Abstract
Non-small-cell lung cancer (NSCLC) is the leading cause of cancer-related death. Developing minimally invasive techniques that can diagnose NSCLC, particularly at an early stage, may improve its outcome. Using microarray platforms, we previously identified 12 microRNAs (miRNAs) the aberrant expressions of which in primary lung tumors are associated with early-stage NSCLC. Here, we extend our previous research by investigating whether the miRNAs could be used as potential plasma biomarkers for NSCLC. We initially validated expressions of the miRNAs in paired lung tumor tissues and plasma specimens from 28 stage I NSCLC patients by real-time quantitative reverse transcription PCR, and then evaluated diagnostic value of the plasma miRNAs in a cohort of 58 NSCLC patients and 29 healthy individuals. The altered miRNA expressions were reproducibly confirmed in the tumor tissues. The miRNAs were stably present and reliably measurable in plasma. Of the 12 miRNAs, five displayed significant concordance of the expression levels in plasma and the corresponding tumor tissues (all r>0.850, all P<0.05). A logistic regression model with the best prediction was defined on the basis of the four genes (miRNA-21, -126, -210, and 486-5p), yielding 86.22% sensitivity and 96.55% specificity in distinguishing NSCLC patients from the healthy controls. Furthermore, the panel of miRNAs produced 73.33% sensitivity and 96.55% specificity in identifying stage I NSCLC patients. In addition, the genes have higher sensitivity (91.67%) in diagnosis of lung adenocarcinomas compared with squamous cell carcinomas (82.35%) (P<0.05). Altered expressions of the miRNAs in plasma would provide potential blood-based biomarkers for NSCLC.
Keywords: diagnosis, lung cancer, microRNA, plasma, qRT-PCR.
Non-small-cell lung cancer (NSCLC) is the most common type of lung cancer and the number-one cancer killer in the United States and worldwide. NSCLC mainly consists of two major histological types: adenocarcinoma (AC) and squamous cell carcinoma (SCC). The disease is frequently diagnosed at advanced stages, resulting in an overall 5-year survival rate of about 14%.1 Conversely, the 5-year survival rate in stage I NSCLC patients who have received effective treatments can be as high as 83%.1 Therefore, finding NSCLC earlier may reduce the mortality.1 The sensitivity of chest X-ray and sputum cytology for early detection of NSCLC is low.2, 3 Bronchoscopy excels at detecting centrally occurring lung tumors. However, it is invasive.4 Although CT provides excellent anatomic information and can noninvasively detect NSCLC earlier at small size, the improved sensitivity is associated with over-diagnosis.1, 2, 3, 4, 5 Therefore, developing minimally invasive techniques by taking advantage of recent developments in molecular genetics for diagnosis of early stage NSCLC is clinically important. Blood plasma is obviously preferred choice for development of such diagnostic markers. Numerous tumor-specific molecular alterations have been identified in plasma/serum and shown the potential as biomarkers in patients with lung cancers.5, 6 However, none of the tested markers thus far had sufficiently achieved the required characteristics for diagnosis of NSCLC.
MicroRNAs (miRNAs) open up a new field for molecular diagnosis of cancer.7, 8, 9, 10, 11, 12, 13 Accumulating reports suggest that circulating miRNAs are present in blood and have the potential as biomarkers for several malignancies including lung cancer.14, 15 For example, serum miRNA signatures were recently identified that could predict survival of NSCLC patients.16 Although the measurement of miRNA expressions in plasma/serum has been postulated as a promising approach in diagnosing lung cancer, the concept requires further investigations to demonstrate its potential clinical application.
Using microarray platforms, we recently identified 12 miRNAs the abnormal expressions of which in lung tumor tissues were associated with early-stage NSCLC.17, 18, 19 Here, we extended our previous research by investigating whether the miRNAs could be detected in plasma and used as minimally invasive biomarkers for NSCLC. We first confirmed the expressions of the miRNAs in paired surgical tissue and plasma specimens from an independent NSCLC cases by using real-time quantitative reverse transcription PCR (Real-time qRT-PCR). We then examined plasma expressions of the miRNAs in a cohort of 58 NSCLC patients and 29 healthy subjects to assess the diagnostic value for NSCLC. The study shows that altered expressions of the miRNAs in plasma might provide potential blood-based biomarkers for NSCLC.
MATERIALS AND METHODS
Patients and Clinical Specimens
The study was divided into two phases. In the first phase, we determined whether the stage I NSCLC-associated miRNAs the aberrant expressions of which could be confirmed in independent surgical tissues, and whether the miRNAs were present in plasma and their changes in plasma were consistent with those in the corresponding tumors. To this end, we obtained the paired lung tumor tissues and noncancerous lung tissues and plasma specimens of 28 stage I NSCLC patients (Table 1) from Baltimore VA Maryland Health Care System (BVAMHCS) and University of Maryland Medical Center (UMMC). The cohort consisted of 18 male and 10 female, 21 white American and seven African American. Their ages ranged from 46 to 79 years. In total, 14 tumors were classified as AC and 14 as SCC. Smoking pack-years of the cases were 38±27. Furthermore, blood samples were collected from 28 age-, race-, gender-, and smoking status-matched healthy individuals, and served as control specimens to assess changes of the miRNAs in plasma of the cancer patients. Sample collection was done in accordance with the protocols approved by the Institutional Review Boards (IRBs) of BVAMHCS and University of Maryland Baltimore. Blood specimens of the cancer cases were collected before the induction of anesthesia, before surgery. None of the patients received preoperative adjuvant chemotherapy or radiotherapy.
