Abstract
1,N2-Etheno(ε)guanine (ε) is formed in DNA as a result of exposure to certain vinyl monomers (e.g. vinyl chloride) or from lipid peroxidation. This lesion has been shown to be mutagenic in bacteria and mammalian cells. 1,N2-ε-G has been shown to block several model replicative DNA polymerases, with limited bypass. Recently an archebacterial DNA polymerase, Sulfolobus solfataricus Dpo4, has been shown to copy past 1,N2-ε-G. In this study we examined the abilities of recombinant, full-length human polymerase (pol) δ and three human translesion DNA polymerases to copy past 1,N2-ε-G. The replicative polymerase pol δ was completely blocked. Pols ι and κ showed similar rates of incorporation of dTTP and dCTP. Pol η was clearly the most active of these polymerases in copying past 1,N2-ε-G, incorporating in the order dGTP > dATP > dCTP, regardless of whether the base 5’ of 1,N2-ε-G in the template was C or T. Pol η also had the highest error frequency opposite 1,N2-ε-G. Analysis of the extended products of the pol η reactions by mass spectrometry indicated only two products, both of which had G incorporated opposite 1,N2-ε-G and all other base pairing being normal (i.e., G:C and A:T). One-half of the products contained an additional A at the 3’ end, presumably arising from a non-informational blunt-end addition or possibly a slipped insertion mechanism at the end of the primer-template replication process. In summary, the most efficient of the four human DNA polymerases was pol η, which appeared to insert G opposite 1,N2-ε-G and then copy correctly. This pattern differs with the same oligonucleotide sequences and 1,N2-ε-G observed using Dpo4, emphasizing the importance of polymerases in mutagenesis events.
Introduction
A number of recent successes have been achieved in the understanding of molecular mechanisms of mutagenesis, particularly in regard to structural and cell biology, but the basic understanding of the events still has major questions (1). Knowledge of cellular outcomes is useful (and indeed critical for practical applications) but is not particularly insightful regarding molecular mechanisms. One of the complicating factors is the plethora of enzyme systems that process damaged DNA, including replication, repair, and other processes such as topoisomerase interactions (2). Our laboratories have focused on studying some of the basic chemistry involved with DNA adducts and the enzymology of replication of damaged DNA, particularly which individual events differ from normal replication processes (1).
1,N2-Etheno(ε)-G1 was originally reported by Leonard’s group (3) and serves as a prototype for the so-called exocyclic DNA adducts, a series of lesions formed by various bis-electrophiles (4). Included in the list of compounds that can generate etheno adducts are the electrophilic oxidation products of industrial vinyl monomers (e.g., vinyl chloride, urethane, etc. (5, 6)), some nitrosamines (7), mucochloric acid (8), and lipid peroxidation products (9–11). 1,N2-ε-G produces misincorporation and mutations in systems involving model DNA polymerases and in bacterial and mammalian cells (12–14). The two added exocyclic carbons block the normal coding “face” involved in Watson-Crick pairing and preclude the usual hydrogen bonding mode (Scheme 1). The frequency of mutations is not particularly high in bacterial and mammalian cells into which the 1,N2-ε-G lesion has been introduced (13, 14). The low frequency of errors may be attributed, at least in part, to the presence of bacterial and mammalian glycosylases that can act on this lesion (15).
Scheme 1.
1,N2-ε-G (R = deoxyribose)
Therefore, 1,N2-ε-G is a useful model for studying DNA polymerase action, because of the inherent significance of the lesion and potential relevance to diseases (16) and as a prototype for other exocyclic guanine adducts (4, 17). Recently we examined the catalytic interaction of 1,N2-ε-G with the archebacterial translesion DNA polymerase Sulfolobus solfataricus Dpo4 in detail (18). The mechanism was studied using typical steady-state kinetics. However, analysis of the sequence of the products formed by full-length extension in the presence of all four dNTPs led to several insights about the complexity of the process (2–4 major products formed, depending on the sequence) and the extent of slippage coupled to insertion. Four crystal structures of reaction steps were obtained, providing support for the proposed pathway (18).
The work with 1,N2-ε-G was extended to four human DNA polymerases, including the replicative DNA polymerase (pol) δ and three of the translesion polymerases (η,ι,κ). Steady-state kinetics and sequence analysis of products indicate that pol η is the most efficient of the human polymerases examined and that the process differs considerably from catalysis by Dpo4.
Experimental Procedures
Materials
Escherichia coli uracil DNA glycosylase (UDG) was purchased from Sigma Chemical Co. (St. Louis, MO).
Unmodified oligonucleotides were purchased from Midland Certified Reagents (Midland, TX). Oligonucleotides containing 1,N2-ε-G were prepared in the Vanderbilt facility from a phosphoramidite derivative of 1,N2-ε-G, using a modification of procedures described previously (18, 19), with HPLC (reversed-phase) purification. Mass spectra and capillary gel electrophoresis of these oligonucleotides have been published elsewhere (18).
Pol δ (20), pol η (20), pol ι (21), and pol κ2 were expressed in baculovirus-infected insect cell systems and purified to electrophoretic homogeneity as described previously. Human proliferating cell nuclear antigen (PCNA) was expressed in E. coli and purified as described earlier (22).
Polymerization Assays and Gel Electrophoresis
A 32P-labeled primer, annealed to either an unmodified or adducted template, was extended in the presence of single dNTPs (Scheme 1). Each reaction was initiated by adding 4 µL of dNTP-Mg2+ solution (final concentrations of 100 µM of each dNTP and 5 mM MgCl2) to a preincubated enzyme•DNA complex (final concentrations of 50 mM Tris-HCl (pH 7.5), 100 nM DNA duplex, polymerase (0.8 to 20 nM, depending on the system), 5 mM dithiothreitol (DTT), 100 µg bovine serum albumin (BSA) mL−1, and 10% glycerol (v/v)) at 37 °C, yielding a total reaction volume of 8 µL.
