Abstract
MicroRNAs are small non-coding RNAs that modulate gene expression at post-transcriptional level, playing a crucial role in cell differentiation and development. Recently, some reports have shown that a limited number of mammalian microRNAs are also involved in anti-viral defense. In this study, the analysis of the hepatitis B virus (HBV) genome by the computer program MiRanda led to the identification of seven sites that are potential targets for human liver microRNAs. These sites were found to be clustered in a 995-bp segment within the viral polymerase ORF and the overlapping surface antigen ORF, and conserved among the most common HBV subtypes. The HBV genomic targets were then subjected to a validation test based on cultured hepatic cells (HepG2, HuH-7 and PLC/PRF/5) and luciferase reporter genes. In this test, one of the selected microRNAs, hsa-miR-125a-5p, was found to interact with the viral sequence and to suppress the reporter activity markedly. The microRNA was then shown to interfere with the viral translation, down-regulating the expression of the surface antigen. Overall, these results support the emerging concept that some mammalian microRNAs play a role in virus-host interaction. Furthermore, they provide the basis for the development of new strategies for anti-HBV intervention.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs that modulate gene expression at post-transcriptional level by targeting mRNAs for degradation or by inhibiting translation (1). At the time of writing, the miRNA database (http://www.mirbase.org) (2) contained ∼1100 human miRNAs which may regulate up to 30% of the protein-coding genes (3). MicroRNAs have been shown to play a prevalent role in development and cell differentiation, maintenance of stem cell character, and apoptosis (4,5). Moreover, several studies indicate that miRNA genes may act as oncogenes or tumor suppressors (6,7).
MicroRNAs are generally transcribed by RNA polymerase II as long precursors, pri-miRNAs, which are processed in the nucleus to ∼70-nt hairpin structures by the enzyme Drosha. These products, pre-miRNAs, are then transported to the cytoplasm and processed into ∼22 nt mature miRNAs by the action of the multi-domain RNase III-like enzyme Dicer (8). Each miRNA duplex is then unwound and the strand with the lower stability in the 5′-end (guide strand) is preferentially selected (9) and incorporated into a large, dynamic multi-protein complex called RNA-induced silencing complex (RISC). The targets of miRNA-loaded RISC are mRNAs presenting a near-perfect sequence complementarity with 2–8 nt in the 5′-portion of the miRNA (the so-called seed region), and additional base pairings with its 3′-region. RISC mediates down-regulation of target gene expression by cleavage or translational inhibition of the target mRNA. The choice between these two modes of action is dictated by the degree of complementarity between miRNA and its target. Near-perfect complementarity produces cleavage of the mRNA, followed by its complete degradation, whereas partial complementarity causes translational inhibition (10). In vertebrates, most of the time, the consequence is an inhibition of translation. In addition to repressing translation, miRNA interactions can lead to deadenylation and decapping, leading to rapid mRNA decay (11).
RNA-mediated post-transcriptional gene silencing can also be triggered by exogenous dsRNA molecules. In plants and invertebrates, viral dsRNA molecules are processed into small interfering RNAs (siRNAs) by Dicer and are incorporated into RISC to target pathogen’s genome and mRNAs for cleavage (12,13). Although undisputed in plants and invertebrates, a defensive role for RNA silencing in vertebrates has not been sufficiently defined. In vertebrates, cell exposure to long dsRNA triggers the interferon response as primary form of innate antiviral defense. This leads to a global shutdown in protein translation, cellular RNA degradation and often the death of virus-infected cells (14). Nevertheless, recent data provide increasing evidence that vertebrate miRNAs may directly affect viral gene expression.
