Abstract
Hfq is a key regulator involved in multiple aspects of stress tolerance and virulence of bacteria. There has been an intriguing question as to how this RNA chaperone achieves two completely opposite functions—annealing and unwinding—for different RNA substrates. To address this question, we studied the Hfq-mediated interaction of fragments of a non-coding RNA, DsrA, with its mRNA target rpoS by using single-molecule fluorescence techniques. These experiments permitted us to observe the mechanistic steps of Hfq-mediated RNA annealing/unwinding at the single-molecule level, for the first time. Our real-time observations reveal that, even if the ring-shaped Hfq displays multiple binding sites for its interaction with RNA, the regulatory RNA and the mRNA compete for the same binding site. The competition makes the RNA-Hfq interaction dynamic and, surprisingly, increases the overall annealing efficiency by properly aligning the two RNAs. We furthermore reveal that when Hfq specifically binds to only one of the two RNAs, the unwinding process dominates over the annealing process, thus shedding a new light on the substrate selectivity for annealing or unwinding. Finally, our results demonstrate for the first time that a single Hfq hexamer is sufficient to facilitate sRNA–mRNA annealing.
INTRODUCTION
During the transfer of genetic information, RNA molecules often interconvert between well-defined structures. Many of these structural changes are involved in the regulation of critical steps of transcription and translation. RNA chaperones are a group of proteins that assist in these structural changes by using either ATP hydrolysis or the binding energy of protein–RNA interactions. Numerous RNA chaperones have been discovered and their biological functions are well-characterized (1). However, their operational mechanism and mechanistic steps are still unclear. The bacterial protein Hfq is a good model system to address these questions (2). Hfq was initially discovered as an Escherichia coli host factor required for bacteriophage Qβ RNA replication (3). The importance of Hfq in other cellular functions became clear from various deleterious phenotypes observed in an hfq-null mutant (4). Subsequent studies revealed that Hfq acts as a post-transcriptional regulator by facilitating the trans-annealing of small noncoding regulatory RNAs (sRNA) to their mRNA targets (5,6), thus resulting in up- or down-regulation of various proteins (7–9). In addition, Hfq is involved in the regulation of RNA decay (10,11) and is a virulence factor in pathogenic bacteria (12,13). Thus, Hfq is a potential target for anti-bacterial drug development. Finally, due to its homology to Sm and Sm-like (L-Sm) proteins, the interest in Hfq increased as a simpler model system for the understanding of the operational mechanisms of Sm and L-Sm proteins in general (14,15).
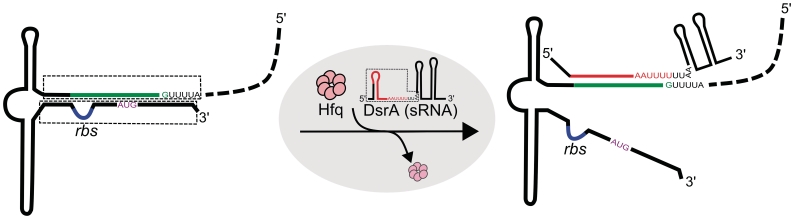
Structural studies revealed that Hfq is a homohexameric toroidal protein, in which uridine-containing sequences bind to the proximal face of the protein torus, while adenine-rich sequences bind to its distal face (16,17). Therefore, Hfq could potentially bind two RNA substrates simultaneously (16,18,19) and thus promote intermolecular interaction by bringing complementary RNA sequences together. However, roles of the proximal/distal RNA binding sites of Hfq in the RNA chaperone activity have not been directly proved, and it is still possible that the periphery of the torus may play a role in Hfq–RNA interactions. A bulk fluorescence resonance energy transfer (FRET) study (20) confirmed that Hfq is an RNA chaperone which enhances the interaction of DsrA (an E. coli regulatory sRNA) with its mRNA target rpoS (encoding σS, the stress and stationary phase transcription factor) by facilitating both the annealing of DsrA and rpoS, and the unwinding of the inhibitory stem of rpoS (Figure 1). However, the detailed mechanisms and the dynamic nature of Hfq activity are still unclear, and basic questions as to the oligomeric state for the active form of the hexameric Hfq is under debate. Furthermore, how Hfq can selectively achieve completely opposite functions—annealing and unwinding—for different RNA substrates is still ambiguous. To address these questions, here we developed single-molecule FRET assays recapitulating the essential aspects of the ternary interaction between DsrA, rpoS and Hfq (21) and observed the mechanistic steps of both RNA annealing and unwinding in real time.
Figure 1.
Regulation of rpoS translation by DsrA. Base pairing of DsrA with the leader sequence of rpoS enhances the translation of rpoS by exposing the ribosome binding site (rbs, blue line) and start codon (AUG, purple) which were previously sequestered in the inhibitory stem structure (21). This process does not happen readily, but requires the RNA chaperone Hfq as a cofactor. Hfq unwinds the inhibitory stem of rpoS and also accelerates the annealing of DsrA and rpoS. To understand the detailed mechanism of RNA annealing and unwinding processes we focused on the core of the RNA–RNA interactions (dashed boxes).
