Abstract
The FtsK translocase pumps dsDNA directionally at ∼5 kb/s and facilitates chromosome unlinking by activating XerCD site-specific recombination at dif, located in the replication terminus of the Escherichia coli chromosome. We show directly that the γ regulatory subdomain of FtsK activates XerD catalytic activity to generate Holliday junction intermediates that can then be resolved by XerC. Furthermore, we demonstrate that γ can activate XerCD-dif recombination in the absence of the translocase domain, when it is fused to XerCD, or added in isolation. In these cases the recombination products are topologically complex and would impair chromosome unlinking. We propose that FtsK translocation and activation of unlinking are normally coupled, with the translocation being essential for ensuring that the products of recombination are topologically unlinked, an essential feature of the role of FtsK in chromosome segregation.
INTRODUCTION
Faithful chromosome duplication and segregation has to precede cell division to maintain genomic stability. The FtsK DNA translocase of Escherichia coli functions in coordinating chromosome unlinking and cell division by using an approximately 200 amino acid N-terminal domain and a linker of roughly 600 amino acids, to facilitate the late stages of cell division (1,2), and a C-terminal DNA translocase domain, of about 500 amino acids, that acts in chromosome unlinking (3,4). FtsK is targeted to the invaginating septum at about the time of completion of DNA replication and likely only accesses chromosomal DNA that has not been segregated in a timely manner; for example, when chromosomal dimers have formed, decatenation is incomplete or completion of replication is delayed (5–7). FtsK-like proteins are highly conserved in eubacteria, where they appear to undertake the same functions as E. coli FtsK, or alternatively function in the transport of DNA into the forespore, during Bacillus subtilis sporulation (8–10).
Characterization of the isolated FtsK translocase domain has shown that it translocates dsDNA rapidly (∼5 kb/s) and directionally in vitro, removing protein and nucleic acid roadblocks as it translocates (11–18). Directionality of translocation is imparted by FtsK preferentially loading at specific DNA sequences, KOPS [GGGNAGGG; (14,17,19)] that are oriented in the chromosome such that any loading events at KOPS will lead to FtsK translocating towards the dif recombination site, contained in the replication termination region (18). FtsK translocation stops when the translocase encounters XerCD bound to dif (13).
In the initial in vitro characterization of FtsK translocation, it was proposed that the ATP hydrolysis-dependent stimulation of XerCD–dif recombination required a direct remodelling of the XerCD–dif synaptic complex in addition to any translocation role (11). Subsequently, it was demonstrated that the C-terminal γ regulatory subdomain of FtsK is responsible for activation, by a direct interaction with XerD (20). Furthermore, in a substrate containing a nick within dif, γ alone could stimulate cleavage of dif by XerD, independently of ATP and could facilitate strand exchange between the nicked substrate and an intact dif site (20). However, no information was forthcoming on the role of ATP hydrolysis and translocation in the recombination between intact dif sites. These studies led to the proposal that a pre-existing nick in the dif DNA provides flexibility in the XerCD–dif nucleoprotein complex, thereby removing the requirement for an ATP-hydrolysis-dependent remodelling during the activation process. Recently, it was shown that the expression of isolated γ domain in cells lacking FtsKc could lead to a low level (∼1%) of recombination between two copies of dif in the chromosome (21). Analysis of the analogous Lactococcus lactis recombination system in E. coli (the recombinase XerS, the L. lactis FtsK homologue and repeats of the difSL site) showed that FtsK translocation stimulated recombination, but γ alone had little effect on recombination at the cognate Lactococcus dif site. The authors, therefore, conclude that there is an effect of FtsK translocation on the DNA that is required to stimulate recombination, in addition to the γ–XerD interaction (21).
Here we demonstrate that FtsK translocation is required to obtain unlinked products of XerCD–dif recombination. An isolated γ subdomain of FtsK, or γ fused to XerCD, can still activate recombination, but the products are now topologically complex and would hinder chromosome segregation. Indeed, there is no obligatory role for ATP hydrolysis during the recombination reaction, as the entire FtsK motor domain is dispensable. However, the XerCD-γ fusion proteins cannot support chromosome dimer resolution (CDR) in the absence of FtsK activity in vivo. Finally we show that, in XerCD–dif recombination reactions, γ stimulates the activation of XerD to form Holliday junction (HJ) intermediates that can then be resolved by XerC.
