Abstract
The Sm protein Hfq binds small non-coding RNA (sRNAs) in bacteria and facilitates their base pairing with mRNA targets. Molecular beacons and a 16 nt RNA derived from the Hfq binding site in DsrA sRNA were used to investigate how Hfq accelerates base pairing between complementary strands of RNA. Stopped-flow fluorescence experiments showed that annealing became faster with Hfq concentration but was impaired by mutations in RNA binding sites on either face of the Hfq ring or by competition with excess RNA substrate. A fast bimolecular Hfq binding step (∼108 M−1s−1) observed with Cy3-Hfq was followed by a slow transition (0.5 s−1) to a stable Hfq–RNA complex that exchanges RNA ligands more slowly. Release of Hfq upon addition of complementary RNA was faster than duplex formation, suggesting that the nucleic acid strands dissociate from Hfq before base pairing is complete. A working model is presented in which rapid co-binding and release of two RNA strands from the Hfq ternary complex accelerates helix initiation 10 000 times above the Hfq-independent rate. Thus, Hfq acts to overcome barriers to helix initiation, but the net reaction flux depends on how tightly Hfq binds the reactants and products and the potential for unproductive binding interactions.
INTRODUCTION
Small non-coding RNA (sRNAs) form gene regulatory networks in bacteria that control the response to cellular stress such as cold shock, oxidative stress, iron levels, sugar metabolism and quorum sensing (1–3). An important class of sRNAs control gene expression by base pairing with complementary sequences in their mRNA targets, inhibiting or activating translation initiation by covering or exposing the ribosome binding site (1,4,5). Binding of sRNAs can also increase degradation of the mRNA by RNase E (5–7).
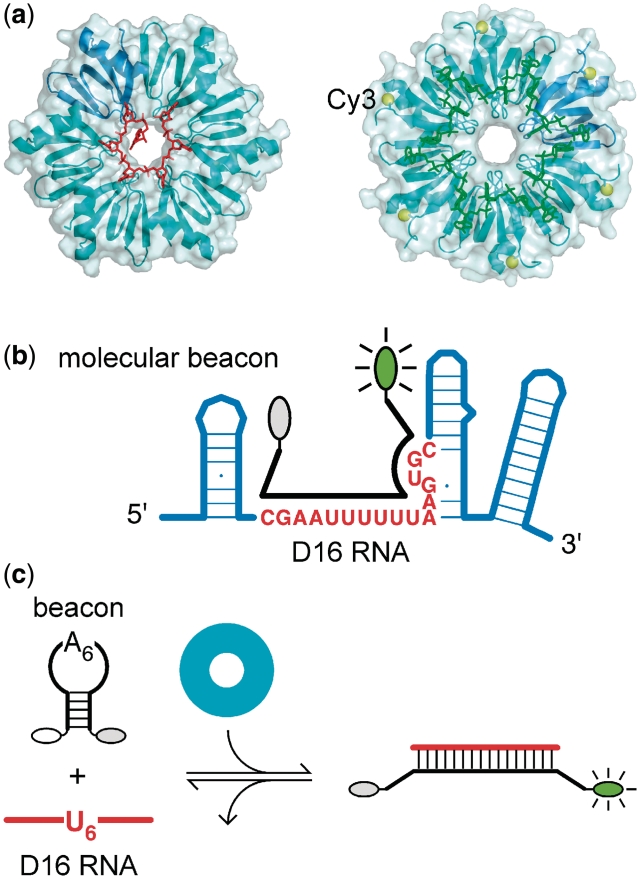
Nearly all sRNAs that act via this antisense mechanism require the RNA chaperone protein Hfq for their regulatory function (8,9). Hfq is an abundant Escherichia coli protein that binds and stabilizes the sRNAs, and facilitates base pairing between sRNAs and their mRNA targets (10). A member of the Sm family of RNA binding proteins, Hfq forms a ring-shaped homohexamer (11) that binds 6 nt of U-rich single-stranded RNA around the inner ‘proximal’ surface of the ring (Figure 1a, left) (12,13). Biochemical experiments (14–16) and a second crystallographic structure showed that the distal face of Hfq binds A-rich sequences, with 3 nt per monomer (Figure 1a, right) (17).
Figure 1.
Molecular beacon assay for RNA annealing and strand exchange. (a) Left, structure of Staphylococcus aureus Hfq in complex with U-rich RNA (red) (13). One monomer is shown in darker blue. Right, structure of E. coli Hfq in complex with A18 (green) (17). E. coli Hfq was modified with Cy3 at position 65 (yellow spheres). (b) 16-nt RNA substrate (D16, red letters) contains the U-rich Hfq binding site of DsrA sRNA. Molecular beacons dMB-D16 or rMB-D16 (black) were modified with 5′-FAM and 3′-DABCYL. The loop of the molecular beacon is exactly complementary to D16 RNA. (c) Annealing of D16 RNA and the beacon produces a rise in fluorescence and is facilitated by Hfq (cyan).
How Hfq facilitates base pairing between complementary sequences in mRNAs and sRNA is not understood. One model is that simultaneous binding of two RNA strands to Hfq (on the same face or opposite faces of the hexamer) enhances base pairing by bringing the RNAs closer together (15,18). Alternatively, Hfq may act primarily by unfolding secondary structures in one or both RNA partners.
To understand how Hfq acts in sRNA regulation, we have investigated how Hfq promotes association of DsrA sRNA with the rpoS mRNA leader, which up-regulates translation of RpoS stress response regulator (19,20). Hfq binds six U’s in DsrA through its proximal RNA binding site (15,21) (Figure 1b). Hfq is also recruited to the rpoS mRNA via an upstream (AAN)n sequence motif that interacts with the Hfqs distal face (22) and is important for regulation of rpoS translation in Escherichia coli by sRNAs (23). A similar interaction has been identified in fhlA mRNA, which is regulated by OxyS sRNA (24).