MATERIALS AND METHODS
Protein purification
Escherichia coli Hfq was over-expressed and prepared as described previously (22), except that to avoid the use of denaturing conditions previously imposed by purification on a polyA sepharose column, this affinity purification step was replaced by a strong anion-exchange chromatography on a Mono Q HR5/5 column (GE Healthcare). The process of purification was as follows: cells from post-induction cultures were resuspended in a buffer containing 20 mM Tris–HCl pH 7.5, 0.5 M NaCl, 10% (v/v) glycerol, 0.1% (v/v) Triton X-100 and a protease inhibitor cocktail (SIGMAFASTTM Protase Inhibitor Cocktail) at 4°C. The suspension was sonicated and lysed cells were cleared by centrifugation at 15 000g for 30 min. The supernatant was heated at 80°C for 15 min, followed by centrifugation at 15 000g for 30 min. DNase I (40 µg/ml) and RNase A (30 µg/ml) were added to the cleared lysate at RT for 1 h. RNAse digestion resulted in weakening of RNA:Hfq complexes, which can be removed by the ion-exchange column (see below). The resulting solution was then applied to a 1-ml Ni2+–NTA column (GE Healthcare). The resin was washed with 20 mM Tris–HCl, pH 7.8, 0.3 M NaCl, 20 mM imidazole and the protein was eluted with a gradient of imidazole (20–500 mM). Finally, to completely remove all traces of RNA bound to Hfq, the protein was further purified by anion exchange chromatography on a Mono Q HR5/5 column (GE Healthcare). Elution was performed with a linear gradient of NaCl, which allows to separate remaining RNA from Hfq. Absence of significant RNA contamination of the protein preparation can be verified by its absorbance spectrum, which has high sensitivity for the detection of nucleic acid contamination, and by the absence of detectable bands upon Sybr green staining of sample-resolved polyacrylamide gel electrophoresis. We stored Hfq at 4°C in 50 mM Tris–HCl pH 7.5, 1 mM EDTA, 10% glycerol and 50 mM NH4Cl.
RNA preparation
The following RNA strands (written from 5′–3′) were purchased from Samchully (South Korea).
rpoS_reg: Cy3/Cy5-AUUUUGAAAUUCGUUACAAGGGGAAAUCCGUAAACCC
rpoS_rbs: CAAGGGAUCACGGGUAGGAGCCACCUUAUGAGUCAGAAU-Cy7
DsrA_core: AACACAUCAGAUUUCCUGGUGUAACGAAUUUUUUAAG-Cy3/Cy5
DsrA_pA: AACACAUCAGAUUUCCUGGUGUAACGAAAAAAAAAAG-Cy3/Cy5
Each strand was labeled with one of the fluorophores indicated above at the amine-modified base at the terminal. The RNA ends opposite from the dye-labeling position were biotinylated as needed. The labeling efficiencies were >90% except the case of Cy5-labeling of DsrA_pA (72%), Cy3-labeling of rpoS_reg (73%) and Cy5-labeling of rpoS_reg (72%).
For the annealing of rpoS_reg, and rpoS_rbs the mixture of Cy3-labeled rpoS_reg (120 μM in H2O, 1 μl), Cy7-labeled rpoS_rbs (70 μM in H2O, 8 μl), NaCl (5 M in H2O, 1 μl) and 10 mM Tris–HCl (pH 8.0, 5 μl) with 50 mM NaCl was slowly cooled down from 90 to 4°C. The rpoS_reg:DsrA_pA duplex and rpoS_reg:DsrA_core duplex annealings were performed in a similar way.
Single-molecule FRET experiments
Polymer-coated quartz slides were prepared as described previously (23). After immobilizing biotinylated RNAs, and adding an imaging buffer containing Hfq and/or complementary RNAs, single-molecule images were obtained in a wide-field total-internal-reflection fluorescence microscope using an electron multiplying charge-coupled device (EM-CCD) camera (iXon DV887ECS-BV, Andor Technology) and a C++ program developed in our laboratory. Measurements were performed at 23°C in an imaging buffer with the following buffer composition: 50 mM Tris–HCl (pH 8.0), 50 mM NaCl and an oxygen scavenger system to slow photobleaching.
To obtain single-molecule time traces, or dwell-times, either 300-ms (DsrA:rpoS annealing), or 1000-ms (rpoS_reg:rpoS_rbs unwinding) of EM-CCD exposure time was used. To obtain FRET histograms, initial 10 data points of single-molecule time traces were collected from short movies with exposure times of 200-ms (DsrA_core:rpoS_reg annealing), 300-ms (rpoS_reg:rpoS_rbs annealing) or initial 20 data points with 400-ms (rpoS_reg:rpoS_rbs unwinding). Matlab, IDL and Origin were used for data analysis.