MATERIALS AND METHODS
Proteins
FtsK50C and FtsK50CΔγ purifications were as described (14). FtsK γ and Lγ are as in (17). XerC and XerD mutant proteins were purified as described (22). The γ3 fusion was generated by cloning three tandem repeats of γ separated by 14 amino acid linkers (GGGSEGGGSEGGSG) into pBAD24 such that the N-terminus had a 6× His-tag. XerCγ and XerDγ were cloned by PCR so that the same 14 amino acid linker was inserted at the C-terminus of the XerC or XerD gene followed by an XbaI restriction site, then the γ domain of FtsK. These constructs were put into pBAD24 for expression, as were wild-type XerC and XerD.
The γ3, XerC, XerCγ, XerD and XerDγ proteins were each expressed in B834 xerD. Cells were grown in LB at 37°C to A600 0.8, supplemented with 0.2% arabinose then grown for a further 3 h. Harvested cells were resuspended in buffer A (50 mM Tris pH 7.5, 500 mM NaCl, 10% glycerol), with 5 mM imidazole. Lysis (Avestin pressure homogeniser) followed by centrifugation at 40 000g for 30 min. Supernatant was loaded onto a 2 ml TALON column, washed with 80 ml buffer A, then eluted with buffer A + 150 mM imidazole. XerD and XerDγ were then dialysed into 25 mM Tris (pH 7.5), 500 mM NaCl, 1 mM EDTA, 1 mM DTT and 50% glycerol and stored at −20°C. XerC and XerCγ were further purified by addition of 0.5 M ammonium sulphate, followed by application to a 1 ml phenyl sepharose HP column (GE Healthcare) in buffer A + 0.5 M ammonium sulphate. Protein was eluted with a gradient to buffer B, 50 mM Tris (pH 7.5), 1 mM EDTA, 50 mM NaCl. Following TALON purification the γ3 protein was diluted 1:1 in 10 mM Tris (pH 7.5), 1 mM EDTA, and loaded onto a 1 ml Q fast flow column (GE Healthcare) in buffer A2 [50 mM Tris (pH 7.5), 1 mM EDTA, 50 mM NaCl, 1 mM DTT] and then eluted with a gradient to buffer A2 + 1 M NaCl.
In vivo recombination reactions
These were carried out as previously described (17). Briefly a strain lacking the C-terminus of FtsK [DS9041, ftsK::cat1, insertion at nucleotide 678 (23)] was transformed with a 2× dif reporter plasmid [pFX142; (11)] and a protein expression vector. Cells were grown overnight with glucose, subcultured into fresh LB and grown to A600 0.5. At this point 0.05% arabinose was added; samples were taken at 1 h intervals and plasmid DNA prepared (Qiagen mini-prep). DNA was then separated on 0.8% agarose in 1× TBE (89 mM Tris–borate, 1 mM EDTA).
In vivo growth competition
Assays were carried out as described previously (18,24). Each protein was constitutively expressed, without induction, from the arabinose promoter of pBAD24. The relative viability defect per cell, per generation was calculated as in (25).
In vitro recombination reactions
Recombination reactions were carried out in recombination buffer: 20 mM Tris (pH 7.5), 5 mM MgCl2, 1.25% PEG 8000, 1.25% PEG 300. Proteins and DNA were mixed on ice, before 2 mM ATP was added and reactions placed at 37°C. Final concentrations of each protein, when present in a reaction, were 40 nM FtsK50C hexamer, 80 nM XerD/XerDγ, 150 nM XerC/XerCγ and 1 µM γ3 or γ. Plasmid DNA (400 ng [∼5 nM] per reaction) was pSI56 (Ip et al., 2003), except for Figure 2, B+C: pFX142. Reactions were stopped after 10 min (unless otherwise stated) by phenol/chloroform extraction, followed by isopropanol precipitation. Subsequently, DNA was electrophoresed directly, digested with EcoRI (New England Biolabs), or nicked with DNaseI (6). Separation was on 0.8% agarose, 1× TBE, at 55V for 16 h. DNA was visualized with Cybr Green using a Fuji FLA-3000.
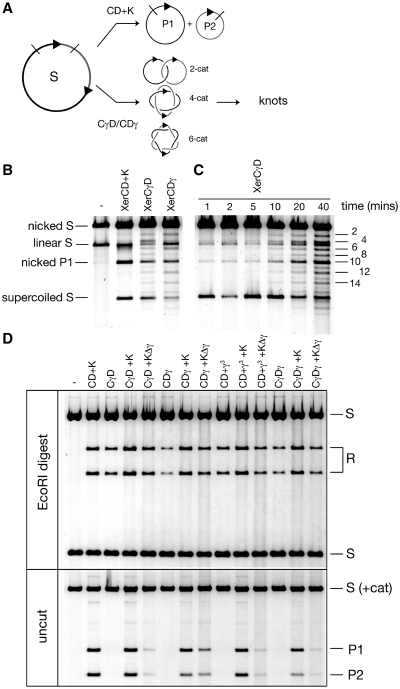
Figure 2.