Table 1. Demographic and clinical characteristics of a cohort of 28 stage I NSCLC cases and 28 healthy individuals.
| 28 NSCLC cases | 28 Healthy smokers | |
|---|---|---|
| Age, median (s.d.) | 65.8 (6.9) | 67 (6.4) |
| Sex | ||
| Female | 10 (35.7%) | 10 (35.7%) |
| Male | 18 (64.3%) | 18 (64.3%) |
| Race | ||
| African American | 7 (25%) | 7 (25%) |
| White | 21 (75%) | 21 (75%) |
| Smoking status | ||
| Pack-years, median (s.d.) | 35.3 (23.2) | 35.8 (25.8) |
| Stage | ||
| All are stage I | 28 (100%) | |
| Histological types | ||
| AC | 14 (50%) | |
| SCC | 14 (50%) | |
Abbreviations: AC, adenocarcinoma; NSCLC, non-small-cell lung cancer; SCC, squamous cell carcinoma.
The second phase of the study was to evaluate the diagnostic value of the miRNAs for NSCLC in plasma. We obtained blood samples from BVAMHCS, UMMC, and NCI-Cooperative Human Tissue Network under the IRB protocols. The specimens were collected from 58 NSCLC patients with various stages and histological types and 29 healthy individuals (Table 2). Among the cancer patients, 29 were African American and 29 were Caucasian and 20 patients were female and 38 were male. In total, 15 had stage I NSCLC, 15 with stage II, 12 with stage III, and 16 with stage IV. Histological diagnosis was made on bronchoscopic biopsy specimens and thoracotomy according to the World Health Organization (WHO) categories.20, 21 The NSCLC stage classification was based on WHO classification and the International Association for the Study of Lung Cancer staging system. There were no significant differences of age, race, gender, and smoking status between the NSCLC patients and healthy individuals. Peripheral blood (10 ml) was drawn from the subjects using standardized phlebotomy procedures in BD Vacutainer spray-coated K2EDTA Tubes (BD, Franklin Lakes, NJ, USA). The blood samples from cancer patients were collected at the time of initial consultation, before definitive surgical management and/or adjuvant therapy. The specimens were processed within 2 h of collection by centrifugation at 1300 × g at for 10 min 4°C. Plasma was transferred to a fresh tube and stored at −80°C until use.
Table 2. Demographic and clinical characteristics of an independent cohort of 58 NSCLC cases and 29 healthy individuals.
| 58 NSCLC cases |
29 Healthy subjects | ||
|---|---|---|---|
| 24 SCC patients | 34 AC patients | ||
| Age, median (s.d.) | 67.4 (8.7) | 68 (8.2) | 66 (7.8) |
| Sex | |||
| Female | 9 (37.5%) | 11 (32.3%) | 10 (34.5%) |
| Male | 15 (62.5%) | 23 (67.7%) | 19 (65.5%) |
| Race | |||
| African American | 12 (50%) | 16 (47.1%) | 7 (24.1%) |
| White | 12 (50%) | 18 (52.9%) | 22 (75.9%) |
| Smoking status | |||
| Pack-years, median (s.d.) | 34.6 (27.8) | 30.6 (21.1) | 30.4 (28.3) |
| Stage | |||
| I | 6 (25%) | 9 (26.6%) | |
| II | 7 (29.2%) | 8 (23.5%) | |
| III | 6 (25%) | 6 (17.6%) | |
| IV | 5 (20.8%) | 11 (32.3%) | |
Abbreviations: AC, adenocarcinoma; NSCLC, non-small-cell lung cancer; SCC, squamous cell carcinoma.
RNA Isolation
RNA was extracted from the tissue specimens by using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) as described in our previous study.17, 18 The mirVana miRNA isolation kit (Ambion) was also utilized to extract RNA from plasma according to the manufacturer's instructions. Briefly, 350 μl plasma was diluted with two volumes of Lysis/Binding Solution (Ambion). miRNA homogenate additive, in volumes of 1/10, was then added in each aliquot. An equal volume of acid/phenol/chloroform (Ambion) was added to each aliquot. The resulting solutions were spun for 10 min at 10 000 × g, and then passed through a mirVana column in sequential 700-μl aliquots. 20 ng of eluted RNA were pretreated with one unit of DNase (Invitrogen Corporation, Carlsbad, CA, USA) and 0.38 μl of RNase inhibitor (Invitrogen Corporation). RNA's purity and concentration were determined from OD260/280 readings using a dual beam UV spectrophotometer (Eppendorf AG, Hamburg, Germany). Integrity of RNA was determined by using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA (with an RNA integrity number >6) underwent in further analysis.