RESULTS
Single-molecule FRET assay for strand-exchange
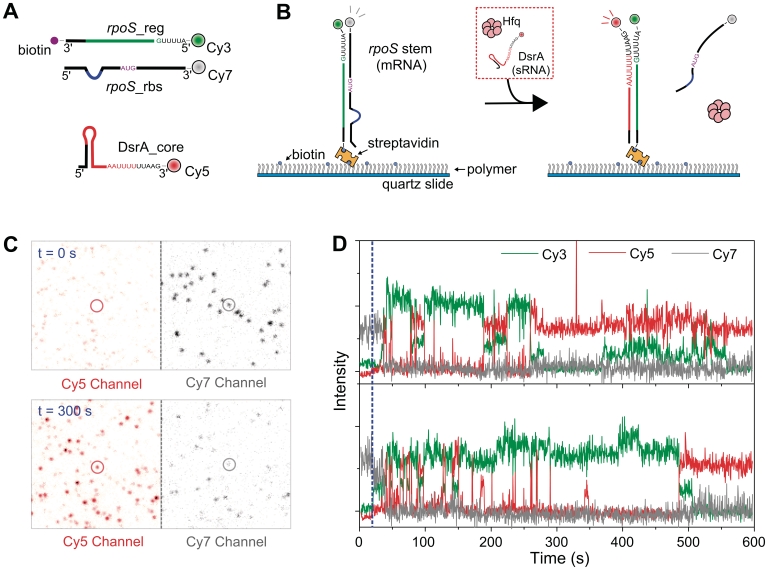
We established a single-molecule FRET assay to emulate the strand-exchange process summarized in Figure 1. We prepared three RNA fragments based on the core parts of the RNA–RNA, and RNA–Hfq interactions: (i) DsrA_core, the 5′-portion of DsrA containing sequences for base-pairing with rpoS and the primary Hfq binding site; (ii) rpoS_reg, the leader sequence of the rpoS inhibitory stem; and (iii) rpoS_rbs, the complementary sequence of rpoS_reg which contains both the ribosome binding site (rbs) of rpoS and its translation start codon (Figure 2A). Each RNA strand was labeled with distinct fluorophores so that high FRET signal occurs in the properly annealed states, and rpoS_reg was additionally biotinylated for immobilization (Figure 2A).
Figure 2.
Strand-exchange experiments. (A) RNA fragments used in this study. The three synthetic RNA fragments contain both the RNA–RNA interaction sequences (red line for DsrA_core, and green line for rpoS_reg), and the main Hfq-interaction sequences (DsrA_core: AAUUUUUUAA and rpoS_reg: AUUUUG). The labeling positions of fluorophores and biotin are shown. (B) Experimental scheme. Pre-annealed rpoS_reg:rpoS_rbs complex was immobilized on a polymer-coated surface. While single-molecule fluorescence images were taken, the buffer containing Cy5-labeled DsrA_core (2 nM) and Hfq (5 nM) was delivered. (C) Single-molecule images before and 300 s after the addition of Hfq and DrsA. The circled molecule corresponds to first trace in (D). (D) Single-molecule time traces of Cy3 (green), Cy5 (red) and Cy7 (gray) intensities. Blue dashed line indicates the Hfq injection point.
To observe the individual steps of the strand-exchange process, we immobilized pre-annealed rpoS_reg:rpoS_rbs complexes on a polymer-coated quartz surface via a streptavidin–biotin interaction, and rapidly delivered a buffered solution containing DsrA_core and Hfq (Figure 2B) while acquiring single-molecule images on a wide-field total-internal-reflection fluorescence microscope (24). Comparison of single-molecule images before the injection of the DsrA_core and Hfq and after incubation for 300 s clearly shows that Hfq induces the displacement of rpoS_rbs from rpoS_reg, and promotes the annealing of DsrA_core to rpoS_reg (Figure 2C), thus demonstrating that our single-molecule strand-exchange assay is properly operating. Representative fluorescence intensity time traces provide the insights into the strand-exchange process (Figure 2D); the unwinding of the rpoS_reg:rpoS_rbs complex (monitored as the disappearance of Cy7 signal) occurs rapidly after the injection of Hfq and DsrA_core (dashed blue line), but the full annealing of DsrA_core and rpoS_reg (monitored from the appearance of stable Cy3/Cy5 FRET) is established after a number of transient and unproductive binding events of DsrA_core (monitored via Cy3/Cy5 FRET spikes). Our observation justifies the hypothesis that the unwinding and annealing of RNA molecules by Hfq are independent processes, and thus can be studied separately.