After 30 min, reactions were quenched with 72 µL of 20 mM EDTA (pH 8.0) in 95% formamide (v/v). Aliquots (3 µL) were separated by electrophoresis on a denaturing gel containing 8.0 M urea and 16% acrylamide (w/v) (from a 19:1 acrylamide:bisacrylamide solution (w/w), AccuGel, National Diagnostics, Atlanta, GA) with 80 mM Tris-borate buffer (pH 7.8) containing 1 mM EDTA. The gel was exposed to a phosphorimager screen (Imaging Screen K, Bio-Rad, Hercules, CA) overnight. The bands (representing extension of the primer) were visualized with a phosphorimaging system (Bio-Rad, Molecular Imager® FX, Hercules, CA) using the manufacturer’s Quantity One Software, Version 4.3.0.
Steady-State Kinetics
Unless indicated otherwise, all polymerase reactions were performed at 37 °C in 50 mM Tris-HCl buffer (pH 7.5) containing 10% glycerol (v/v), 5 mM DTT, and 100 µg BSA mL−1. For unmodified and modified templates, the molar ratios of primer/template to enzyme were at least 100:1 and 20:1 respectively, and the reactions were done at ten dNTP concentrations (usually reaction time of 10 min).
Liquid Chromatography (LC)-Mass Spectrometry (MS)/MS Analysis of Oligonucleotide Products from Pol η Reactions
Pol η reactions were performed at 37 °C for 1.5 h in 50 mM Tris-HCl buffer (pH 7.8) containing 25% glycerol (v/v), 5 mM DTT, 50 mM NaCl, 5 mM MgCl2, and 100 µg BSA mL−1. The reactions were done with 13 µM oligonucleotide substrate, 0.8 µM pol η, and the four dNTPs at 0.5 mM each, in a final reaction volume of 150 µL. The reaction was terminated by extraction of excess dNTPs using a “spin column” (Bio-Spin 6 chromatography column, Bio-Rad). To the above filtrate (200 µL), concentrated Tris-HCl and DTT solutions were added to restore the initial concentrations, and UDG solution was added (20 units). The reaction was incubated at 37 °C for 4 h to hydrolyze the uracil residue on the extended primer. The final reaction mixture was then heated at 95 °C for 1 h in the presence of 0.25 M piperidine, followed by removal of solvent by lyophilization. The dried residues were dissolved in 100 µL of H2O for the MS analysis.
MS was performed on a DecaXP ion trap instrument (ThermoFinnigan, San Jose, CA) in the Vanderbilt facility. Separation of oligonucleotides was carried out with a YMC ODS-AQ column (2.0 × 250 mm, 120 Å). Buffer A contained 10 mM NH4CH3CO2 (pH 7.0) and 1% CH3CN (v/v); Buffer B contained 10 mM NH4CH3CO2 (pH not adjusted) and 95% CH3CN (v/v). The following gradient program was used with a flow rate of 0.2 mL min−1: 0–3 min, 100% A; 3–25 min, linear program to 30% B (v/v); 25–28 min, linear program to 100% B; 28–40 min, hold at 100% B; 40–41 min, linear program to 100% A; 41–50 min, hold at 100% A (for next injection). The desired oligonucleotide products were eluted at 13 min. Samples were infused using an autosampler, with a 20 µL aliquot withdrawn from a 100-µl reaction. Electrospray (ES) conditions were: source voltage, 3.4 kV; source current, 80 µA; sheath gas flow rate setting, 29; auxiliary sweep gas flow rate setting, 10; capillary voltage, −47 V; capillary temperature, 320 °C; tube lens voltage, −16 V. MS/MS conditions were: normalized collision energy, 35%; activation Q, 0.250; time, 30 ms; 1 scan. Product ion spectra were acquired over the m/z 250–2000 range. The abundant ions from LC-MS spectra were selected for collision-induced dissociation (CID) analysis, and the cut-off was set >15% of the most abundant ion. When more than one ion came from a single species, the peak responding to the doubly charged parent ion was chosen for fragmentation analysis. The calculations of the CID fragmentations of oligonucleotide sequences were done using a program linked to the Mass Spectrometry Group of Medicinal Chemistry at the University of Utah (http://medlib.med.utah.edu/masspec/).
Results
Primer Extension Studies
The initial studies were done with four recombinant full-length DNA polymerases, all expressed in baculovirus systems and purified to electrophoretic homogeneity. Pol δ was used with PCNA, in that strong PCNA effects have been seen for bypass of 8-oxo-7,8-dihydro G with bovine pol δ (23); however, we have not found such effects with pol η, ι, or κ and any DNA adducts, at least with relatively short templates (20, 21).2
Pol δ was completely blocked at 1,N2-ε-G in this system (Figure 1A). Of the three human translesion polymerases used, only pol η fully extended the primer (Figure 1B). Pol ι and pol κ showed some 1-base incorporation (Figures 1C and 1D).
Figure 1.
Extension of 32P-labeled 13-mer paired with an 18-mer template containing G or 1,N2-ε-G at position 14 (Table 1). Z = T (base 5’ of G*). All incubations were done with 100 nM primer template and the indicated concentration of the polymerase. PCNA (400 nM) was present in the case of pol δ. The lengths of the bands are indicated.
Steady-state Kinetic Assays with Individual Nucleoside Triphosphates
The activity with pol δ was too low for detailed analysis. Other assays were done following preliminary experiments with 1 mM concentrations of individual dNTPs (Tables 2–4).
Table 2.
Steady-state Kinetic Parameters for One-base Incorporation by Human Pol ηa
| template | dNTP | Km (µM) | kcat(s−1) |
kcat/Km (mM−1 s−1) |
f (misinsertion frequency)b |
|---|---|---|---|---|---|
| C | 0.91 ± 0.35 | 0.23 ± 0.02 | 250 | (1) | |
| 5' – | |||||
| A | 110 ± 40 | 0.26 ± 0.03 | 2.4 | 0.0096 | |
| 3' –GTA– | |||||
| G | 9.1 ± 2.0 | 0.058 ± 0.003 | 6.4 | 0.026 | |
| C | 18 ± 5 | 0.021 ± 0.001 | 1.2 | (1) | |
| 5' – | |||||
| A | 15 ± 5 | 0.059 ± 0.004 | 3.9 | 3.3 | |
| 3' –G*TA– | |||||
| G | 9.3 ± 2.3 | 0.056 ± 0.003 | 6.0 | 5.0 | |
| C | 30 ± 2 | 0.015 ± 0.0003 | 0.5 | (1) | |
| 5'– | |||||
| A | 44 ± 5 | 0.12 ± 0.004 | 2.7 | 5.4 | |
| 3' –G*CA– | |||||
| G | 4.0 ± 1.1 | 0.017 ± 0.001 | 4.3 | 8.6 | |
G*: 1, N2 -εG
f = (kcat/Km)wrong dNTP/(Kcat/Km)correct dNTP
Table 4.