The ability of Hfq to make multiple interactions with its natural RNA substrates complicates the study of its mechanism. To understand how Hfq accelerates the formation of RNA double helices, apart from its interactions with the upstream (AAN)4 site in rpoS mRNA, we designed an unstructured 16 nt RNA substrate containing the U-rich Hfq-binding site in DsrA (D16; Figure 1b) (25). Molecular beacons complementary to D16 RNA were derivatized with fluorescein (6-FAM) on the 5′-end and DABCYL on the 3′-end, such that base pairing with D16 RNA produces a large increase in fluorescence intensity (Figure 1c). An advantage of this assay is that the increase in beacon fluorescence directly reports base pairing with the D16 substrate RNA. In contrast to FRET assays for RNA annealing (26), this method does not detect ternary complexes in which two unpaired strands are bridged by Hfq.
We previously showed that Hfq increases the rate of RNA annealing up to 100-fold and that Hfq-dependent annealing is fastest when at least one strand binds Hfq weakly (25). The inverse correlation between binding affinity and annealing kinetics, plus the observation that high Hfq concentrations inhibit association of DsrA and rpoS mRNA, led us to propose that Hfq must dissociate from the incipient double helix during annealing (27). In this model, Hfq cycles on and off its RNA substrates during each round of annealing and strand exchange, with the net reaction flux depending on the strength of competing Hfq–RNA interactions and the stability of the product RNA duplex.
Here, we use D16 RNA and Cy3-labeled Hfq to measure the kinetics of individual steps in the Hfq-dependent RNA annealing reaction. We show that Hfq binds nucleic acid strands rapidly (∼108 M−1s−1), forming an unstable ternary complex from which Hfq dissociates rapidly, most likely before annealing is complete. Mutations that weaken RNA binding to the distal and proximal faces of Hfq reduce its RNA annealing activity as expected, consistent with RNA binding to both sides of Hfq.
MATERIALS AND METHODS
Molecular beacons and RNA oligomers
Oligoribonucleotides (Invitrogen) were purified by 8% PAGE and dissolved in water. D16: 5′ rCGAAUUUUUUAAGUGC; R16: 5′ rGCACUUAAAAAAUUCG. Molecular beacons and 5′ carboxyfluorescein D16 (D16-FAM) were synthesized and purified by reverse phase HPLC (Trilink Biotechnologies) as previously described (25). dMB-D16: 5′ (6-FAM) dCCAGGGCACTTAAAAAATTCGCCTGG (C6-NH-DABCYL) 3′; rMB-D16: 5′ (6-FAM) rCCAGGGCACUUAAAAAAUUCGCCUGG (C6-NH-DABCYL) 3′; dMB-R16 5′ (6-FAM) dCCCCTCGAATTTTTTAAGTGCAGGGG (C6-NH-DABCYL) 3′. Concentrations were determined by absorption at 260 nm; D16 ε260 = 172.3 OD µ−1mol−1; R16 ε260 = 187.7 OD µ−1mol−1; dMB-D16 ε260 = 289.6 OD µ−1mol−1; rMB-D16 ε260 = 295.6 OD µ−1mol−1; dMB-R16 ε260 = 283.4 OD µ−1mol−1.
Hfq purification
His-tagged E. coli Hfq and its K56A and Y25D variants were expressed from plasmids kindly provided by A. Feig (Wayne State University), and purified as previously described (15). Wild-type (untagged) Hfq was over-expressed as previously described (11) and purified using a Hi-Trap Co2+ column as previously described (23). Natural and his-tagged Hfq preparations had similar activities on oligonucleotide substrates, and were free of cellular RNA based on the relative absorption at 260 and 280 nm. Hfq-Cy3 was prepared by substituting Ser 65 in wild-type Hfq with cysteine (QuikChange; Strategene). Hfq:S65C was treated with Cy3 maleimide (GE Healthcare) according to the manufacturer’s protocol and re-purified by Hi-Trap Co2+ chromatography (23). The extent of labeling was estimated from absorbance at 280 and 550 nm and was typically 20–30%. Variations in the extent of labeling did not measurably change the observed binding kinetics.
Beacon hybridization kinetics
The hybridization kinetics in TNK buffer (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 50 mM KCl) or HB buffer (10 mM Tris–HCl pH 7.5, 50 mM NH4Cl, 0.2 mM EDTA, 2% glycerol) at 30°C was measured using an Applied Photophysics SX 18MV stopped-flow spectrometer as previously described (25). Reactions typically contained 50 nM molecular beacon (final), 100 nM target RNA and 50–830 nM Hfq6 as stated in the text and figure legends. Excitation was at 496 nm and the emission intensity was recorded using a 515-nm cutoff filter.
The normalized change in fluorescence, ΔF(t), was fit to a double exponential rate equation,
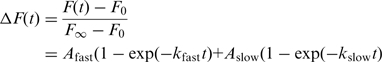 |
(1) |
and the observed rate constants for three or more trials were averaged. For the purposes of simulation, absolute amplitudes of annealing reactions in the presence of Hfq were determined from the decrease in equilibrium fluorescence intensity, when 50 nM dMB-D16 plus 50 nM D16 was titrated with Hfq (Supplementary Figure S1). The fluorescence intensity without Hfq was assumed to represent 26.8 nM dMB-D16•D16, the amount of beacon–RNA duplex predicted by the 19.6 nM dissociation constant in TNK buffer (25). A linear plot of Kd apparent for beacon–RNA hybridization versus Hfq concentration, 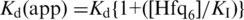 , yielded KI = 138 nM Hfq6 (Supplementary Figure S1).
, yielded KI = 138 nM Hfq6 (Supplementary Figure S1).
Fluorescence anisotropy
Hfq binding to D16-FAM RNA was determined from polarization measurements as described earlier (25). Two complexes were detected with dissociation constants of 23 and 600 nM Hfq6 in TNK buffer at 30°C (25). Similar measurements with dMB-D16 lacking the 3′-DABCYL modification yielded Kd = 1.4 µM Hfq6 in TNK, 30°C for the DNA beacon.
