Abstract
Lentiviral vectors are effective tools for gene transfer and integrate variable numbers of proviral DNA copies in variable proportions of cells. The levels of transduction of a cellular population may therefore depend upon experimental parameters affecting the frequency and/or the distribution of vector integration events in this population. Such analysis would require measuring vector copy numbers (VCN) in individual cells. To evaluate the transduction of hematopoietic progenitor cells at the single-cell level, we measured VCN in individual colony-forming cell (CFC) units, using an adapted quantitative PCR (Q-PCR) method. The feasibility, reproducibility and sensitivity of this approach were tested with characterized cell lines carrying known numbers of vector integration. The method was validated by correlating data in CFC with gene expression or with calculated values, and was found to slightly underestimate VCN. In spite of this, such Q-PCR on CFC was useful to compare transduction levels with different infection protocols and different vectors. Increasing the vector concentration and re-iterating the infection were two different strategies that improved transduction by increasing the frequency of transduced progenitor cells. Repeated infection also augmented the number of integrated copies and the magnitude of this effect seemed to depend on the vector preparation. Thus, the distribution of VCN in hematopoietic colonies may depend upon experimental conditions including features of vectors. This should be carefully evaluated in the context of ex vivo hematopoietic gene therapy studies.
Keywords: Lentivirus, CD34+ cell, Q-PCR, method, hematopoietic cells, CFC
Introduction
Genetically corrected hematopoietic stem cells (HSC) can be used as an alternative to allogenic HSC transplantation for the correction of several types of inherited diseases.1 Different vectors have been used for the gene modification of HSC, but recombinant HIV-1-derived (rHIV) lentiviral vectors (LV) appear to be promising for gene-modification of the long-term repopulating HSC population, as shown in several preclinical models2, 3, 4 and in clinical trials in man.5 Thus, novel clinical applications of rHIV vectors are actively developed, including for instance a treatment of Wiskott–Aldrich syndrome (WAS). A LV encoding the WAS protein (WASP) is being developed to treat this life-threatening X-linked primary deficiency.6
Depending on the experimental conditions, rHIV vectors can transduce variable percentages of cells and integrate variable numbers of copies of proviral DNA into the genome of target cells. Indeed, it has been shown that human HSC are permissive to the integration of multiple copies of rHIV. As high as six integrations per cell have been detected on average in populations of human hematopoietic cells engrafted in the bone marrow of immuno-incompetent mice.7 The stable insertion of genes in hematopoietic progenitor cells has a significant impact as these cells will transmit their genomic heritage to a considerable number of cells given their proliferation and differentiation potential. The biological potency of the vector is expected to correlate positively with the frequency of transduced cells and also with the number of integration per cell unless transgene silencing is observed, as suggested in some studies.8 At the same time, genotoxicity related to the number of vector insertions per cell can result from inappropriate transgene expression in cells9 or from effects of elements contained in the integrated cassette,10 thus requiring a strict control of the number of gene insertion per cell. It is therefore important to determine the distribution of vector copies in the infected cell population at the single cell level to assess the efficacy and safety, that is, the therapeutic window of integrative vectors.
Several methods have been used to measure the transduction of human hematopoietic cells with LV but there are few reports of validated techniques to determine vector copy distribution in a population from single-cell measures. In earlier studies, average number of vector copies have been measured in cell populations using Southern blot detection of vector sequences, calibrated on dilutions of genomic DNA and a housekeeping gene .11 More recently, quantitative PCR (Q-PCR) has been used to provide more precise measures and the technique can be calibrated on dilutions of a plasmid bearing both vector and genomic sequences to determine average vector copy number (VCN) in a human cell population transduced with a rHIV LV.12 Such average values does not provide any indication on the initial frequency of transduced cells in the target population nor does it estimate the numbers of integrations in individual transduced cells. Hematopoietic progenitor cells have the ability to form colonies arising from a single cell when cultured at low cell density in semi-solid medium. After a culture period of about 2 weeks in methylcellulose, individual colonies can be picked and the genomic DNA of the cells can be extracted from these colony-forming cells (CFC) with proteinase K and phenol/chloroform to determine the presence or absence of vector by PCR and agarose gels, thus, determining the frequency of transduced hematopoietic progenitor cells in the initially-infected population of cells.13 Protocols for the quantification of VCN in CFC by Q-PCR have been reported recently but not validated with experimental data.14 Here, we have developed a simplified method relying on a single-step genomic DNA extraction and a duplex Q-PCR method to quantify VCN in individual CFC. We have validated this approach experimentally. Transduced and cloned human cell lines were generated as controls and used to demonstrate the feasibility, sensitivity and reproducibility of this protocol. Measures of VCN in CFC have been used to evaluate various conditions for the transduction of hematopoietic progenitor cells with LV encoding the green fluorescence protein (GFP) reporter transgene (GFP-LV) or WASP (WASP-LV). Results show that the frequency of transduced CFC and the distribution of VCN in these cells could be augmented by repeated infection. This approach should be useful to optimize rHIV transduction protocols and to verify vector safety.
Results
Generation of controlled human cell lines to measure rHIV vector integration
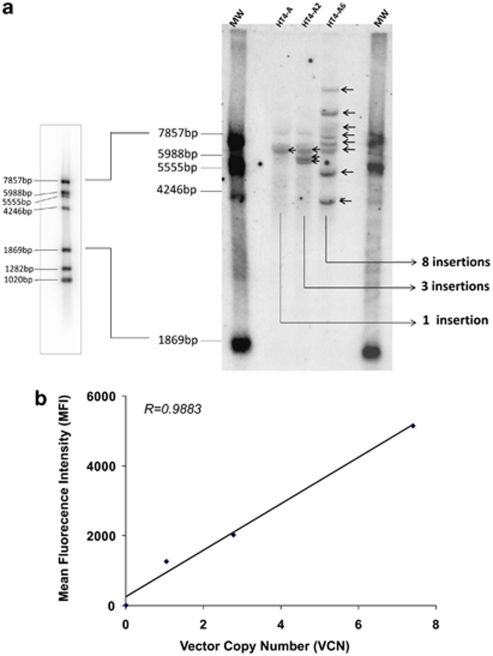
Transduced human cells containing a known amount of rHIV integration can serve as control materials to analyze VCN in cells. A panel of such control cells was generated by infecting the human fibrosarcoma HT1080 cell line with a GFP-LV, then selecting and characterizing stably-transduced single-cell clones with variable numbers of VCN. HT1080 cells were used because of their rapid growth and few chromosomal changes from the diploid karyotype (http://www.lgcstandards-atcc.org/ and Ref. 15). Following transduction with the vector and two rounds of single-cell cloning, a single HT1080 cell clone HT4-A was first selected and analyzed. Vector insertion sites were determined with the vector integration tag analysis (VITA) technique described elsewhere16 showing a single vector insertion in chromosome 19 (Table 1). Subsequently, this clone was re-infected with the GFP-LV and two daughter clones displaying different levels of expression of GFP were selected by single-cell cloning. These HT4-A2 and HT4-A6 cells showed, respectively, three and eight vector insertions in various genomic locations, including the initial insertion in chromosome 19. The number of vector insertion sites in the HT4-A, HT4A-2 and HT4-A6 was confirmed by Southern-blot analysis using a single-restriction enzyme (XbaI) to reveal the LV integration banding pattern. As shown in Figure 1a, we observed one band in clone HT4-A, three bands in HT4-A2 and eight bands in clone HT4-A6. With the slight over-exposition of the blot, a very weak band appears at about 8 kb in all HT4 clones and corresponds to a nonspecific hybridation in the stringency conditions used in the experiment.