Steady-state Kinetic Parameters for One-base Incorporation by Human Pol κa
| Template | dNTP | Km (µM) | kcat(s−1) |
kcat/Km (mM−1 s−1) |
f (misinsertion frequency)b |
|---|---|---|---|---|---|
| 5' – | C | 3.6 ± 0.5 | 0.065 ± 0.002 | 18.1 | 1 |
| 3' –GTA– | T | 2900 ± 900 | 0.14 ± 0.03 | 0.048 | 0.0027 |
| 5' – | C | 230 ± 50 | 0.0045 ± 0.0003 | 0.020 | 1 |
| 3' –G*TA– | T | 2600 ± 700 | 0.052 ± 0.008 | 0.020 | 1 |
G*: 1, N2-ε-G
f = (kcat/Km)wrong dNTP/(kcat/Km)correct dNTP
With pol η, both A and G were preferably misincorporated, compared to C (Table 2). T was misincorporated the least. One possibility is that incorporation is actually opposite the next base 5’ of 1,N2-ε-G, based upon our experiences with this adduct and Dpo4 in the same sequences (18), and therefore both oligonucleotide templates shown in Table 1 were used C, T). The results (Table 2) indicate that the reactions with 1,N2-ε-G (and the G:A and G:G mispairs) proceeded with relatively good kcat values but Km values were unfavorable. One guide to catalytic efficiency is kcat/Km, the catalytic specificity parameter. With either C or T 5’ of 1,N2-ε-G, the order of incorporation was G > A > C.
Table 1.
Oligonucleotides Used in This Study
| 13-mer | 5′ GGGGGAAGGATTC |
| 18-mer | 3′ CCCCCTTCCTAAGG*ZACT |
G* = G or 1, N2-ε-G
Z = T or C
Incorporation with pol ι was much less efficient and only dTTP was misincorporated (Table 3), in contrast to pol η (Table 2). Only the template containing T was analyzed (Table 3). Interestingly, the parameter kcat/Km for dCTP incorporation opposite 1,N2-ε-G by pol ι was only about 6-fold less than opposite G, compared with pol η (about 200–500 fold), indicating only some tolerability with this lesion to pol ι, in spite of inherently less efficiency than with pol η.
Table 3.
Steady-state Kinetic Parameters for One-base Incorporation by Human Pol ιa
| template | dNTP | Km (µM) | kcat(s−1) |
kcat/Km (mM−1 s−1) |
f (misinsertion frequency)b |
|---|---|---|---|---|---|
| 5' – | C | 150 ± 40 | 0.26 ± 0.02 | 1.7 | (1) |
| 3' –GTA– | T | 770 ± 130 | 0.12 ± 0.01 | 0.16 | 0.094 |
| 5' – | C | 72 ± 18 | 0.020 ± 0.001 | 0.28 | (1) |
| 3' –G*TA– | T | 980 ± 240 | 0.26 ± 0.03 | 0.27 | 0.96 |
G*: 1, N2-εG
f = (kcat/Km)wrong dNTP/(kcat/Km)correct dNTP
The results with pol κ (Table 4) were similar to pol ι (dTTP incorporation). The efficiencies were even lower. The difference of kcat/Km of dCTP incorporation opposite 1,N2-ε-G (vs. G) with pol κ was largest (900-fold) among the polymerases tested, indicating the least tolerability to this lesion.
Analysis of Pol η Extension Products
With prolonged incubation conditions at lower concentrations of template/primer DNA substrate it was possible to extend the primer to what appeared to be a mixture of full-length product and a further 1-base extension, as judged by the results of gel electrophoresis studies (Figures 2, 3). The results appeared to be similar regardless of whether C or T was 5’ of the 1,N2-ε-G (Figure 3).
Figure 2.
Extension of primers beyond 1,N2-ε-G:G and 1,N2-ε-G:A pairs by pol η. X denotes 1,N2-ε-G in parts A and B. The primer strand contained a 32P label at the 5’-end. (A) The incubation included 125 nM primer:template complex (shown at bottom of each experiment), 50 nM pol η, and a 62.5 µM mixture of all four dNTPs. At times varying up to 30 min, aliquots were analyzed for extension of the 32P-labeled primer. (B) Incubations were done with the indicated primer-template complexes for a fixed time (30 min) as a function of dNTP concentration. The incubations were done with 100 nM primer-template complex (shown at bottom of each experiment), 40 nM pol η, and the indicated concentrations of a mixture of all four dNTPs. In both parts A and B, aliquots of the reaction mixture were analyzed by gel electrophoresis and phosphoimaging. The lengths of the oligonucleotides are shown at the sides of the electrophoretograms.
Figure 3.
Prolonged extension of a 13-mer primer opposite 18-mer templates containing G or 1,N2-ε-G by pol η. (A) Experimental design and results of experiments done with 32P-labeled primer and analysis of products by gel electrophoresis. The concentration of primer/template complex was 10 nM, and the concentrations of pol η are indicated. The reactions proceeded for 60 min at 37 °C. (B) Results of analysis of products (made under conditions similar to part A, without 32P) by LC-MS/MS. The gap in the primer strand opposite the A was generated by the UDG cleavage of a uracil originally placed at this position. See Figures 4–6 for details of the LC-MS/MS analysis.
The “single base” incorporation studies (Table 2) indicated that pol η had a preference for inserting dNTPs in the order G > A > C. However, this information does not indicate which bases will be incorporated in further extensions. Pol η could extend primers with either G or A placed opposite the 1,N2-ε-G adduct (Figure 2A). When the extension was examined as a function of the concentration of the added dNTP mixture (Figure 2B), the results were ambiguous. Although the tendency appeared to be that the primer with an A opposite 1,N2-ε-G was extended more readily (and an effort to quantitate total products to yield a “kcat” and “Km” was consistent with this view, i.e. ~ 5-fold greater catalytic efficiency), the patterns for extension of the primer beyond A and G (paired with 1,N2-ε-G) were qualitatively very different. Extension beyond the G tended to yield mainly +1 and fully extended (and 19-mer products, i.e. blunt-end additions), but with A opposite 1,N2-ε-G more intermediate length products accumulated (Figure 2B). Similar results were obtained with the two primers (A and G at 3′ end) and the template having a C 5′ of the 1,N2-ε-G (results not shown).