(A) Schematic of recombination reactions; FtsK dependent recombination gives exclusively free products (P1 + P2) whereas the XerCγ or XerDγ fusion proteins produce mainly catenated products with a small amount of free P1 + P2. Catenated products up to six crossings (6-cat) are shown but higher forms are apparent in the gels. Second recombination events on the catenanes can produce knotted products, both twist and torus knots depending upon the synaptic complexity. DNA supercoiling is not shown for clarity. (B) Recombination reactions (20 min) with the indicated proteins were nicked with DNaseI in the presence of ethidium, to reveal the presence of catenated/knotted recombination products. Both fusion proteins produce catenanes, whereas FtsK (K) with XerCD (CD) does not. Nicking was not complete leaving some supercoiled plasmid substrate (supercoiled S). The smaller product, P2, was present lower down the gel but is not shown here. (C) Timecourse of recombination with XerCγD proteins. Reactions were nicked as previously. Catenanes (as labelled; the 10-crossing catenane co-migrates with nicked P1) appear at early time points and become stronger upon incubation, concomitant with the appearance of knotted products (weaker unlabelled bands, interdigitated with the catenanes). (D) Recombination reactions with the indicated proteins were either digested with EcoRI to show clearly the amount of recombination (upper panel) or electrophoresed without cutting (lower panel) to show the level of free circle product. Supercoiled catenated products were not resolved from the supercoiled substrate in the lower gel.
Analysis of knots and catenanes from XerCγD reactions
Reactions containing 800 ng DNA were nicked, electrophoresed, visualized with ethidium bromide, and the individual catenane or knot bands excised separately from the gel. DNA was recovered (Qiagen kit), and subsequently cut with EcoRI, and electrophoresed again (0.8% agarose, 1× TBE). After visualization with Sybr Green, catenanes showed the recombinant restriction pattern whereas knots were largely the parental pattern.
Isolation and analysis of Holliday junctions
Recombination reactions were supplemented with the indicated concentration of peptide WRWYCR that inhibits HJ processing. After 10 min, DNA was phenol/chloroform extracted and precipitated with isopropanol. It was cut with EcoRI, treated with calf intestinal phosphatase and separated on 0.7% agarose in 1× TBE + 0.3 µg/ml ethidium bromide, at 60V for 16 h. HJs were excised and purified (Qiagen kit). DNA was then 5′-end labelled using T4 PNK and γ32P ATP. A portion of this DNA was then further digested with ScaI for 5 h. DNA was isopropanol precipitated and resuspended in a buffer containing 50 mM NaOH, 1 mM EDTA, and heated to 55°C for 5 min immediately prior to separation on 1% agarose in 50 mM NaOH, 1 mM EDTA, for 14 h at 30V. DNA was then transferred to Hybond N+ membrane (GE Healthcare) and visualized on a Fuji FLA-3000.
RESULTS
The isolated γ domain of FtsK is sufficient to activate recombination
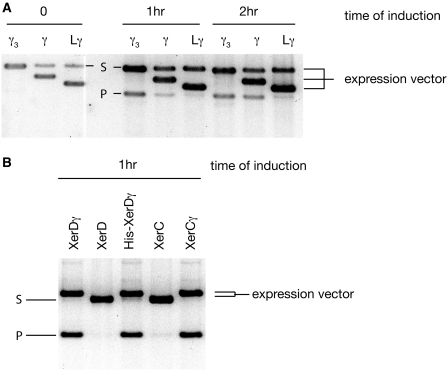
Previous results have indicated that ATP hydrolysis by FtsK was necessary to stimulate recombination between pairs of dif sites in vitro (11,14,26). In order to test whether this is the case in vivo, a strain lacking the C-terminus of FtsK (FtsKC−) was transformed with a reporter plasmid harbouring two dif sites in direct repeat (pFX142), along with a vector bearing the isolated FtsKγ domain under control of an inducible promoter. If recombination occurs then two smaller circular products are formed by a deletion reaction from the circular reporter plasmid. Cells were cultured in the presence of glucose to repress expression from the promoter before addition of inducer. Three different versions of the γ domain were tested: the isolated γ domain used for crystallization (17); Lγ, which also has the linker sequence that attaches the N-terminus of γ to the motor domain β; and γ3, with three repeats of γ, each separated by 14 amino-acid linkers. After addition of inducer for each promoter, samples were taken at 1 h and 2 h time points and the DNA extracted and analysed (Figure 1A). Recombination products are seen when all three γ variants were expressed. γ3 was more proficient in inducing recombination than isolated γ, which in turn was more efficient than Lγ. Although the exact levels of protein expression were not investigated in these experiments, we believe that they are likely to be very similar, because each construct is based on the same FtsKγ domain, expressed from the same promoter. Further, in over-expression tests, each form of the γ protein was expressed to a similarly high level upon induction. We conclude that γ is sufficient to stimulate recombination between dif sites in vivo, and activation of XerCD–dif recombination can be completely separated from the DNA translocation activity of the FtsK motor. The topology of the recombination products is not revealed by these assays, as, in vivo, TopoIV can unlink catenanes to produce free DNA circles.