RNA unwinding occurs when a poly-U site exists only in one of the two RNAs
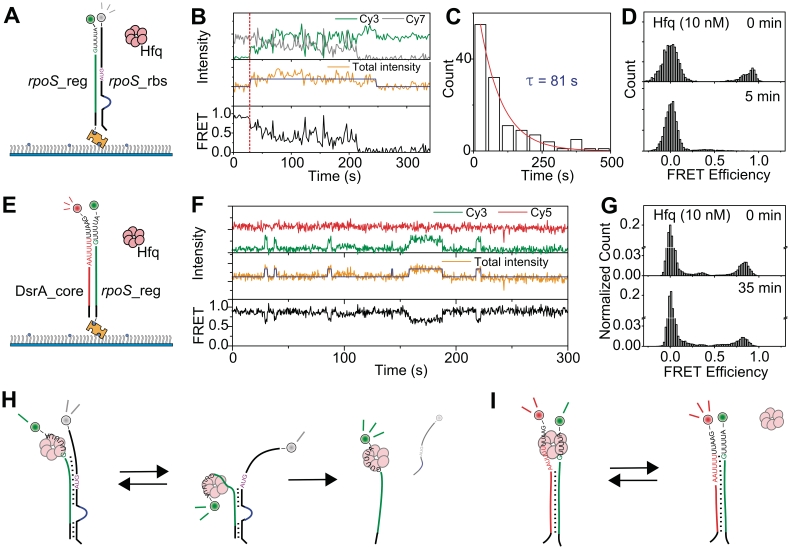
To investigate the detailed mechanism of rpoS_reg:rpoS_rbs complex unwinding by Hfq, we added Hfq to the detection chamber where rpoS_reg:rpoS_rbs complexes were immobilized (Figure 3A). An example of fluorescence intensity time traces in Figure 3B show that Hfq binding to RNA substrates can be monitored by intensity jumps of the sum signal (middle). This technique, termed protein induced fluorescence enhancement (PIFE), has been used to study the translocation of molecular motors on nucleic acids (25,26). Consistent with the conclusion, intensity jumps became more frequent with increasing Hfq concentration while the dwell time of the high intensity state remained the same (Supplementary Figure S1). In addition to the binding/dissociation events of Hfq, Figure 3B shows that Cy3–Cy7 FRET rapidly fluctuates upon Hfq binding, and eventually disappears (bottom). The unwinding time histogram gave 81 s of rpoS_reg:rpoS_rbs dissociation time (Figure 3C), and the unwinding was almost complete after incubation of 5 min (Figure 3D). From these observations, we infer that Hfq binding to an rpoS_reg:rpoS_rbs complex induces a partial unwinding of the complex and the competition of Hfq and rpoS_rbs for the interaction with rpoS_reg causes FRET fluctuations (Figure 3H). The model that a complete unwinding of the complex spontaneously follows due to the reduced stability of rpoS_reg:rpoS_rbs complex upon Hfq binding is consistent with previous data showing that Hfq destabilizes the secondary structure of rpoS (18,21).
Figure 3.
Unwinding mechanism of Hfq. (A) Experimental scheme of rpoS_reg:rpoS_rbs unwinding. Pre-annealed rpoS_reg:rpoS_rbs was immobilized on a polymer-coated quartz surface, and Hfq (10 nM) was added into the detection chamber. (B) Example traces of fluorescence intensities (top: green for Cy3 and gray for Cy7; middle: orange with blue for the sum of Cy3 and Cy7) and corresponding FRET (bottom: black). Blue lines in the middle panel were added as an eye-guide. The red dashed line indicates the injection point of Hfq. (C) Unwinding time of rpoS_reg:rpoS_rbs complex after the binding of Hfq. (D) FRET histograms before the injection of Hfq (top) and after 5 min incubation with Hfq (bottom). (E) Experimental scheme of DsrA_core:rpoS_reg unwinding. DsrA:rpoS_reg complex was immobilized on a polymer-coated quartz surface and Hfq (10 nM) was added into the detection chamber. (F) Representative fluorescence intensity and FRET time traces of the experiment described in (E). (G) FRET histograms of the experiment described in (E) before the injection of Hfq (top), and after the incubation with Hfq for 35 min (bottom). Model of rpoS_reg:rpoS_rbs unwinding by Hfq (H), and that of DsrA:rpoS_reg unwinding (I).
Next, we performed the same experiments with the pre-annealed DsrA_core:rpoS_reg complex (Figure 3E). Representative time traces of fluorescence intensities and FRET (Figure 3F) show that the binding of Hfq (monitored from the intensity jumps in the middle) induces a partial unwinding of the complex (monitored from the FRET drops occurring simultaneously with the Hfq binding), but complete unwinding of the annealed DsrA_core:rpoS_reg complex was very rare in this case (Figure 3F); even after incubation for 35 min, most of DsrA_core:rpoS_reg complexes remain intact (Figure 3G), which is well contrasted to the rapid unwinding of the rpoS_reg:rpoS_rbs complex after the binding of Hfq. We speculate that the DsrA_core:rpoS_reg complex does not proceed to a complete unwinding because both RNAs have Hfq-binding sites allowing Hfq holds both RNAs simultaneously in this case (Figure 3I).
