Abstract
The transcription of ribosomal RNA (rRNA) is critical to life. Despite its importance, ribosomal DNA (rDNA) is not included in current genome assemblies and, consequently, genomic analyses to date have excluded rDNA. Here, we show that short sequence reads can be aligned to a genome assembly containing a single rDNA repeat. Integrated analysis of ChIP-seq, DNase-seq, MNase-seq and RNA-seq data reveals several novel findings. First, the coding region of active rDNA is contained within nucleosome-depleted open chromatin that is highly transcriptionally active. Second, histone modifications are located not only at the rDNA promoter but also at novel sites within the intergenic spacer. Third, the distributions of active modifications are more similar within and between different cell types than repressive modifications. Fourth, UBF, a positive regulator of rRNA transcription, binds to sites throughout the genome. Lastly, the insulator binding protein CTCF associates with the spacer promoter of rDNA, suggesting that transcriptional insulation plays a role in regulating the transcription of rRNA. Taken together, these analyses confirm and expand the results of previous ChIP studies of rDNA and provide novel avenues for exploration of chromatin-mediated regulation of rDNA.
INTRODUCTION
The transcription of ribosomal RNA (rRNA) is a critical process for all cells, accounting for up to 80% of all cellular RNA production (1). Deficiencies in rRNA transcription lead to reduced ribosome biogenesis, altered cellular growth and increased cell death (2). Highlighting its central importance in cellular function, dysregulation of rRNA biogenesis has been implicated in many human diseases (3): Treacher Collins syndrome (4), Diamond–Blackfan anemia (5), 5q-syndrome (6), cartilage-hair hypoplasia (7), Shwachman–Diamond syndrome (8), dyskeratosis congenita (9), CHARGE syndrome (10) and cancers with amplification of the c-Myc oncogene, the product of which is a positive regulator of rRNA transcription (11). Accordingly, rRNA transcription is tightly regulated at many levels, including that of chromatin structure.
The diploid human genome contains 400 copies of a 43-kb rDNA unit tandemly arrayed in nucleolar organizer regions (NORs) on the five acrocentric chromosomes. Each unit contains ∼13.3 kb of sequence encoding the 28S, 5.8S and 18S rRNAs (hereafter referred to as the ‘coding region’) and a non-coding intergenic spacer (IGS) containing an enhancer, spacer promoter and the core promoter of the adjoining rDNA repeat (12). Transcriptionally active rDNA is euchromatic, hypomethylated at CpG sites and marked with histone modifications generally associated with transcriptionally active nucleoplasmic genes (i.e., H3K4me3 and H3K9ac). Transcriptionally silent rDNA is heterochromatic, hypermethylated and marked with repressive histone modifications (i.e. H3K27me3 and H4K20me3) (12,13). The coding sequence of each transcriptionally active rDNA unit is transcribed by RNA polymerase I (Pol I) into a pre-ribosomal RNA (pre-rRNA), containing the sequences encoding the 18S, 5.8S, and 28S rDNA species (14,15). The mature rRNA species are generated through a complex series of RNase cleavages and chemical modifications and subsequently assembled into ribosomes (16,17).
The recent development of ChIP-seq (18) has allowed for rapid assessment of protein occupancy throughout the genome. However, because rDNA is not included in reference genome assemblies to which sequence reads are normally aligned, virtually no ChIP-seq studies reported to date have analyzed rDNA. In this report, we show that short-sequence reads generated from ChIP-seq experiments can be accurately aligned to genome assemblies containing rDNA. We present the locations of nine histone modifications at rDNA in multiple cell types, confirming and expanding the results of previous ChIP studies that have focused primarily on the promoter. Results from analysis of MNase-seq, DNase-seq and RNA-seq data are consistent with a model wherein the coding region of rDNA is contained within nucleosome-depleted open chromatin that is highly transcriptionally active. We also present the first ChIP-seq analysis of human UBF and RPA116 (the second largest subunit of RNA Pol I), validating their distribution along rDNA and demonstrating extensive nucleoplasmic chromatin association of UBF. We also report the association of CTCF with the rDNA spacer promoter, suggesting the presence of an insulator element. Taken together, our results provide a high-resolution map of chromatin structure at rDNA and provide a reference for future studies of chromatin-mediated regulation of rDNA.
MATERIALS AND METHODS
Cell culture, siRNA knockdown, and gene expression analysis
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 50 µg/ml gentamicin at 37°C, 5% CO2. K562 cells were cultured in RPMI medium 1640 supplemented with 10% FBS and 50 µg/ml gentamicin at 37°C, 5% CO2. For UBF knockdown, HEK293T cells were transfected with a control or UBF siRNA SmartPool (Dharmacon). Cells were harvested 48 h after transfection for analysis. UBF depletion was assayed by western blot (rabbit anti-UBF, Santa Cruz #9131, 1:1000 and rabbit anti-tubulin, ICN BioMedicals, 1:5000). RNA was extracted using TRIzol (Invitrogen) and cDNA was prepared using the High-Capacity cDNA Archive Kit (ABI). We selected 11 genes with significant UBF binding at their TSS in both HEK293T and K562 cells (ATG2A, C10ORF140, CNOT4, IPP, KDELR1, MED26, PCBP2, PRCC, PSMD3, PSMD14, SETD2) to analyze in HEK293T cells following UBF knockdown. TaqMan probes (ABI) were used to assay the expression of each gene on a GeneAmp 7300 real-time thermal cycler (ABI). GAPDH was used as endogenous control for all reactions.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described (19). For polymerase chain reaction (PCR), triplicate wells were performed for each primer set using Sybr Green (ABI) on an ABI 7300 real-time thermal cycler. Relative enrichment was calculated using the ΔΔCt method. Primer details are listed in Supplementary Table S5. Following successful ChIP–PCR, 30 µl ChIP or 500 ng input DNA was used to prepare sequencing libraries as described (19). Sequencing of ChIP and input libraries was performed on an Illumina Genome Analyzer II at the Case Western Reserve University Genomics Core. We obtained the following unique read numbers: HEK293T RPA116: 14 103 573; HEK293T UBF, 15 473 371; HEK293T input: 20 913 306; K562 UBF: 8 801 725; K562 input: 12 188 805. Antibodies used for ChIP were rabbit anti-RPA116 (a gift from Ingrid Grummt, 5 µl/ChIP), rabbit anti-UBF (H-300, Santa Cruz #9131, 5 µg/ChIP), mouse anti-UBF (F-9, Santa Cruz #13125, 5 µg/ChIP), rabbit anti-H3K4me1 (Abcam #8895, 8µg/ChIP), and rabbit anti-CTCF (Millipore #07-729, 10 µl/ChIP). The ChIP-seq data sets generated in this publication have been deposited to the SRA (SRA027342).
