Abstract
High dispersal rates between patches in spatially structured populations can impede diversification and homogenize diversity. These homogenizing effects of dispersal are likely to be enhanced by coevolving parasites that impose strong selection on hosts for resistance. However, the interactive effects of dispersal and parasites on host diversification have never been tested. We used spatially structured, experimental populations of the bacterium Pseudomonas fluorescens, cultured with or without the phage SBW25Ф2 under three levels of dispersal (none, localized or global), and quantified diversity in terms of evolved bacterial colony morphologies after approximately 100 bacterial generations. We demonstrate that higher levels of colony morphology richness evolved in the presence of phage, and that dispersal reduced diversity most strongly in the presence of phage. Thus, our results suggest that, while parasites can drive host diversification, host populations coevolving with parasites are more prone to homogenization through dispersal.
Keywords: adaptive radiation, experimental evolution, antagonistic coevolution, geographical mosaic theory, metapopulation
1. Introduction
Spatial population structure can contribute to genetic diversification by allowing divergent evolutionary trajectories to be followed in different patches. This may arise by chance through the stochastic nature of allele frequency fluctuations in small population patches [1–3], or through the localizing of ecological interactions favouring local adaptation [4,5]. High dispersal rates can cause migration load through genetic swamping (the replacement of locally selected alleles with alleles common in the rest of the metapopulation [6]) and further impede local adaptation in spatially structured populations by homogenizing genetic variation [7] or selective trajectories between patches [8,9]. Such patterns are commonly inferred from negative correlations between measures of adaptive divergence and gene flow [9]. To our knowledge, the only direct empirical test has used spatially structured populations of the bacterium Escherichia coli cultured with and without population mixing, where as expected, diversity only evolved in the absence of population mixing [4] (although the effect of spatial structure on diversity may be somewhat dependent upon the mechanism maintaining diversity, cf. [10]). While parasites can directly drive divergence in hosts if, for example, multiple resistance mechanisms are available [11–13], coevolving host populations could be more prone to the homogenizing of selection by dispersal because parasites often impose strong selection on hosts for resistance. However, the interactive effects of parasites and dispersal on the diversification of hosts have not been studied.
We used an experimental evolution approach to test the effects of dispersal on host diversification in spatially structured laboratory populations of the bacterium Pseudomonas fluorescens and its naturally associated phage SBW25Ф2 [14]. In the absence of phage, P. fluorescens readily diversifies in laboratory microcosms, forming three ecologically distinct classes distinguishable by their niche preference and colony morphology: smooth (SM) inhabits the liquid broth and has a smooth-colony morphology, wrinkly spreader (WS) inhabits the air–broth interface and has a wrinkled-colony morphology, and fuzzy spreader (FS) inhabits the bottom of the microcosm and has a fuzzy-colony morphology [15]. These phenotypes are heritable, and diversity is stably maintained by negative frequency-dependent selection. Hence, to a large extent, such diversification is deterministic, with similar morphotypes occurring in all replicate populations. However, introduction of phage SBW25Ф2 into populations of P. fluorescens inhibits adaptive radiation within populations by reducing density, thus weakening diversifying selection mediated by resource competition [16,17]. However, between populations, coevolutionary trajectories are divergent and distinct morphotypes tend to dominate in different replicate populations, depending on which genetic background is the first to obtain a resistance mutation. In addition, phage select for novel colony morphologies not seen in the absence of phage, notably mucoids [16,17].
2. Material and methods
Each population was propagated in 64 wells on a 96-well microtitre plate (i.e. an eight-well by eight-well grid), each well containing 100 µl of King's B (KB) liquid media. Eighteen replicate phage-free spatially structured populations were initiated with approximately 1.7 × 106 bacterial cells per well and 18 replicate phage-containing populations were initiated with approximately 1.7 × 106 bacterial cells and 170 viral particles per well using a 96-pin replicator. These densities were chosen to be equivalent to the starting population densities previously used in experiments with this system [14]. Populations were propagated by serial transfer for 12 transfers (every 2 days, 1 µl of each well was used to inoculate a fresh well using a 96-pin replicator) under one of the following dispersal regimes: global—all patches were pooled and homogenized prior to transfer; localized—the contents of all eight wells in each row were pooled and homogenized prior to even-numbered transfers, the contents of all eight wells in each column were pooled and homogenized prior to odd-numbered transfers; and none—no between well mixing occurred prior to transfer. Diversity was measured by determining the morphologies of 100 random colonies on KB agar plates [15,18,19]. Population diversity was calculated as the complement of the Simpson index [20]:
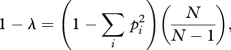 |
where pi is the proportion of the ith morph and N is the total number of colonies sampled. This measure is the probability that two randomly selected colonies are morphologically different. Six colony morphology variants were observed: SM, two distinct WSs (WS1 and WS2), FS, mucoids (MUs), opaques (OPs) and swarming spreaders (SSs).