Figure 1.
(A) Timecourse of in vivo recombination of a plasmid with two dif sites in direct repeat. DNA was prepared from cells at the indicated times after induction. Three different constructs expressing variants of isolated FtsKγ domain (γ) cloned into various plasmid backgrounds were used (hence the size differences of the expression vectors). Recombination yields the product P after induction of γ. The smaller, non-replicating product migrates further in the gel and becomes lost from the population over time, so has not been shown. Note that the γ3 expression vector migrates at the same position as the unrecombined reporter plasmid. (B) In vivo recombination following induction of the Xer recombinases and their fusions to γ. DNA was sampled 1 h after induction of each protein. The expression vectors and reporter plasmid migrate at similar positions, but the larger recombination product is clearly visible in the lanes where the fusion proteins are expressed. The stimulation of recombination by the γ fusions to XerC and XerD was so efficient that significant recombination was also observed at the zero time point (not shown) because of leaky expression of the proteins.
The ability of isolated γ to stimulate recombination at dif was also tested in vitro. Equal concentrations of purified γ or γ3 proteins were added to recombination reactions containing XerCD and dif sites. The dif substrates were either on plasmids (11,27), or on short radiolabelled fragments (14). In both cases addition of isolated FtsK γ domain was sufficient to stimulate Xer-mediated recombination at dif (Figure 2D and data not shown). Nevertheless, appreciable levels of recombination required a high concentration of γ or γ3, for reasons unknown.
XerCγ and XerDγ fusion proteins are active in recombination at dif in the absence of FtsK
In order to test recombination activity with stoichiometric levels of γ to the XerCD recombinases, fusion proteins, with a 14 amino acid linker coupling the C-terminus of the recombinase to γ, were produced, giving XerCγ and XerDγ. These were expressed from the arabinose promoter of pBAD24 in a FtsKC− strain containing a two-dif reporter plasmid as above. DNA samples taken 1 h after induction show almost complete conversion of substrate to recombinant deletion product in the XerCγ and XerDγ fusions (Figure 1B). Controls expressing wild-type XerC or XerD from the same promoter failed to give recombination in the FtsKC− background, as expected. Thus fusion of γ to the C-terminus of either Xer recombinase allowed recombination at dif independent from the translocase motor of FtsK.
In order to determine whether Xer-mediated recombination in the presence of these fusion proteins required the appropriate partner recombinase to be present, the fusion constructs were expressed in a strain lacking FtsKC and either XerC or XerD. Recombination was only observed in strains where the XerCγ or XerDγ fusion protein had its appropriate partner recombinase present (data not shown), indicating that recombination still required both XerC and XerD proteins but could be stimulated by γ covalently attached to either.
Crystal structures of several tyrosine recombinases have revealed that the recombinase C-termini interact with the partner recombinases to control catalytic activity (28–30). In order to confirm that the activation of recombination seen with the fusion proteins was specific to the γ domain, a variety of other fusions were constructed at the C-terminus of either XerC or XerD. These failed to show any recombination in the absence of FtsK (data not shown).
Activation of Xer-mediated recombination at dif in the absence of FtsK yields complex topological products
XerCγ and XerDγ fusion proteins were purified and their recombination activity tested in vitro. Both fusion proteins, when provided with wild-type partner recombinase, were competent in stimulating high levels of recombination between dif sites on a supercoiled plasmid substrate (Figure 2B and D), as well as on short oligonucleotide substrates (data not shown), without the addition of FtsK.
Following recombination, DNA was nicked prior to electrophoresis to reveal the topology of the products. XerCD–FtsK mediated recombination between two dif sites on the same plasmid gives rise to exclusively free circular products (11) (Figure 2A and B). Recombination with the fusion proteins in the absence of FtsK, however, produced mainly catenated products, though with some free circles also present (Figure 2B). The same was true for stimulation of XerCD by addition of γ3 (Figure 2D; data not shown). This is the pattern of recombination products expected from random collision of XerCD–dif sites in a supercoiled plasmid, and mimics the recombination products seen with either Cre- or FLP-mediated deletion from a plasmid (27,31). Accumulation of recombination products was relatively slow in the absence of FtsK; a timecourse of recombination stimulated by XerCγ and XerD showed a slow gradual increase in catenated product over time (Figure 2C), whereas the majority of XerCD–FtsK recombination occurs within the first few minutes (data not shown). At longer times XerCγD reactions showed the presence of odd numbered, knotted products. These were confirmed to be the result of second rounds of recombination, since isolated catenanes and knots showed recombinant and parental restriction patterns, respectively.