The gel results (Figure 2) did not provide clear answers as to what bases are incorporated into primers, and a further kinetic analysis of enzyme efficiencies would require multiple experimental settings and still not yield definitive predictions. We have previously analyzed primer extension products (including those past 1,N2-ε-G (13)) by modified Maxam-Gilbert sequencing methods and found these unsatisfactory. Recently we have applied an approach of substituting uracil for thymidine in the primer and then cleaving the product with UDG (18). The products (< 10-mers) are short enough for LC-MS/MS analysis using CID in an ion-trap instrument (24). With some assumptions about the variability of ES MS responses, it is possible to estimate the fractions of different components of mixtures (18).
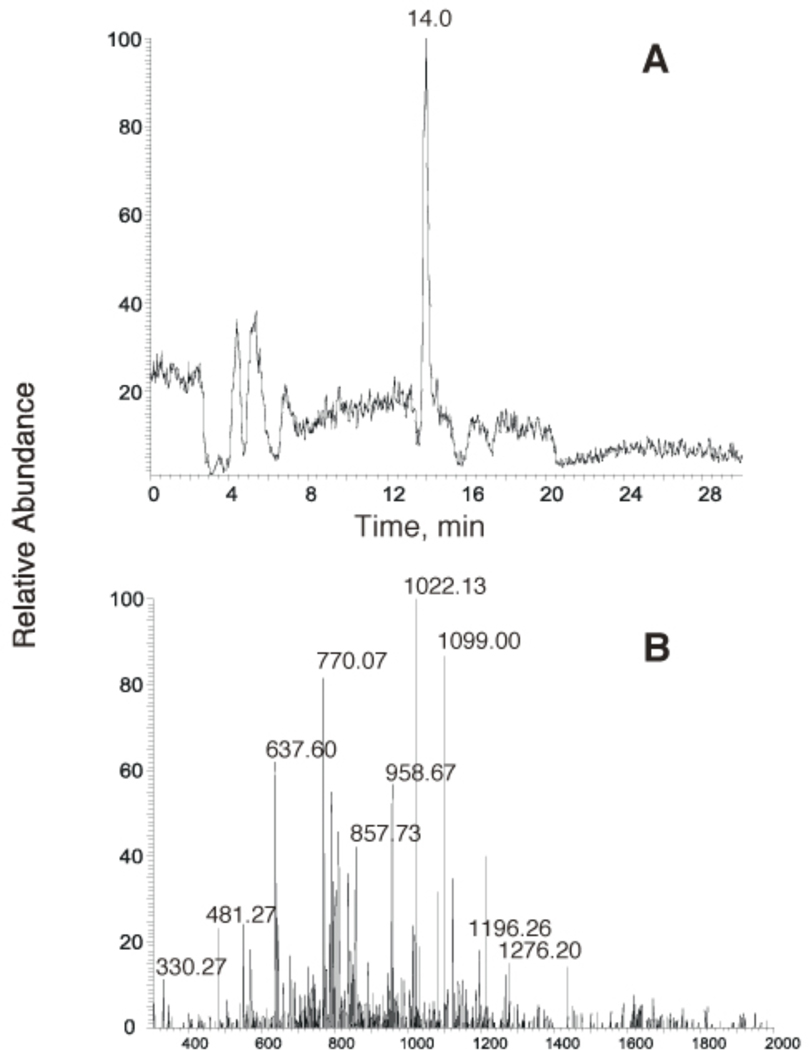
The LC-MS/MS approach was applied to the products generated by extension of the primer by pol η (Figure 3B). The products were eluted from HPLC together with the hydrolyzed primer at tR 13 min (Figure 4). The two ions at m/z 814.9 and 1086.4 were −4 and −3 ions from the primer (5’-GGGGGAAGGAp-3’) following UDG digestion. Thus, these two ions were not chosen for CID analysis. Three other ions, at m/z 732.3, 836.7, and 842.5, were considered further as potential products. With the assumption that these ions are −2, −3, or −4 ions, the corresponding molecular mass can be calculated, and only ions at m/z 732.3 and 836.7 can generate reasonable composition results based on the Oligo Composition Calculator. For example, if the ion at m/z 836.6 is an M-3H species, the molecular mass will be 2518.2. When the molecular mass of 2518.2 was applied to the Composition Calculator, two hits were obtained with a one mass unit cut-off (± 1) (a two mass unit cut-off (± 2) was also applied in some cases). One possibility was 1 C, 2 T’s, 3 A’s, and 2 G’s plus one phosphate at the 5’-end. Reconstruction of this composition gives a candidate sequence as 5’-pTCGATGA-3’ plus an extra A. The final choice of the sequence 5’-pTCGATGAA-3’ offered the best match of the CID spectra, especially the doubly-charged “a-B ion” at m/z 1022.1. Using the same strategy, the m/z 732.3 ion generated the sequence of another product 5’-pTCGATGA-3’ (see CID spectra in Figures 5 and 6 and calculated CID values in the Supporting Information, Tables 1S and 2S).
Figure 4.
Analysis of the pol η extension product (Figure 3) containing the oligonucleotide fragments obtained by UDG hydrolysis. (A) Total ion current (TIC) chromatogram. (B) ES mass spectrum of the peak eluted at 13.0 min in part A.