Hfq binding kinetics
Association of Hfq with D16 RNA was measured by stopped-flow spectroscopy as described above. 50 nM D16-FAM was mixed with 0.3–1 µM Hfq-Cy3 (monomer) in TNK, 30°C. Excitation was at 490 nm; formation of the complex was measured by the loss of donor intensity at 515 nm using a 520 ± 10 nm band pass filter; for very fast transitions, a 515 nm cut-off filter was used to improve the signal. The change in fluorescence signal was followed for 0.2 or 60 s, to capture fast and slow phases of binding. The FRET efficiency was calculated from the change in donor fluorescence, EFRET = [1−(FDA/FD)], in which FDA and FD are the donor emission in the presence and absence of the acceptor, respectively. The binding constant calculated from the change in EFRET in the presence of Hfq-Cy3 acceptor was similar to that obtained from anisotropy measurements. Addition of unlabeled RNA to Hfq-Cy3 did not change the Cy3 emission spectrum.
Release of Hfq was measured by pre-binding 100 nM D16-FAM and 0.5 µM Hfq-Cy3 (monomer) ≥5 min, then challenging the complex with 100 nM R16 RNA. Data were collected over 1 and 20 s (split time base), and the initial increase in FAM emission (≤0.2 s) fit to a single-exponential rate equation. Similar results were obtained by fitting the entire 20 s to a double exponential rate equation. Amplitudes were Afast = 40% and Aslow = 60%.
Strand-exchange
The kinetics of strand exchange was measured as described above for annealing, except that 50 nM dMB-D16 and 50 nM D16 were pre-equilibrated at 30°C, with or without 50 nM Hfq6, then mixed with 50 nM R16 in the stopped-flow spectrometer. Similar results were obtained when Hfq was added to the syringe containing R16.
Simulations
The binding kinetics of Hfq-Cy3 and D16-FL RNA were fit to the three-state mechanism in Scheme I using Berkeley Madonna, and the entire range of data was simulated using average values for microscopic rate constants for each step to determine how well they recapitulated the observed macroscopic rate constants and equilibrium binding constant. For simplicity, both D16-Hfq complexes were assumed to have the same FRET value.
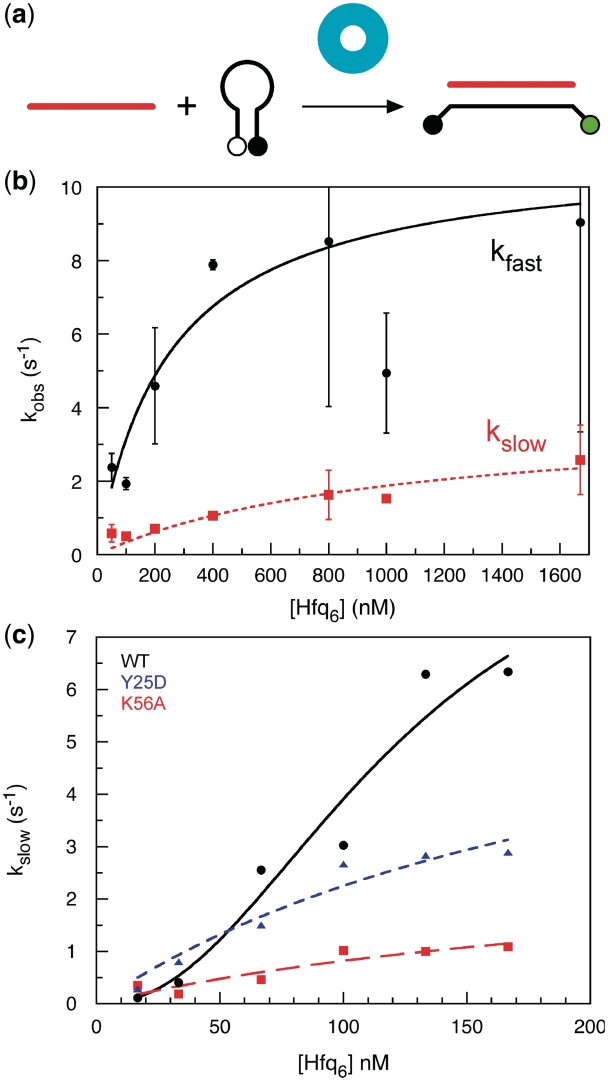
Scheme I.
Simulated kinetics of D16-dMB-D16 hybridization in TNK buffer were compared with experimental progress curves with 50–800 nM D16 and 0–600 µM Hfq6. Stopped flow traces were scaled to equilibrium concentrations of DB product as described above. Rate constants for annealing in absence of Hfq (kon = 3.4 • 105 M−1s−1, koff = 6.7 • 10−4 s−1) and for Hfq binding were measured experimentally [this work and (25)]. The rate of Hfq release was estimated from RNA competition experiments.
RESULTS
Hfq-dependence of annealing rates
To understand how Hfq accelerates base pairing between nucleic acid strands, we used D16 RNA and its complementary beacon to measure the annealing kinetics by stopped-flow fluorescence (Figure 2a) (25). The loop of the molecular beacon was the exact complement of D16 RNA and contains 6 A’s; the stem of beacon was designed to maintain low fluorescence in the absence of target without strongly inhibiting target binding. Because RNA binding and annealing by Hfq decreases with salt concentration, reactions were performed in moderate salt (TNK; 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 50 mM KCl).
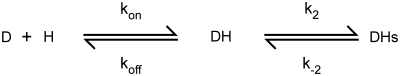
Figure 2.