Data sets
Data for histone modifications, CTCF, and a corresponding input from K562, HUVEC, H1-hESC, and NHEK cells were obtained from the ENCODE Broad Histone track of the UCSC Genome Browser. DNase-seq data from K562, HUVEC, H1-hESC and NHEK cells were obtained from the ENCODE Duke/UNC/UT Open Chromatin track of the UCSC Genome Browser. Short nucleolar RNA-seq data from K562 cells were obtained from the ENCODE CSHL small RNA-seq track of the UCSC Genome Browser. MNase-seq data from CD4 + T cells (20) and CTCF ChIP-seq data from mouse ES cells (21) were obtained from the SRA (SRA000234 and SRX000540, respectively).
Sequencing data alignment and analysis
Because rDNA is not included in the human genome assembly, we created a custom build of HG18. We removed the unsequenced bases near the centromere of chromosome 13 and added a full, non-repeat masked human rDNA repeat (GenBank accession no. U13369), yielding ‘rDNA_chr13’. A custom HG18 assembly containing rDNA_chr13 rather than chromosome 13 was constructed with bowtie-build (22). We designated this HG18 build ‘HG18_plus_rDNA’. A similar genome build was constructed for mouse, wherein a full, non-repeat masked mouse rDNA repeat (GenBank accession no. BK000964) was added to chromosome 12 of the MM8 assembly. This build was designated ‘MM8_plus_rDNA’.
Data sets were aligned to our custom assemblies with Bowtie (22), allowing two mismatches per read. Prior to alignment, non-unique reads were removed from each FASTQ file. During alignment, reads with more than one reportable alignment were discarded using the ‘-m 1’ option. Peaks were detected with F-seq (23). Fragment size was set to 200 for all analyses except DNase, for which it was 0. We analyzed sequenced input samples as described above and subtracted the signal at each base from the corresponding base of the ChIP data using R. Input subtraction was performed prior to all analyses of ChIP-seq data in this study.
For detection of RPA116 and UBF peaks throughout the whole genome, we used Sole-Search (24). ChIP-seq and corresponding input datasets, with non-unique reads removed, were aligned to HG18 without rDNA using Bowtie, allowing only unique alignments. Bowtie alignment files were converted to TagAlign format and uploaded to the SoleSearch web server, using an FDR of 0.001. Detailed information on UBF peaks is given in Supplementary Data 1.
To compare nucleosome occupancy at the coding region to the IGS of rDNA, we obtained the median MNase-signal in 100-bp windows along the whole rDNA repeat. We averaged the median signal for the coding region (windows 1–133, representing 0–13.3 kb of the rDNA repeat) and IGS (windows 134–430, representing 13.4–43 kb of the rDNA repeat). The median signals of the coding region and IGS were compared by t-test.
Correlation analysis
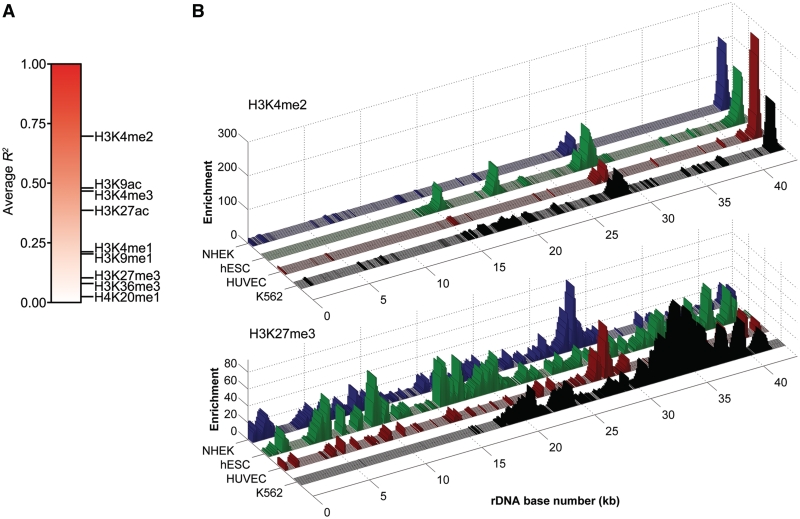
To obtain data points for correlations, the rDNA was divided into 100-bp windows and the median signal for each window was determined. To compare modifications, DNase hypersensitivity, RPA116, UBF and CTCF at rDNA between cell types, least-squares regression analysis was performed in R. To generate an average correlation score for each histone modification, we averaged the R2 values for all cell-type comparisons done for a given modification. Three-dimensional plots of histone modifications at rDNA were generated with MatLab. See Supplementary Table S1 for a complete list of R2 values determined by these analyses.
To assess rDNA co-occupancy of histone modifications within a cell type, the rDNA was windowed to 100 bp as above and pairwise comparisons between all pairs of histone modifications for a given cell type were performed in R. Matrices of the pairwise correlation scores were then plotted as heatmaps using the gplots R package (http://cran.r-project.org/web/packages/gplots/index.html).
Analysis of nucleoplasmic UBF peaks
To determine the distribution of significant peaks with respect to RefSeq genes, we used the Location-Analysis feature of the ChIP-seq Tool Set (http://chipseq.genomecenter.ucdavis.edu/cgi-bin/chipseq.cgi), which uses the UCSC known genes list to define the start and end of genes. UBF peaks <2 kb from a ReqSeq TSS were binned into the TSS category. Peaks in exons or introns >2 kb downstream of the TSS in RefSeq genes were placed in the exon and intron categories, and peaks >2 kb upstream of a RefSeq TSS or otherwise outside of a RefSeq gene were considered intergenic. Detailed location analysis results are included in Supplementary Data 1.
To determine UBF signal at all TSSs in the human genome, the complete list of RefSeq human genes was downloaded from the UCSC Genome Browser and merged to a file containing all UBF binding sites throughout the genome. The median input-normalized UBF signal ±5 kb of each TSS in 200-bp windows was determined. TSSs were sorted ascending by gene name and then descending by median UBF signal intensity. The list was filtered to include only unique records, removing duplicate TSSs and retaining the TSS with the highest median signal for a given gene. Signals were then Z-score transformed, sorted descending by average signal intensity for each TSS, and heatmapped with Java TreeView.
