder W, Urquhart W, Ung H, Yoshioka K. The role of cyclic nucleotide-gated ion channels in plant immunity. Molecular Plant. 2011 doi: 10.1093/mp/ssr018. (Doi: 10.1093/SSR018, in press) [DOI] [PubMed] [Google Scholar]
- Ougham H, Hortensteiner S, Armstead I, Donnison I, King I, Thomas H, Mur L. The control of chlorophyll catabolism and the status of yellowing as a biomarker of leaf senescence. Plant Biology. 2008;10:4–14. doi: 10.1111/j.1438-8677.2008.00081.x. [DOI] [PubMed] [Google Scholar]
- Plieth C, Trewavas AJ. Reorientation of seedlings in the Earth's gravitational field induces cytosolic calcium transients. Plant Physiology. 2002;129:786–796. doi: 10.1104/pp.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah BW, Leopold AC. Deferral of leaf senescence with calcium. Plant Physiology. 1973;52:236–239. doi: 10.1104/pp.52.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta. 1989;975:384–394. [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. The Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd JJ, Franklin-Tong VE. Calcium signalling in plants. Cellular and Molecular Life Sciences. 1999;55:214–232. doi: 10.1007/s000180050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Storey R, Leigh RA. Processes modulating calcium distribution in citrus leaves. An investigation using X-ray microanalysis with strontium as a tracer. Plant Physiology. 2004;136:3838–3848. doi: 10.1104/pp.104.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzberg D, Kirshner B, Rav-David D, Elad Y, Granot D. Botrytis cinerea induces senescence and is inhibited by autoregulated expression of the IPT gene. European Journal of Plant Pathology. 2008;120:289–297. [Google Scholar]
- Urquhart W, Gunawardena AHLAN, Moeder W, Ali R, Berkowitz GA, Yoshioka K. The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Molecular Biology. 2007;65:747–761. doi: 10.1007/s11103-007-9239-7. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence associated genes in response to stress and hormone treatment. Plant Molecular Biology. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE8. 2007 doi: 10.1371/journal.pone.0000718. e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Heath MC. Role of calcium in signal transduction during the hypersensitive response caused by basidiospore-derived infection of the cowpea rust fungus. The Plant Cell. 1998;10:585–598. doi: 10.1105/tpc.10.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Kachroo P, Tsui F, Sharma SB, Shah J, Klessig DF. Environmentally-sensitive, SA-dependent defense response in the cpr22 mutant of Arabidopsis. The Plant Journal. 2001;26:447–459. doi: 10.1046/j.1365-313x.2001.2641039.x. [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF. The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. The Plant Cell. 2006;18:747–763. doi: 10.1105/tpc.105.038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta WN, Siegelbaum SA. Structure and function of cyclic nucleotide-gated channels. Annual Review of Neuroscience. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.