Addition of FtsK, but not FtsKΔγ, leads to the majority of recombination giving free products
Recombination reactions were initiated as above using combinations of the fusion proteins and wild-type recombinase proteins, and additionally either FtsK50C, or FtsK50CΔγ was added. FtsK50CΔγ can translocate efficiently, but lacks the γ domain and therefore cannot activate XerCD-dif (13). At the concentrations used both FtsK50C and FtsK50CΔγ have been shown to load and to translocate efficiently on DNA by a triplex displacement assay (12,13). Since the Xer-mediated recombination could be carried out by the fusion proteins independently of direct FtsK activation, these assays allowed the dissection of the contribution of DNA translocation by the FtsK motor activity to the overall topology of the products.
Reactions were stopped after 5 min, and the DNA was either cut with EcoRI so that the overall level of recombination could easily be assessed, or electrophoresed without treatment, so that the amount of free circle recombination products could be seen (Figure 2D). It is clear that each combination of fusion proteins, or the addition of γ3 to wild-type XerCD proteins, produce recombination. Further, each combination yields roughly similar levels of product. Additionally, when FtsK50C was present, there was a reproducible moderate increase in recombination efficiency (Figure 2D, upper panel). Strikingly, the majority of the recombination products were free circles in the presence of FtsK50C rather than the catenated products seen in the absence of FtsK (Figure 2D, lower panel). When FtsK50CΔγ was added to the reactions then only a modest increase in the level of free recombination products was seen. This was true of the situations where either or both Xer-γ fusion proteins were used, and when wild-type XerCD proteins were supplemented with γ3, even though recombination in these reactions could progress without the need for the γ domain attached to FtsK. Therefore, it was concluded that DNA translocation by the FtsK motor protein leads to simplification of topology, but that this was only efficient when the motor still had the γ domain attached.
XerC-γ and XerD-γ can function in chromosome dimer resolution only in the presence of active FtsK
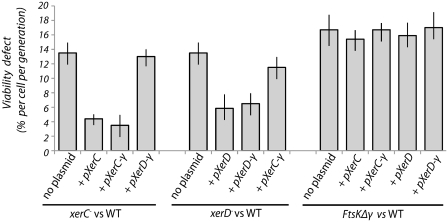
The ability of the Xer-γ fusion proteins to function in CDR was tested using growth competition experiments (Figure 3). Strains deficient in CDR show a reduced fitness when co-cultured with an otherwise isogenic strain that is proficient in CDR (5,18,24). In every generation, roughly 15% of cells form a chromosome dimer, which, if unresolved, causes the cell to filament, and eventually die (25,32). Thus, there is a growth defect associated with the inability to carry out CDR when compared to a wild-type strain over time.
Figure 3.
XerC-γ and XerD-γ function in vivo, but cannot carry out CDR without FtsK. Histograms show the viability defect per cell, per generation, calculated from the competition assay of the indicated mutant strain (carrying xerC-, xerD- or ftsKΔγ mutations) versus the wild-type isogenic reference strain AB1157, both in the absence and presence of indicated plasmid (encoding XerC, XerD, XerC-γ or XerD-γ).
It was found that a plasmid expressing the XerC-γ fusion could complement a xerC mutant as effectively as a plasmid carrying wild-type XerC (Figure 3). Similarly, XerD-γ was observed to complement a xerD mutant as well as wild-type XerD. Complementation is not complete with either plasmid as there is still a growth defect with respect to wild-type, but both plasmids reduce the defect from ∼13% to between 3 and 6%. The failure to restore wild-type growth levels could be due to protein expression levels in these cells: proteins were constitutively expressed from the arabinose promoter without induction, and are likely to vary from the wild-type levels of expression. However, it was clear that the fusion proteins performed as well as wild-type when expressed from plasmids in the complementation assay. Therefore, each fusion protein is seen to be proficient for CDR in an otherwise wild-type background. Further tests showed that the fusion proteins were not toxic to the cells at this level of production (data not shown), so it seems likely that the failure of both wild-type and fusion proteins to fully complement the mutant phenotype could be due to limiting protein levels in these cells.