Gene ontology analysis was performed using the PANTHER Classification System (25). For each cell type, a list of genes with significant UBF peaks <2 kb from their TSS was uploaded to PANTHER. Lists were then analyzed using the Biological Process, Pathways, and PANTHER Protein Class options. The Homo sapiens gene list was used as the background gene list. Results of PANTHER analyses are included as Supplementary Data 2. Motif analysis was performed with the Cis Element Annotation System (CEAS) (26) and detailed results are included in Supplementary Data 3. Analysis of overlap between HEK293T and K562 peaks was performed with the GFF-Overlap feature of the ChIP-seq Tool Set.
Comparison of UBF ChIP-seq data to expression data
Publically available expression data were downloaded from GEO. Accession numbers are as follows: HEK293T, GSE21092 and K562, GSE8832. Data sets were generated with the Affymetrix GeneChip Human Genome U133 Plus 2.0 array platform. Replicates were normalized to one another using the RMA method (27) included in the affy R package and averaged. For genes represented by multiple probes, the probe with the highest average expression value was retained for analysis. Expression values for three categories of genes were obtained: all genes, genes with a significant UBF peak <2 kb from their TSS as determined by SoleSearch, and genes without substantial UBF binding at their TSS. Statistical significance between groups was assessed using t-tests. P-values were multiplied by two to account for the two comparisons made against the genes with a significant SoleSearch peak.
RESULTS
Alignment of high-throughput sequencing data to rDNA
There are ∼400 copies of rDNA in the average mammalian genome arranged in variable orientations on several chromosomes (1). Sequencing of rDNA loci was not performed during the sequencing of the human genome (28,29), and thus current genome assemblies do not contain rDNA. Despite the high copy number of rDNA, each individual unit is similar in repetitiveness to the human genome as a whole [48.88% for rDNA versus 50% for the whole genome (18)]. Therefore, in principle, alignment of short sequence reads to rDNA should not be any more problematic than alignment of reads to other regions of the genome. We implemented the following pipeline for analysis of ChIP-seq data. First, the complete sequence of one rDNA unit was added to the human genome assembly (HG18). We reasoned that if reads were aligned to rDNA alone, out of the context of the whole genome assembly, reads derived from elsewhere in the genome with sequences similar to those occurring in rDNA might be forced to align to rDNA, resulting in false positives. The rDNA sequence was added to the proximal tip of chromosome 13, on which rDNA is endogenously located, so that ChIP-seq signals corresponding to rDNA could be easily compared to those on nucleoplasmic chromatin. We call this assembly HG18_plus_rDNA. To further reduce false positives we discarded non-unique reads and reads aligning to more than one region of the genome. Finally, to mitigate the effects of systematic biases that might be present in the data, ChIP-seq signals were normalized against control input DNA from each cell type. Importantly, input libraries prepared and sequenced by independent labs were analyzed and found to be similar to one another, suggesting that the stability of the rDNA in a given cell type is not likely to affect the signal output (Supplementary Figure S1). We note that this analysis pipeline, which incorporates multiple filtering steps, is highly conservative and inherently designed to retain only robust rDNA signals that are likely genuine. In addition, with the exception of one region located 2–5 kb into the repeat, the sequence of the rDNA locus is of sufficient uniqueness to avoid false negatives. However, the limitation of this approach is that the signal obtained at rDNA is an aggregate of signal at all rDNA repeats and does not discriminate between transcriptionally active and repressed rDNA.
Distribution of histone modifications at rDNA
We analyzed publically available ChIP-seq data from K562 cells generated by the ENCODE consortium. K562, a lymphoblastoid chronic myelogenous leukemia line, was first selected for analysis because these cells are grown under specific cell culture guidelines established by the ENCODE consortium, thus minimizing experimental variability due to culture conditions. In total, we analyzed nine histone modifications, six generally associated with transcriptional activation (H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K27ac and H3K36me3) and three generally associated with transcriptional repression (H3K9me1, H3K27me3 and H4K20me1). Consistent with previous analysis of rDNA by standard ChIP, we detected strong enrichment of the active modifications H3K4me2/3 and H3K9ac at the rDNA promoter, just upstream of the transcription start site (TSS) (Figure 1A and Supplementary Figure S2). This region also shows enrichment of H3K4me1 and H3K27ac, marks that have not previously been analyzed at rDNA. Enrichment of active modifications was also detected within the IGS of rDNA, at a site located ∼28–29 kb into the rDNA repeat (Figure 1A and Supplementary Figure S2). H3K4me1 ChIP-PCR was used to verify enrichment at this region (Supplementary Figure S3). Little or no enrichment of active modifications was detected in the coding region. This is an interesting observation, as it has been previously speculated that the coding region of active rDNA repeats is nucleosome-poor or completely free of nucleosomes (30–33). In general, enrichment of active modifications is more punctate than repressive modifications, which are broadly distributed along the IGS and sometimes within the coding region of rDNA (i.e. H3K9me1; Figure 1A and Supplementary Figure S2). Interestingly, H3K36me3, a mark generally associated with the bodies of transcriptionally elongating genes, is virtually absent from the coding region of the rDNA. While the significance of this finding is currently unclear, the results may suggest that the function of H3K36me3 differs between the nucleoplasm and nucleolus.
Figure 1.
Distribution of histone modifications at rDNA. (A) Patterns of histone modifications at rDNA in K562 cells. A schematic representation of the human rDNA repeat is shown above the ChIP-seq plots. UCE: upstream control element; CPE: core promoter element; ETS: external transcribed spacer; ITS: internal transcribed spacer; SAL box: transcription terminator region with SAL repeats; SP: spacer promoter. The coding region spans ∼0–13.3 kb of the rDNA repeat, with the remainder made up by the non-coding IGS. Shown below the rDNA repeat are patterns of histone modifications at rDNA in K562 cells as determined by ChIP-seq. Plots for HUVEC, H1-hESC and NHEK are presented in Supplementary Figure S2. (B) Correlation heatmap of pairwise comparisons between median signals for all histone modifications at rDNA in K562 cells. Heatmaps for HUVEC, H1-hESC and NHEK are presented in Supplementary Figure S4. (C) Normalized tag density for histone modifications. Normalized tag density was determined by dividing the average intensity of the three highest peaks at rDNA by the average intensity of the 100 highest peaks along chromosome 13.