Annealing rate increases with Hfq concentration. (a) Scheme as in Figure 1. (b) Observed annealing rates were measured by stopped-flow fluorescence in TNK buffer at 30°C, using 50 nM dMB-D16, 100 nM D16 RNA and 50–1600 nM Hfq hexamer. Individual progress curves were fit to Equation (1) (‘Materials and Methods’ section). Error bars show the standard deviation between trials; the small amplitude of the fast phase >600 nM Hfq6 accounts for the greater uncertainty in the rate constants. The increase in kobs was fit to a single-site binding isotherm, with [Hfq]1/2 = 250 ± 80 nM (kfast, black circles) and 1.0 ± 6 µM Hfq (kslow, red squares). (c) Effect of Hfq mutations on annealing activity. Distal face mutation Y25D (blue triangles) decreases activity less than proximal face mutation K56A (red squares). Experiments as above but with 100 nM D16 RNA in HB buffer, both of which increase overall activity. Rate constants for the slow phase are shown. Data for the wild-type Hfq were fit to a cooperative binding model.
Hfq strongly increased the rate of strand association. In the absence of Hfq, 50 nM D16 and 50 nM dMB-D16 at 30°C base pair slowly, with kfast = 0.016 s−1 and kslow = 0.0066 s−1 (25). In 600 nM Hfq6, these rates increased 100-fold, to 1.6 and 0.42 s−1, respectively (Figure 2b). Control reactions confirmed that Hfq does not unfold the beacon in the absence of target, as previously observed (25).
The slow and fast observed annealing rates appeared to saturate ∼1 µM Hfq6, and the amplitude of the fast rate decreased, suggesting that Hfq binding becomes rate-limiting (Figure 2b). Fluorescence anisotropy measurements showed that the Hfq hexamer binds the U-rich D16 RNA with a dissociation constant of 23 nM in TNK buffer (25). However, 300 nM Hfq6 was needed to reach half-maximal velocity under these conditions, more than predicted from the binding affinity of D16 RNA but less than predicted from the Kd for the DNA beacon, which was 1.4 µM (data not shown). These results are consistent with the need for Hfq to interact with both strands in order to accelerate annealing.
Requirement for interactions with both faces of Hfq
A U-rich sequence in DsrA sRNA binds the proximal face of Hfq, while an upstream A-rich motif in rpoS mRNA interacts with the distal face (15,22). To determine if our oligonucleotide substrates interact with both proximal and distal faces of Hfq, annealing reactions with the DNA beacon were repeated with Hfq variants in which one or the other RNA binding site was mutated. Reactions with Hfq mutants were done in low salt Hfq buffer (HB; 10 mM Tris–HCl pH 7.5, 50 mM NH4Cl, 0.2 mM EDTA, 2% glycerol) to enhance substrate binding and annealing (25).
The mutation K56A, which disrupts binding of DsrA to the proximal face of Hfq (15), strongly reduced annealing of D16 RNA (Figure 2c, red squares), as expected. The mutation Y25D, which reduces the affinity of A-rich RNA for the distal face of Hfq (15), also had a moderate effect on annealing activity (Figure 2c, blue triangles). Thus, both RNA binding sites contributed to the ability of Hfq to facilitate base pairing between D16 RNA and the DNA beacon.
Hfq binds single-stranded RNA rapidly
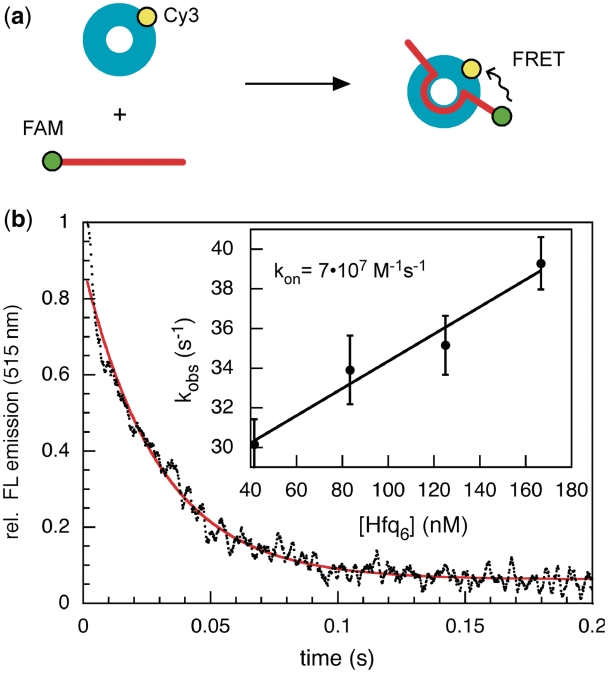
To determine how rapidly Hfq interacts with short RNAs, the binding kinetics were measured via energy transfer (FRET) from fluorescein-labeled D16 (D16-FAM) to Cy3-labeled Hfq (Figure 3a). The FAM donor emission decreased upon Hfq-Cy3 binding with an initial rate of 30–40 s−1 (Figure 3b), corresponding to an apparent second order rate constant for hexamer binding kon = 6.9 ± 0.9 • 107 M−1s−1 (Figure 3b inset). Thus, Hfq binds D16 RNA much faster than D16 hybridizes with its complementary beacon under our standard conditions (100 nM D16, 50 nM dMB-D16, 50 nM Hfq6).
Figure 3.
Rapid RNA binding kinetics of Hfq. (a) FRET assay for Hfq binding. 50 nM D16-FAM RNA (red) was mixed with 0.25–1 µM Cy3-labeled Hfq monomer (cyan). (b) The decrease in FAM emission due to FRET was detected using a 515 nm cut-off filter (black) and fit to a single exponential rate equation over 0.2 s (red line). The linear increase in kobs with [Hfq6] (inset) yielded kon ≥ 6.9 ± 0.9 • 107 M−1s−1. Error bars show the standard deviation among five trials. Due to fewer data at the shortest times, the on-rate may be slightly faster than reported here. A slower binding phase (∼0.2 s−1) was detected when the reaction was followed for 60 s (Supplementary Figure S3).
The off-rate predicted from the binding kinetics was koff = 27 s−1 (Supplementary Figure S3), 20 times faster than the dissociation rate of 1.6 s−1 predicted from the equilibrium constant of 23 nM Hfq6. When the Hfq binding reactions were followed for 60 s, however, we also observed a slow phase (0.15–0.2 s−1) that accounted for 60% of the total change in donor intensity. The slow binding phase, which did not become faster with Hfq concentration (Supplementary Figure S3), suggested that Hfq–RNA complexes undergo an additional conformational change that stabilizes the complex (Scheme I).