Quantification of miRNA Expression by qRT-PCR
The reverse transcription (RT) reaction was carried out with TaqMan MicroRNA RT Kit (Applied Biosystems) according to the manufacture's instruction. An amount of 1.33 μl of cDNA solution was amplified in 20 μl of mixture. Quantitative PCR was then performed on a Bio-Red IQ5 Multi-color RT-PCR Detection System (Bio-Red, Hercules, CA, USA). All assays were undertaken in triplicates, and one no-template control and two interplate controls were carried along in each experiment. Expression levels of the miRNAs were calculated using comparative cycle threshold (Ct) method.17, 18, 19 Ct values of the target miRNAs were normalized in relation to that of miR-16. As explained later, miR-16 was used as an internal control gene. The fold change was calculated using the equation 2−ΔΔCt.
Selection of Internal Control Gene for Quantification of Plasma miRNAs
RNA was first purified from plasma of 20 NSCLC patients and 20 healthy subjects. Synthesized Caenorhabditis elegans miRNA, cel-miR-238 (Integrated DNA Technologies, Coralville, IA, USA) that did not exist in the human genomes, was spiked into each RNA sample with a concentration of 100 fmol/μl. qRT-PCR was undertaken in the same amount of RNA of each sample to determine Ct values of miR-16, RNU6B, and cel-miR-238. Ct values of miR-16 and RNU6B were compared with that of cel-miR-238 that was used to normalize the difference of experimental efficiency among different samples. To further evaluate the stability of miR-16 and RNU6B in plasma, plasma from five healthy individuals was first incubated with an equal volume of 2 × denaturing solution (Ambion) to inhibit endogenous RNase activity. The synthesized cel-miR-238 was spiked into each sample, which was subsequently divided into three parts. The first aliquot of each sample was processed for RNA extraction on day 1. The last two aliquots were stored in 4°C for 3 and 7 days, respectively, before being processed for RNA isolation. The Ct values of the two human endogenous genes and cel-miR-238 were simultaneously determined by qRT-PCR.
Specificity and Sensitivity of qRT-PCR Assay for Quantification of the miRNAs
Plasma was obtained from 10 healthy individuals and divided into four parts in 350 μl for each, respectively. The first aliquot from each specimen was processed on day 1 for isolating RNA, whereas others were stored at −20°C and processed on day 3, 7, and 30. qRT-PCR was performed in the RNA samples to determine whether the miRNAs were stably present and reproducibly detectable in plasma. To further assess stability of plasma miRNAs, plasma specimens from another five healthy subjects were split in three parts, respectively. The first two parts of each sample were incubated with RNase A (Qiagen) at different concentrations (0.1 and 1 mg/ml) for 24 h, respectively. The third part of each specimen was treated and served as controls. In addition, the synthesized cel-miR-238 was spiked into the plasma samples treated with RNase A (Qiagen) at different concentrations and the specimens without the treatment. The expressions of the endogenous miRNAs and spiked miRNA were measured in the specimens by qRT-PCR in parallel. Moreover, plasma of another five healthy subjects was divided into three aliquots. The first aliquot of each sample was subjected to five repeated cycles of freeze thawing, whereas other two aliquots were stored in −80°C freezer for 3 and 7 days, respectively. Expression of the endogenous miRNAs was also measured in these specimens. To determine specificity of qRT-PCR assay for quantification of miRNAs in plasma, the synthesized cel-miR-238 was diluted in RNA isolated from five denatured plasma specimens by 10 orders of magnitude, respectively. qRT-PCR with primers specific to cel-miR-238 was performed in the samples. The plasma samples that were not spiked with the synthesized cel-miR-238 were also tested in parallel and used as controls. Furthermore, to assess the ability of qRT-PCR for specifically amplifying human miRNA genes, two members of a human miR-486 family, miR-486-3p and miR-486-5p, were synthetically generated (Integrated DNA Technologies). The synthesized miR-486 variants were diluted in DEPC water and then polyadenylated and reversely transcribed. Each of the miR-486 variants was subjected to two independent qRT-PCR reactions, wherein each reaction there were PCR primers specific to only one of the two variants. To determine the sensitivity and dynamic range of qRT-PCR for quantification of the plasma miRNAs, RNA was first extracted from plasma of five healthy individuals and then diluted by 10 orders of magnitude in DEPC water. Expressions of the 12 miRNAs were assessed by qRT-PCR in the serially diluted samples. All the experiments were performed in triplicate and repeated three times.