To address the significance of the ChIP-seq signals at rDNA, we compared the intensity of peaks at the rDNA to peaks on nucleoplasmic chromatin. One might predict, since sequence reads were aligned to a genome assembly containing only one copy of the rDNA, that signals at rDNA would appear inflated relative to nucleoplasmic chromatin. However, there are a number of unknown factors that could influence the signal at rDNA, including but not limited to ChIP efficiency, the proportion of active versus repressive rDNA loci, the copy number of rDNA in a given cell type, and the scaling of signals at high-copy sequences during peak detection. Nevertheless, we would expect genuine ChIP-seq signals at rDNA to be at least as intense as nucleoplasmic signals. Indeed, the intensities of all histone signals at rDNA were equal to or higher than those at nucleoplasmic genes (Figure 1C). Similar results were found for ChIP-seq signals analyzed in additional cell types.
Visual inspection of the data shows that the active marks tend to show similar profiles, an observation that is also apparent for the repressive marks. To more systematically address this in an unbiased fashion, the data were median-smoothed in 100-bp windows, and correlation scores between all nine histone modifications were calculated. The correlation scores were then hierarchially clustered and plotted in a heatmap. The results clearly show that, with the exception of H3K36me3, the active and repressive modifications separate into two distinct groups (Figure 1B and Supplementary Figure S4). These data indicate that overall, active histone marks are distributed similarly along the rDNA in K562 cells, and that repressive marks tend to correlate more with one another than with active marks.
Cell-type specificity of histone marks at rDNA
We next investigated whether the distribution of histone modifications at rDNA in K562 cells is similar to that in other cell types, including HUVEC (human umbilical vein endothelial cells), H1-hESC (human embryonic stem cells), and NHEK (normal human epidermal keratinocytes). Similar to K562 cells, enrichment of the active marks H3K4me1/2/3 and H3K9ac was detected at the promoter of rDNA and at the 28–29 kb site located in the IGS. HUVEC and NHEK cells also show enrichment of H3K27ac at the TSS (data not available for H1-hESC). H1-hESC shows similar enrichment of H3K4me1/2/3 and H3K9ac at the promoter and IGS, but shows additional peaks within the IGS at ∼15 and ∼20 kb. With respect to the repressive histone modifications, there is less similarity to K562, and in fact, each cell type shows a fairly specific pattern of enrichment (Supplementary Figure S2). Overall, the data indicate that each histone mark shows some level of cell-type specificity, though cluster analyses reveal distinct groupings of active and repressive marks in each cell type that are similar to K562 (Supplementary Figure S4).
Visual inspection of the data suggests that the distribution of active marks tends to be more consistent among the four cell types than repressive marks. To test this more systematically, we performed pairwise linear regression analyses between all four cell types for each modification and plotted the average R2 values on a color scale (Figure 2A). The results indicate that the three active modifications associated with the promoter and IGS of rDNA (H3K4me2/3 and H3K9ac) are most conserved between cell types. H3K27ac and H3K4me1, marks that localize to gene enhancer elements on nucleoplasmic chromatin, are the next most conserved. This scenario is reminiscent of that in the nucleoplasm, where marks associated with promoters tend to be invariant between cell types while marks associated with enhancers display cell-type specificity (34). The repressive marks and H3K36me3 show the lowest degree of correlation between cell types. Three-dimensional plots of H3K4me2 and H3K27me3 ChIP-seq data in all cell lines analyzed are presented to illustrate this point (Figure 2B).
Figure 2.
Comparison of rDNA histone modifications in multiple cell types. (A) Gradient plot representing average correlation scores for each histone modification at rDNA. (B) Three-dimensional representations of H3K4me2 and H3K27me3 ChIP-seq data, illustrating the general correlation of active modifications and lack of correlation of repressive modifications between cell types.
Chromatin accessibility and transcription of rDNA
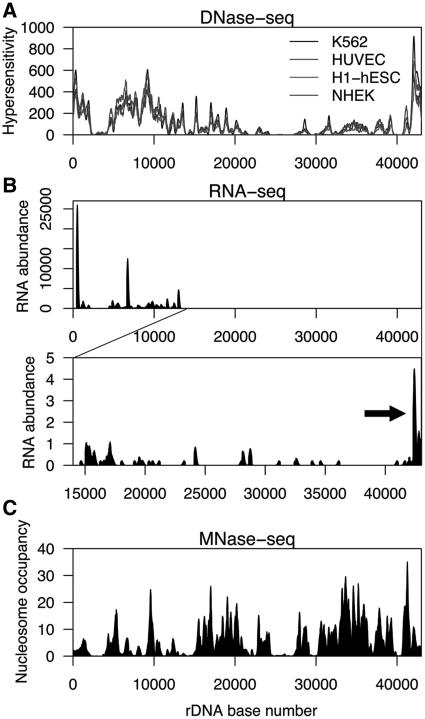
rDNA has an exceptionally high transcriptional output, producing up to 80% of all RNA in a cell (1). This fact suggests that the chromatin of rDNA is readily accessible to the Pol I machinery and therefore likely to be open. The results of DNase-seq analysis, which measures accessibility of chromatin based on sensitivity to DNase I digestion, indicate that the coding region of rDNA, parts of the IGS, and the promoter region are in an open state (Figure 3A). DNase hypersensitivity was also highly correlated between cell types (R2 = 0.88–0.97; Supplementary Table S1).
Figure 3.
Chromatin accessibility, transcription, and nucleosome occupancy at rDNA. (A) Profile of DNase hypersensitivity at rDNA in K562, HUVEC, H1-hESC and NHEK cells. The DNA sequence of the region at 2–5 kb contains large stretches of high identity to other genomic regions, and thus reads matching this region would have been discarded during alignment. Therefore, the reduction in signal at 2–5 kb is likely a false negative. (B) Nucleolar small RNA-seq profile of rDNA in K562 cells. The lower panel shows a zoomed-in view of rDNA from ∼14–43 kb with a reduced y-axis to show low-abundance RNAs. RNA-seq signal corresponding to pRNA is indicated by an arrow. (C) Profile of nucleosome occupancy at rDNA in CD4 + T cells as determined by MNase-seq.