Table 1. Characterization of vector copies and vector insertion sites in HT1080 clones.
|
Clones |
VCN by Q-PCR (Taqman) |
Insertion site analysis by VITA (n=2) |
||||
|---|---|---|---|---|---|---|
| n | VCN Ave±s.d. (extremes) | Nb IS | Size(bp)_Chrom(no.)_Position | Gene | Description function | |
| HT4-A | 37 | 1.1±0.2 (0.74–1.5) | 1 | 107_19_13093976 | BTBD14B | Transcriptional regulator |
| 285_8_81041615 | MRPS28 | Mitochondrial ribosomal protein | ||||
| HT4-A2 | 16 | 2.79±0.65 (1.9–3.6) | 3 | 183_14_71097682 | SIPA1L1 | Signal-induced proliferation-associated protein |
| 107_19_13093976 | BTBD14B | Transcriptional regulator | ||||
| 164_3_178970571 | No gene | |||||
| 136_17_10245972 | MYH8 | Myosin heavy chain 8, skeletal muscle | ||||
| 114_17_44346886 | UBE2Z | Ubiquitin-conjugating enzyme E2Z | ||||
| HT4-A6 | 17 | 7.13±1.32 (5.3–8.7) | 8 | 330_17_77173163 | NPLOC 4 | Nuclear protein localisation 4 |
| 63_12_61459846 | PPM 1H | Protein phosphatase 1H | ||||
| 68_7_19590033 | No gene | |||||
| 62_7_101494262 | CUX-1 | Cut-like homeobox 1a | ||||
| 107_19_13093976 | BTBD14B | Transcriptional regulator | ||||
Abbreviations: Q-PCR, quantitative-PCR; s.d., standard deviation; VCN, vector copy numbers; n, number of Q-PCR experiments.
The bold data represent the identical insertion that is expected to be found in the three different cell lines.
Figure 1.
Characterization of the HT4-A, HT4-A2 and HT4-A6 clones. (a) Southern Blot on XbaI-digested genomic DNA from the HT1080 clones. The probes bands are the XbaI fragments released from an internal region to the vector and the flanking genomic DNA. (b) Correlation between VCN obtained by Q-PCR and MFI obtained by flow cytometry in the three clones.
In previous studies, we have used a duplex Q-PCR for comparative amplification of vector-specific sequences (human immunodeficiency virus or woodchuck hepatitis post-transcriptional regulatory element) versus a human cellular gene (the human albumin gene (ALB) for which two copies are present per cell) to determine the number of rHIV vector insertions in human cells.12 The reaction is calibrated by a standard curve made by serial dilutions of a plasmid carrying a single copy of vector bearing WPRE and of the ALB sequence (see supplementary Table S1). Analysis of the genomic DNA from HT4-A, HT4-A2 and HT4-A6 cell clones by such Q-PCR protocol determined 1.1±0.2, 2.8±0.7 and 7.1±1.3 VCN in repeated experiments (Table 1). These results were consistent with the determination of vector insertions by VITA and by Southern blot in each of the clones. The numbers of vector copy also correlated strongly with the expression of the integrated GFP transgene in the different cell clones as determined by the mean fluorescence intensity of the cells in flow cytometry (Figure 1b).
Such concurring results from VITA, Southern blot, Q-PCR and flow cytometry characterize these three cell lines, which can be used for analytical purposes as a panel of control cells carrying a known range of rHIV vector integrations.
Validation of the Q-PCR method to determine VCN in hematopoietic progenitor cells
Hematopoietic progenitor cells are endowed with sufficient proliferation potential so that single progenitor cells can form visible colonies when cultured in semi-solid medium in the presence of cytokines. The transduction of hematopoietic progenitor cells can therefore be assessed at the clonal level by measuring VCN in a single unit of CFC in which all cells originate from a single progenitor cell. This prompted us to assess if the duplex Q-PCR technique that we employed previously could be used to quantify VCN in CFC. One technical challenge is that CFC contain small numbers of cells (between 1000–10 000 cells) requiring specific DNA extraction method, therefore requiring an evaluation of the sensitivity, feasibility and robustness of the approach.
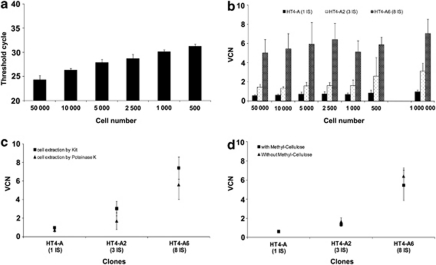
First, we determined if the sensitivity of our Q-PCR protocol would be adequate in this desired cell number range. Serial dilutions of the standard curve plasmid showed the expected amplification of HIV versus ALB or WPRE versus ALB sequences with as little as 102 copies of plasmid corresponding to 8.26 × 10−7 ng of DNA per reaction (see supplementary Table S1). The amplification of HIV or WPRE sequences was considered to be equivalent. Considering the size of the plasmid in relation to the size of the human genome and the amount of DNA per cell, this level would be compatible with the amplification of the integrated proviral vector DNA in approximately 100 cells. The sensitivity of the Q-PCR is therefore in principle adequate for the analysis of CFC.