Statistical Analysis
To determine whether genes were statistically differentially expressed between the paired tumor tissues and normal tissues, we expected the acceptable number of false positives to be 1.0, fold difference between the two types of samples at 2.0, s.d. of the gene measurements on the base-two logarithmic scale at 0.7, and desired power at 0.9. Given 12 miRNAs needed to be evaluated, at least nine specimens for each tissue type were required to achieve the statistical criteria.22 To define an optimal miRNA marker panel for distinguishing cancer patients from normal controls, a case–control study was designed that consisted of lung cancer cases and cancer-free individuals. The area under receiver operating characteristic curve (AUC) of H0 (the null hypothesis) was set at 0.5. H1 represented the alternative hypothesis; accordingly, at least 28 subjects were required in each category to show a minimum difference of interest between an AUC of 0.75 versus an AUC of 0.5 with 80% power at the 5% significance level.23 Therefore, the two sets of cohorts of cases and controls recruited in the present study provided enough statistical power to analyze the miRNAs.
Pearson's correlation analysis was applied to determine association of miRNA expressions of miRNAs in tumor tissues with those in the paired plasma specimens. The analysis was also applied to assess relationship between plasma miRNA expressions and demographic characteristics of the patients and healthy individuals. Clinicopathological diagnoses were used as reference standards to decide sensitivity and specificity of the genes. The receiver–operator characteristic curve and AUC analyses were used to determine accuracy of each miRNA and cut-off value of each miRNA in a specimen with a given specificity rate.24 Logistic regression models with constrained parameters and similar to least absolute shrinkage and selection operator were applied to select an optimal panel of potential biomarkers.25 All analyses, including correlation coefficient, Wilcoxon test, logistic regression, ANOVA, and Student's t-test, were performed using log-transformed data. All P-values shown were two sided, and a P-value of <0.05 was considered statistically significant.
RESULTS
The Changes of the NSCLC-associated miRNAs were Reproducibly Validated in an Independent Set of Surgical Tissues
We previously used microarray-based techniques to analyze primary tumor tissues and identified 12 miRNAs the expression changes of which were associated with stage I NSCLC. The miRNAs include miR-21, 126, 145, 139, 182, 200b, 205, 210, 375, 429, 486-5p, and 708. To determine whether aberrations of the miRNAs could be confirmed by a different approach in independent tumor tissues, we assessed expressions of the miRNAs by qRT-PCR in 28 stage I NSCLC tissue and the paired noncancerous lung tissue specimens. All of the miRNAs displayed more than two2-fold higher or lower expression in the tumor tissues, as compared with the corresponding noncancerous tissues (all P<0.001) (Table 3). The observations were consistent with the previous findings,17 suggesting that the identified miRNAs by microarray platforms could be well confirmed by qRT-PCR in a different set of surgical specimens.
Table 3. miRNAs differentially expressed in 28 stage I NSCLC tissues versus the paired noncancerous lung tissue specimens by qRT-PCR.
| miRNAs | Log2 (fold)* (mean±s.d.) | P-value |
|---|---|---|
| miR-21 | 2.67265789±0.26879567 | 0.00072756 |
| miR-126 | −2.95674568±0.23674323 | 0.00028312 |
| miR-145 | −2.46785613±0.20874515 | 0.00052668 |
| miR-139 | −2.13567129±0.19585334 | 0.00015854 |
| miR-182 | 2.21567349±0.29864376 | 0.00026994 |
| miR-205 | 2.86871452±0.25647823 | 0.00057825 |
| miR-200b | 2.15668178±0.23456782 | 0.00032678 |
| miR-210 | 2.32567489±0.30124675 | 0.00076148 |
| miR-375 | 2.84519747±0.25784738 | 0.00057838 |
| miR-429 | −2.00934864±0.30032546 | 0.00023757 |
| miR-486-5p | −2.85638753±0.27545613 | 0.00043221 |
| miR-708 | 2.26721137±0.31145759 | 0.00054983 |
Abbreviation: NSCLC, non-small-cell lung cancer.
Median fold changes were determined by Wilcoxon test of paired data.
Selection of an Internal Control Gene for Quantification of Plasma miRNAs
To find an internal control that can reliably quantify expressions of the target miRNAs in plasma, we evaluated miR-16 and RNU6B in plasma of NSCLC and healthy subjects for the Ct values of qRT-PCR. The two genes have previously been proposed as potential internal normalization controls for miRNA quantification.26 To normalize the difference of experimental efficiency among different samples, we performed qRT-PCR in the same amount of RNA or fixed volume of RNA eluted from a given volume of starting plasma (350 μl). Furthermore, we compared Ct values of endogenous miR-16 and RNU6B with that of spiked cel-miR-238. No significant difference was observed on freshly prepared plasma for Ct values of miR-16 (P=0.464) and RNU6B (P=0.418) between cancer patients and healthy controls (Supplementary Figure 1). We further measured the genes in the samples prepared at different time points. Ct values of miR-16 did not change after prolonged storage at 4°C for 7 days (P>0.05, Supplementary Figure 2A). However, RNU6B exhibited less stability than miR-16, because the Ct values of RNU6B on the samples that were stored for 7 days were significantly higher than that on the samples prepared on day 1 (P=0.03) (Supplementary Figure 2B). Furthermore, mean Ct value of miR-16 was statistically lower than that of RNU6B in the same RNA samples tested on day 1 (19.2±1.2 versus 29.6±2.9, P<0.05). Altogether, the observations indicated that miR-16 displayed higher stability and abundance than did RNU6B in plasma. Therefore, miR-16 was used as the normalization control to determine expressions of the 12 miRNAs in the present study.