We next assessed the levels of transcripts emanating from rDNA using small RNA-seq data from K562 nucleoli. As expected, extremely high levels of RNA corresponding to the coding region of rDNA were detected. It is also known that ∼150 nt rDNA promoter-associated RNAs (pRNAs) are involved in rDNA silencing. These transcripts are produced from a spacer promoter located ∼2 kb upstream of the core rDNA promoter (35–37). We therefore adjusted the y-axis scale of the RNA-seq data to look for pRNA and other potential low-abundance rDNA transcripts. We detected RNA signal corresponding to pRNA at the promoter region (Figure 3B), as well as weak signals throughout the IGS. Notably, one such signal is located ∼28–29 kb into the repeat, at the same location where high enrichment of active histone marks was detected (Figure 1A and Supplementary Figure S2). We next assessed the strand specificity of the mapped RNA-seq reads. In total, 75.5% (335 816/444 558) of nucleolar RNA-seq reads mapped uniquely to rDNA. Of these, only 16 mapped to the negative strand. We therefore conclude that, under normal cellular conditions, transcription of rDNA predominantly occurs in the sense direction. These results are consistent with previous studies assessing the strandedness of rDNA transcription (38).
Nucleosome occupancy of rDNA
The IGS of both active and inactive rDNA loci contains nucleosomes (12). However, whether the coding region of active rDNA repeats contains nucleosomes or is nucleosome-free is controversial (33). We assessed nucleosome occupancy of rDNA using data obtained by high-throughput sequencing after micrococcal nuclease digestion (MNase-seq) in CD4 + T cells (20) (Figure 3C). On average, median signal across the coding region was ∼2.2-fold lower than that across the IGS of the locus (3.19 versus 7.01, P = 1.18 × 10−9). We therefore conclude that, overall, the coding region of rDNA has reduced nucleosome occupancy relative to the IGS. However, because active and inactive rDNA repeats were sampled together in the MNase-seq assay, we cannot discern whether the coding region of active rDNA repeats is completely nucleosome-free and the residual signal is due to nucleosome occupancy only at inactive rDNA loci, or if the coding regions of both active and inactive copies of rDNA are nucleosome-depleted relative to the IGS.
ChIP-seq analysis of Pol I chromatin association
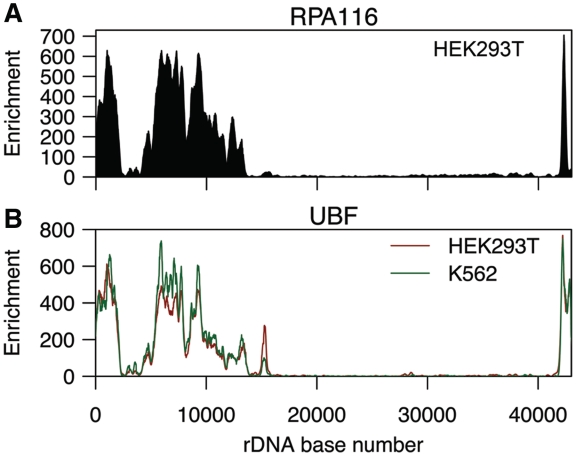
Transcription of rRNA is mediated by Pol I and its associated basal transcription machinery (14). ChIP-seq of RPA116, the second-largest subunit of the Pol I, showed high enrichment of Pol I across the promoter and coding region of rDNA (Figure 4A). This result is consistent with previous studies (39,40) and was validated by ChIP-PCR (Supplementary Figure S5A and B). We also detected 31 RPA116 peaks on non-rDNA chromatin. Many of these nucleoplasmic peaks were located close to centromeres, a known source of false positives in ChIP-seq experiments (41). Most of the nucleoplasmic peaks also appeared irregular, with a jagged, discontinuous distribution of tags rather than the smooth gradations in tag density seen at legitimate peaks. We therefore conclude that the nucleoplasmic signals are artifacts, and that Pol I is exclusively nucleolar.
Figure 4.
ChIP-seq analysis of Pol I and UBF. (A) ChIP-seq profile of the RPA116 subunit of Pol I at rDNA in HEK293T cells. (B) ChIP-seq profile of UBF at rDNA in HEK293T and K562 cells.
ChIP-seq analysis of UBF chromatin association
UBF (upstream binding factor) maintains the rDNA promoter in an open state and is necessary for the formation of the Pol I preinitiation complex (1,40,42). ChIP-seq of UBF revealed substantial enrichment in the promoter and coding region of rDNA (Figure 4B), consistent with previous work (39,40) and confirmatory ChIP-PCR assays (Supplementary Figure S5C and D). The distributions of UBF and RPA116 were highly similar to one another in HEK293T cells (R2 = 0.87). UBF binding in HEK293T and K562 was also strongly correlated (R2 = 0.94), as was HEK293T UBF and K562 RPA116 binding (R2 = 0.88).