In 2005, Voinnet and collaborators for the first time demonstrated that a mammalian miRNA, miR-32, has an antiviral role (15). MiR-32 was shown to target the open reading frame 2 of the primate foamy virus type 1 (PFV-1), thereby inhibiting viral mRNA translation and restricting the accumulation of the retrovirus in cultured human cells. Other cellular miRNAs, miR-196 and miR-448, were found to be up-regulated by interferon beta and capable of inhibiting hepatitis C virus (HCV) replication (16). MicroRNAs miR-24 and miR-93 have been shown to target vesicular stomatitis virus (VSV) and protect mice against VSV infection (17). Interactions between cellular miRNAs and human immunodeficiency virus (HIV) also exist. One study has shown that miR-28, miR-125b, miR-150, miR-223 and miR-382 are overexpressed in resting T4 lymphocytes and are able to target sequences in the 3′-end of HIV-1 RNA, thus silencing almost all viral messengers (18). Neutralizing these cellular miRNAs in T4 cells from patients with HIV under highly active antiretroviral therapy increased by 10-fold the in vitro efficiency of virus isolation. These observations strongly argue for a role of these cellular miRNAs in HIV latency. Very recently, miR-125b has been shown to affect the replication of human papillomavirus (19). A database of computationally predicted viral targets for mammalian miRNAs has also been established (20). Overall, these data indicate that some cellular miRNAs are a part of the host’s innate antiviral defense (21). However, it has also been proposed that miRNAs are among the host molecules that viruses co-opt to suppress their own replication to evade immune elimination and establish a persistent infection (22).
In this study, we focused our interest on human hepatitis B virus (HBV), a widely spread hepatotropic virus that causes persistent infections (23). We first performed an in silico analysis of the HBV genome looking for potential target sites for human miRNAs, then these targets were validated by assay methods based on cultured hepatic cells. This work led to the identification of hsa-miR-125a-5p as an effective suppressor of the expression of HBV surface antigen.
MATERIALS AND METHODS
Materials
MiRIDIAN miR-125a-5p mimic, miRIDIAN miR-125a-5p hairpin inhibitor, and their controls with unrelated sequences were from Dharmacon. DNA oligonucleotides were obtained from Invitrogen. The psiCheck-2 vector was purchased from Promega.
Computational analyses
Computer program miRanda (24) was used to scan the genomes of HBV subtypes ayw, adw and adr (GeneBank accession numbers X65257.1, X02763.1, AY123041.1, respectively) for the presence of target sites for the human miRNAs listed in miRBase (http://www.mirbase.org/) (2). Search was conducted with the following cut-off parameters: minimal free energy of binding less than −12 kcal/mol and miRanda score >140. Alignment of the DNA sequences of the viral genomes was performed with ClustalW.
DNA constructs
The HBV genomic segments that were supposed to be targeted by human miRNAs, whose size ranged from 18 to 29 bp, were obtained by chemical synthesis of complementary oligonucleotides containing upstream XhoI and EcoRV restriction sites and a downstream NotI site. Each couple of oligonucleotides was annealed, 5′-phosphorylated with T4 polynucleotide kinase, and ligated into XhoI and NotI sites of psiCheck-2. Screening of recombinant clones was performed by digestion with EcoRV. Selected constructs were then sequenced to confirm their identity. Control plasmids with inverted or mutated sequences were obtained by the same approach.
Cell cultures and transfections
Human hepatoblastoma HepG2 cells were cultured in RPMI 1640 containing 10% fetal bovine serum, 2 mM l-glutamine, 50 U/ml penicillin and 100 µg/ml streptomycin; cell line HuH-7 was cultured in DMEM containing 10% fetal bovine serum, 2 mM l-glutamine and the same antibiotics; PLC/PRF/5 cells were cultured as HuH-7 except that medium contained 0.1 mM non-essential amino acid. The day before transfection, cells were trypsinized and seeded in medium without antibiotics in 12-well plates. Transfections were performed with cells at 80–90% of confluence by using 3 µl of Lipofectamine 2000 (Invitrogen) for 1 µg of nucleic acids, as described by the manufacturer. HepG2, HuH-7 and PLC/PRF/5 were transfected with 0.2, 0.05 and 0.1 µg of reporter plasmid, respectively. After 6 h, transfection mix was replaced with complete medium.
Assays
Assays were performed 48 h after transfection. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega), as described (25). Briefly, cells were washed with phosphate buffered saline and lysed chemically. Lysates were then centrifuged to remove cellular debris and 20 µl of the supernatants were loaded into an automated luminometer (Turner Biosystems). Then the instrument performed a sequential auto-injection of 100 µl of Luciferase Assay Reagent II (substrate for firefly luciferase) and 100 µl of Stop and Glow Reagent (stop solution for firefly luciferase containing the substrate for Renilla luciferase). The mean of the luciferase activities measured for 10 s each were used to calculate ratios between Renilla and firefly luciferases. The extracellular HBV surface antigen (HBsAg) produced by PLC/PRF/5 cells was quantitated by an enzyme-linked immunosorbent assay (Alpha Diagnostic International) according to the manufacturer’s protocol. Each experiment of transfection and subsequent assays was performed at least three times. Reported data are the mean ± SD and statistical significance was assessed with Student’s t-test. Surface antigen mRNA was quantitated by RT–PCR with primers binding upstream (5′-TCCTCTGGGATTCTTTCCCGA-3′) and downstream (5′-CACTGCATGGCCTGAGGATG-3′) of the target site for hsa-miR125a-5p. Hsa-miR-125a-5p was quantitated by RT-PCR using the TaqMan miRNA assays kit from Applied Biosystems, as described (26).