3. Results
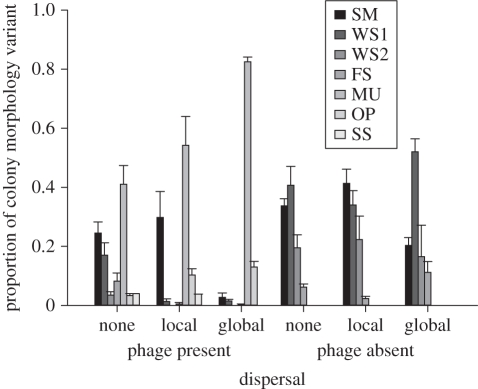
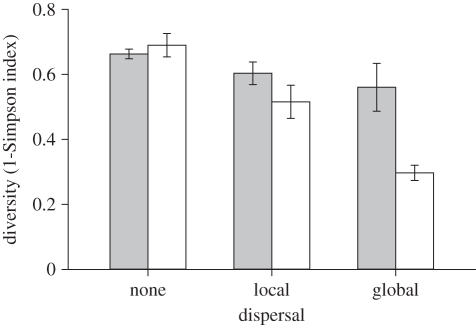
Highest bacterial colony morphology richness was observed in the presence of phage under no dispersal (figure 1, dispersal × phage interaction, F2,30 = 9.23, p < 0.001; simple effect of phage: no dispersal, F1,30 = 29.99, p < 0.001; localized dispersal, F1,30 = 1.38, p > 0.05; global dispersal, F1,30 = 0.15, p > 0.05). Up to seven distinct colony morphologies were observed to coexist within these spatially structured populations, including several that were never observed in the absence of phage (figure 1; e.g. MUs, OPs and SSs). In the presence of phage, increasing dispersal led to the domination of populations by MUs (figure 1; simple effect of dispersal with phage: F2,30 = 20.34, p < 0.001). However, no such decline in colony morphology richness with increasing dispersal was observed in the absence of phage; in all cases, the same four colony morphotypes were always observed, regardless of dispersal regime (figure 1; simple effect of dispersal without phage: F2,30 = 0.41, p > 0.05). Dispersal and phage, therefore, had interactive effects on Simpson index diversity (figure 2, dispersal × phage interaction, F2,30 = 5.91, p < 0.01), such that dispersal decreased diversity in the presence of phage, but had no effect on diversity in the absence of phage (simple effect of dispersal: with phage, F2,30 = 21.45, p < 0.001; without phage, F2,30 = 1.47, p > 0.05).
Figure 1.
Colony morphology richness. Bars represent mean proportion of colony morphology variant (±s.e.m.) within populations under each treatment after 12 transfers (n = 6).
Figure 2.
Colony morphology diversity. Bars represent mean diversity measured as the complement of the Simpson index (±s.e.m.) in the absence (grey) and presence (white) of phage after 12 transfers (n = 6).
4. Discussion
Our results suggest that dispersal decreases diversity much more strongly in the presence of coevolving parasites, and furthermore, that parasites can both promote and impede host divergence in spatially structured populations depending upon the scale of dispersal. Without dispersal, phage imposed selection for the evolution of novel colony morphologies that were never observed in the absence of phage. Taken together with previous experiments on this system [16,17], it is likely that different morphologies evolved in different patches owing to chance variation in resistance mutations (and linked genetic variation) between patches. These diverse colony morphologies were able to coexist at a ‘regional’ population level owing to spatial structure. Increasing dispersal reduced diversity in the presence of phage owing to the domination of populations by MU colony morphology variants. This presumably occurred as a result of the homogenization of coevolutionary trajectories between patches. No such decline in diversity with increasing dispersal was observed in the absence of phage. Ecological metacommunity models and experiments emphasize that high rates of dispersal can homogenize diversity through enhancing regional competitive exclusion [21–23]. Our experiments suggest that the homogenizing effect of dispersal is likely to be even stronger when combined with a source of strong selection, such as that imposed by shared parasites (and other natural enemies) on hosts.