Second, we determined whether we could amplify small amounts of cellular genomic DNA material under conditions compatible with the study of CFC. Extraction of genomic DNA from CFC was performed by a single-step proteinase K lysis, which is reportedly successful with small numbers of cells.17 The effects of cell number and of the extraction conditions were tested. Genomic DNA was obtained from decreasing numbers of HT4-A, HT4-A2 and HT4-A6 cells after extraction with proteinase K in the presence or not of methylcellulose. As shown in Figure 2a, the cycle threshold (CT) for the ALB sequence increased correspondingly to the reduction in cell sample size. The lowest amount of genomic DNA that could provide an interpretable signal with a CT of 31.2±0.5 (n=94) corresponded to 100 cells, thus confirming the range of predicted sensitivity of the Q-PCR. The determination of VCN from variable amounts of genomic DNA obtained from each of the clones showed similar values over the range of cell-equivalent tested (Figure 2b). However, lower VCN values than expected were obtained from the Q-PCR amplification of small amounts of genomic DNA extracted with proteinase K from HT4-A, HT4-A2 and HT4-A6 cells (Figure 2b). Under these conditions, the copy number was inferior by about 30–40% from the expected value. With the six cell dilutions shown in Figure 2b, clone HT4-A gave an average of 0.7±0.1 VCN (range 0.6–0.8), which is 30% less than expected value of 1; HT4-A2 gave an average of 1.7±0.4 VCN (range 1.3–2.5), which is 40% less than expected value of 3; and HT4-A6 gave an average of 5.7±0.5 VCN (range 5–6.4), which is 30% less than expected value of 8. To understand the origin of this suboptimal accuracy, we investigated several parameters. Copy numbers measured from genomic DNA obtained from large numbers of cells with a commercial DNA extraction kit were close to the expected values (VCN 0.97, 3.12 and 7.1 for each of the three clones) (Figure 2b controls on the right side of graph), demonstrating the accuracy of the Q-PCR amplification step in itself when it is performed on optimal material. The quality of the genomic DNA was therefore examined. The proteinase K extraction method was found to give lower VCN values than a commercial DNA extraction kit even with large numbers of cells, although this lower trend was not found to be statistically significant (Figure 2c). The presence or absence of methylcellulose with the cells did not significantly impact the values of VCN in the clones (Figure 2d). Thus, the genomic DNA extraction step in itself appears to be a critical parameter for the accuracy of VCN determination in CFC by Q-PCR. The advantage of the rapid single-step proteinase K procedure must therefore be mitigated by an underestimation of VCN values by 30–40%. However, with this limitation defined, the approach remains useful and provides dose-dependent results in order to evaluate the distribution of VCN at the clonal level.
Figure 2.
Determination of VCN by Q-PCR. (a) The sensitivity of the Q-PCR was evaluated by measuring CT values (average of duplicate measures) for the amplification of the albumin gene sequences in decreasing numbers of control cells. Q-PCR was carried out from 1/6 of the total genomic DNA extracted from 500–50 000 cells. (b) VCN in the HT4-A, HT4-A2 and HT4-A6 clones were measured after proteinase K lysis extractions of separate preparations of cells ranging from 500–50 000 cells per condition. The Q-PCR was performed on 1/6 of the extracted genomic DNA as in Figure 2a. The control indicated on the right side of the graph corresponds to Q-PCR on genomic DNA obtained from 1 × 106 cells extracted with a commercial Promega kit. (c) Comparison of VCN values obtained on the three clones using genomic DNA extracted either by proteinase K lysis from a number of cells inferior to 5 × 104 or using a commercial kit and 1 × 106 cells per extraction. (d) Comparison of VCN values obtained by Q-PCR from the three clones using genomic DNA extracted by proteinase K lysis in the presence (5 μl per condition) or absence of methylcellulose.
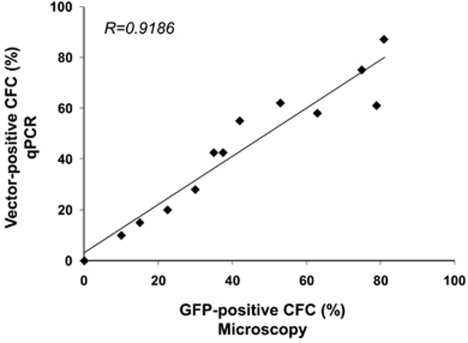
Third, we used this Q-PCR technique to evaluate the transduction of cord blood CD34+ hematopoietic progenitor cells with various concentrations of the GFP-LV by comparing the number of vector copies in relation to transgene expression in individual CFC. Infected CD34+ cells were seeded in methylcellulose and after 2 weeks, colonies were scored under epifluorescence microscopy to determine GFP expression. Genomic DNA was extracted from each colony with proteinase K to measure vector copies by duplex Q-PCR. The colonies were scored positive for vector when they displayed a specific signal even though VCN values could be lower than one copy per cell (cut-off value 0.1). Under these conditions, a positive correlation (r2=0.92; n=13) was found between the frequency of CFC expressing the GFP protein and scoring positive for vector by Q-PCR (Figure 3). There were only 4±7% of CFC scored positive by microscopy that had no detectable vector integration by qPCR and inversely, only 6±5% of CFC scored negative by microscopy that gave a positive signal by qPCR.
Figure 3.
Correlation between GFP-positive CFC observed by microscopy and vector-positive CFC evaluated by Q-PCR (n=13 experiments).
It has been proposed that the probability of transduction of single hematopoietic cells in a preparation can be estimated from the average mean transduction rate using the Poisson distribution analysis.18 Assuming that each cell has equivalent probability of being transduced and that the distribution of events is not modified by cell culture, then the transduction rate can predict the percentage of cells receiving at least one copy of vector. The transduction rate can be estimated from the mean average copy number per cell in a whole population of cells. In a series of experiments, CD34+ cells were infected with the GFP-LV at 2 × 108 IG per ml giving mean average VCN of 1.4±0.4 in the whole population (n=3 experiments) and in this set of experiments, the presence of vector was measured by Q-PCR on CFC (Table 2 and Figure 5a). This showed 70% vector-positive individual CFC (Table 2 and Figure 5a), which is close to the expected transduction efficiency of 65% calculated from the Poisson distribution as per Ref. 18. Similar findings were made with another vector. In six independent transductions of CD34+ cells with a WASP-LV vector, we measured a mean average value of 0.4±0.1 vector copies per cell in the cultured whole cell population (data not shown). In one of these six experiments, the transduction frequency of CFC was found to be 40% by Q-PCR (Table 2), which is also very close to an expected 35% transduction frequency on the basis of Poisson distribution.
Table 2. Effects of vector concentration on the transduction of CFC.
| LV vectors | LV concentration (IG ml−1) | Number of CFC tested | Vector-positive CFC (%) | Median VCN within transduced CFC (range) |
|---|---|---|---|---|
| GFP-LV (ultracentrifuged) n=3 experiments | 2 × 107 | 120 | 48 | 0.9 (0.1–4.9) |
| 20 × 107 | 120 | 70 | 1.2 (0.1–12.7) | |
| WASP-LV (ultracentrifuged) n=2 experiments | 5 × 107 | 78 | 27 | 1.1 (0.2–4.4) |
| 10 × 107 | 73 | 32 | 0.9 (0.1–7.1) | |
| WASP-LV (chromatography-purified) n=2 experiments | 5 × 107 | 143 | 40 | 0.5 (0.1–11.9) |
| 10 × 107 | 40 | 50 | 0.7 (0.2–4.5) |
Abbreviations: CFC, colony-forming cell; GFP-LV, green fluorescence protein-lentiviral vector; VCN, vector copy numbers.