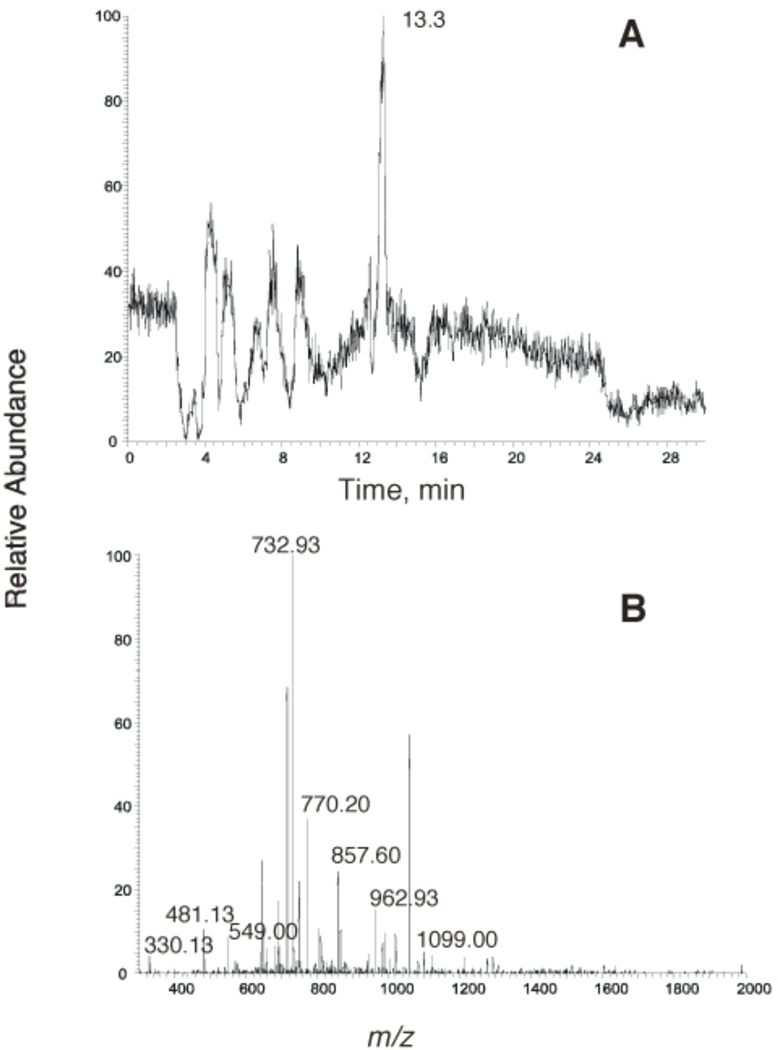
Figure 5.
Analysis of the longer pol η extension product (Figure 3) using LC-MS/MS. (A) Trace for ion m/z 836.7. (B) CID mass spectrum of ion m/z 836.7.
Figure 6.
Analysis of the shorter pol η extension product (Figure 3) using LC-MS/MS. (A) Trace for ion m/z 732.3. (B) CID mass spectrum of ion m/z 732.3.
The results (Figures 4–6) indicate an equimolar mixture of a full-length product and the same product extended to include an additional A. The apparent lengths of the products are consistent with the gel electrophoresis experiment (Figure 3A). In both of the products only a G was observed opposite 1,N2-ε-G, with fidelity of base incorporation occurring after this (Figure 3B).
Discussion
In this work, three major approaches were used to examine the selectivity of recombinant human DNA polymerases to process 1,N2-ε-G in DNA: (i) run-on gel analysis with all four dNTPs (Figure 1), (ii) steady-state kinetic analysis of 1-base incorporation with individual dNTPs (Tables 2–4), and (iii) LC-MS/MS analysis of the full-length product of extension by pol η. The first two approaches provide independent support for the dominant role of pol η, at least among the polymerases available here. The latter two approaches provide evidence that insertion of G opposite 1,N2-ε-G is a dominant process for pol η (and that the putative 1,N2-ε-G:G pair is preferentially extended, with apparently little slippage beyond the mispair).
In previous studies with Dpo4 (18, 26) and this work (Figures 3–6) we have found LC-MS/MS analysis to be a very powerful method of analyzing the sequences of oligonucleotide extension products and revealing insight into the events involved in replication past DNA adducts (Scheme 2). Critical to the success is the post-polymerase cleavage of the product to generate fragments small enough to use conventional ion trap mass spectrometers for analysis of the fragments carrying limited charge (i.e. −2 or −3). Some mass spectrometers may be capable of achieving good fragmentation results with longer oligonucleotides, although our limit with the DecaXP ion trap instrument is ~10 residues. We routinely use uracil and UDG for the post-reaction cleavage, even if a mispair must be introduced (although it should have some distance from the primer-template junction). We have tried using 8-oxo-7,8-dihydroG and 8-oxo-7,8-dihydroG glycosylase for cleavage but with limited success. The method has also been applied to Dpo4 products derived from polycyclic hydrocarbon epoxide N6-A adducts.3
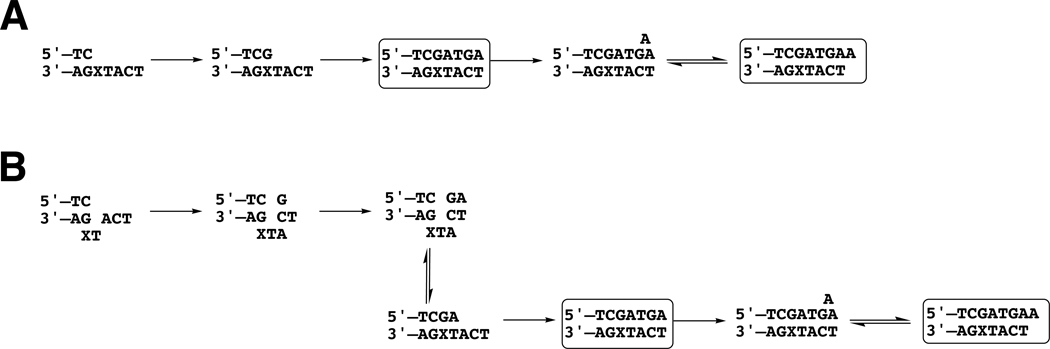
Scheme 2.
(A) Proposed Mechanism of Polymerization Past 1,N2-ε-G. (B) Alternate Mechanism for Incorporation of First Two Residues.a
aX indicates 1,N2-ε-G. Identified products are indicated with boxes.