The three-state binding model in scheme I described the observed Hfq binding kinetics well, using the values for kon and koff above and assuming k2 = 0.5 s−1 and k−2 = 0.05 s−1 (Supplementary Figure S3). Step 2 was slower than the fast annealing rate under the same conditions and thus must not be required for strand association, although it may contribute to the slow phase of annealing. Although scheme I does not account for differences in the two RNA binding sites on Hfq or hexamer stability, a conformational change in Hfq prior to RNA binding did not explain the observed kinetics.
Rapid Hfq release during annealing
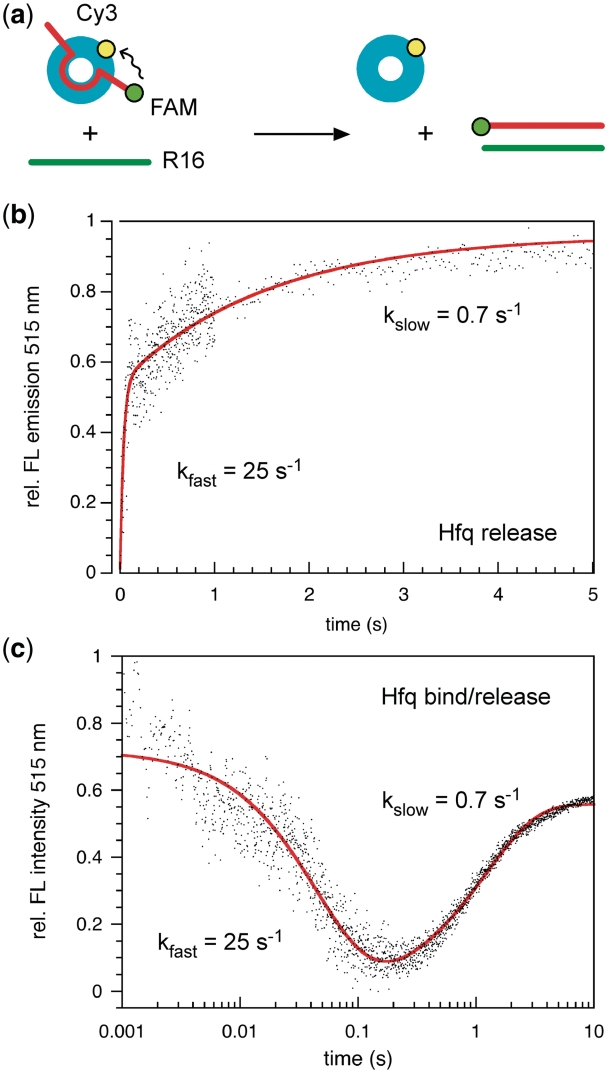
As the initial D16-Hfq complex dissociates in less than a second and the DNA beacon also interacts weakly with Hfq, we expected the Hfq–RNA-beacon ternary complex to exist only transiently in our reactions. To measure the rate of Hfq release from its substrates during the annealing cycle, D16-FAM RNA was pre-bound to Hfq-Cy3 to form a high FRET complex. Release of Hfq and the increase in donor intensity due to loss of FRET was triggered by the addition of RNA complementary to D16 (R16) (Figure 4a). The initial rate of Hfq release was 25 ± 1 s−1 (Figure 4b), comparable to the Hfq dissociation rate but much faster than the observed annealing rate reported by opening of the molecular beacon (Figure 2b). This can be explained if Hfq dissociates from one or both nucleic acid strands before annealing is complete. We also observed a loss of FRET at 0.7 s−1, which was on the same time scale as the slow phase of D16 binding (Figure 4b), and presumably reflects dissociation of more stable Hfq-D16 complexes.
Figure 4.
Hfq binding and release during annealing. (a) Hfq displacement was measured by pre-mixing 100 nM D16-FAM (red) with 0.5 µM Hfq-Cy3, and challenging the D16-FAM • Hfq-Cy3 complex with 100 nM complementary RNA (R16, green line). (b) Release of Hfq results in a loss of FRET and increase in FAM emission at 515 nm. Data (black) were fit to a double exponential equation (red curve), yielding observed rate constants of 25 ± 1 s−1 and 0.7 ± 0.2 s−1 (four trials). (c) Ternary complex formation. R16 RNA (100 nM) was pre-mixed with 0.5 μM Hfq-Cy3, and challenged with 100 nM complementary D16-FAM RNA. Binding of D16-FAM to the R16 • Hfq-Cy3 complex results in FRET and decreased FAM emission at 515 nm; subsequent release of Hfq lowers FRET and increases FAM emission. Data were fit to a double exponential (red curve); observed rate constants were 19 ± 1 s−1 and 0.86 ± 0.03 s−1 (five shots).
To obtain further evidence for Hfq release during or after annealing, Hfq-Cy3 was pre-equilibrated with unlabeled R16 RNA and then mixed with D16-FAM. An initial rise in FRET (or decrease in donor fluorescence), which we attribute to the formation of a ternary complex with kobs ∼20 s−1, was followed by a loss of FRET at ∼0.9 s−1, corresponding to dissociation of Hfq. This is likely not due to a change in the Cy3 environment, as we also observed a change in FAM anisotropy θ in the absence of Cy3 consistent with the expected changes in molecular weight. When R16 was added to D16-FAM•Hfq complexes (θ ∼ 0.12), we observed a short-lived increase in FAM anisotropy to θ ≥ 0.14, consistent with a higher molecular weight ternary complex (data not shown). This spike in fluorescence anisotropy quickly relaxed to θ ∼ 0.07, close to the value of the D16–R16 duplex and larger than that of free D16 (0.033).