These experiments are also related in Figure 5 (see Figure legend).
Altogether, these results experimentally validate the feasibility of using Q-PCR to quantify the frequency of transduction in single CFC.
Application of the Q-PCR on CFC to evaluate the effects of vector concentration and number of hits on the transduction of hematopoietic progenitor cells
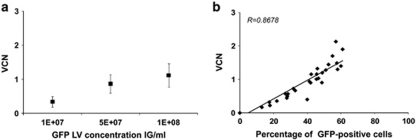
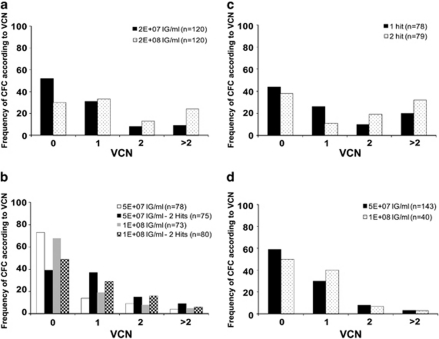
Various parameters, such as the concentration of vector and the number of hits of vector, can be adapted to optimize the infection of hematopoietic progenitor cells with LV. Increasing concentrations of a GFP-LV augmented the percentage of GFP-expressing cells in the bulk population of cultured CD34+ cells as expected19 and augmented the mean average VCN in the population (Figure 4a), thus, resulting in a good correlation between transgene expression and cell transduction (Figure 4b). At the individual progenitor cell level, increasing the concentration of LV increased the frequency of transduced CFC (Table2 and Figure 5a). In addition, this also modified the distribution of VCN in the progenitor cell population as shown by increased median values or range of VCN when using higher vector concentrations (Table 2). The frequency of CFC containing greater than two vector copies per cell was also augmented as represented by categories of frequency according to VCN in Figure 5a. Yet, in the conditions tested, the majority of transduced CFC only integrated one copy of vector per cell.
Figure 4.
(a) Transduction of CD34+ cells with increasing concentrations of a GFP-LV tested from 0.1 to 10 × 107 IG ml−1 in eight independent experiments. Results show the mean average VCN determined on the cells expanded in liquid culture in the presence of cytokines for 2 weeks after genomic DNA kit extraction (mean±s.d.). (b) Correlation between average VCN and the frequency of GFP expression measured by FACS in 40 experiments in which various GFP-LV vector concentrations were tested.
Figure 5.
Effects of experimental conditions on the frequency of transduction and on the distribution of vector copies in the population. CFC with VCN values comprised between 0 and 0.1 were categorized as 0; those comprised between 0.1 and 1.1 were categorized as 1; those comprised between 1.2 and 2.1 were categorized as 2 and those with VCN superior to 2.1 were categorized as >2. Bars represent average percentage of CFC in each category over the total number of CFC analyzed. The number of CFC analyzed is indicated between brackets for each graph. (a) Transduction with an ultracentrifuged GFP-LV using various concentrations of vector. Results represent data pooled from three separate transduction experiments. (b) Transduction with an ultracentrifuged WASP-LV using several concentrations of vectors and either one or two consecutive infections (hits). Results represent data pooled from two separate transduction experiments. (c) Transduction with the GFP-LV at concentration of 2 × 108 IG ml−1 given once or twice. Results represent data pooled from two separate transduction experiments. (d) Transduction with two batches of chromatography purified WASP-LV using two concentrations. Data from one experiment.
The effects of multiple rounds of infection on CFC transduction were analyzed. CD34+ cells were infected once for 6 h or twice in a consecutive manner, by adding the same concentration of vector for a second time. A second hit of infection with the GFP-LV had little impact on the frequency of vector-positive CFC as seen in Table 3, but instead, it shifted the median of VCN values from 1.4 to 2.2. This effect is also illustrated in Figure 5c where higher percentages of CFC are found in categories of cells with VCN equal or superior to two. The range of VCN obtained after infection with the GPF-LV is very broad, even with one hit. In contrast, performing a second hit with the WASP-LV, increased the frequency of transduced CFC but did not modify the median or range of VCN values (Table 3). However, it increased the percentage of CFC with two copies of vector (Figure 5b) without significant impact on higher categories. Thus, these results show that different types of LV have the potential to behave differently according to the experimental conditions. With the WAS-LV tested here, the transduction efficiency could be more effectively augmented by two consecutive hits of vector rather than doubling the vector concentration.
Table 3. Effects of repeated infection on the transduction of CFC.
| LV vectors | Number of hits | Number of CFC tested | Vector-positive CFC (%) | Median VCN within transduced CFC (range) |
|---|---|---|---|---|
| GFP-LV | 1 | 78 | 56 | 1.4 (0.1–14.5) |
| 2 | 79 | 62 | 2.2 (0.1–20.9) | |
| WASP-LV | 1 | 151 | 29 | 0.9 (0.1–7.1) |
| 2 | 155 | 53 | 1 (0.1–6.5) |
Abbreviations: CFC, colony-forming cell; GFP-LV, green fluorescence protein-lentiviral vector; VCN, vector copy numbers.
These experiments are also related in Figure 5 (see Figure legend). Vectors were tested at the concentration of 2 × 108 IG ml−1 (GFP-LV) or 0.5-1 × 108 IG ml−1 (WASP-LV) and data were pooled from two separate transductions with each vector.
Application of the Q-PCR on CFC to evaluate the activity of a purified WASP-LV
In the perspective of clinical gene therapy studies for WAS, we have developed a large-scale purification process for the WASP-LV using chromatography and membrane steps.20 Preparations of chromatography-purified WASP-LV were generated during this development phase and were previously reported.21 Two batches of chromatography-purifed WASP-LV were tested here to determine the ability to transduce individual CFCs in comparison with the same vector concentrated by ultracentrifugation. Comparable levels of transduction were found between these two types of preparations as shown in Table 2. Approximately 40–50% of the CFC were transduced with the chromatography-purified vector compared with about 30% with the ultracentrifuged vector, but the median of VCN were slightly higher in the latter (0.9–1.1) than the former (0.5–0.7). Approximately 90% of the CFC that were transduced with the chromatography-purified WASP-LV had less than two vector copies per cell as shown in Figure 5d (with both concentrations tested, 3% of CFC have >2 VCN and 7–8% have two VCN). Thus, contrary to what observed with the GFP-LV, increasing the concentration of the WASP-LV improved the transduction frequency, yet, had little effect on VCN augmentation per cell. This suggests that a plateau of transduction was probably reached with the highest value of 108 IG per ml of this preparation of LV. In a non-mutually exclusive manner this also suggests that different preparations of LV may have different infectivity properties for hematopoietic progenitor cells.