Gel electrophoresis experiments provided only partial answers as to what residues were incorporated in primer extension studies. Analysis of the pol η insertion reaction (Table 2) indicated a preference for G > A, but the analysis of extension beyond G and A (Figure 2) did not provide a clear answer regarding preferences. LC-MS indicated that full extension products contained mainly G opposite 1,N2-ε-G, followed by a high fidelity extension (Figure 3). Consideration of all of the results indicates that analysis of events using single dNTPs does not necessarily lead to an accurate prediction of the overall pattern of incorporation, probably due to the complexity of the polymerase reaction and influences of neighboring bases that may not be accounted for in experiments with individual steps. One issue that can be raised is whether the extension of a primer by ~5 bases is a realistic approximation of the biological situation, in that the general view of translesion DNA polymerases is that they do not remain on the DNA for very many cycles (Figure 1) and are replaced by pol δ (or pol ε) (27). Thus, the question can be raised as to whether the experiments presented in Table 2 or Figure 3 are more representative o the cellular situation. This point was addressed by Yang et al. (28) in in vitro experiments with α-hydroxypropano G adducts, and the conclusion was that the translesion polymerase needed to copy ≥ 7 nucleotides past the adduct before pol δ resumed synthesis.
S. solfataricus Dpo4 and eukaryotic pol η have been compared with regard to their similar catalytic properties (29, 30). For instance, both enzymes catalyze high efficiency and high fidelity incorporation of dCTP opposite 8-oxo-7,8-dihydroG (26, 31–33). However, their activities with 1,N2-ε-G differ considerably. Both polymerases have the ability to proceed past the lesion with some efficiency ((18) and Figure 1). However, even with the same oligonucleotide sequences (18), Dpo4 produces a variety of products dominated by a mode in which 1,N2-ε-G is skipped, producing a −1 frameshift. When Dpo4 does insert directly opposite 1,N2-ε-G, the choice is dATP (18). In contrast, human pol η inserts dGTP opposite 1,N2-ε-G and then copies with high fidelity and apparently without frameshifts (Scheme 2A). The simplest conclusion is that the insertion of G is direct; an alternate 3-base slippage mechanism can be proposed (Scheme 2B) but we have no evidence for or against this. The addition of an extra A at the end in part of the product could be the result of either (i) non-instructional blunt-end ligation or (ii) a slippage/insertion mechanism (Scheme 2).
Even in the case of pol η, polymerization past 1,N2-ε-G is considerably retarded compared to G (Fig. 1). In the best case, the rate of pol η-catalyzed insertion of the preferred dNTP (dGTP) opposite 1,N2-ε-G is only ~2 % as efficient (on the basis of kcat/Km) as for insertion of dCTP opposite G (Table 2). An interesting point to note (Table 2) is that the efficiency for insertion of dGTP opposite 1,N2-ε-G is similar to that for insertion opposite G. Thus, one conclusion is that 1,N2-ε-G has no more coding information than G in this context. One possibility is that 1,N2-ε-G:G or G:G pairing can be explained by a Hoogsteen pair, in that conventional Watson-Crick pairing is impossible. However, such pairing must remain speculative in the absence of structural data. No evidence for Hoogsteen pairing with pol η was obtained in a comparative study with pol ι using N2,N2-dimethyl G (21).
Previously we reported studies on the bypass of 1,N2-ε-G by more replicative polymerases (12). E. coli pol I (Klenow fragment) exonuclease− appeared to insert both G and C opposite 1,N2-ε-G and to produce −1 and −2 frameshifts, although no quantitation was done. E. coli pol II (exonuclease−), however, inserted A, T, and C. With the Klenow fragment, the best rate of incorporation (kcat/Km) opposite 1,N2-ε-G was lower than incorporation of dCTP opposite G by a factor of 2 × 10−6 (12) (compared to 2 × 10−2 for pol η, Table 2). Comparisons with pol T7− (12) are not useful in that the primer-template pair in the earlier study (12) was too short to be used proficiently (34, 35).
Our results on the kinetic parameters and base selectivity (opposite 1,N2-ε-G) by pol η cannot necessarily be further generalized to different sequence contexts. The kcat/Km values for incorporation of incorrect bases such as dATP and dGTP opposite 1,N2-ε-G (Table 2) were not very different from those opposite unmodified G and thus the preferential incorrect incorporation (dGTP > dATP) opposite 1,N2-ε-G by pol η might not be a direct coding effect of 1,N2-ε-G but possibly related to an effect of the sequence context used here (Scheme 2B). As do other Y-family DNA polymerases, pol κ shows a large effect of the sequence context (even in the case of a one base difference adjacent to the template base) on polymerization efficiency and pol η might also. Therefore we cannot extrapolate our result to a different template such as the sequence having just a purine base (instead of a pyrimidine) at the position 5’ of 1,N2-ε-G. Most other studies on DNA adducts using human polymerases, including Y-family polymerases, have been focused on a single lesion (in one sequence context) with one polymerase and the results are often fragmented. Therefore, it is quite difficult to compare the kinetic parameters of different polymerases directly. Possible explanations of the polymerase specificity on each lesion can be the capability of Watson-Crick (or Hoogsteen) base-pairing, location (groove-side or intercalating), shape, size, hydrophobicity, and stacking of the lesion in the active site of each polymerase. No single mechanism explains all of the effects because of no available information in the structure of each human polymerase with various adducts. Pol η seems to have versatile bypass ability across a variety of lesions including T-T dimers and 8-oxo-7,8-dihydroguanine, among the Y-family polymerases, whereas pol ι seems to have limited ability on some lesions and pol κ seems to have a very limited role in incorporation opposite lesions but a considerable role as a proficient mismatch (or the lesion:base pair) extender (27).
Work with some other exocyclic G adducts (1,N2-propanoG derivatives) has been interpreted in the context of a mechanism in which pol ι inserts opposite the adduct and then pol κ extends the primer strand (36–38). Although the structures of 1,N2-ε-G and an acrolein adduct (36) differ, considerable incorporation of dTTP opposite the adduct was observed in both cases (Table 3). In the case of 1,N2-ε-G, we did not try a mixture of pol ι and κ to extend the primer product further (Figure 1). However, the conclusion that pol η is the most proficient to these polymerases under consideration in inserting opposite 1,N2-ε-G is not affected, in that the steady-state kinetic paramaters clearly favor pol η in this case (Tables 2–4). Another issue in comparing our results with those presented by Washington et al. (36) and Wolfle et al. (37, 38) is that the kcat and kcat/Km parameters we measured for dCTP incorporation opposite (unmodified) G are one to two orders of magnitude higher for all these translesion polymerases (Tables 2–4 and (20, 21)),2 although the effects of differences in sequences may contribute to the discrepancy.