RESULTS AND DISCUSSION
Prediction of target sites for human microRNAs within HBV genome
HBV is characterized by a circular DNA genome of 3.2 kb which is only partially double-stranded (27). This is replicated through an RNA intermediate, the pregenomic RNA, which is produced by transcription of the viral DNA by cellular RNA polymerase. Pregenomic RNA is then converted into the viral DNA genome by the HBV polymerase, a multifunctional enzyme with reverse transcriptase, DNA-dependent DNA polymerase and RNase H activities (28).
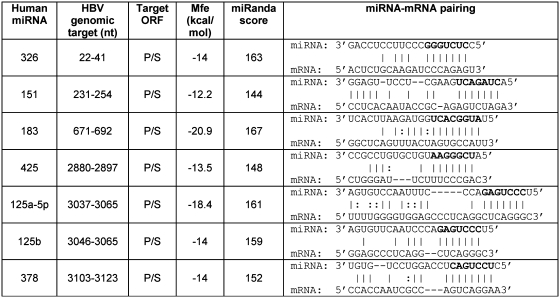
The computational analysis was performed with the computer program miRanda, scanning plus strands of the genomes of the most common HBV subtypes (ayw, adw and adr) for the presence of target sites for human miRNAs. Restriction of the results to (i) those targets that are conserved in the three HBV subtypes, (ii) those miRNA–mRNA pairings with a perfect matching of the seed region (2–8 nt of miRNA), and (iii) those microRNAs that are known to be expressed in liver, the natural site of infection, or in cultured hepatic cells (7,30), yielded seven promising target sites. They were found to be clustered in a 995-bp segment within the viral polymerase (P) ORF and the overlapping surface antigen (S) ORF, as detailed in Figure 1. The results of this search suggested that human hepatic cells may use these miRNAs to down-regulate the expression of HBV P and S genes.
Figure 1.
A computational analysis of hepatitis B virus (HBV) genome by miRanda (24) yielded seven sites that are potential targets for human miRNAs. Polymerase ORF (P) comprises nucleotides 2309–1625; the overlapping surface antigen ORF (S) comprises nucleotides 2848–839. MicroRNAs 125a-5p and 125b are products of different genes but have similar sequences thus targeting related sites. The seed region of miRNAs is marked in bold. Nucleotide numbering is referred to HBV subtype ayw (GeneBank entry X65257.1).
Testing of HBV genomic targets for responsiveness to human hepatic miRNAs
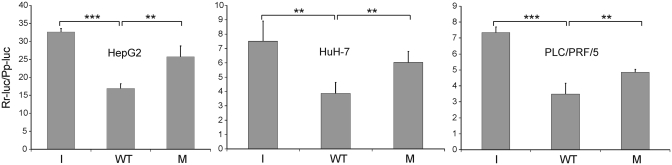
The most common methods for miRNA target validation are based on reporter gene constructs transfected in cultured mammalian cells (29). The seven HBV genomic segments containing putative target sites for human miRNAs were separately cloned downstream of the Renilla luciferase open reading frame contained in the psiCheck-2 vector. The recombinant plasmids were then transfected in cultured HepG2 cells, and luciferase activity was measured 48 h after transfection. The rationale for using this assay is that the binding of a given cellular miRNA to the viral target, transcribed together with the luciferase coding sequence, will repress reporter protein production thereby reducing luciferase activity compared to a control. In these experiments, controls were represented by psiCheck-2 vectors that did not contain any HBV sequences or contained inverted sequences. It should be noted that psiCheck-2 also contains a secondary Photinus (firefly) luciferase cassette designed to be an intraplasmid transfection normalization reporter. Thus when using the psiCheck-2 vector, the Renilla luciferase signal can be normalized to the firefly luciferase signal allowing very accurate measurements of the expression level of the reporter. Under these experimental conditions, the HBV genomic segment 3037–3065, supposed to be targeted by hsa-miR-125a-5p, inhibited markedly the expression of the reporter in HepG2 cells. The activity registered with the wild-type 3037–3065 HBV segment was in fact 48% lower than that obtained with the control plasmid containing the inverted sequence (Figure 2, left panel). The inhibition values obtained with the other six HBV genomic segments were lower