Transduction of hematopoietic cell subsets
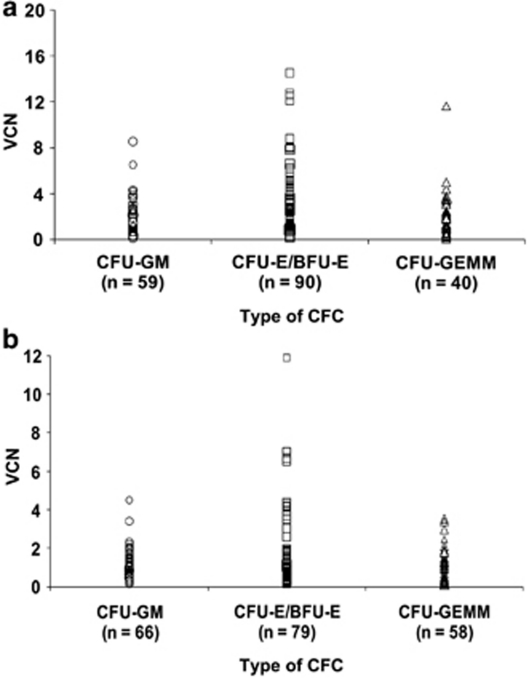
To assess the transduction of the different types of hematopoietic progenitor cells, we examined the effects of the GFP-LV and WASP-LV on the VCN values in each type of hematopoietic colony, combining all experimental conditions tested for each vector. This showed that the VCN were within a similar range in colony-forming units (CFU)-granulocyte, monocyte (GM) and CFU-granulocyte, erythrocyte, monocyte, megakaryocyte (GEMM) colonies and in majority inferior to four copies per cell as 83% of the CFC were transduced with the GFP-LV (Figure 6a) and 96% of the CFC were transduced with the WASP-LV (Figure 6b). However, it is also remarked that both GFP and WASP LV generate higher VCN values in erythroid colonies (colony-forming unit, erythroid (CFU-E)/burst-forming unit, erythoid (BFU-E)) than in myeloid and/or mixed colonies (CFU-GEMM). The difference between VCN values in erythroid colonies versus other categories alone or combined, is statistically significant (P<0.05, Student's t test).
Figure 6.
Distribution of the VCN in the different type of the CFC (CFU-GM, CFU-mix and BFU-E) from CD34+ cells transduced by (a) GFP-LV or (b) by WASP-LV.
Discussion
We herein describe and validate an analytical method to measure rHIV VCN in human CFC, providing experimental data on the transduction of hematopoietic progenitor cells, in particular with a relevant WASP vector.
The method described in this paper is simple and rapid, comprising a single-step extraction of genomic DNA followed by a duplex Q-PCR to amplify the vector and cellular sequences simultaneously. This simplicity presents an advantage over previously-published protocols that analyze the presence of gene transfer vectors in hematopoietic colonies with protocols combining cell lysis, DNA extraction with phenol chloroform or isopropanol, PCR amplification and agarose gene analysis.4, 13 More recent protocols combine these DNA extraction methods with Q-PCR analysis,14 but to our knowledge, without being validated experimentally. Thus, we herein show that a simple protocol can be used and is sufficiently sensitive to reliably determine the frequency of transduced CFC according to expected values. Following transduction of CD34+ cells with a GFP-LV, there is a good correlation between the frequency of PCR-positive CFC and the expression of the transgene. In addition, the transduction frequency of CFC is coherent with values calculated from the average copy number in the bulk population of CD34 cells using Poisson's distribution of single events.18 The precision of the method is comparable with that of Q-PCR performed in standard conditions. Indeed, comparable standard deviations are found on the three control cell lines whether using genomic DNA extraction kits and large amounts of cell material or in conditions mimicking those used for CFC (see Figures 2b and c). The method is therefore simple, sensitive and precise. However, the simplicity of the method must be mitigated by a suboptimal accuracy. Testing the three control characterized cell lines in the same conditions as CFC, we find a 30–40% underestimation of VCN values compared with the expected values. This underestimation is not caused by insufficient quantities of genomic DNA as the Q-PCR is sensitive within the range of cells analyzed. The limitation is probably caused by insufficient quality of the genomic DNA, which prevents the optimal amplification of vector-specific sequences, but the presence of methylcellulose can be excluded as a factor. In spite of this suboptimal accuracy, VCN results obtained with this simple method provide coherent results. Indeed, the VCN obtained on the three control cell lines reflect the expected range of rHIV copies inserted in these cells. The distribution of VCN in individual CFC is consistent with expected values from an idealized Poisson's distribution.18 Theoretical calculations predict that when a mean vector copy number is inferior to two in a cell population, then among transduced cells the majority of individual cells should contain one copy of vector. This distribution was obtained with the GFP-LV or with the WASP-LV and indeed we measured that the majority (about 60 %) of the CFC contained one copy. Also, with mean transduction rates inferior to one, an idealized distribution would predict that less than 10% of individual cells should contain more than two copies per cell and this is also what we measured in experiments using the chromatography-purified WASP-LV. Thus, in spite of a slight underestimation of the VCN values in CFC, the distribution of cells according to VCN categories appears to be consistent with expected data from theoretical calculations. To take into account this possible underestimation, one could apply a corrective factor on the basis of the 30–40% underestimation that was observed with the three control cell lines using this technique. Altogether, our data show that at this point, the Q-PCR method is sufficiently sensitive, precise and acceptably accurate to provide a meaningful measure of the frequency of vector-positive CFC and the distribution of vector copies at the clonal level within a population of hematopoietic progenitor cells.
Further effort outside the scope of this article are needed to improve the accuracy of this technique by optimizing the DNA extraction step or the performance of amplification of rHIV sequences in proteinase K-extracted DNA. In addition, this simple technique could be adapted to high-throughput analysis to obtain data on large numbers of clones. Although the use of CFC is a very practical manner to obtain clones of hematopoietic cells, it is strongly biased for cells of the myelo-erythroid lineage. Further development could be envisioned with the analysis of single cell sorted cultures, which could enable the evaluation of transduction at the single cell level in various hematopoietic cell lineages.
Measuring the number of vector copies in individual target cells is important to assess the therapeutic window of integrative vectors. Their biological potency but also their inherent risk of genotoxicity is related to the number of vector copies integrated per cell. Determining the VCN in hematopoietic progenitor cells in relation to gene expression in these cells or their progeny, could be used to gauge the biological activity of the integrated cassette and may reveal potential occurrence of gene silencing as suggested by some studies.8 In order to evaluate the genotoxic potential of vectors, the Q-PCR on CFC method could be used in combination with an analysis of vector insertion sites. Three characterized human cell lines, which we have been derived from HT1080 cells, could be relevant controls for both types of analytical measures. These cells contain known numbers of rHIV integration, which are in the range of events expected to occur following transduction of HSC. In addition, these cells contain known sites of rHIV integration, which could also serve as references for quality control and validation of protocols aiming to identify vector insertion sites.