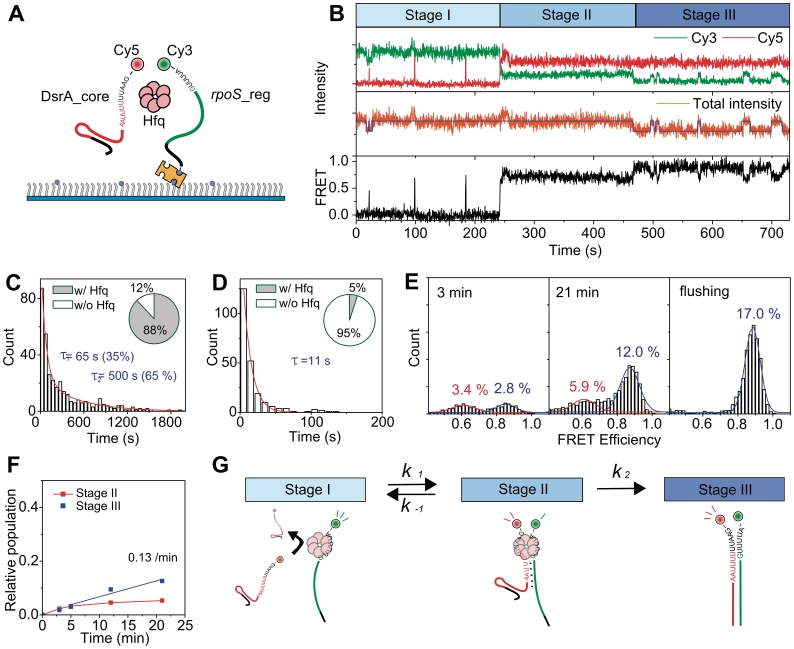
Annealing of DsrA and rpoS occurs through three distinct steps
To study the detailed mechanism of DsrA:rpoS annealing, we added a buffer containing DsrA_core and Hfq to a detection chamber where rpoS_reg is immobilized (Figure 4A). In Figure 4B, representative time traces of fluorescence intensities and corresponding FRET are shown. Under our experimental conditions, the FRET jumps (binding of DsrA_core) were not observed in the absence of Hfq, confirming a critical role of Hfq in this DsrA:rpoS interaction. Figure 4B also shows that the annealing process of DsrA_core and rpoS_reg can be classified into three distinct stages. In stage I, brief binding of DsrA_core (monitored from Cy3/Cy5 FRET spikes) to rpoS_reg:Hfq complex occurred repeatedly while the binding of Hfq with rpoS_reg (monitored from the intensity jumps of sum signal) was maintained for hundreds of seconds (Figure 4C). Under our experimental conditions, 88% of rpoS_reg molecules in stage I were in a Hfq-bound state (inset of Figure 4C). In stage II, the ternary complex of rpoS_reg, DsrA_core and Hfq is stably maintained for much longer time than the ternary complex formation events in stage I (Supplementary Figure S2). Molecules in stage II were observed to either regress back to stage I after the dissociation of DsrA_core, or to proceed to stage III. In stage III, the DsrA_core:rpoS_reg interaction is maintained even after the dissociation of Hfq, confirming the identity of stage III as the fully annealed state. It is noticeable that, compared to stage I, the dissociation time of Hfq is appreciably reduced in stage III (Figure 4D), and the portion of RNA molecules bound to Hfq is decreased (inset of Figure 4D), an observation consistent with the model of an efficient recycling of Hfq (21).
Figure 4.
Annealing of DsrA_core and rpoS_reg. (A) Schematic diagram of the experiment. Cy3-labeled rpoS_reg was immobilized on a polymer-coated quartz surface, and Cy5-labeled DsrA_core (5 nM) and Hfq (2 nM) were added into the detection chamber. (B) Representative time traces of single-molecule FRET experiments. The same color coding as in Figure 3f was used. The annealing process are classified into three distinct stages (stages I–III). (C) Binding lifetime of Hfq to rpoS_reg in stage I, and the portion of Hfq-bound rpoS_reg (inset). (D) Binding lifetime of Hfq to DsrA_core:rpoS_reg complex in stage III, and the portion of Hfq-bound DsrA_core:rpoS_reg complex (inset). (E) Evolution of FRET histograms after the delivery of DsrA_core and Hfq. Three FRET states were detected; a zero-FRET state corresponding to the absence of DsrA_core was omitted here for clarity. The last histogram was obtained after washing out the detection chamber at 24 min with a high-salt buffer (300 mM NaCl) to dissociate Hfq proteins. (F) Kinetics of product accumulation of DsrA_core:rpoS_reg. (G) A proposed model of the DsrA_core:rpoS_reg annealing by Hfq.