The Expressions of the NSCLC-associated miRNAs were Reliably Measurable in Plasma
To determine whether the 12 miRNAs are present in plasma, we measured their expressions in plasma of 10 healthy individuals by qRT-PCR. All tested miRNAs had ≤32 Ct values in each sample, indicating that the miRNAs existed in plasma and were readily detectable. We next investigated the stability of the endogenous miRNAs in archived plasma samples. Aliquots of the plasma specimens were stored at −20°C for 1, 3, 7, and 30 days, respectively. Bioanalyzer showed that there was increasing degradation of total RNA illustrated by gradually decreased heights of 18S and 28S peaks from day 1 to day 30 (Supplementary Figure 3A). However, there was no effect on expression levels of the miRNAs determined by qRT-PCR in the same specimens (Supplementary Figure 3B). Because plasma has high levels of RNase activity, we further measured expressions of the miRNAs in the samples treated with or without RNase A. The abundance of the endogenous miRNAs in the plasma samples with the different treatments was fairly equable (all P>0.05). Conversely, the spiked synthesized cel-miR-238 was almost not detected in the plasma samples (Supplementary Figure 4). The observation suggests that the endogenous miRNAs in plasma possess considerable resistance to the enzymatic cleavage of RNase A. However, the synthesized cel-miR-238 that was added in the samples was not resistant to either endogenous RNase A or exogenous RNase A in human plasma. In addition, aliquots of plasma subjected to five repeated cycles of freeze thawing and those that were stored at −80°C for 3 and 7 days did not display different expressions of the miRNAs. The findings suggest that the plasma miRNAs have adequate resistance to freeze–thaw action. Altogether, the 12 endogenous miRNAs are present in a stable form and consistently measurable in plasma.
To define specificity of qRT-PCR for quantification of miRNAs in plasma, synthesized cel-miR-238 was mixed with the total RNA isolated from denatured plasma specimens. The cel-miR-238 was detectable in the mixed samples. However, cel-miR-238 was not observed in the human RNA that was not spiked with the synthesized miRNA. Furthermore, there was excellent linearity between the cel-miR-238 input and the Ct values of qRT-PCR. In addition, qRT-PCR exhibited a dynamic range of at least eight orders of magnitude, and was able to detect as low as 10 copies of the cel-miR-238 in each reaction, and the correlation coefficient was 0.996 for the synthesized miRNA (Supplementary Figure 5). To further determine whether qRT-PCR assay can specifically amplify the target miRNAs, the synthetic miR-486-5p or miR-486-3p was subjected to two independent qRT-PCR reactions, wherein each reaction there were PCR primers specific to only one of the two genes. Non-specific amplification was found while amplification only of the appropriate gene that matched the specific primers was observed (Supplementary Figure 6). The findings imply that the qRT-PCR assay is highly specific and can discriminate miRNAs that are the members from the same gene family. Therefore, the miRNAs could be specifically detected in plasma.
To determine the sensitivity of qRT-PCR assay for detection of the 12 miRNA, RNA isolated from plasma was diluted in DEPC water by 10 orders of magnitude and then tested for expression of each miRNA. There was excellent linearity between the RNA input and the Ct values for qRT-PCR. Furthermore, the assay had a dynamic range of at least eight orders of magnitude, and was capable of detecting as few as 10 copies of the miRNAs per PCR reaction, and the correlation coefficient was at least 0.992 for the 12 miRNAs (Supplementary Figure 7). Taken together, the miRNAs could be accurately and robustly measured in blood plasma by qRT-PCR.
Abnormal Expressions of Five miRNAs in Plasma were Consistent with those in the Paired Tumors of Stage I NSCLC Patients
To determine relationship of the miRNA expressions in plasma with those in primary tumor tissues, the miRNAs were tested by qRT-PCR in plasma of 28 stage I NSCLC patients and 28 age-, race-, gender-, and smoking status-matched healthy individuals. A miRNA gene is considered to have positive change when ratio of its expression level in plasma from cancer patients versus that in pooled plasma of the matched normal subjects was either ≥2.0 or ≤0.5 (absolute fold change ≥2.0). Using the cut-off level, miR-21, 210, and 182 exhibited increased plasma expressions in 50.00–63.64% cancer patients whose tumors had elevated expressions of the same three genes (Table 4). On the contrary, miR-126 and 486-5p displayed decreased plasma expression levels in 59.09–68.18% cancer patients whose tumors had reduced expressions of the two miRNAs. Furthermore, Pearson's correlation analysis showed that there were significant correlations between the expression levels of the five miRNAs in plasma and those in the paired tumors (all r≥0.85, all P≤0.01). However, another seven genes, including miR-139, 145, 205, 200b, 375, 429, and 708, did not display considerable changes in plasma of the NSCLC patients whose tumors showed either higher or lower expressions of the genes, as compared with those in the plasma samples of the healthy controls. These findings implied that plasma samples and the corresponding primary cancer tissues showed similar tendencies concerning the expressions of miR-21, 126, 182, 210, and 486-5p. The level of the genes in plasma might reflect those in the tumors of the same cancer patients. Therefore, the five miRNAs could be candidate plasma biomarkers for NSCLC and lead to the next step of the study.