Following transduction of CD34+ cells with the two different LVs tested in our study, revealed a slight bias in the distribution of VCN in the different types of progenitor cells. BFU-E seemed to integrate more copies than CFU-GM or CFU-GEMM. As erythroid cells arise from primitive cells such as CFU-GEMM, this would indicate that the experimental conditions favoured the preferential transduction of erythroid-restricted progenitor cells. This is not unexpected as the transduction medium contained stem cell factor (SCF) and thrombopoietin (TPO), which may provide a strong erythroid progenitor cell stimulus. Alternatively, we cannot exclude that the measure of VCN is not as accurate or precise in erythroid colonies as in other cells. Erythroid-restricted colonies are composed of cells that start to compact their chromatin in the course of their development. Further studies are needed to address this point more thoroughly, in particular, in the context of gene therapy studies targeting erythroid progenitor cells.
There is a strong interest in optimizing the conditions for ex vivo hematopoietic gene transfer in the perspective of experimental or clinical applications. Several previous studies have optimized parameters such as cell concentration, cytokines, medium, timing, concentration of vector, multiplicity of infection or number of transduction hits with rHIV vectors. In many cases, such studies were performed with GFP-encoding LV, relying on transgene expression levels8, 19, 22 but providing little to no information on the amount of vector integrated in target cells or the frequency of the targeted population. In this paper, we show that the Q-PCR analysis of CFC is applicable to evaluate the infectivity of CD34+ cells with LV in a transgene-independent manner. Increasing the concentration of infectious vector or repeating the infection were two different but effective ways to augment the level of transduction as measured by the percentage of vector-positive cells and by the distribution of VCN within transduced cells. In some experiments, repeating the infection seemed more effective than increasing the vector concentration to augment the percentage of vector-positive colonies as seen in Figure 5b. This indicates that clonogenic cells could become more permissive to rHIV transduction during the ex vivo culture. Further optimization could be undertaken to increase this effect and improve the levels of transduction. On the other hand, these results reveal that two consecutive hits can augment the frequency of progenitor cells with more than two copies per cell. Therefore a repeat transduction strategy may become toxic to a fraction of the cells and this should be carefully tested. The effect of repeated hits on high VCN was clearly seen with the ultracentrifuged vectors but not with the chromatography-purified batches. This information cannot be obtained from gene expression analyses based on average numbers. To our knowledge, this is the first time that transduction efficiency is documented from quantitative measures of VCN at the single-cell level. Thus, our results strongly suggest that different preparations could behave differently in pharmacological terms, for infection of hematopoietic progenitor cells resulting in variable levels of vector copies per cell. The process of transduction is complex, but the mechanisms behind the observed differences in VCN may involve viral binding or entry as well as proviral integration capacity.
In the perspective of clinical gene therapy studies for WAS, we have developed a large-scale purification process for rHIV LV as described in Ref. 21, and more extensively in Ref. 20. The chromatography-purified WASP-LV has showed an acceptable safety profile in preclinical tests, notably with respect to hematopoietic progenitor cell survival and differentiation, as confirmed here. The CFC transduction levels obtained with this batch of vector were similar to those obtained with the same WASP-LV purified by ultracentrifugation. Transduction is effective (about 40–50% of the CFC can be transduced), whereas providing only a low number of copies (60–85% of CFC contain 1 copy per cell and >90% of the CFC contain equal or less than two copies per cell), even after repeat infection. If we take into account that there is possibly an underestimation factor for VCN in CFC and apply a 30–40% correction, then >90% of the cells would have no more than three copies per cell. Thus, although a probability exists that some cells will integrate high VCN, our data suggest that the chromatography-purified WASP-LV provides a relatively safe level of transduction of hematopoietic cells.
In conclusion, the analysis of vector copy number in individual CFC with Q-PCR determines the frequency and the distribution of vector copies in the population of hematopoietic progenitor cells that were initially targeted by the vector. It is clear that these values are influenced by the experimental transduction conditions and by the type of vector tested. Such data are important to optimize preclinical and clinical transduction protocols with LVs for ex vivo hematopoietic gene therapy applications.
Materials and methods
Generation and titration of lentiviruses
The PGK-GFP/VSVg rHIV vector (GFP-LV) or w1.6hWASP/VSVg rHIV vector (WASP-LV) were produced by transient quadri-transfection of 293T cells and were purified by ultracentrifugation as previously reported.12 In some experiments a WASP-encoding LV produced similarly by transient transfection was concentrated and purified through a series of chromatography and membrane steps as reported.20 Vectors were titered as infectious genomes (IG) per ml on HCT116 cells using duplex Q-PCR as previously described12 and were also titered for p24 levels using an ELISA (Perkin Elmer, Waltham, MA, USA). In some experiments, vectors encoding the GFP were titered by flow cytometry instead, and results were multiplied by a factor 2 to match with IG values, as determined by repeated comparisons between the two titration methods (data not shown). Vector batches used in the study are listed in supplementary Table S2.
Clones derived from HT1080 cells
The fibrosarcoma HT1080 cells originated from ATCC (CCL-121, American Type Culture Collection, Manassas, VA, USA) were grown in DMEM supplemented with glutamine and antibiotics, and containing 10% fetal calf serum. HT1080 cells were transduced with a rHIV encoding the GFP under control of the human phosphor-glycerate kinase promoter kindly provided by Dr L Naldini (Tiget, Milan, Italy). Expression of GFP was measured by flow cytometry. Using the same culture medium, the cells were first cloned by fluorescence-activated cell sorting and then a second cloning was performed by limit dilution culture. One clone (HT4-A) containing one copy was identified and further transduced with the same vector to generate subclones. Such re-infected HT4-A cells were then cloned by limit dilution and wells were screened by fluorescent microscopy and flow cytometry to identify cells presenting different mean fluorescence intensity for GFP expression. Two daughter clones (HTA-A2 and HT4-A6) expressing different levels of fluorescence were selected and characterized by Q-PCR, Southern blot and sequencing.