than 15% and did not reach statistical significance (data not shown). On the other hand, the level of inhibition obtained with the 3037–3065 target is quite impressive since the assay method reveals the effect of a single endogenous miRNA on the translation of an abundant mRNA transcribed from the strong SV40 early enhancer/promoter of the reporter plasmid. It should also be noted that miR-32, miR-24 and miR-93, endowed with antiviral activity toward PFV-1 and VSV (15,17), were shown to inhibit the expression of the reporter by a similar extent (∼50%). As a further control of the effect of hsa-miR-125-5p on the viral target, three mutations, C3058A, C3060G and G3063A, were introduced in the reporter plasmid, within the region of complementarity to the seed region of the miRNA. When the mutated construct was transfected in HepG2 cells, the reporter activity was found to be significantly higher than that obtained with the wild-type construct (Figure 2, left panel), providing a strong evidence for a direct interaction between hsa-miR-125a-5p and the viral target. It should be noted that hsa-miR-125a-5p is one of the best-scoring miRNAs predicted to bind to HBV mRNAs (Figure 1). The reason why the other six miRNAs identified by MiRanda were not found to affect the reporter activity is unknown. One possible explanation is that their target sites may have extensive secondary structures that prevent miRNA binding. Otherwise it should be hypothesized that the false-positive rate of MiRanda is higher than expected.
Figure 2.
Interfering activity of hsa-miR-125a-5p in cultured hepatic cells. HepG2, HuH-7 and PLC/PRF/5 cells were transfected with the luciferase-based reporter plasmid psiCheck-2 containing the HBV 3037–3065 genomic segment belonging to the polymerase/surface antigen ORF (WT). Other plasmids contained a control DNA with inverted sequence (I), or a mutated viral sequence (M) characterized by 3 nt substitutions (C3058A, C3060G and G3063A) in the region of complementarity to the seed region of the miRNA. Luciferase activity was recorded 48 h after transfection. Luciferase activities registered with the wild-type viral DNA (WT) were always lower than those observed with the control DNA with inverted sequence (I), indicating that the endogenous hsa-miR-125a-5p can interact with the viral sequence. P-values at Student’s t-test were *P < 0.05, **P < 0.01 or ***P < 0.001. Rr-luc, Renilla reniformis luciferase; Pp-luc, Photinus pyralis (firefly) luciferase.
Hsa-miR-125a-5p has been reported to be expressed in other hepatic cell lines also, such as HuH-7 and PLC/PRF/5 (7,16,30,31). Testing was then extended to these cells, revealing similar or slightly higher levels of silencing activity (Figure 2, center and right panels). It should be noted that the expression level of the reporter gene in these cell lines is lower than that registered in HepG2 cells, revealing a different activity of the SV40 early enhancer/promoter in these cells. Overall, these data indicate that the 3037–3065 segment HBV genome is sensitive, at the mRNA level, to the silencing effect of hsa-miR-125a-5p expressed in hepatic cells.
Silencing activity of hsa-miR-125a-5p
Little is known about the cellular targets of hsa-miR-125a-5p. This miRNA has been reported to down-regulate the expression of ERBB2 and ERBB3 oncogenes in cultured breast cancer cells (32) and to inhibit the expression of p53 gene (33). In both reports, the interaction of the miRNA with its target was validated by a luciferase reporter assay yielding levels of inhibition of gene expression ≤40%, even if testing was performed with high miRNA concentrations, either produced by an exogenous expression cassette or by transfection of a synthetic miRNA mimic.