Phages are thought to impose pervasive and intense selection in almost all bacterial communities [24]. Recent work demonstrates that soil phage populations are locally adapted to their bacterial hosts on spatial scales similar to those employed in our experiments (i.e. centimetre scales) [25]. This highlights that bacteria-phage coevolution plays an important diversifying role in nature; our findings suggest that such populations may be particularly prone to homogenization through dispersal. However, extrapolation from simple laboratory models to natural systems should be made with great caution. We therefore highlight two important caveats. First, the rates of dispersal used in these experiments were relatively high compared with those observed in some natural systems [26]. Our dispersal regimes consisted of mass migration events at each transfer, with each growth period between transfers allowing approximately 10 bacterial generations. This equated to m (i.e. proportion of immigrants per patch per generation [26,27]) values of approximately 0.10 for global dispersal and approximately 0.09 for localized dispersal. Second, bacteria and phage were dispersed at equal rates in our experiments. In some host–parasite associations such congruent patterns of host and parasite gene flow are observed [28], while in others either the host [29] or the parasite [30,31] have relatively greater levels of gene flow. Our findings may, therefore, be limited to systems where dispersal rates are relatively high and the parasite relies upon the host for its dispersal, as is the case for contact-transmitted parasites, or where co-dispersal of host and parasite is driven by an external factor such as a prevailing wind or aquatic current.
In summary, we demonstrate that parasites and dispersal have interactive effects on the evolution and maintenance of host diversity in spatially structured host populations. We provide empirical evidence that parasites can drive host divergence, by selecting for novel resistance phenotypes, and that these may coexist through spatial population structuring. However, we also show that selection imposed by coevolving parasites can enhance the diversity-homogenizing effects of dispersal, by selecting for highly resistant phenotypes across all patches.
Acknowledgements
This work was funded by a NERC studentship to T.V. and a Royal Society project grant to A.F. We are grateful to the anonymous reviewers for valuable comments on previous versions of the article.
References
- 1.Rozen D. E., Habets M. G., Handel A., de Visser J. A. 2008. Heterogeneous adaptive trajectories of small populations on complex fitness landscapes. PLoS ONE 3, e1715. 10.1371/journal.pone.0001715 (doi:10.1371/journal.pone.0001715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain K., Krug J. 2007. Deterministic and stochastic regimes of asexual evolution on rugged fitness landscapes. Genetics 175, 1275–1288 10.1534/genetics.106.067165 (doi:10.1534/genetics.106.067165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Visser J. A., Rozen D. E. 2005. Limits to adaptation in asexual populations. J. Evol. Biol. 18, 779–788 10.1111/j.1420-9101.2005.00879.x (doi:10.1111/j.1420-9101.2005.00879.x) [DOI] [PubMed] [Google Scholar]
- 4.Habets M. G., Rozen D. E., Hoekstra R. F., de Visser J. A. 2006. The effect of population structure on the adaptive radiation of microbial populations evolving in spatially structured environments. Ecol. Lett. 9, 1041–1048 10.1111/j.1461-0248.2006.00955.x (doi:10.1111/j.1461-0248.2006.00955.x) [DOI] [PubMed] [Google Scholar]
- 5.Venail P. A., MacLean R. C., Bouvier T., Brockhurst M. A., Hochberg M. E., Mouquet N. 2008. Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities. Nature 452, 210–214 10.1038/nature06554 (doi:10.1038/nature06554) [DOI] [PubMed] [Google Scholar]
- 6.Lenormand T. 2002. Gene flow and the limits to natural selection. Trends Ecol. Evol. 17, 183–189 10.1016/S0169-5347(02)02497-7 (doi:10.1016/S0169-5347(02)02497-7) [DOI] [Google Scholar]
- 7.Gandon S., Michalakis Y. 2002. Local adaptation, evolutionary potential and host–parasite coevolution: interactions between migration, mutation, population size and generation time. J. Evol. Biol. 15, 451–462 10.1046/j.1420-9101.2002.00402.x (doi:10.1046/j.1420-9101.2002.00402.x) [DOI] [Google Scholar]
- 8.Garant D., Forde S. E., Hendry A. P. 2007. The multifarious effects of dispersal and gene flow on contemporary adaptation. Funct. Ecol. 21, 434–443 10.1111/j.1365-2435.2006.01228.x (doi:10.1111/j.1365-2435.2006.01228.x) [DOI] [Google Scholar]