Due to low unwinding rate of the DsrA_core:rpoS_reg complex (Figure 3G), the fraction of stage III molecules gradually increases, as shown in time-lapse FRET histograms after the injection of Hfq and DsrA_core (Figure 4E). The last histogram of Figure 4E was obtained after washing out the chamber with high salt buffer (300 mM NaCl) at 24 min, in order to induce a complete dissociation of Hfq (27). In these histograms, the high FRET state (x-axis; E = 0.9) is solely contributed by the molecules in stage III after Hfq release, while the middle FRET state (E = 0.6) is contributed by the molecules in stage II as well as by the stage III molecules with bound Hfq. The fact that the high FRET population of the last histogram is identical within error to the sum of the middle and high FRET populations just before the flushing of Hfq supports the interpretation that the two FRET states correspond to either the partially or the fully annealed states. From the time-lapse FRET histograms, we determined the populations of molecules in stages II and III as functions of the incubation time (Supplementary Data). The time-evolution of stages II and III populations thus obtained shows that the population of stage III molecules linearly increases while the stage II population roughly remains a constant from 5 to 21 min (Figure 4F).
The observations in Figure 4B–F can be summarized as a reaction diagram of Hfq-mediated DsrA:rpoS annealing (Figure 4G), where the first transition (stages I–II) is reversible and approaches a steady-state relatively fast in our experimental condition, and the second transition (stages II–III) is almost irreversible. In the reaction diagram, the apparent forward rate of the first transition (k1) is a parameter depending on the Hfq and DsrA_core concentrations, opposed by the reverse rate of the first transition (k−1) and the forward rate of the second transition (k2). Because the stage II population is highly heterogeneous, the kinetic parameters in Figure 4G (k1, k−1 and k2) could not be uniquely determined from dwell-time analysis. However, an effective value of k2 could be determined as 0.13 min−1 from the steady-state approximation in the range of 5–21 min of (Figure 4F, Supplementary Data).
DsrA and rpoS compete for the same RNA binding site of Hfq
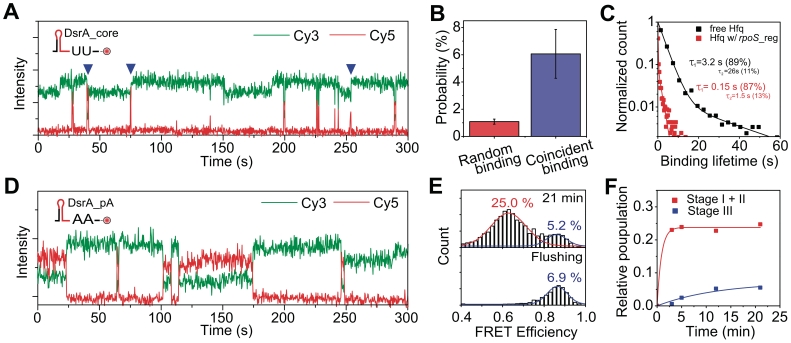
Considering that a biological role of Hfq is to anneal DsrA and rpoS, the formation of many unproductive DsrA:Hfq:rpoS ternary complexes in stage I is intriguing. Hence we asked why the ternary complex in stage I is so unstable. By investingating time traces in stage I, we noticed that many of the brief binding events of DsrA_core (monitored from Cy3–Cy5 FRET in Figure 5A) happen simultaneously (blue triangles in Figure 5A) with the binding or dissociation of Hfq proteins (monitored from intensity jumps of Cy3 signal in Figure 5A). The coincident probability between Hfq binding/dissociation and DsrA_core binding was several times larger than what is expected from a random event (Figure 5B, Supplementary Data). One model to explain the high probability of coincidence is that the two RNAs dynamically compete for the same RNA-binding site of Hfq. According to this competition model, the brevity of DsrA_core binding in stage I should not represent an intrinsic instability of the DsrA_core:Hfq interaction, but rather a hindrance of DsrA-binding caused by the existence of rpoS_reg pre-loaded on Hfq.
Figure 5.
Competition of DsrA_core and rpoS_reg for the same proximal RNA binding site. (A) Representative intensity time traces of DsrA_core:rpoS_reg annealing. Cy5-labeled DsrA_core (2 nM), and Hfq (1 nM) were added in a rpoS_reg-immobilized detection chamber. Frequent coincidence of DsrA_core binding and Hfq dissociation/association events are indicated (blue triangles). (B) Comparison of the random binding and coincident binding probabilities. (C) Normalized histograms of DsrA_core binding lifetime to Hfq loaded with rpoS_core (red), and to free Hfq (black). (D) Representative intensity time traces of the experiment in which Cy5-labeled DsrA_pA (5 nM) and Hfq (2 nM) were added to a rpoS_reg-immobilized detection chamber. Ternary complex is much more stable compared to the case in (A). (E) FRET histogram after 21 min incubation (top), and after flushing with 300 mM NaCl solution at 24 min (bottom). (F) Product accumulation kinetics of DsrA_pA:rpoS_reg.