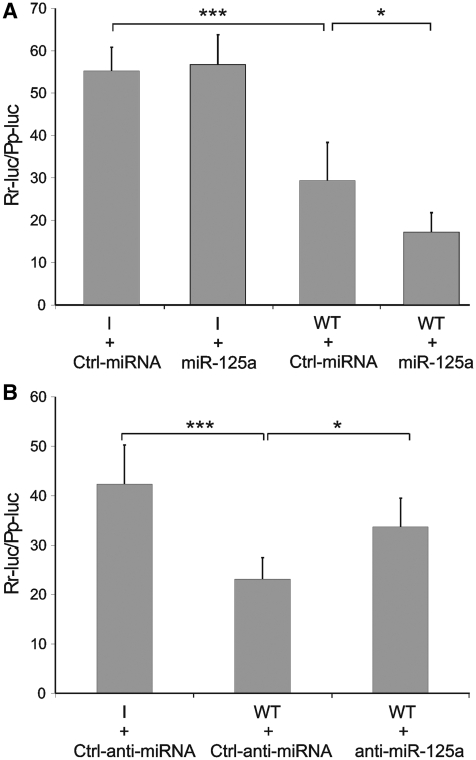
The anti-HBV activity of hsa-miR-125a-5p was then inquired by the use of a synthetic miRNA-mimic and an antisense inhibitor. Transfection of hsa-miR-125a-5p mimic in HepG2 cells increased the reporter-silencing effect of the endogenous miRNAs to 69% (Figure 3A). An antisense miRNA inhibitor was then used to knockdown the endogenous hsa-miR-125a-5p. When the inhibitor was transfected in HepG2 cells, the reporter-silencing activity of the endogenous miRNAs was significantly reduced (Figure 3B). These data clearly indicate that the silencing effect of cellular miRNAs toward the HBV genomic segment 3037–3065 can be attributed to hsa-miR-125a-5p.
Figure 3.
The silencing effect of hsa-miR-125a-5p in HepG2 cells can be modulated by a synthetic miRNA mimic (A) or inhibitor (B). Cells were transfected with the reporter plasmid psiCheck-2 containing the wild-type HBV fragment (WT) or a control DNA with inverted sequence (I), along with 10 nM hsa-miR-125a-5p mimic (miR-125a, A) or 50 nM hsa-miR-125a-5p inhibitor (anti-miR-125a, B). Further controls were a miRNA mimic (Ctrl-miRNA) or inhibitor (Ctrl-anti-miRNA) with unrelated sequences. Luciferase activity was recorded 48 h after transfection. P-values are indicated as described in Figure 2.
At this stage of the work we became aware of some interesting results from a study on miRNA expression profiles in cultured hepatic cells. By using a miRNA detection technique based on microarrays, it was found that 11 miRNAs, including hsa-miR-125a-5p are expressed in the HBV-producing cell line HepG2.2.15 at levels at least 3-fold higher than in its parent cell line HepG2 (31). These results raise the possibility that the expression of hsa-miR125a-5p is inducible and correlated to the viral infection. In this regard, it has very recently reported that the expression in HepG2 cells of HBV X protein increases the expression level of hsa-miR125a-5p (34).
Effect of hsa-miR-125a-5p on viral translation
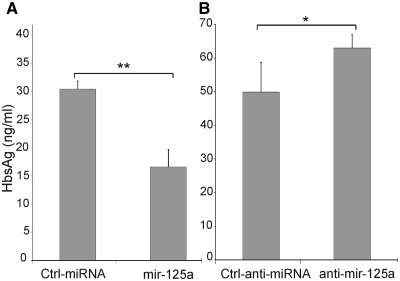
The assays described above are based on the silencing effect of hsa-miR-125a-5p on the translation of a chimeric mRNA containing the Renilla luciferase coding sequence and a segment of HBV mRNA. The data collected clearly indicated that hsa-miR-125a-5p is able, at physiological concentration, to interact effectively with the viral sequence thereby silencing the luciferase expression. However, it was not known whether the same miRNA could be able to interfere with the translation of the natural HBV mRNAs. This point was then addressed by the use of the human hepatoma cell line PLC/PRF/5 that contains several copies of HBV DNA integrated in its genome. This cell line does not produce infectious HBV particles but expresses the HBV S gene into the mature hepatitis B surface antigen (HBsAg) in 22-nm spherical particles that are secreted in the culture medium (35). In this system, cells were transfected with the synthetic hsa-miR-125a-5p mimic and the effect on viral translation of the surface antigen mRNA was assessed by a diagnostic ELISA assay for HBsAg. Transfection of the mimic induced a 46% decrease of the expression of HBsAg (Figure 4A). This level of inhibition is remarkable if we consider that there is a background level of uninhibited translation due to preexisting expression of the endogenous target gene and to the fact that the exogenous miRNA-mimic is not expected to transfect all cells. In a control experiment, transfection of the mimic was found to increase the average cellular content of hsa-miR-125a-5p by ∼3-fold. RT–PCR was then used to quantitate the surface antigen mRNA in PLC/PRF/5 cells transfected with the hsa-miR-125a-5p mimic or with the control miRNA. This assay indicated that hsa-miR-125a-5p does not affect the concentration of the target mRNA, revealing that its silencing effect is due to translation inhibition rather than mRNA cleavage. Overall, these results clearly indicate that hsa-miR-125a is able to interfere with the viral translation of surface antigen mRNAs, thus reducing the amount of HBsAg.