Since the Xer-γ fusion proteins were proficient in recombination in vitro without FtsK (Figure 2), co-culture assays were carried out in a FtsKΔγ background to see if the fusion proteins would be proficient in CDR without FtsK activity (Figure 3). The results show that there is no survival advantage in this background to expressing the fusion proteins, presumably because CDR is not effective. This situation parallels that seen when the loxP site replaces dif, and the Cre recombinase is expressed; recombination itself at loxP is not FtsK dependent-, but CDR still requires a functional FtsK (18,24).
FtsK-dependent Xer recombination at dif is initiated by XerD
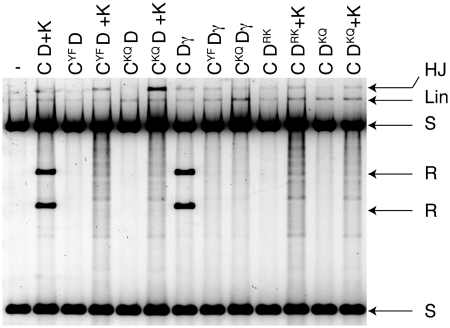
It has previously been shown that XerD can make HJs at dif in the presence of FtsK, when the catalytic mutant XerCY275F was used (11). This was interpreted as meaning that during a recombination reaction at dif stimulated by FtsK, which XerD carries out the first pair of strand exchanges, and then XerC resolves the junction in a second pair of strand exchanges. However, when combinations of the XerCγ or XerDγ fusion proteins, or FtsK50C, were used with various other catalytic mutants of XerC or XerD, it was found that both XerC and XerD could exchange a pair of strands to produce HJs, and so it was ambiguous as to which recombinase normally carried out the first exchanges (Figure 4). The level of HJ that is trapped as an intermediate in these reactions is low, as expected, as HJ can be resolved back to parental conformation by the active recombinase.
Figure 4.
Reactions with recombinase catalytic mutants reveal that HJ can be formed by either XerC or XerD mediated strand exchange at dif. Recombination reactions with combinations of proteins as indicated, were cut with EcoRI to reveal recombinant product and HJ. Many combinations tested produced some level of HJ indicating that either recombinases could potentially carry out the first pair of strand exchanges, even when FtsK50C (K) or γ were present in the reaction. Reactions with FtsK50C have a background smearing upon digestion, but this does not affect the interpretation. Proteins: CYF = XerCY275F, CKQ = XerCK172Q, DRK = XerDR148K, DKQ = XerDK172Q.
There is a problem with using catalytic mutants of tyrosine family integrase proteins; the catalytic potential of partner recombinases are closely interlinked by relatively subtle allosteric changes in the interacting proteins, so that only one pair of cleavages can occur at any one time. Thus, inactivating one recombinase at a recombination site with mutants in the catalytic site could potentially inappropriately activate the partner recombinase (22,33).
Therefore, to investigate which pair of strands is usually exchanged upon stimulation of XerCD-dif by γ, catalytically wild-type proteins, or the γ fusion protein derivatives thereof, were used. The initial pair of strand exchanges forms a HJ, which can be specifically trapped using small peptides (34). Therefore a peptide was titrated into a recombination reaction on a plasmid substrate with two dif sites (Figure 5A), and does indeed show an accumulation of HJs. Upon restriction digestion the resulting HJ migrates slowly in the gel, and can be isolated and analysed. Using 1 µM peptide, HJs from reactions were isolated following EcoRI digestion. These were then labelled at the 5′-ends with 32P. Strand exchange by either XerC or XerD gives diagnostic fragment sizes when the HJs were further digested with another restriction enzyme, ScaI. Individual strand sizes were analysed in denaturing agarose gels (Figure 5B). As has been demonstrated before (35), XerC can generate HJs at dif in the absence of FtsK, which gives a background level of XerC-mediated strand exchange in all reactions. This level is apparent in the reactions with only XerCD present (Figure 5B). Upon addition of FtsK50C, or either fusion protein, or the γ3 protein to these reactions, the diagnostic XerD exchange product was seen in addition to this XerC-mediated background. This indicates that upon stimulation by the FtsKγ domain, XerD does indeed carry out the first pair of strand exchanges at dif, and the same effect is seen whether the γ is attached to FtsK, or either recombinase protein or even isolated γ. This confirms the earlier assumption, made using catalytic mutants, that in a XerCD–FtsK reaction at dif, XerD initiates recombination and forms a HJ intermediate that is subsequently resolved by XerC (11). It also confirms that the covalent fusion proteins appear to recombine dif by the same order of strand exchanges as when FtsK stimulates the reaction, and that these fusions use the normal reaction mechanism.