To test this competition model, we measured binding lifetime of free Hfq to DsrA_core as follows. We immobilized a Cy3-labeled DsrA_core on a polymer-coated quartz surface, added Hfq into the detection chamber, and monitored the fluorescence signals of Cy3 (Supplementary Figure S1A). As before, the binding of Hfq could be monitored as intensity jumps of Cy3 signal (Supplementary Figure S1B). Dwell-time analysis of Hfq binding shows that the binding lifetime of DsrA to a free Hfq is much longer than when Hfq is pre-loaded by rpoS_reg (Figure 5C), consistent with the competition model. We also observed that, reciprocally, the binding lifetime of rpoS_reg to Hfq is reduced when Hfq is occupied by DsrA_core (Supplementary Figure S3).
The competition is advantageous for efficient annealing of DsrA and rpoS
Previous biochemical and structural studies revealed that Hfq has two RNA binding sites: the proximal site which prefers U-rich sequences (17,19), and the distal site which prefers A-rich sequences (16,19). Both RNA fragments used here have U-rich sequences (at the 3′-end of DsrA_core and at the 5′-end of rpoS_reg). Thus, it is a reasonable hypothesis that the proximal RNA-binding site is the competition site. To see what happens when the two RNAs are targeted to different binding sites in Hfq, we prepared a DsrA mutant, DsrA_pA, in which the U-rich sequence at the 3′-end of DsrA_core was replaced with an A-rich sequence. As a consequence, DsrA_pA is expected to primarily bind to the distal RNA binding site of Hfq (16,19). Consistent with the hypothesis, control experiments with a distal side mutant of Hfq (Y25A) showed that binding stabilities of DsrA_core and rpoS_reg was not reduced while binding lifetime of DsrA_pA was reduced (Supplementary Figure S4). When we added Cy5-labeled DsrA_pA and Hfq to a detection chamber where Cy3-labeled rpoS_reg was immobilized, we could still observe a transient formation of ternary complexes (Supplementary Figure 5D), but in this case the binding lifetime of DsrA_pA on rpoS_reg:Hfq complex was increased by 100-folds compared to the case of DsrA_core (Supplementary Figure S5).
Despite the significant increase of the ternary complex stability in DsrA_pA, the relative population of the high FRET state of mutant DsrA_pA is reduced compared to that of DsrA_core (Figure 5E, top). Strikingly, high-salt flushing to remove Hfq shows that most of the middle FRET population is disrupted after the dissociation of Hfq (Figure 5E, bottom). Thus we conclude that the annealing efficiency of DsrA_pA and rpoS_reg is reduced despite of the greatly increased ternary complex ratio (Figure 5F). These observations indicate that the RNA-RNA interaction is rarely established (Figure 5F, blue) even though the ternary complex is rapidly formed (Figure 5F, red) and maintained for a much longer time. Because the replacement of the U-rich sequence with A-rich sequence does not cause any appreciable reduction in the stability of DsrA_pA:rpoS_reg complex (Supplementary Figure S6), the difference of the annealing efficiencies was not caused by the reduced stability of the RNA complex.
Observations in Figure 5A–F support the competition of the two RNAs for the proximal RNA binding site of Hfq. However, the competition does not seem to be limited to the proximal binding site, because the binding lifetime of rpoS_reg was still reduced several fold when Hfq was loaded with DsrA_pA (Supplementary Figure S7). Therefore, we conclude that the major competition site of DsrA and rpoS binding is on the proximal side, but non-specific interaction on the periphery of the torus, which is highly positively charged, is also important for RNA-binding (28).
We note that the non-specific RNA interaction itself does not support an efficient RNA annealing because when rpoS_rbs was added together with Hfq to the rpoS_reg-immobilized chamber, we still observed frequent transient binding events of rpoS_rbs, but we could not observed any appreciable annealing (Supplementary Figure S8). We speculate that when one of the two RNAs binds to Hfq only via non-specific interactions, the annealing of the two RNAs is inefficient due to a very low probability of the proper alignment of the two RNA for annealing.