Table 4. Prevalence of altered miRNA expressions in the paired tumor and plasma samples from 28 stage I NSCLC patientsa.
| miRNAs | No. of subjects with altered expressions of miRNA (%) |
|
|---|---|---|
| Tumor, no. positive (%) | Plasma, no. positive (%)b | |
| miR-21 | 28/28, (100) | 19/28, (67.86) |
| miR-126 | 28/28, (100) | 18/28, (64.29) |
| miR-182 | 28/28, (100) | 17/28, (60.71) |
| miR-210 | 28/28, (100) | 16/28, (57.14) |
| miR-486-5p | 28/28, (100) | 20/28, (71.43) |
Abbreviation: NSCLC, non-small-cell lung cancer.
Expression of the miRNAs in each sample was calculated by using comparative cycle threshold (Ct) method with the equation 2−ΔΔCt. Ct values of the target miRNAs were normalized in relation to that of miR-16.
A miRNA gene is considered to have positive change when ratio of its expression level in plasma from cancer patients versus that in pooled plasma samples of the matched normal controls was either ≥2.0 or ≤0.5 (absolute fold change ≥2.0).
A Panel of miRNAs was Optimized as Plasma-based Biomarkers for NSCLC
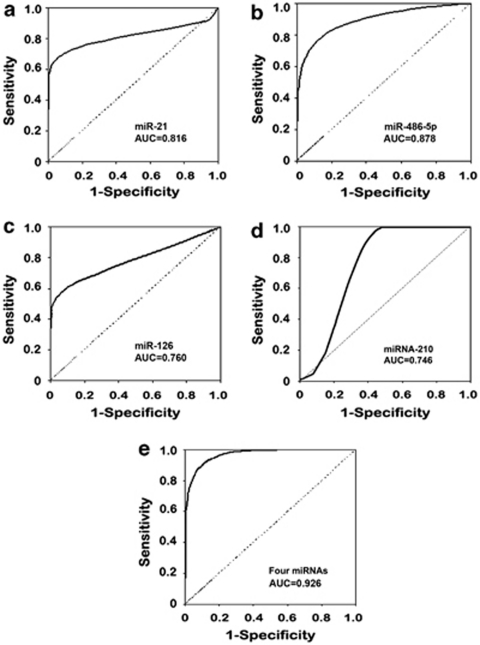
To evaluate diagnostic value of the five miRNAs in plasma for NSCLC, plasma expressions of the genes were measured by qRT-PCR in a cohort of 58 NSCLC patients and 29 healthy individuals. Overall, expression levels of miR-21, 210, and 182 in cancer patients were statistically higher than those in healthy subjects (all P<0.001) (Supplementary Table 1). In contrast, miR-126 and miR-486-5p were present at lower plasma levels in NSCLC patients than in the healthy subjects (all P<0.001). The individual miRNAs exhibited AUC values of 0.662–0.878 in distinguishing NSCLC patients from the healthy individuals, revealing 51.72–84.48% sensitivity and 65.52–82.76% specificity (Supplementary Table 2). On the basis of the AUC values, a logistic regression model with the best prediction was defined that consisted of miR-21, 126, 210, and 486-5p. As shown in Figure 1, the composite panel of the four miRNAs produced 0.926 AUC that was higher than those of individual genes used alone (all P<0.05). Given a specificity of 96.55%, the four miRNAs used in conjunction revealed a sensitivity of 86.22% in differentiating NSCLC patients from the healthy subjects. Furthermore, Pearson's correlation analysis indicated that the estimated correlations among expression levels of the four miRNAs were low (all r<0.50, all P>0.05), implying that plasma expressions of the four miRNAs were complementary to each other. The data further supported that the combined analyses of the genes outperformed a single one used alone for diagnosis of NSCLC.
Figure 1.
Receiver–operator characteristic (ROC) curve analysis of expression levels of the four miRNAs in plasma of 58 patients diagnosed with NSCLC and 29 healthy individuals. The area under the ROC curve (AUC) for each miRNA conveys its accuracy in differentiating NSCLC patients from the healthy subjects in terms of sensitivity and specificity. The individual genes produce 0.662–0.878 AUC values (a–d), being significantly lower than 0.926 AUC by the four genes genes used in combination as a marker panel (e) (all P<0.05).
The four miRNAs used in combination had higher diagnostic sensitivity in more advanced stages (II–IV) as compared with stage I NSCLC (P<0.05) (Table 5). However, combining the four genes still produced 73.33% sensitivity and 96.55% specificity in distinguishing stage I NSLCLC patients from the healthy subjects. In addition, the diagnostic sensitivity of the genes used together was significantly higher for ACs (91.67%) than SCCs of the lungs (82.35%) (P<0.05). Finally, there was no association between the changes of the genes with age, gender, ethnic group, and smoking status of the participants (all P>0.05).