Southern Blot
The number of rHIV insertions in HT4, HT4-A2 and HT4-A6 cells was determined by Southern blot using genomic DNA extracted by chloroform/phenol extraction with AutoGen extractor, NA2000 (Geneworx, Blénod lès Pont à Mousson, France) after proteinase K (Invitrogen, Cergy Pontoise, France) lysis. A measure of 40 μg of genomic DNA of each cell sample was digested with 70 units of XbaI (Gibco BRL/Invitrogen, Cergy Pontoise, France) at 37 °C over night and the digested DNA samples were subjected to an 0.8% agarose gel electrophoresis. The DNA fragments in the gel were transferred onto a Nylon membrane (Amersham, Piscataway, NJ, USA). The membrane was hybridized with 50% deionized formamide over night at 42 °C with a 2185 bp fragment of the integrative lentiviral DNA obtained by AflII-BamHI digestion of the plasmid pCCLsincppthPGK-GFP-WPRE and labeled with [α-32P]dATP using the Prime-it Random Primer Labeling kit (Stratagene, Lyon, France). The final wash was preformed with 0.1 × SSC-1% SDS at 68 °C for 1 h. After 10 days of exposure on a phosphor screen (Molecular Dynamics PhosphorImager System; GE Healthcare Bio-Sciences, Piscataway, NJ, USA), radioactive bands were revealed by the Storm system (GE-Healthcare Bio-Sciences).
Analysis of vector genomic insertion sites
rHIV insertion sites were determined following NlaIII digestion of the genomic DNA of HT4-A, HT4-A2 and HT4-A6 cell clones as described previously.16 Sequencing reactions were performed using Big Dye terminator sequencing chemistry (Applied Biosystems, Carlsbad, CA, USA) from the M13 forward or M13 reverse primers and run on a 377-XL Applied Biosystems automated sequencer. Junctions obtained were matched on the human genome using the BLAT program (UCSC human Genome Working draft (March 2006 (NCBI36/hg18). The description of the human transcripts was obtained from ftp://hgdownload.cse.ucsc.edu.
Human CD34+ cells source, transduction and culture
Umbilical cord blood progenitor CD34+ cells were obtained by immunomagnetic selection (Miltenyi Biotec, Paris, France) from mononuclear cell fractions of cord blood samples obtained from uncomplicated births at Hopital Louise Michel, Evry, France, in compliance with French national bioethics law. Cells were first pre-activated by culturing overnight 5 × 104 cells in 0.2 ml of X-vivo20 medium (Lonza, Levallois Perret, France) supplemented with 50 U ml−1 penicillin, 50 μg ml−1 streptomycin and 2 m-glutamine (Gibco BRL/Invitrogen), SCF (25 ng ml−1), Flt-3 ligand (50 ng ml−1), TPO (25 ng ml−1), IL-3 (10 ng ml−1) (R&D Systems, Lille, France). Pre-activated cells were then infected with LV using different concentration of vectors ranging from 2 × 107 IG per ml to 2 × 108 IG per ml for 6 h in the presence of polybrene (6 μg ml−1). When cells were re-infected, the second hit of vector was added overnight. At the end of the transduction step, cells were washed and either plated in semi-solid medium for CFC assays or cultured in the presence of cytokines for in vitro expansion. For in vitro expansion, the transduced CD34+ cells were seeded at 5 × 104 cells per ml in 24-well flat-bottom plates in X-vivo20 medium supplemented with 10% fetal calf serum (Gibco), penicillin/streptomycin, -glutamine and the same recombinant cytokines as used for transduction. Fresh medium was added every 3 days. Cultures were incubated at 37 °C 5% CO2 for 14 days.
CFC assays
CFC assays were performed in duplicate by plating 1000 transduced or untransduced cells per ml of Methocult (H4434), a complete methylcellulose medium supplemented with human cytokines (Stem Cell Technologies, Vancouver, CA, USA) according to the manufacturer's instructions. After 2 weeks of culture, 5% CO2, 37 °C in humid atmosphere, CFU-E/BFU-E, CFU-GM and CFU-GEMM were counted by inverse microscopy using standard visual criteria.
Q-PCR and analysis of CFC
Well-isolated colonies were aspirated carefully with a pipette tip under the miscroscope and the cells were suspended into 100 μl of phosphate-buffered saline in a 96-well U-bottom plates. The plates were centrifuged at 1500 r.p.m. for 10 min. Green fluorescent (GFP+) colonies were identified by fluorescence microscopy. Medium was aspirated and the cells pelleted from each CFC unit were suspended in 10 μl of phosphate-buffered saline. Genomic DNA was extracted from these pellets using proteinase K lysis consisting in adding 20 μl of lysis buffer (0.3 m Tris HCl, pH 7.5; 0.6 m CaCl2; 1.5 % Glycerol; 0.675% Tween-20; and 0.3 mg ml−1 Proteinase K) to each well and incubating the plate at 65 °C for 30 min, 90 °C for 10 min and at 4 °C to end the reaction for a minimum of 10 min. After lysis, 30 μl of water was added to each well to obtain 60 μl of genomic DNA preparation. In all experiments using this technique, the Q-PCR was always carried out from 10 μl of this genomic DNA preparation (that is, amplifying 1/6 of the extracted material), and in duplicate.
The Q-PCR consisted of a duplex detection of WPRE or HIV sequences normalized to ALB, as described for titrations. The probes were conjugated to FAM for HIV or WPRE sequences and to VIC for Albumin. Amplification reactions (25 μl) contained 10 μl of genomic DNA and 15 μl of TaqMan buffer (Absolute Q-PCR Rox Mix, ABgene AB-1139/B), 0.1 μ primers (forward and reverse), 0.1 μ TaqMan probe and consisted of 40 cycles at 95 °C (15 s) then 60 °C (1 min) on an ABI PRISM 7700 sequence detector (Applied Biosystems). Standard amplification curves were obtained by serial dilutions of the pRRLcpptPGKGFP-WPRE-Alb plasmid containing the appropriate sequences in cis from the vectors and ALB gene. All PCR measures were performed at least in duplicate. All Q-PCR experiments on CFC include samples from untransduced CFC as negative controls.
Data were edited using the Primer Express software (Applied Biosystems). Data were interpreted in the linear portion of the standard curve. Linear regression coefficient of the standard curve should be >0.99. In this portion of the curve the ratio between HIV/ALB of the plasmid standard is equivalent to 1±0.2 (n=28). The detection's threshold is determined with this standard. When the CT of Albumin is superior to 32 cycles, the ratio HIV/ALB is different from the value of 1±0.2 defined in the linear portion of the curve. The duplicate CT should vary by less than 0.5 CT. The ALB and HIV or WPRE CT of H2O should locate between 35 and 40. HIV CT of untransduced cells should be like H2O. ALB CT of untransduced cells and transduced samples should be around 23–28. The amount of WPRE/ALB or HIV/ALB is equimolar to the amount of pRRLcpptPGKGFP-WPRE-Alb plasmid. However, because in diploid cells, there are two molecules of ALB per cell, the number of integrated VCN per cell is determined by multiplying the ratio WPRE/ALB or HIV/ALB by two. Results of VCN inferior to 0.1 copy per cell were considered to be negative.
Preparation of genomic DNA from cells other than CFC for Q-PCR
Genomic DNA from cell lines or from bulk CD34+ were extracted with the ‘Wizard genomic DNA purification kit' (Promega Corporation, Madison, WI, USA) or with a single-step proteinase K lysis as described above.