Together with the annealing experiments, the results above are consistent with transient binding of two single-strands to Hfq, followed by dissociation of Hfq from the RNA duplex. Although excess unlabeled D16 RNA also competed away the FRET signal from the D16-FAM•Hfq-Cy3 complex, equimolar concentrations of complementary R16 were sufficient to trigger Hfq dissociation. The rate of strand exchange, which was measured by displacement of the beacon from D16 RNA in the presence of R16 RNA, was 0.08 s−1 in 50 nM Hfq6 (Supplementary Figure S4). Therefore, once a stable duplex has formed, further strand exchange is slow under these conditions, presumably because Hfq binds the D16•R16 duplex poorly (25).
RNA inhibition of Hfq annealing
The experiments above show that D16 RNA and the dMB-D16 beacon bind and unbind Hfq rapidly, compared to the observed annealing rate, and that Hfq is likely released from the RNA before the DNA–RNA hybrid has completely formed. If annealing involves a ternary complex between Hfq and two complementary nucleic acids, these and other results (26) suggest that the reaction rate depends on the amount of each RNA that initially binds Hfq.
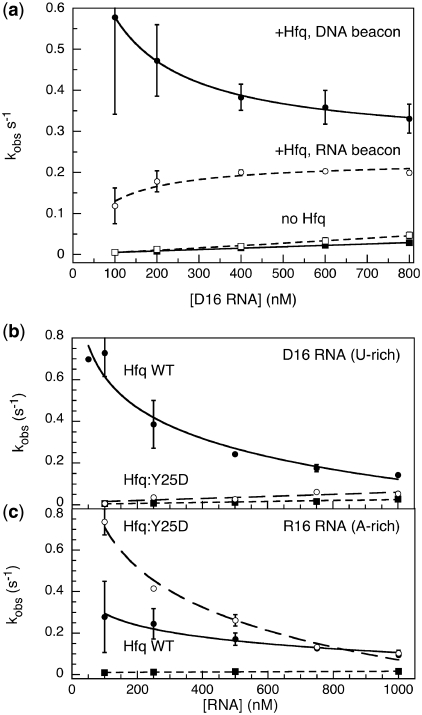
We attempted to measure the second order rate constant for forming the ternary complex, by conducting annealing experiments with increasing concentrations of D16 RNA. Surprisingly, excess D16 reduced the observed annealing rate with the DNA beacon (filled circles; Figure 5a). The annealing rate increased with D16 concentration in the absence of Hfq as expected (filled squares; Figure 5a). One explanation for these results is that two molecules of D16 RNA bind Hfq, inhibiting association of the DNA beacon with Hfq, and thus lowering the apparent annealing rate. In agreement with this possibility, annealing of the RNA beacon rMB-D16, which binds Hfq more tightly, was not inhibited by excess D16 RNA (open circles; Figure 5a).
Figure 5.
Inhibition of Hfq-dependent annealing by excess RNA. (a) Annealing kinetics of molecular beacons in TNK buffer, with 100–800 nM D16 RNA. Solid symbols and lines, DNA beacon dMB-D16; open symbols and dashed lines, RNA beacon rMB-D16. Circles, +50 nM Hfq6; data were fit to a single-site binding isotherm, with [D16]1/2 = 90 nM. Squares, no Hfq; data were fit to a line, yielding kon = 3.4 • 104 and 5.8 • 104 M−1s−1 for DNA and RNA beacons, respectively (25). Error bars represent the standard deviation among 3–4 trials. (b) Comparison of wild-type and Y25D Hfq. Reactions as in (a) but in HB buffer. Filled circles, 50 nM wild-type Hfq6; open circles, 50 nM Y25D Hfq6; filled squares, no Hfq. Fitted curves as in (a). (c) Reactions as in (b), but with R16 A-rich RNA and its complementary beacon dMB-R16. Symbols as in (b).
We next asked whether the ability of D16 to inhibit annealing depends on its U-rich sequence, by comparing the annealing kinetics of D16 with dMB-D16 with annealing of the A-rich R16 RNA with its complementary beacon dMB-R16 (filled circles, Figure 5b and c). Annealing of R16 was three times slower than annealing of D16 when the RNA concentration was 100 nM, suggesting that R16 cannot form active ternary complexes as easily as D16. However, excess R16 also inhibited the reaction, presumably by competition with its dMB-R16 beacon for access to Hfq (Figure 5c).
Finally, we asked whether interactions with the distal face of Hfq are important for RNA inhibition, by comparing the ability of D16 and R16 RNA to inhibit the annealing of Y25D Hfq (open circles, Figure 5b and c). As expected, the Y25D Hfq was less active than wild-type Hfq over the entire range of D16 concentrations. The DNA beacons may partly interact with the distal face of the hexamer, consistent with a recent report that distal face mutations abolish DNA binding by Hfq (28). Hfq:Y25D was more active than wild-type Hfq in 100 nM R16, although this reaction was also inhibited by excess R16 (open circles, Figure 5c). One explanation for this result is that the dMB-R16 DNA beacon also interacts with the distal face of Hfq. The Y25D mutation lessens competitive binding to the distal face by R16, which contains 6 A’s, increasing the fraction of ternary complexes that productively engage both RNA and beacon.
DISCUSSION
Although Hfq promotes the association of sRNAs with their mRNA targets (11,29), how Hfq accelerates strand annealing is not understood. Using an unstructured 16-nt RNA containing the U6 Hfq binding site from DsrA as a model substrate, we find that Hfq binds the RNA rapidly, but that a fraction of Hfq complexes are unstable, releasing single or double-stranded RNA in ∼0.1 s. Fender et al. (30) recently proposed that RNAs are actively displaced from Hfq by invasion of a second RNA; preliminary data are consistent with this possibility although the binding kinetics on short RNAs are ≥10 times faster than reported by Fender et al. Our results also suggest that nascent duplexes are released from Hfq before the helix has completely formed, consistent with our previous conclusion that Hfq must cycle off its binding site in DsrA sRNA during annealing with rpoS mRNA (27). Thus, Hfq ternary complexes are short-lived in the absence of additional binding motifs that anchor Hfq to the RNA.