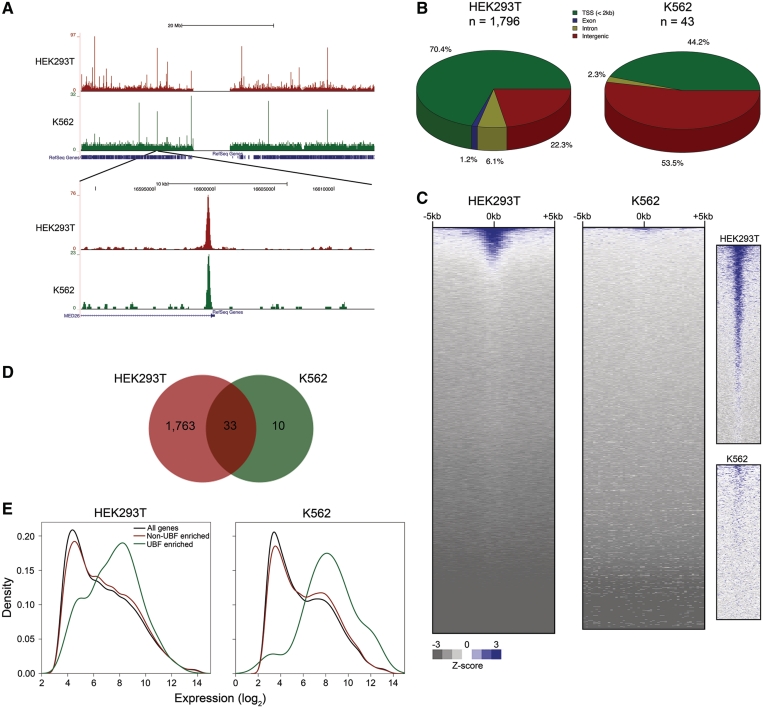
UBF has previously been shown to bind the CCND1 promoter and activate β-catenin-responsive reporter genes, suggesting a non-nucleolar function for UBF in Wnt signaling (43,44). We detected robust enrichment of UBF on nucleoplasmic chromatin in HEK293T (1796 peaks) and K562 (43 peaks) cells (Figure 5A, Supplementary Data 1). The association of UBF with nucleoplasmic chromatin was confirmed by standard ChIP-PCR with two different antibodies (Supplementary Figure S6). We also verified specificity by performing UBF ChIP-PCR following knockdown of UBF (Supplementary Figure S7). In HEK293T cells, 70.4% of the UBF peaks were located within 2 kb of a RefSeq TSS, 22.3% were intergenic, and 7.3% were intragenic. A smaller fraction of UBF sites (44.2%) in K562 were located near TSSs, while the majority (53.5%) of sites were located in intergenic regions (Figure 5B). Analysis of UBF binding to all TSSs in the genome revealed that UBF was bound to ∼10% and ∼1% of all TSSs in HEK293T and K562, respectively, suggesting that some UBF TSS binding events are slightly below the FDR threshold used (Figure 5C). Of the UBF peaks detected in K562 cells, 33/43 (76.7%) overlapped with those detected in HEK293T. (Figure 5D).
Figure 5.
UBF is associated with nucleoplasmic chromatin. (A) UCSC Genome Browser view of UBF binding on human chromosome 19 in HEK293T and K562 cells. A zoomed-in view is shown in the lower panel. (B) Distribution of UBF binding sites with respect to RefSeq genes. (C) UBF signal ± 5 kb of all unique TSSs in the human genome in descending order of average signal intensity. The panels on the right show a zoomed-in view of the topmost area of each heatmap, showing details of UBF signal in each cell type. (D) Venn diagram indicating overlap of UBF binding sites in HEK293T and K562 cells. (E) Density histograms of gene expression levels for all genes, genes with significant UBF peaks <2 kb from their TSS, or genes with low or no UBF binding at their TSS. HEK293T UBF enriched versus all genes, P = 1.85 × 10−28; HEK293T UBF enriched versus non-enriched genes, P = 3.29 × 10−21; K562 UBF enriched vs. all genes, P = 0.0013; K562 UBF enriched versus non-enriched genes, P = 0.0031 by t-test.
We next correlated the UBF ChIP-seq data to gene expression. The results indicate that genes containing high levels of UBF at the TSS are generally expressed at relatively high levels (Figure 5E). Transcription factors and genes involved in developmental processes and nucleobase metabolism were significantly over-represented among genes with a significant UBF peak at their TSS in HEK293T (Supplementary Data 2). Despite the low number of UBF sites in K562 cells, gene ontology analysis revealed that transcription factors and nucleobase metabolism genes were significantly over-represented (Supplementary Data 2). Genes encoding nucleolar and ribosomal proteins were not over-represented among UBF-bound genes in either cell type. Interestingly, in HEK293T cells, genes involved in Wnt signaling were significantly over-represented among UBF-enriched genes, in support of previous studies suggesting a role for UBF in β-catenin signaling (43,44). Motif analysis revealed 150 and 56 transcription factor binding motifs within significant UBF peaks in HEK293T and K562, respectively (Supplementary Data 3). The sequence logos and functions associated with the five most significantly enriched motifs in HEK293T and K562 are shown in Supplementary Tables 2 and 3, respectively. These results raise the possibility that UBF collaborates with a variety of cofactors to regulate nucleoplasmic transcription.
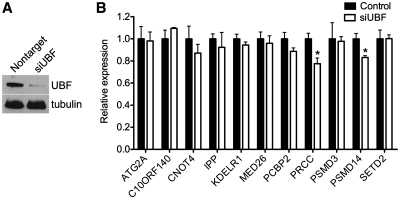
To test whether binding of UBF to nucleoplasmic genes has a functional effect, we knocked down UBF with siRNA (Figure 6A) and quantified transcript levels of 11 UBF-bound genes. Two of the 11 genes (PRCC and PSMD14) showed a modest, but significant decrease in expression 48 h after UBF knockdown in HEK293 cells (Figure 6B). No genes were increased upon UBF knockdown. These data suggest that UBF functions to modestly increase expression of a subset of nucleoplasmic genes. Further studies are required to determine whether the effect of UBF on nucleoplasmic transcription is generally modest, if other targets are more dramatically affected upon UBF depletion, or if complete loss of UBF has a more dramatic effect.
Figure 6.
UBF regulates nucleoplasmic gene transcription. (A) Western blot showing depletion of UBF protein 48 h after siRNA transfection in HEK293T cells. (B) Expression analysis of nucleoplasmic genes 48 h after UBF siRNA transfection in HEK293T cells. Error bars represent mean + SEM (n = 3). *, P < 0.05 by t-test.