DISCUSSION
It has been intriguing how RNA chaperones manifest two completely opposite functions—annealing and unwinding—in a substrate-specific manner. We focused on the Hfq RNA chaperone activity on E. coli DsrA and rpoS. Using single-molecule FRET and synthesized RNA fragments from DsrA and rpoS, we observed that Hfq-dependent annealing proceeds through three separable stages—transient unproductive binding events, partial annealing and full annealing. Analysis of binding kinetics revealed that Hfq helps the annealing of the two RNAs by using the proximal RNA-binding site. The competition of the two RNAs to occupy the same RNA binding site of Hfq resulted in many unproductive formations of transient ternary complexes, but surprisingly the competition was advantageous for the efficient annealing of the two RNAs; when rpoS and DsrA fragments are designed to use different RNA binding sites of Hfq, the annealing efficiency was dramatically decreased. Therefore, the distal binding site of Hfq may not directly involved in annealing of DsrA and rpoS. Note that, despite belonging to the same protein family, Sm protein mechanisms probably do not bind RNA on their distal face (29). We infer that when the two RNAs compete for the same binding site, there is a higher probability that complementary sequences of two RNAs are properly aligned compared to the case when the two RNAs use different Hfq binding sites (proximal/distal). This interpretation is consistent with Hfq as local concentration enhancer, without the need to consume ATP (30). Furthermore, there have been debates on the oligomeric state of the active form of Hfq, and our finding demonstrates that a single Hfq hexamer can facilitate the DsrA–rpoS annealing process.
We need to emphasize that our observation is not contradictory to the recent observation that the poly-A region in the leader sequence of rpoS greatly enhances the annealing efficiency of DsrA and rpoS (18,31) because the A-rich region of rpoS can contribute to the recruitment of DsrA and Hfq while the U-rich region is actually used in the annealing process. Therefore, we propose the following model for Hfq-mediated rpoS–sRNA interactions. First, by using the two binding sites, Hfq enhances the local concentration of two RNAs by forming a stable ternary complex. Then, by using the same RNA binding site, Hfq drives eventual annealing of two RNAs by placing two RNAs in close proximity.
Finally, while the annealing activity of Hfq is manifested when two interacting RNAs have poly-U sites as in DsrA and rpoS fragments, RNA unwinding activity is manifested when only one RNA molecule has a specific Hfq binding site as in the inhibitory stem of rpoS (21). The unwinding of the RNA complex is initiated because the binding of Hfq to the RNA complex induces a partial unwinding of the complex, resulting in a reduced stability of the RNA.
In conclusion, our study has revealed for the first time that both the rpoS and DsrA compete for the same binding site of Hfq, making the RNA–Hfq interaction more dynamic, but also surprisingly greatly increasing the overall annealing efficiency. We also show that when Hfq specifically binds to only one of two RNAs, in the inhibitory stem of rpoS, the unwinding process dominates over the annealing process, thus shedding a new light on the substrate selectivity for annealing or unwinding.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Creative Research Initiatives (Physical Genetics Laboratory, 2009-0081562); World Class University project (R31-2009-100320); CEA, CNRS and University Paris 7. Funding for open access charge: Creative Research Initiatives (Physical Genetics Laboratory, 2009-0081562).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are very indebted to B. Cayrol for his help in preparing Hfq protein, to R. Roy and T. Ha who contributed to this work at early stage, and to R.A. Lease, R. Roy and C. Joo for critical reading of the article. W. Hwang thanks S. Bae, J. Lee and H. Uhm for the assistance in aligning the optical setup.
REFERENCES
- 1.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 2.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Qβ ribonucleic acid. J. Biol. Chem. 1972;247:824–821. [PubMed] [Google Scholar]
- 4.Tsui H-CT, Leung H-CE, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq Protein Increases OxyS RNA Interaction with Target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 6.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq. A Bacterial Sm-like Protein that Mediates RNA-RNA Interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect Biol. 2010 doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caron MP, Lafontaine DA, Masse E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 2010;7:140–144. doi: 10.4161/rna.7.2.11056. [DOI] [PubMed] [Google Scholar]
- 12.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. RNAs: regulators of bacterial virulence. Nat. Rev. Microbiol. 2010;8:857–866. doi: 10.1038/nrmicro2457. [DOI] [PubMed] [Google Scholar]
- 14.Arluison V, Derreumaux P, Allemand F, Folichon M, Hajnsdorf E, Regnier P. Structural Modelling of the Sm-like Protein Hfq from Escherichia coli. J. Mol. Biol. 2002;320:705–712. doi: 10.1016/s0022-2836(02)00548-x. [DOI] [PubMed] [Google Scholar]
- 15.Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell Mol. Biol. 2009;272:149–189. doi: 10.1016/S1937-6448(08)01604-3. [DOI] [PubMed] [Google Scholar]
- 16.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq- RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Arluison V, Mura C, Guzman MR, Liquier J, Pellegrini O, Gingery M, Regnier P, Marco S. Three-dimensional structures of fibrillar Sm proteins: Hfq and other Sm-like proteins. J. Mol. Biol. 2006;356:86–96. doi: 10.1016/j.jmb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S, Lee J, Hohng S. Single-molecule three-color FRET with both negligible spectral overlap and long observation time. PLoS ONE. 2010;5:e12270. doi: 10.1371/journal.pone.0012270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc. Natl Acad. Sci. USA. 2007;104:12610–12615. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 31.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.