Figure 4.
Hsa-miR-125a-5p interferes with viral expression of surface antigen. PLC/PRF/5 cells were transfected with 10 nM hsa-miR-125a-5p mimic (miR-125a, A) or 50 nM hsa-miR-125a-5p inhibitor (anti-miR-125a, panel B). Controls were miRNA mimic (Ctrl-miRNA) or inhibitor (Ctrl-anti-miRNA) with unrelated sequences. Forty-eight hours after transfection HBsAg was quantitated by an ELISA test. P-values are indicated as described in Figure 2.
In a final experiment, the synthetic miRNA inhibitor was transfected into the PLC/PRF/5 cells to knock-down hsa-miR-125a-5p. This induced a significant increase in the expression of HBsAg (Figure 4B), providing a direct evidence that the interaction between the selected miRNA and the viral mRNA occurs naturally in PLC/PRF/5 cells, leading to a reduced expression of HBsAg.
The HBV 3037–3065 genomic segment, targeted by hsa-miR-125a-5p, encodes amino acid residues 244–252 of polymerase and 64–72 of surface antigen. The active site of HBV polymerase has not been fully characterized since a three-dimensional structure of the enzyme has not been determined yet. Based on its primary structure and mutational studies (36), the encoded amino acids fall within a polymerase region, the spacer (residues 178–336) that is far from the YMDD catalytic motif. However, it cannot be ruled out the possibility that the encoded residues contribute to the folding and functionality of the enzyme. In this regard, it has been reported a lamivudine-resistant HBV variant showing an amino acid substitution in the spacer region (37). On the other hand, the encoded residues of the surface antigen fall within the extracellular pre-S1 domain of the protein that is responsible for receptor binding on hepatocytes. Furthermore, they overlap to an amino acid segment (residues 1–75) that has been shown to be essential for infectivity (38). This may pose constrains for the virus not to mutate this region. In this regard, we performed a BLAST search of nucleotide databases looking for sequences homologous to the HBV 3037–3065 genomic segment. This search yielded over 4000 entries identical to the query sequence and belonging to HBV strains and/or isolates of all subtypes (ayw, adw, adr, ayr and adyw). This finding confirms that the HBV target site for binding of hsa-miR-125a-5p is very well conserved.
CONCLUSIONS
Human HBV infection has a wide range of clinical outcomes, from self-limited asymptomatic infection to fulminant hepatitis and death (23,39). In addition, chronic infection is closely correlated with the development of hepatocellular carcinoma (40). Although new infections are preventable through vaccination, new antiviral agents are being sought for the treatment of the over 300-million affected individuals worldwide (41). This work shows that hsa-miR-125a-5p, a microRNA expressed in human liver, is able to down-regulate the expression of HBV S gene thus reducing the amount of secreted HBsAg. This effect will likely counteract viral replication, since it is known that (i) the expression levels of HBsAg are correlated with viral titer, and (ii) experimental knockdown of HBsAg leads to suppression of HBV replication (42). If these observations are confirmed by experiments in vivo, then it is possible to hypothesize that the mild outcome of HBV infection observed in many individuals is not only due to the well-known immune response but also to an RNA-mediated defense operated by hepatic cells. In this scenario, delivery of synthetic hsa-miR-125a-5p mimics or treatments that modulate the expression of cellular hsa-miR-125a-5p could represent useful strategies in developing new anti-HBV therapeutics. Finally, the determination of the expression levels of hsa-miR-125a-5p in different individuals may also be useful in studying susceptibility to infection.