Table 5. Diagnostic performance of the composite panel of the four miRNAs in a cohort of 58 NSCLC patients and 29 healthy subjects.
| Sensitivity, % | Specificity, % | |
|---|---|---|
| All cases | 86.22 (50/58) | 96.55 (28/29) |
| Different histological types | ||
| SCC | 82.35 (28/34) | 96.55 (28/29) |
| AC | 91.67 (22/24)a | 96.55 (28/29) |
| Cases with different stages | ||
| I | 73.33 (11/15) | 96.55 (28/29) |
| II | 86.67 (13/15)b | 96.55 (28/29) |
| III | 91.67 (11/12)b | 96.55 (28/29) |
| IV | 93.75 (15/16)b | 96.55 (28/29) |
Abbreviations: AC, adenocarcinoma; NSCLC, non-small-cell lung cancer; SCC, squamous cell carcinoma.
Difference of sensitivity of the four miRNAs used together in diagnosis of SCC and AC, P<0.05.
Comparison of stage I with II, III, and IV, respectively, regarding diagnostic sensitivity of the four miRNAs. All P<0.05.
DISCUSSION
In the present study, we reproducibly validated our previously identified NSCLC-associated miRNAs in independent tumor tissues by using qRT-PCR assay. We further showed that the miRNAs were stably present and readily measurable in plasma. We finally demonstrated that a panel of plasma miRNAs would provide potential biomarkers that could differentiate NSCLC patients from healthy individuals.
Upregulation of miR-21 has been found in numerous human cancers.27, 28, 29, 30 miR-21 was recently classed as an oncomir the overexpression of which could lead to tumor development and progression.29 Furthermore, the measurement of miR-21 expression in plasma might be useful in diagnosis of pancreatic cancers.31 In addition, elevated serum miRNA-21 levels in patients with metastatic hormone-refractory prostate cancer could be a predictive factor for the efficacy of docetaxel-based chemotherapy.32 Moreover, a high correlation of miRNA expression level of a panel of six miRNAs, including miR-21, was found between breast tumors and paired sera.33 Additionally, miR-21 was significantly over-expressed in specimens of cancer patients compared with those in normal controls. miR-21 could be one of blood-based biomarkers for breast cancer diagnosis, grading and prognosis.33 The data produced from our present study imply that plasma miR-21 could serve as a biomarker for diagnosis of lung cancer. miR-486-5p was found to be underexpressed in eight types of human tumors, including lung cancer.34 The observation from the current study using plasma is consistent with the previous finding, and further suggests that miR-486-5p might be a potential plasma-based biomarker for lung cancer. miR-126 has been considered as a tumor suppressor in human carcinogenesis, and its decreased expression is associated with metastasis of breast cancers.35 Furthermore, the reduced expression of miR-126 was recently found to be closely associated with clinicopathological features of breast cancer.33 Moreover, by analyzing a miRNA profile of SCC samples of Chinese lung cancer patients, Yang et al identified that miR-126 was one of the miRNAs that were downregulated in the tumors compared with noncancerous lung tissues.36 Furthermore, enforced expression of miR-126 in gastric cancer cells inhibited cell growth by inducing cell-cycle arrest in G0/G1 phase, migration and invasion in vitro, as well as tumorigenicity and metastasis in vivo. In addition, miR-126 might have a tumor-suppressor role in tumorigenesis by targeting an adapter protein, Crk.37 Our current study shows that measuring reduced plasma expression of miR-126 that could distinguish NSCLC patients from control subjects. miR-210 can regulate the hypoxic response of tumor cells and tumor growth.38 Elevated expression of miR-210 was significantly associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer.10 Furthermore, increased miR-210 expressions in serum could be useful as one of markers for patients with diffuse large B-cell lymphoma and pancreatic ductal AC.31, 39 Our present observation further suggests that miR-210 might be a potential plasma marker for NSCLC.
Because tobacco use is the major cause of NSCLC, most of the previously identified lung cancer-associated molecular genetic changes are related to the smoking status.1, 5 The use of such alterations as biomarkers might produce over-diagnosis. Interestingly, plasma expressions of the four miRNA genes are independent of smoking pack-years of the participants. Although our result appears to be promising, there are several limitations in this study. First, because the sample size of the two cohorts was too small and resulting sensitivity (73.33%) was too low for diagnosis of stage I NSCLC. Further launching the miRNAs in large cohorts and independent studies is clearly required. Furthermore, the gene expressions had more close associations with AC compared with SCC of the lungs. However, due to the small size of the samples, whether the miRNAs could be used as specific markers for AC remains to be investigated. Second, the 12 candidate miRNAs chosen in the present study were based on our previous microarray analysis data on primary lung cancer.17 We will evaluate lung tumor-associated miRNAs defined from other groups to identify additional plasma-based miRNAs that can be added to the current ones so that the diagnostic efficacy of the approach could be improved. Third, the plasma miRNA expressions were more related to advanced stages of NSCLC compared with stage I NSCLC, suggesting the potential as prognostic markers for the disease. However, we were not able to assess plasma expression changes of the miRNAs before and after surgery, and their prognostic value in predicting outcome of NSCLC in the present study, because blood samples were only obtained from the lung cancer cases before they received surgery, and follow-up information of the patients was not yet available.