One important conclusion from our results is that Hfq brings RNA strands together at 108 M−1s−1, 10 000 times faster than the protein-independent rate of 3.4 • 104 M−1s−1. In doing so, Hfq overcomes entropic and electrostatic barriers to helix formation. A second conclusion from our results is that Hfq appears capable of forming dynamic complexes that exchange RNA rapidly (∼0.1 s), and less dynamic complexes (∼20 s) that bind the RNA more tightly. Thus, the chaperone activity of Hfq may depend on which RNA binding partners are available at any particular moment.
Model for RNA annealing by Hfq
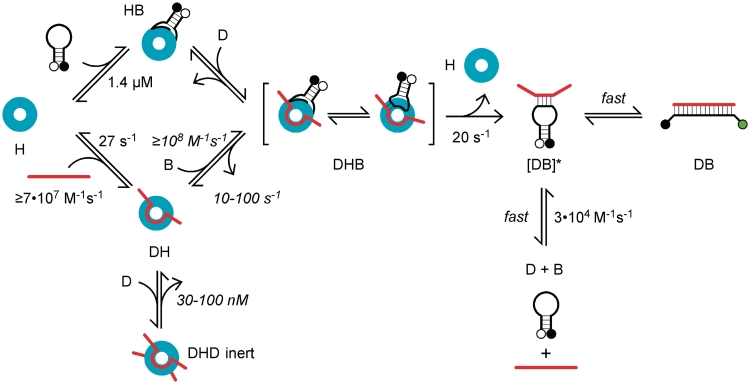
We used the kinetics of oligonucleotide annealing and Hfq binding to construct a working model for the mechanism of Hfq-dependent RNA annealing (Figure 6). The model was evaluated by simulations of experimental data, and was qualitatively successful in predicting the change in the annealing kinetics and reaction endpoints with variations in D16 or Hfq concentration (Supplementary Figure S5). Although the annealing kinetics was best approximated by the mechanism and rate constants in Figure 6, we cannot exclude mechanisms involving two Hfq hexamers, or in which only one strand binds Hfq and the second strand joins the complex via base pairing with the first strand. Our working model does not recapitulate the slow phase of the reaction or Hfq saturation, indicating there are additional Hfq interactions to be discovered. Nonetheless, its basic features, such as the rapid formation of the ternary complex and the release of Hfq from the duplex, were robust to variations on the mechanism.
Figure 6.
Model for RNA annealing by Hfq. Mechanism based on experiments with D16 RNA and a DNA beacon. Hfq6 (H, cyan) binds D16 RNA (D, red) to form the DH complex, with Kd = 23 nM at 30°C in TNK buffer. The DNA beacon (B, black) binds Hfq (HB) more weakly. The DH or HB complexes transiently form a ternary complex DHB, from which Hfq release produces an unstable intermediate [DB]* that either zippers into the fluorescent product DB or dissociates (D + B). With Hfq, initial strand association is ≥108 M−1 s−1, compared with the background rate of 3.4 • 104 M−1 s−1. D16 RNA competes with the DNA beacon for binding, producing an inert complex DHD. Apparent equilibrium constants predicted by simulations of the kinetic data were consistent with the effect of Hfq on the RNA–beacon binding equilibrium (KI = 138 nM Hfq6) and the decrease in the annealing kinetics with D16 (∼80 nM D16). Experimentally measured rate constants are in plain text; simulated values are in italics.
RNA binding
First, based on FRET measurements of D16–Hfq interactions (Figure 3 and Scheme I), we assumed that D16 RNA binds Hfq (DH) with an association rate constant of 7 • 107 M−1s−1 and dissociates at 27 s−1. The second Hfq-D16 binding step was too slow to lead directly to annealing and was modeled as a side reaction. We also assumed the DNA beacon and D16 RNA can bind Hfq in any order (top and bottom paths, Figure 6). However, the DNA beacon binds Hfq much more weakly than D16 RNA (Kd ∼ 1.4 µM Hfq6; koff ∼ 100 s−1) and is presumably the less traveled path.
RNA inhibition
In the next step of our model, the Hfq–RNA complex (DH) recruits a second strand of nucleic acid, forming either a reactive ternary complex with the beacon (DHB), or an inert complex with two strands of D16 RNA (DHD). Formation of a DHD complex best explained why annealing was slower when excess RNA was added to the reaction (Figure 5), and is plausible given the low sequence specificity of RNA binding to the distal face of Hfq (17). However, the RNA inhibition could also be explained by models in which the beacon must compete with D16 for binding to the proximal face of Hfq (31).
Ternary complex
In order for Hfq to facilitate strand annealing, the three components must physically interact at some point during the reaction (DHB; Figure 6). Evidence for ternary complexes in our experiments comes from sequential binding and release of D16 RNA from Hfq in the presence of complementary RNA (Figure 4c), and the associated change in the anisotropy of the fluorescently labeled complexes (data not shown).
The ternary complexes are unstable, as Hfq dissociates from D16 RNA at 20–30 s−1, with or without a complementary strand. The transient and dynamic nature of the ternary complexes with oligonucleotide substrates was confirmed by our simulations of the annealing kinetics. Regardless of the exact mechanism used to simulate the data, the experimental annealing rates could only be reproduced by assuming ternary complexes form with a second order rate constant ≥108 M−1s−1, 10 000 times over the background rate of helix formation. Dissociation of D16 from Hfq ranged from 10–100 s−1, in agreement with the experimental dissociation rate of 27 s−1. Simulated values for Hfq release after the first annealing step were at least 15 s−1, also in agreement with the experimental value of ∼25 s−1.
Annealing and turnover
After the ternary complex is formed, we propose that the nucleic acid strands are released as a high energy intermediate, in which some inter-strand base pairs have already formed (DB*; Figure 6). This intermediate either rapidly zippers into the fluorescent duplex (DB) or dissociates (D + B). Although binding is fast, annealing is inefficient, as the beacon or RNA can also dissociate from Hfq before the strands begin to base pair. Thus, annealing depends on the probability of initiating a helix during the lifetime of the ternary complex.