In vivo work with 1,N2-ε-G (E. coli) showed apparent incorporation A and T opposite the residue, although the analysis was not designed to score complex mutations (13). In a study involving stable chromosomal integration of 1,N2-ε-G in Chinese hamster ovary cells (14), the number of simple base pair substitutions was limited and many complex mutations were found. However, the dominant base pair change at the site of the lesion was a G to A transition, implying insertion of dTTP opposite 1,N2-ε-G. The discrepancy with our present results might be due to the several possible reasons. One possibility is that DNA substrates with different sequences and lengths can affect and change the subtle base preference and efficiency in nucleotide incorporation opposite 1,N2-ε-G by the individual polymerases we tested here. The earlier work and more recent information about translesion polymerases raises the question of which translesion DNA polymerases are expressed (and operative) in cultured Chinese hamster ovary cells (and other cultured cells). Apparently these cells have not been analyzed with regard to which translesion polymerases are expressed. This is a general problem in understanding mutation biology, with all adducts. Other possible reason for the limited concordance between the apparent mutations in the Chinese hamster ovary cells is that some translesion polymerases with higher activity and different preferences are more active than the three polymerases studied here in different sequences and longer DNA strands. One alternate approach to establishing better discernment of which translesion polymerases are functional with specific DNA adducts is to utilize cells from transgenic “knock out” lines or animals, or the use of small RNA interference systems.
In conclusion, pol η was the most active of the four DNA polymerases tested with 1,N2-ε-G and was very miscoding (Scheme 2, Figure 1, Tables 2–4). The specific biochemical mechanisms for the preferred insertion of G opposite 1,N2-ε-G and proficient (and high fidelity) extension beyond the 1,N2-ε-G:G mispair are unclear. The behavior is considerably distinct from that of another translesion polymerase, S. solfataricus Dpo4. The results emphasize the importance of the individual mechanisms of DNA polymerases in determining incorporation events. Knowledge of thermodynamic preferences for pairing is of limited predictive value, particularly involving rare or unfavorable events such as interactions with bases whose coding potential is severely restricted.
Supplementary Material
Acknowledgment
This work was supported in part by National Institutes of Health research grants R01 ES010375 (F.P.G.), R01 ES011331 (C.J.R.), and P30 ES000267 (F.P.G., C.J.R). H.Z. was supported by a Merck Research Fellowship. A.K.G. was supported by T32 ES007028. We thank K. Trisler for assistance in preparation of the manuscript.
Footnotes
Abbreviations: BSA, bovine serum albumin; CID, collision-induced dissociation; DTT, dithiothreitol; ε, etheno; ES, electrospray; LC, liquid chromatography; MS, mass spectrometry; PCNA, proliferating cell nuclear antigen; pol, (DNA) polymerase; TIC, total ion current; UDG, uracil DNA glycosylase.
Choi, J-Y., Angel, K. C., and Guengerich, F. P., submitted for publication.
Zang, H., Chowdhury, G., Angel, K. C., Harris, T. M., and Guengerich, F. P., submitted for publication.
Supporting Information Available. Tables with calculated CID fragments of the primer products (obtained after UDG hydrolysis) 5’-pTCGATGA-3’ and 5’-pTCGATGAA-3’.
References
- 1.Guengerich FP. Effects of carcinogen-bound DNA on individual DNA polymerases. Chem. Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed. Washington, D. C.: Am. Soc. Microbiol. Press; 2006. [Google Scholar]
- 3.Sattsangi PD, Leonard NJ, Frihart CR. 1,N2-Ethenoguanine and N2,3-ethenoguanine. Synthesis and comparison of the electronic spectral properties of these linear and angular triheterocycles related to the Y bases. J. Org. Chem. 1977;42:3292–3296. doi: 10.1021/jo00440a020. [DOI] [PubMed] [Google Scholar]
- 4.Singer B, Bartsch H. Exocyclic DNA Adducts in Mutagenesis and Carcinogenesis. Vol. 150. Lyon, France: International Agency for Research on Cancer Scientific Publications; 1999. [Google Scholar]
- 5.Guengerich FP, Kim D-H, Iwasaki M. Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem. Res. Toxicol. 1991;4:168–179. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- 6.Guengerich FP, Persmark M, Humphreys WG. Formation of 1,N2- and N2,3-ethenoguanine from 2-halooxiranes: isotopic labeling studies and isolation of a hemiaminal derivative of N2-(2-oxoethyl)guanine. Chem. Res. Toxicol. 1993;6:635–648. doi: 10.1021/tx00035a008. [DOI] [PubMed] [Google Scholar]
- 7.Hecht SS, Young-Sciame R, Chung FL. Reaction of α-acetoxy-N-nitrosopiperidine with deoxyguanosine: oxygen-dependent formation of 4-oxo-2-pentenal and a 1,N2-ethenodeoxyguanosine adduct. Chem. Res. Toxicol. 1992;5:706–712. doi: 10.1021/tx00029a018. [DOI] [PubMed] [Google Scholar]
- 8.Kronberg L, Sjöholm R, Karlsson S. Formation of 3,N4-ethenocytidine, 1,N6-ethenoadenosine, and 1,N2-ethenoguanosine in reactions of mucochloric acid with nucleosides. Chem. Res. Toxicol. 1992;5:852–855. doi: 10.1021/tx00030a019. [DOI] [PubMed] [Google Scholar]
- 9.Sodum RS, Chung FL. 1,N2-Ethenodeoxyguanosine as a potential marker for DNA adducts formation by trans-4-hydroxy-2-nonenal. Cancer Res. 1988;48:320–323. [PubMed] [Google Scholar]
- 10.Morinello EJ, Ham AJL, Ranasinghe A, Sangaiah R, Swenberg JA. Simultaneous quantitation of N2,3-ethenoguanine and 1,N2-ethenoguanine with an immunoaffinity/gas chromatography/high-resolution mass spectrometry assay. Chem. Res. Toxicol. 2001;14:327–334. doi: 10.1021/tx0002076. [DOI] [PubMed] [Google Scholar]