- 9.Rasanen K., Hendry A. P. 2008. Disentangling interactions between adaptive divergence and gene flow when ecology drives diversification. Ecol. Lett. 11, 624–636 10.1111/j.1461-0248.2008.01176.x (doi:10.1111/j.1461-0248.2008.01176.x) [DOI] [PubMed] [Google Scholar]
- 10.Saxer G., Doebeli M., Travisano M. 2009. Spatial structure leads to ecological breakdown and loss of diversity. Proc. R. Soc. B 276, 2065–2070 10.1098/rspb.2008.1827 (doi:10.1098/rspb.2008.1827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank S. A. 1993. Evolution of host parasite diversity. Evolution 47, 1721–1732 10.2307/2410216 (doi:10.2307/2410216) [DOI] [PubMed] [Google Scholar]
- 12.Brockhurst M. A., Buckling A., Rainey P. B. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc. R. Soc. B 272, 1385–1391 10.1098/rspb.2005.3086 (doi:10.1098/rspb.2005.3086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press [Google Scholar]
- 14.Buckling A., Rainey P. B. 2002. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. Lond. B 269, 931–936 10.1098/rspb.2001.1945 (doi:10.1098/rspb.2001.1945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rainey P. B., Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72 10.1038/27900 (doi:10.1038/27900) [DOI] [PubMed] [Google Scholar]
- 16.Buckling A., Rainey P. B. 2002. The role of parasites in sympatric and allopatric host diversification. Nature 420, 496–499 10.1038/nature01164 (doi:10.1038/nature01164) [DOI] [PubMed] [Google Scholar]
- 17.Brockhurst M. A., Rainey P. B., Buckling A. 2004. The effect of spatial heterogeneity and parasites on the evolution of host diversity. Proc. R. Soc. Lond. B 271, 107–111 10.1098/rspb.2003.2556 (doi:10.1098/rspb.2003.2556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buckling A., Kassen R., Bell G., Rainey P. B. 2000. Disturbance and diversity in experimental microcosms. Nature 408, 961–964 10.1038/35050080 (doi:10.1038/35050080) [DOI] [PubMed] [Google Scholar]
- 19.Buckling A., Rainey P. B. 2002. The role of parasites in sympatric and allopatric diversification. Nature 420, 496–499 10.1038/nature01164 (doi:10.1038/nature01164) [DOI] [PubMed] [Google Scholar]
- 20.Simpson E. H. 1949. Measurement of diversity. Nature 163, 688. 10.1038/163688a0 (doi:10.1038/163688a0) [DOI] [Google Scholar]
- 21.Mouquet N., Miller T. E., Daufresne T., Kneitel J. M. 2006. Consequences of varying regional heterogeneity in source-sink metacommunities. Oikos 113, 481–488 10.1111/j.2006.0030-1299.14582.x (doi:10.1111/j.2006.0030-1299.14582.x) [DOI] [Google Scholar]
- 22.Mouquet N., Loreau M. 2003. Community patterns in source-sink metacommunities. Am. Nat. 162, 544–557 10.1086/378857 (doi:10.1086/378857) [DOI] [PubMed] [Google Scholar]
- 23.Matthiessen B., Mielke E., Sommer U. 2010. Dispersal decreases diversity in heterogeneous metacommunities by enhancing regional competition. Ecology 91, 2022–2033 10.1890/09-1395.1 (doi:10.1890/09-1395.1) [DOI] [PubMed] [Google Scholar]
- 24.Weinbauer M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181 10.1016/j.femsre.2003.08.001 (doi:10.1016/j.femsre.2003.08.001) [DOI] [PubMed] [Google Scholar]
- 25.Vos M., Birkett P. J., Birch E., Griffiths R. I., Buckling A. 2009. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325, 833–833 10.1126/Science.1174173 (doi:10.1126/Science.1174173) [DOI] [PubMed] [Google Scholar]
- 26.Slatkin M. 1985. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16, 393–430 10.1146/annurev.ecolsys.16.1.393 (doi:10.1146/annurev.ecolsys.16.1.393) [DOI] [Google Scholar]
- 27.Wright S. 1931. Evolution in Mendelian populations. Genetics 16, 97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulvey M., Aho M. J., Lydeard C., Leberg P. L. 1991. Comparative population structure of a parasite (Fascioloides magna) and its definitive host. Evolution 45, 1628–1640 10.2307/2409784 (doi:10.2307/2409784) [DOI] [PubMed] [Google Scholar]
- 29.Delmotte F., Bucheli E., Shykoff J. A. 1999. Host and parasite population structure in a natural plant-pathogen system. Heredity 82, 300–308 10.1038/sj.hdy.6884850 (doi:10.1038/sj.hdy.6884850) [DOI] [PubMed] [Google Scholar]
- 30.Davies C. M., Webster J. P., Kruger O., Munatsi A., Ndamba J., Woolhouse M. E. 1999. Host–parasite population genetics: a cross-sectional comparison of Bulinus globosus and Schistosoma haematobium. Parasitology 119, 295–302 10.1017/S0031182099004722 (doi:10.1017/S0031182099004722) [DOI] [PubMed] [Google Scholar]
- 31.Dybdahl M. F., Lively C. M. 1996. The geography of coevolution: comparative population structures for a snail and its trematode parasite. Evolution 50, 2264–2275 10.2307/2410696 (doi:10.2307/2410696) [DOI] [PubMed] [Google Scholar]