Helix formation by Hfq
The physical mechanism by which Hfq stimulates the initiation of RNA duplexes remains unknown. Many sRNA–mRNA pairs that depend on Hfq for their function contain U-rich and A-rich sequences that recruit each RNA partner to an opposite face of the Hfq hexamer. Thus, one possibility is that Hfq promotes sRNA–mRNA base pairing by simply co-localizing two strands of RNA, either to the same or opposite face of the hexamer (15,26). Hfq may additionally lower electrostatic barriers to helix initiation (32).
The fluorescent probes do not reveal how our substrates interact with Hfq or with each other in the ternary complex. Both strands may bind the proximal face, stabilizing initial base pairs between them. On the other hand, the effects of the Y25D mutation suggest that interactions with the distal face are needed to productively engage the DNA beacon, regardless of the sequence of its loop. It has been reported that Hfq binding to DNA is impaired by mutations on Hfqs distal face and C-terminus (28). However, it is not evident from existing structures how short RNAs bound to opposite faces of the Hfq hexamer would reach each other. Alternatively, RNA binding to the distal face may contribute to activity by stabilizing the hexamer or inducing a conformational change in Hfq. Our data also do not rule out interactions between two Hfq hexamers.
In all plausible mechanisms, the net velocity of the annealing reaction was exquisitely sensitive to the concentration (and turnover) of ternary complex. Our simulations predict a Michaelis constant for the ternary complex between 70 and 700 nM for our substrates, in rough agreement with the increase in annealing rate with Hfq6 concentration. Thus, the ability of Hfq to rapidly bind and release different substrates is central to its function as an RNA chaperone, as noted for other RNA chaperone proteins (18).
Comparison with natural RNAs
In contrast with the oligomeric substrates used here, which contain a single stretch of U’s (D16) or A’s (R16), natural sRNAs and their mRNA targets often contain additional binding sites for Hfq that may modulate its intrinsic annealing activity. The (AAN)n motif that recruits Hfq to the rpoS mRNA is >60 nt upstream of the sRNA complementary region (22). By interacting with the distal face of Hfq, this motif could tether Hfq to the rpoS mRNA while allowing DsrA sRNA on the proximal face to engage its complement. Alternatively or in addition, the (AAN)n motif may transfer Hfq away from DsrA during the annealing reaction, ensuring that the antisense duplex remains in place (22). Since our 16 bp substrates are fully complementary and lack this (AAN)n motif, Hfq dissociates from the D16•R16 duplex completely, explaining why the ternary complexes are unstable.
Hfq likely has additional functions on natural RNAs that contribute to sRNA regulation, apart from the intrinsic annealing activity studied here. Biochemical probing (22,33,34) and single-molecule FRET experiments (35) show that Hfq acts in part by melting the mRNA secondary structure, making it more accessible to an incoming sRNA. In addition, some mRNAs may contact both faces of Hfq; in this case sRNA binding would presumably displace the mRNA segment bound to the proximal face (24).
Dynamic versus stable Hfq complexes
An unexpected finding was that D16 RNA forms Hfq complexes with 0.04 and 20 s lifetimes, respectively. Two-step binding explains how Hfq can bind D16 with a Kd of 23 nM, yet attain annealing rates of 10 s−1 or more. As D16 has little or no secondary structure, the second step is not likely due to unfolding of D16. While at present we can only speculate on the physical differences between these complexes, a reorganization of interactions within the hexamer or a change in the number of subunits which contact the RNA (30) are possibilities.
Regardless of the structure of the complexes, the possibility that Hfq can switch between stable and dynamic RNA binding modes has important implications for its biological function. Hfq binds certain transcripts such as rpsO mRNA with a picomolar dissociation constant (36), while other RNAs such as DsrA are bound more loosely (21). If specific RNA interactions or protein partners change the conformation of Hfq complexes, they may be able to switch its behavior from chaperone to stable binder.
In summary, we find that Hfq can greatly accelerate RNA base pairing, and appears to do so by overcoming barriers to helix initiation. The net flux of the annealing reaction depends on rapid formation of ternary complexes, release of Hfq from the products, and competition with unproductive complexes containing non-complementary RNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01 GM46686). Funding for open access charge: National Institute of General Medical Sciences.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Toby Soper and Kevin Doxzen for help with Hfq purification.
REFERENCES
- 1.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harbor Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altuvia S, Wagner EG. Switching on and off with RNA. Proc. Natl Acad. Sci. USA. 2000;97:9824–9826. doi: 10.1073/pnas.97.18.9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaberdin VR, Blasi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 6.Caron MP, Lafontaine DA, Masse E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 2010;7:140–144. doi: 10.4161/rna.7.2.11056. [DOI] [PubMed] [Google Scholar]
- 7.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 10.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq Protein Increases OxyS RNA Interaction with Target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 12.Senear AW, Steitz JA. Site-specific interaction of Qbeta host factor and ribosomal protein S1 with Qbeta and R17 bacteriophage RNAs. J. Biol. Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- 13.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Haseth PL, Uhlenbeck OC. Interaction of Escherichia coli host factor protein with oligoriboadenylates. Biochemistry. 1980;19:6138–6146. doi: 10.1021/bi00567a029. [DOI] [PubMed] [Google Scholar]
- 15.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, Waldsich C, von Haeseler A, Schroeder R. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 2010;38:3794–3808. doi: 10.1093/nar/gkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 19.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS ONE. 2010;5:e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Updegrove TB, Correia JJ, Galletto R, Bujalowski W, Wartell RM. E. coli DNA associated with isolated Hfq interacts with Hfq's distal surface and C-terminal domain. Biochim. Biophys. Acta. 2010;1799:588–596. doi: 10.1016/j.bbagrm.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 30.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopkins JF. Ph.D. Thesis. Baltimore: Johns Hopkins University; 2007. Secondary structure rearrangements monitored by fluorescent probes in RNA folding and RNA-protein interactions. [Google Scholar]
- 32.Woodson SA. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010;7:1–10. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.