- 11.Lee SH, Arora JA, Oe T, Blair IA. 4-Hydroperoxy-2-nonenal-induced Formation of 1,N2-etheno-2'-deoxyguanosine adducts. Chem. Res. Toxicol. 2005;18:780–786. doi: 10.1021/tx0497088. [DOI] [PubMed] [Google Scholar]
- 12.Langouët S, Müller M, Guengerich FP. Misincorporation of dNTPs opposite 1,N2-ethenoguanine and 5,6,7,9-tetrahydro-7-hydroxy-9-oxoimidazo[1,2-a]purine in oligonucleotides by Escherichia coli polymerases I exo− and II exo−, T7 polymerase exo−, human immunodeficiency virus-1 reverse transcriptase, and rat polymerase β. Biochemistry. 1997;36:6069–6079. doi: 10.1021/bi962526v. [DOI] [PubMed] [Google Scholar]
- 13.Langouët S, Mican AN, Müller M, Fink SP, Marnett LJ, Muhle SA, Guengerich FP. Misincorporation of nucleotides opposite 5-membered exocyclic ring guanine derivatives by Escherichia coli polymerases in vitro and in vivo: 1,N2-ethenoguanine, 5,6,7,9-tetrahydro-9-oxoimidazo[1,2-a]purine, and 5,6,7,9-tetrahydro-7-hydroxy-9-oxoimidazo[1,2-a]purine. Biochemistry. 1998;37:5184–5193. doi: 10.1021/bi972327r. [DOI] [PubMed] [Google Scholar]
- 14.Akasaka S, Guengerich FP. Mutagenicity of site-specifically located 1,N2-ethenoguanine in Chinese hamster ovary cell chromosomal DNA. Chem. Res. Toxicol. 1999;12:501–507. doi: 10.1021/tx980259j. [DOI] [PubMed] [Google Scholar]
- 15.Saparbaev M, Langouët S, Privezentev CV, Guengerich FP, Cai H, Elder RH, Laval J. 1,N2-Ethenoguanine, a mutagenic DNA adduct, is a primary substrate of Escherichia coli mismatch-specific uracil-DNA glycosylase and human alkyl-N-purine DNA glycosylase. J. Biol. Chem. 2002;277:26987–26993. doi: 10.1074/jbc.M111100200. [DOI] [PubMed] [Google Scholar]
- 16.Nair J, Barbin A, Velic I, Bartsch H. Etheno DNA-base adducts from endogenous reactive species. Mut. Res. 1999;424:59–69. doi: 10.1016/s0027-5107(99)00008-1. [DOI] [PubMed] [Google Scholar]
- 17.VanderVeen LA, Druckova A, Riggins JN, Sorrells JL, Guengerich FP, Marnett LJ. Differential DNA recognition and cleavage by EcoRI dependent on the dynamic equilibrium between two forms of the malondialdehyde-deoxyguanosine adducts. Biochemistry. 2004;44:5024–5033. doi: 10.1021/bi0472898. [DOI] [PubMed] [Google Scholar]
- 18.Zang H, Goodenough AK, Choi J-Y, Irminia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4. Analysis and crystal structures of multiple base-pair substitution and frameshift product with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 19.Goodenough AK, Kozekov ID, Zang H, Choi J-Y, Guengerich FP, Harris TM, Rizzo CJ. Site-specific synthesis and polymerase bypass of oligonucleotides containing 6-hydroxy-3,5,6,7-tetrahydro-9H-imidazo[1,2-a]purinone, an intermediate in the formation of 1,N2-etheno-2'-deoxyguanosine. Chem. Res. Toxicol. 2005;18:1701–1714. doi: 10.1021/tx050141k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J-Y, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J. Mol. Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 21.Choi J-Y, Guengerich FP. Kinetic analysis of inefficient and error-prone bypass at bulky N2-guanine DNA adducts by human DNA polymerase i. J Biol Chem. 2006;281 doi: 10.1074/jbc.M600112200. in press. [DOI] [PubMed] [Google Scholar]
- 22.Einolf HJ, Guengerich FP. Kinetic analysis of nucleotide incorporation by mammalian DNA polymerase δ. J. Biol. Chem. 2000;275:16316–16322. doi: 10.1074/jbc.M001291200. [DOI] [PubMed] [Google Scholar]
- 23.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion opposite 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase δ. J. Biol. Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 24.Ni J, Pomerantz SC, Rozenski J, Zhang Y, McCloskey JA. Interpretation of oligonucleotide mass spectra for determination of sequence using electrospray ionization and tandem mass spectrometry. Anal. Chem. 1996;68:1989–1999. doi: 10.1021/ac960270t. [DOI] [PubMed] [Google Scholar]
- 25.Christian NP, Reilly JP, Mokler VR, Wincott FE, Ellington AD. Elucidation of the initial step of oligonucleotide fragmentation in matrix-assisted laser desorption/ionization using modified nucleic acids. J. Am. Soc. Mass. Spectrom. 2001;12:744–753. doi: 10.1016/S1044-0305(01)00228-8. [DOI] [PubMed] [Google Scholar]
- 26.Zang H, Irminia A, Choi J-Y, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. High fidelity and efficient incorporation of dCTP opposite 7,8-dihydro-8-oxo-deoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 27.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 28.Yan I-Y, Miller H, Wang Z, Frank EG, Ohmori H, Hanaoka F, Moriya M. Mammalian translesion DNA synthesis across an acrolein-derived deoxyguanosine adduct. Participation of DNA polymerae η in error-prone synthesis in human cells. J. Biol. Chem. 2003;278:13989–13994. doi: 10.1074/jbc.M212535200. [DOI] [PubMed] [Google Scholar]
- 29.Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang ZG, Woodgate R. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 30.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic pol eta. Nucl. Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 32.Carlson KD, Washington MT. Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase η. Mol. Cell Biol. 2005;25:2169–2176. doi: 10.1128/MCB.25.6.2169-2176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG Lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 35.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo− and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 36.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Human DNA polymerase ι promotes replication through a ring-closed minor-groove adduct that adopts a syn conformation in DNA. Mol. Cell Biol. 2005;25:8748–8754. doi: 10.1128/MCB.25.19.8748-8754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell Biol. 2006;26:381–386. doi: 10.1128/MCB.26.1.381-386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.