The insulator-binding protein CTCF associates with rDNA
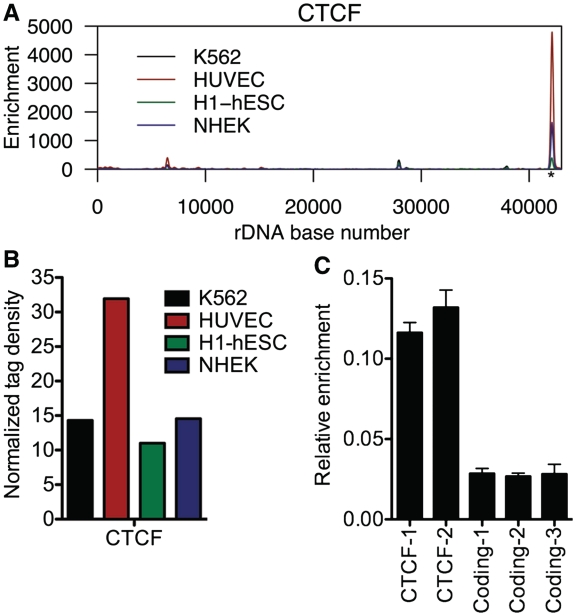
Because rDNA is organized into tandemly repeated arrays (NORs), it stands to reason that there is a mechanism to prevent leaky transcription between adjacent repeats. A candidate for such a mechanism could be an insulator element. Insulator elements generally function to demarcate discrete transcriptional units and prevent inappropriate transcription (45). A large proportion of conserved DNase hypersensitive sites overlap with CTCF, a well-characterized insulator-binding protein (46). Additionally, CTCF has been shown to localize to the nucleolus and repress rRNA transcription (47). We therefore aligned CTCF ChIP-seq data from K562, HUVEC, H1-hESC and NHEK cells to HG18_plus_rDNA and found that CTCF was highly enriched at the 3′ end of rDNA, at the spacer promoter (Figure 7A). The binding pattern of CTCF was highly consistent across multiple cell types (R2 = 0.76–0.99; Supplementary Table S1), suggesting that this CTCF binding site serves an important, conserved function. We also aligned CTCF ChIP-seq data from mouse ES cells to a build of the mouse genome containing an rDNA repeat and observed CTCF binding at the spacer promoter of rDNA (Supplementary Figure S8). Notably, an independent laboratory has also recently observed CTCF binding to human and mouse rDNA at a site similar to that observed in our analyses, further validating our findings (48).
Figure 7.
The insulator-binding protein CTCF is associated with rDNA. (A) ChIP-seq profile of CTCF at rDNA in K562, HUVEC, H1-hESC and NHEK cells. The putative CTCF motif is indicated by an asterisk. (B) Normalized tag density scores for CTCF at rDNA. (C) ChIP-PCR validation of CTCF rDNA association in K562 cells using two primer sets under the CTCF ChIP-seq peak. Error bars represent mean + SD for triplicates.
We searched the sequence under the CTCF peak in human and mouse rDNA for consensus binding motifs using the CTCF Binding Site Database (CTCFBSDB) (49). Three of four positional weight matrices used by CTCFBSDB yielded scores indicating a suggestive match for a CTCF consensus binding site in human rDNA (Supplementary Table S4A), and all positional weight matrices yielded suggestive scores for a CTCF site in mouse rDNA (Supplementary Table S4B). Normalized tag density for CTCF was ∼11–32, suggesting that CTCF is present at many copies of rDNA (Figure 7B). ChIP-PCR confirmed the association of CTCF with rDNA in K562 cells (Figure 7C). Combined with our analysis of DNase-seq data demonstrating hypersensitivity at this location (Figure 3A), these results suggest that the 3′ of rDNA, in addition to promoting the transcription of rRNA, also acts as an insulator element to demarcate the boundaries of each rDNA repeat and/or repress rRNA transcription. This model is supported by a previous study demonstrating CTCF-mediated transcriptional repression of rDNA in human cells (47). However, as this study was under review, a report was published demonstrating that CTCF promotes association of UBF, Pol I, and active histone marks with the rDNA spacer promoter in mouse and human cells (48). Therefore, CTCF may act in a context-dependent manner to regulate rDNA transcription in both the positive and negative directions.
DISCUSSION
The rDNA has posed significant obstacles to genomic analysis and has thus far been analyzed only by standard ChIP and other biochemical techniques. Here, we integrated ChIP-seq, DNase-seq, MNase-seq and RNA-seq data sets to assemble a high-resolution map of chromatin structure at the human rDNA locus that verifies and expands the results of previous rDNA ChIP studies. Additionally, our results complement those of previous genome-wide ChIP-seq studies by focusing on a critically important region of the genome that has thus far not been analyzed by this method. We present four novel findings. First, histone modifications at rDNA are located not only at the promoters, but also at sites within the IGS. Second, the distributions of active modifications are more similar within and between different cell types than repressive modifications. Third, UBF, primarily a nucleolar transcription factor, binds to many sites on nucleoplasmic chromatin. Fourth, the insulator binding protein CTCF associates with rDNA, at a site situated between the spacer and core promoters, suggesting that transcriptional insulation plays a role in regulation of rRNA transcription.
The significance of the peak of active modifications present in multiple cell types at the ∼28–29 kb site within the IGS is not yet clear. The sequence composition of this region is unremarkable, with a ∼55% GC content and 3.8% of bases masked due to the presence of a simple repeat. This region also contains a stretch of DNA similar to a clone hypomethylated in sperm but not somatic tissues (50). It is possible that this region harbors an as yet uncharacterized transcription unit. Another possibility is that this region harbors a novel functional element associated with active transcription, such as an enhancer. Enhancers are generally marked with H3K4me1/2, located distal to TSSs, and usually hypersensitive to DNase digestion (34). The 28–29 kb IGS site has these same characteristics. Further studies such as reporter assays using this region of rDNA could be useful in addressing this further. It is important to note that these two possibilities are not mutually exclusive, as it has recently been shown that active enhancers can produce short transcripts (51).
UBF, well known as a regulator of rRNA transcription, is generally thought to be a nucleolus-specific factor. However, previous studies have suggested that UBF may function outside the nucleolus as a mediator of β-catenin signaling (43,44). We find that UBF is enriched throughout the genome, suggesting that UBF may function in a broader range of cellular processes than previously appreciated. We show that siRNA-mediated depletion of UBF leads to a modest decrease in the expression of two nucleoplasmic genes, PRCC and PSMD14. While the majority of cellular UBF is likely to function in rRNA transcription, we propose that a small amount of UBF may have an extranucleolar transcriptional role.
There are limitations associated with this study. First, because we have included only a single rDNA repeat in our genome assembly, all sequences aligning to rDNA will ‘pile up’ at the single copy of rDNA; thus, the signals we observe are an aggregate of signals at all rDNA copies immunoprecipitated in each ChIP experiment. We are therefore unable to determine how many copies of rDNA contain a particular histone mark or are bound by a given protein. Another limitation of this study is that the data sets we have analyzed likely contain a mixed population of active and inactive rDNA repeats, and thus, the data represent an average of these two populations. Therefore, we cannot definitively conclude whether the distinct patterns of active and repressive histone marks seen occur on independent repeats or if they coexist on the same repeat. However, previous studies combining ChIP with methylation-sensitive restriction digest (ChIP-chop) and bisulfite sequencing have demonstrated that marks associated with transcriptional activation (H3K4me, H3K9ac and H4ac) tend to associate with DNA-hypomethylated, ostensibly active repeats, while repressive modifications (H3K9me, H4K20me) associate with hypermethylated, silent repeats (10,12,52–54). We therefore suggest that the distinct patterns of modifications we observe occur on independent repeats.