Acknowledgments
We are very grateful for technical help from Khalil Seye and Gregory Cedrone, and thank Nicolas Laroudie and the vector production group for providing vector batches and in particular the chromatography-purifed R&D batches. We also thank Guillaume Sirantoine for sequencing the vector insertion sites, Daniel Stockholm and William Vainchenker for their input with methods and Thierry Larmonnier and Lucia Braga-Vacherie from the Genethon Biological Resources Center for help with cell line banking. We are also very grateful to the staff of the Maternité de l'hôpital Louise Michel, Evry, France for providing umbilical cord samples and to AFM (Association Francaise contre les myopathies) and the EC HEALTH-FP6 integrated project CONSERT for sponsoring this project.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Gene Therapy website (http://www.nature.com/gt)
Supplementary Material
References
- Fischer A, Cavazzana-Calvo M. Gene therapy of inherited diseases. Lancet. 2008;371:2044–2047. doi: 10.1016/S0140-6736(08)60874-0. [DOI] [PubMed] [Google Scholar]
- Piacibello W, Bruno S, Sanavio F, Droetto S, Gunetti M, Ailles L, et al. Lentiviral gene transfer and ex vivo expansion of human primitive stem cells capable of primary, secondary, and tertiary multilineage repopulation in NOD/SCID mice. Nonobese diabetic/severe combined immunodeficient. Blood. 2002;100:4391–4400. doi: 10.1182/blood.V100.13.4391. [DOI] [PubMed] [Google Scholar]
- Enssle J, Trobridge GD, Keyser KA, Ironside C, Beard BC, Kiem HP. Stable marking and transgene expression without progression to monoclonality in canine long-term hematopoietic repopulating cells transduced with lentiviral vectors. Hum Gene Ther. 2010;21:397–403. doi: 10.1089/hum.2009.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods NB, Fahlman C, Mikkola H, Hamaguchi I, Olsson K, Zufferey R, et al. Lentiviral gene transfer into primary and secondary NOD/SCID repopulating cells. Blood. 2000;96:3725–3733. [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Galy A, Roncarolo MG, Thrasher AJ. Development of lentiviral gene therapy for Wiskott Aldrich syndrome. Expert Opin Biol Ther. 2008;8:181–190. doi: 10.1517/14712598.8.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods NB, Muessig A, Schmidt M, Flygare J, Olsson K, Salmon P, et al. Lentiviral vector transduction of NOD/SCID repopulating cells results in multiple vector integrations per transduced cell: risk of insertional mutagenesis. Blood. 2003;101:1284–1289. doi: 10.1182/blood-2002-07-2238. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hangoc G, Campbell TB, Goodman M, Tao W, Pollok K, et al. Identification of parameters required for efficient lentiviral vector transduction and engraftment of human cord blood CD34(+) NOD/SCID-repopulating cells. Exp Hematol. 2008;36:947–956. doi: 10.1016/j.exphem.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AH, Sadelain M. The genetic engineering of hematopoietic stem cells: the rise of lentiviral vectors, the conundrum of the ltr, and the promise of lineage-restricted vectors. Mol Ther. 2007;15:445–456. doi: 10.1038/sj.mt.6300060. [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knoss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamedali A, Moreau-Gaudry F, Richard E, Xia P, Nolta J, Malik P. Self-inactivating lentiviral vectors resist proviral methylation but do not confer position-independent expression in hematopoietic stem cells. Mol Ther. 2004;10:249–259. doi: 10.1016/j.ymthe.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Charrier S, Dupre L, Scaramuzza S, Jeanson-Leh L, Blundell MP, Danos O, et al. Lentiviral vectors targeting WASp expression to hematopoietic cells, efficiently transduce and correct cells from WAS patients. Gene Ther. 2007;14:415–428. doi: 10.1038/sj.gt.3302863. [DOI] [PubMed] [Google Scholar]
- Ailles L, Schmidt M, Santoni de Sio FR, Glimm H, Cavalieri S, Bruno S, et al. Molecular evidence of lentiviral vector-mediated gene transfer into human self-renewing, multi-potent, long-term NOD/SCID repopulating hematopoietic cells. Mol Ther. 2002;6:615–626. [PubMed] [Google Scholar]
- Schuesler T, Reeves L, Von Kalle C, Grassman E. Copy number determination of genetically-modified hematopoietic stem cells. Methods Mol Biol. 2009;506:281–298. doi: 10.1007/978-1-59745-409-4_19. [DOI] [PubMed] [Google Scholar]
- Chen TR, Hay RJ, Macy ML. Intercellular karyotypic similarity in near-diploid cell lines of human tumor origins. Cancer Genet Cytogenet. 1983;10:351–362. doi: 10.1016/0165-4608(83)90092-4. [DOI] [PubMed] [Google Scholar]
- Mantovani J, Charrier S, Eckenberg R, Saurin W, Danos O, Perea J, et al. Diverse genomic integration of a lentiviral vector developed for the treatment of Wiskott-Aldrich syndrome. J Gene Med. 2009;11:645–654. doi: 10.1002/jgm.1346. [DOI] [PubMed] [Google Scholar]
- Rook MS, Delach SM, Deyneko G, Worlock A, Wolfe JL. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am J Pathol. 2004;164:23–33. doi: 10.1016/S0002-9440(10)63092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehse B, Kustikova OS, Bubenheim M, Baum C. Pois(s)on--it's a question of dose. Gene Ther. 2004;11:879–881. doi: 10.1038/sj.gt.3302270. [DOI] [PubMed] [Google Scholar]
- Haas DL, Case SS, Crooks GM, Kohn DB. Critical factors influencing stable transduction of human CD34(+) cells with HIV-1-derived lentiviral vectors. Mol Ther. 2000;2:71–80. doi: 10.1006/mthe.2000.0094. [DOI] [PubMed] [Google Scholar]
- Merten OW, Charrier S, Laroudie N, Fauchille S, Dugué C, Jenny C, et al. Large-scale manufacture and characterisation of a lentiviral vector produced for ex vivo gene therapy application Hum Gene Ther 2010. Nov 2. [Epub ahead of print] PMID: 21043787. [DOI] [PubMed]
- Denard J, Rundwasser S, Laroudie N, Gonnet F, Naldini L, Radrizzani M, et al. Quantitative proteomic analysis of lentiviral vectors using 2-DE. Proteomics. 2009;9:3666–3676. doi: 10.1002/pmic.200800747. [DOI] [PubMed] [Google Scholar]
- Millington M, Arndt A, Boyd M, Applegate T, Shen S. Towards a clinically relevant lentiviral transduction protocol for primary human CD34 hematopoietic stem/progenitor cells. PLoS One. 2009;4:e6461. doi: 10.1371/journal.pone.0006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.