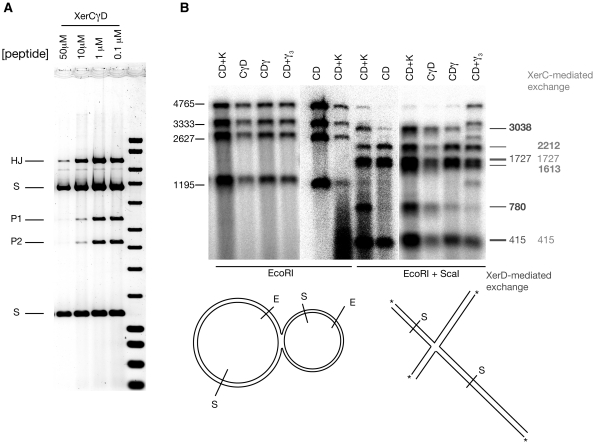
Figure 5.
(A) Recombination in the presence of peptide WRWYCR traps HJs. Recombination was carried out in the presence of the indicated concentration of peptide and subsequently cut with EcoRI so that HJs migrate slowly. (B) Denaturing alkali gels allow determination of exchanged strands in isolated HJs. Isolated HJs were 5′-end labelled at each EcoRI cut site. Subsequently, some of the DNA was then further digested with ScaI, and samples were then denatured and electrophoresed. The relative positions of each site are shown diagrammatically below the gel. Sizes of the four strands resulting from EcoRI digestion are shown alongside (left). The expected sizes of top strand exchange (XerC-mediated) and bottom strand exchange (XerD-mediated) are shown on the right. Two strand sizes (3038 and 780) are specific for XerD mediated exchange (shown in bold), while XerC mediated exchange produces two different diagnostic product sizes (2212 and 1613, also in bold). The other strand sizes (415 and 1727) are common to both events. Note that there is always a background of XerC-mediated exchange, which can be estimated from the CD alone lane. However, upon stimulation by γ (in any form) the level of XerD-mediated exchange is greatly increased. Note that there was partial digestion by ScaI of the XerCD + γ3 reaction (far right lane) so that the four bands seen with EcoRI digestion are still present.
DISCUSSION
Bacterial chromosome dimer resolution and decatenation by XerCD recombination at the chromosomal dif site requires the FtsK DNA translocase, with its γ domain stimulating recombination (11,17,20,35). Here we show that stimulation of recombination by the γ domain can be completely uncoupled from the ATPase and DNA translocase activities of the FtsK motor domain. Therefore, we can rule out models in which supercoil induction by the translocating motor, or direct protein remodelling by the ATPase activity of FtsK are involved in the recombination reaction. Addition of γ alone, or its covalent fusion to either (or both) recombinase proteins, leads to efficient recombination. We also show directly that upon interaction with γ, XerCD switches its catalytic activity so that XerD carries out the first pair of strand exchanges to produce a HJ that is subsequently resolved by XerC.
Exactly how the XerD–γ interaction activates recombination at dif by stimulating XerD to carry out the first pair of strand exchanges is unknown. Based on structural models proposed for the Cre tetramer and subsequently validated for both Flp and λ-Int recombinases, one pair of recombinases is active and the other pair is inactive at any given time. Activity of a recombinase pair is determined by the DNA bend direction and protein–protein interactions (Figure 6) (20,28–30). In order to swap which pair of recombinases in tetramer catalyses the first pair of exchanges the entire complex would need to be remodelled so that the DNA bend directions are changed and the protein–protein interactions are also changed, seemingly involving the breaking and reforming of all the protein–protein or protein–DNA interfaces in the complex. Therefore, it is attractive to think that the interaction of γ with XerD occurs prior to synapsis to reconfigure the XerCD complex so that XerD is poised to be active in catalysis. When the second XerCD–dif is encountered then a synapse would be produced where both XerDs in the tetramer are in the active configuration. It is unclear at the moment whether both XerDs require interaction with γ, or whether a single XerD/γ interaction would be sufficient for this.
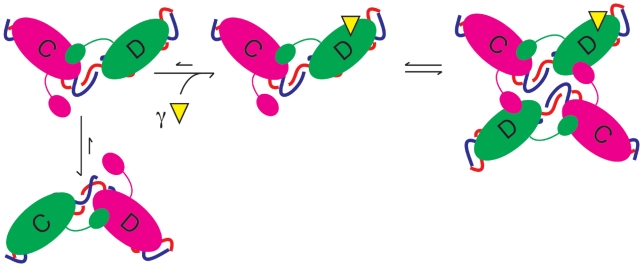
Figure 6.