FUNDING
Funding for open access charge: Second University of Naples and Regione Campania (L5/2002).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Roberto Mazzanti and Dr Ornella Fantappiè of Azienda Ospedaliero-Universitaria Careggi, Florence, Italy for providing the PLC/PRF/5 cells.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths-Jones S, Saini HK, van Dongen S, Bateman A, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 9.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 11.Eulalio A, Huntzinger E, Izaurralde F. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 14.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 19.Nuovo GJ, Wu X, Volinia S, Yan F, di Leva G, Chin N, Nicol AF, Jiang J, Otterson G, Schmittgen TD, et al. Strong inverse correlation between microRNA-125b and human papillomavirus DNA in productive infection. Diagn. Mol. Pathol. 2010;19:135–143. doi: 10.1097/PDM.0b013e3181c4daaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu PW, Lin LZ, Hsu SD, Hsu JB, Huang HD. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res. 2007;35:D381–D385. doi: 10.1093/nar/gkl1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009;37:1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan VS, Drake A, Chen J. Virus-specific host miRNAs: antiviral defenses or promoters of persistent infection? Trends Immunol. 2009;30:1–7. doi: 10.1016/j.it.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin R, Locarnini S. Treatment of chronic hepatitis B: current challenges and future directions. Rev. Med. Virol. 2003;13:255–272. doi: 10.1002/rmv.393. [DOI] [PubMed] [Google Scholar]
- 24.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potenza N, Moggio L, Milano G, Salvatore V, Di Blasio B, Russo A, Messere A. RNA interference in mammalian cells by RNA-3′-PNA chimeras. Int. J. Mol. Sci. 2008;9:299–315. doi: 10.3390/ijms9030299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, Turner M, Cooper MD. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int. Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 28.Crouch RJ. Ribonuclease H: from discovery to 3D structure. New Biol. 2005;2:771–777. [PubMed] [Google Scholar]
- 29.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Zhao JJ, Wang CM, Li MY, Han P, Wang L, Cheng YQ, Zoulim F, Ma X, Xu DP. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin. Med. J. 2009;122:10–14. doi: 10.3901/jme.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Christofer CB. Coordinate Suppression of ERBB2 and ERBB3 by Enforced Expression of Micro-RNA miR-125a or miR-125b. J. Biol. Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Gao JS, Tang X, Tucker LD, Quesenberry P, Rigoutsos I, Ramratnam B. MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 2009;583:3725–3730. doi: 10.1016/j.febslet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Lu Y, Toh ST, Sung W, Tan P, Chow P, Chung AYF, Jooi LLP, Lee CGL. Lethal-7 is down-regulated by the hepatitis B virus X protein and targets signal transducer and activator of transcription 3. J. Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 35.MacNab GM, Alexander JJ, Lecatsas G, Bey EM, Urbanowicz JM. Hepatitis B surface antigen produced by a human hepatoma cell line. Br. J. Cancer. 1976;34:509–515. doi: 10.1038/bjc.1976.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yatsuji H, Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S, Iwao E, Fujimoto Y, Ochi H, et al. Emergence of a novel lamivudine-resistant hepatitis B virus variant with a substitution outside the YMDD motif. Antimicrob. Agents Chemother. 2006;50:3867–3874. doi: 10.1128/AAC.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Duff Y, Blanchet M, Sureau C. The pre-S1 and antigenic loop infectivity determinants of the hepatitis B virus envelope proteins are functionally independent. J. Virol. 2009;83:12443–12451. doi: 10.1128/JVI.01594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoulim F. Therapy of chronic hepatitis B virus infection: inhibition of the viral polymerase and other antiviral strategies. Antiviral Res. 1999;44:1–30. doi: 10.1016/s0166-3542(99)00056-x. [DOI] [PubMed] [Google Scholar]
- 41.Feld J, Lee JY, Locarnini S. New targets and possible new therapeutic approaches in the chemotherapy of chronic hepatitis B. Hepatology. 2003;38:545–553. doi: 10.1053/jhep.2003.50389. [DOI] [PubMed] [Google Scholar]
- 42.Xiangji L, Feng X, Qingbao C, Weifeng T, Xiaoqing J, Baihe Z, Feng S, Hongyang W, Mengchao W. Knockdown of HBV surface antigen gene expression by a lentiviral microRNA-based system inhibits HBV replication and HCC growth. J. Viral Hepat. 2010 doi: 10.1111/j.1365-2893.2010.01346.x. doi:10.1111/j.1365-2893.2010.01346.x [Epub ahead of print, 19 July 2010] [DOI] [PubMed] [Google Scholar]