Although showing altered changes in tumor tissues, miR-139, 145, 205, 200b, 375, 429, and 708 did not display abnormal plasma expressions in stage I lung cancer patients. The explanations for the discrepancy remain unclear. One possible reason might be that NSCLC develops from complex and multistep processes. Although showing dysregulation within lung tumors, the changes in plasma of the cancer-related miRNAs might occur at comparable late stage of the disease. Therefore, the genes could not show considerable expression differences in plasma between patients with early-stage NSCLC and healthy controls. However, future investigation of possible mechanism underlies the discrepancy is needed.
In conclusion, we demonstrated that the expressions of the miRNAs identified from surgical tumor tissues could readily and specifically be measured in plasma. More importantly, detection of a panel of the miRNAs in plasma would potentially be used as a minimally invasive diagnostic tool for NSCLC, including ACs of the lungs. Nonetheless, further validating the utility of the potential biomarker in an independent cohort is required.
Acknowledgments
This work was supported in part by American Cancer Society Research Scholar Grant, National Cancer Institute (NCI) Grants CA-135382, Clinical Innovator Award from Flight Attendant Medical Research Institute and an exploratory research grant from Maryland Stem Cell Research Fund (FJ).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
Supplementary Material
References
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Flehinger BJ, Melamed MR, Zaman MB, et al. Early lung cancer detection: results of the initial (prevalence) radiologic and cytologic screening in the Johns Hopkins Study. Am Rev Respir Dis. 1984;130:549–554. doi: 10.1164/arrd.1984.130.4.549. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Franklin WA, Gazdar AF, et al. Early detection of lung cancer: clinical perspectives of recent advances in biology and radiology. Clin Cancer Res. 2001;7:5–22. [PubMed] [Google Scholar]
- Toloza EM, Harpole L, McCrory DC. Noninvasive staging of non-small cell lung cancer: a review of the current evidence. Chest. 2003;123:137S–146S. doi: 10.1378/chest.123.1_suppl.137s. [DOI] [PubMed] [Google Scholar]
- Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- Galasso M, Elena Sana M, Volinia S. Non-coding RNAs: a key to future personalized molecular therapy. Genome Med. 2010;2:12. doi: 10.1186/gm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;6:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Foekens JA, Sieuwerts AM, Smid M, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Patnaik SK, Kannisto E, Knudsen S, et al. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- Lebanony D, Benjamin H, Gilad S, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- Yu L, Todd NW, Xing L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Todd NW, Yu L, et al. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;8:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar SP, Brambilla C, Pugatch B.Squamous cell carcinomaIn: Travis WD, Brambilla E, Muller-Hermelink HK (eds).Pathology and Genetics, Tumours of the Lung, Pleura, Thymus and Heart IARC Press: Lyon; 200526–30. [Google Scholar]
- Colby TV, Noguchi M, Henschke C.AdenocarcinomaIn: WD Travis, E Brambilla, HK Muller-Hermelink (eds).Pathology and Genetics, Tumours of the Lung, Pleura, Thymus and Heart IARC Press: Lyon; 200535–44. [Google Scholar]
- Sahai H, Kurshid A. Formulae and tables for the determination of sample sizes and power in clinical trials for testing differences in proportions for the two-sample design: A review. Statistics in Medicine. 1996;15:1–21. doi: 10.1002/(SICI)1097-0258(19960115)15:1<1::AID-SIM134>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Dodd LE, Pepe MS. Partial AUC estimation and regression. Biometrics. 2003;59:614–623. doi: 10.1111/1541-0420.00071. [DOI] [PubMed] [Google Scholar]
- Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford: New York; 2003. [Google Scholar]
- Tian GL, Fang HB, Liu Z, et al. Regularized (Bridge) Logistic Regression for Variable Selection Based on ROC Criterion. Statistics and Its Interface. 2009;2:493–502. [Google Scholar]
- Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Pezzolesi MG, Platzer P, Waite KA, et al. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. Am J Hum Genet. 2008;82:1141–1149. doi: 10.1016/j.ajhg.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Si ML, Wu H, et al. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- du Rieu MC, Torrisani J, Selves J, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56:603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila Pa) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Yang LF, Zhu Y, et al. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2010;14:1768–1773. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- Wang F, Zheng Z, Guo J, et al. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586–593. doi: 10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- Navon R, Wang H, Steinfeld I, et al. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;25:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcón C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li X, Yang Q, et al. The role of microRNA in human lung squamous cell carcinoma. Cancer Genet Cytogenet. 2010;2:127–133. doi: 10.1016/j.cancergencyto.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Feng R, Chen X, Yu Y, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Huang X, Ding L, Bennewith KL, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;6:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;5:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.