Taken together, our analyses provide the first high-resolution picture of chromatin structure at rDNA. Our results provide novel insight into a region not previously studied by ChIP-seq and serve as a reference for further studies of chromatin-mediated regulation of rDNA. Future studies focusing on the regulatory potential of the rDNA CTCF binding site and the peak of active modifications ∼28–29 kb into the rDNA repeat will be particularly informative in delineating novel modes of chromatin-level regulation at this critical region of the genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institute of General Medical Sciences (5T32GM008613-14 to G.E.Z.); the National Institute of Child Health and Development (R01HD056369 to P.C.S.); National Human Genome Research Institute (5R01HG004722 to P.C.S.). Funding for open access charge: National Institute of Child Health and Development (R01HD056369); National Human Genome Research Institute (5R01HG004722).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ingrid Grummt for the RPA116 antibody. We also thank Batool Akhtar-Zaidi, Greg Crawford, Terry Furey, and Iris Gonzalez for helpful discussions during the course of this project and Stephanie Doerner for helpful comments on the manuscript.
REFERENCES
- 1.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Opferman JT, Zambetti GP. Translational research? Ribosome integrity and a new p53 tumor suppressor checkpoint. Cell Death Differ. 2006;13:898–901. doi: 10.1038/sj.cdd.4401923. [DOI] [PubMed] [Google Scholar]
- 3.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon J, Trainor P, Dixon MJ. Treacher Collins syndrome. Orthod. Craniofac. Res. 2007;10:88–95. doi: 10.1111/j.1601-6343.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 5.Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes P-E. Impaired ribosome biogenesis in Diamond–Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridanpää M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 8.Ganapathi KA, Austin KM, Lee C-S, Dias A, Malsch MM, Reed R, Shimamura A. The human Shwachman–Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007;110:1458–1465. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokai I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 10.Zentner GE, Hurd EA, Schnetz MP, Handoko L, Wang C, Wang Z, Wei C, Tesar PJ, Hatzoglou M, Martin DM, et al. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum. Mol. Genet. 2010;19:3491–3501. doi: 10.1093/hmg/ddq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 12.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 13.Grummt I. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum. Mol. Genet. 2007;16:R21–R27. doi: 10.1093/hmg/ddm020. [DOI] [PubMed] [Google Scholar]
- 14.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 15.Russell J, Zomerdijk JCBM. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henras A, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life. Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 18.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein–DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schones DE, Cui K, Cuddapah S, Roh T-Y, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blahnik KR, Dou L, O'Geen H, McPhillips T, Xu X, Cao AR, Iyengar S, Nicolet CM, Ludascher B, Korf I, et al. Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res. 2010;38:e13. doi: 10.1093/nar/gkp1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–W554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 29.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 30.Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sogo JM, Ness PJ, Widmer RM, Parish RW, Koller T. Psoralen-crosslinking of DNA as a probe for the structure of active nucleolar chromatin. J. Mol. Biol. 1984;178:897–919. doi: 10.1016/0022-2836(84)90318-8. [DOI] [PubMed] [Google Scholar]
- 32.Lucchini R, Pauli U, Braun R, Koller T, Sogo JM. Structure of the extrachromosomal ribosomal RNA chromatin of Physarum polycephalum. J. Mol. Biol. 1987;196:829–843. doi: 10.1016/0022-2836(87)90408-6. [DOI] [PubMed] [Google Scholar]
- 33.Conconi A, Widmer RM, Koller T, Sogo J. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 34.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 35.Sylvester JE, Gonzalez IL, Mougey EB. In: The Nucleolus. Olson M, editor. Georgetown, TX: Landes Bioscience; 2003. pp. 58–73. [Google Scholar]
- 36.Mayer C, Schmitz K-M, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 37.Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gagnon-Kugler T, Langlois F, Stefanovsky V, Lessard F, Moss T. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell. 2009;35:414–425. doi: 10.1016/j.molcel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 39.O'Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002;22:657–658. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nix D, Courdy S, Boucher K. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rRNA chromatin. Epigenetics. 2009;4:374–382. doi: 10.4161/epi.4.6.9449. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Wu A, Sun H, Drakas R, Garofalo C, Cascio S, Surmacz E, Baserga R. Functional significance of type 1 insulin-like growth factor-mediated nuclear translocation of the insulin receptor substrate-1 and β-catenin. J. Biol. Chem. 2005;280:29912–29920. doi: 10.1074/jbc.M504516200. [DOI] [PubMed] [Google Scholar]
- 44.Grueneberg DA, Pablo L, Hu K-Q, August P, Weng Z, Papkoff J. A functional screen in human cells identifies UBF2 as an RNA polymerase II transcription factor that enhances the β-catenin signaling pathway. Mol. Cell. Biol. 2003;23:3936–3950. doi: 10.1128/MCB.23.11.3936-3950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 46.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torrano V, Navascues J, Docquier F, Zhang R, Burke LJ, Chernukhin I, Farrar D, Leon J, Berciano MT, Renkawitz R, et al. Targeting of CTCF to the nucleolus inhibits nucleolar transcription through a poly(ADP-ribosyl)ation-dependent mechanism. J. Cell Sci. 2006;119:1746–1759. doi: 10.1242/jcs.02890. [DOI] [PubMed] [Google Scholar]
- 48.van de Nobelen S, Rosa-Garrido M, Leers J, Heath H, Soochit W, Joosen L, Jonkers I, Demmers J, van der Reijden M, Torrano V, et al. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenet. Chromat. 2010;3 doi: 10.1186/1756-8935-3-19. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao L, Zhou M, Cui Y. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 2008;36:D83–D87. doi: 10.1093/nar/gkm875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X-Y, Loflin PT, Gehrke CW, Andrews PA, Ehrlich M. Hypermethylation of human DNA sequences in embryonal carcinoma cells and somatic tissues but not in sperm. Nucleic Acids Res. 1987;15:9429–9449. doi: 10.1093/nar/15.22.9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y, Schmitz K-M, Mayer C, Yuan X, Akhtar A, Grummt I. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat. Cell Biol. 2009;11:1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 54.Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS ONE. 5:e12175. doi: 10.1371/journal.pone.0012175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.