Schematic diagram of activation of XerD by FtsKγ. The model is based upon numerous crystal structures where the direction of DNA bend determines which monomer is active (shown in green) or inactive (magenta) upon duplex DNA. In the absence of FtsKγ (yellow triangle) the equilibrium is skewed towards the XerC-active state. Upon interaction of XerD with FtsKγ, the equilibrium changes so that the XerD-active state is now favoured. This state then can make a productive synapse, wherein XerD exchanges the first pair of strands to form a HJ, which is subsequently resolved by XerC.
Based upon the Cre model stated above, evidence suggests that there is an equilibrium between the XerC- and XerD-active conformations, or directions of DNA bends, which in an intact dif site is skewed towards the XerC-active state (Figure 5). It was shown that, in vivo, XerC forms and resolves HJs in ‘futile’ cycles at the dif site (35), independently of FtsK activity. In vitro, suicide substrates based on the dif sequence with nicks in one or other strand help to alter the equilibrium to favour one or other recombinase becoming active (36); a nick in the top strand activates XerC mediated cleavage even further. XerD-mediated cleavage is only seen, albeit at a low level compared to XerC-mediated cleavage, when a nick is placed in the bottom strand. Notably, this XerD-mediated cleavage activity is further stimulated by the presence of γ (20). Finally, HJ formation mediated by XerD is only seen when γ is present, separately or as a fusion protein (Figure 3) (11,20). So, it seems that γ interacting with XerD alters this equillibrium to favour the XerD-active conformation. A γ–XerD interaction before synapsis could also imply an active role of translocation in bringing the synapse together, and could in some as yet unknown way, aid in producing simple topology products.
Alternatively, if FtsK encounters a ready-synapsed pair of XerCD–dif sites, it must be able at this stage to activate only simple synapses for recombination, a form of topological filter, so that only simple products are formed. It is clear that addition of γ or the XerC-γ/XerD-γ fusion proteins allows recombination from complex synapses, so any topological filter would have to result from translocating FtsK. Perhaps the disassembly/reassembly of the synapse to allow XerD activation could be involved in this—maybe only simple synapses are able to withstand the process and survive to re-assemble in a simple XerD-active synapse?
Whatever the nature of the interaction between γ and XerD, it seems clear that having γ associated with an active translocation motor is key to the production of simple, unlinked products. This is supported by the finding that the XerC-γ and XerD-γ fusion proteins can only support chromosome dimer resolution in living cells when there is a functional FtsK present. This could be because FtsK is required to efficiently synapse two sites as a result of its translocation activity, so that in the absence of FtsK activity, dif recombination is too inefficient to effectively support CDR. Alternatively it could be because the recombination at dif in the absence of FtsK produces complex, catenated products that cannot be segregated.
We conclude by noting that the XerCD recombination machinery can use two very different mechanisms to ensure topological selectivity during unlinking by site-specific recombination. During chromosome dimer resolution and unlinking, the action of FtsK ensures the products of recombination are unlinked; this recombination cannot distinguish inter- and intra-molecular recombination and can therefore act to decatenate monomeric chromosomes as well as resolve dimeric chromosomes to monomers (6). By comparison, the action of XerCD on recombination sites present in natural plasmids is restricted to intramolecular events by the use of accessory DNA sequences and proteins that ensure that recombination is restricted to a synaptic complex of defined topology (23). Such topological filters are used by various unrelated DNA processing systems to provide biological selectivity.
FUNDING
Funding for open access charge: Wellcome Trust Program Grant WT083469.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank J. Graham for valuable discussions, A. Segall for short peptides and S. Colloms for technical advice.
REFERENCES
- 1.Dubarry N, Possoz C, Barre FX. Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol. Microbiol. 78:1088–1100. doi: 10.1111/j.1365-2958.2010.07412.x. [DOI] [PubMed] [Google Scholar]
- 2.Lesterlin C, Pages C, Dubarry N, Dasgupta S, Cornet F. Asymmetry of chromosome Replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet. 2008;4:e1000288. doi: 10.1371/journal.pgen.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 4.Crozat E, Grainge I. FtsK DNA translocase: the fast motor that knows where it's going. Chembiochem. 2010;11:2232–2243. doi: 10.1002/cbic.201000347. [DOI] [PubMed] [Google Scholar]
- 5.Bigot S, Corre J, Louarn JM, Cornet F, Barre FX. FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol. Microbiol. 2004;54:876–886. doi: 10.1111/j.1365-2958.2004.04335.x. [DOI] [PubMed] [Google Scholar]
- 6.Grainge I, Bregu M, Vazquez M, Sivanathan V, Ip SC, Sherratt DJ. Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 2007;26:4228–4238. doi: 10.1038/sj.emboj.7601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy SP, Chevalier F, Barre FX. Delayed activation of Xer recombination at dif by FtsK during septum assembly in Escherichia coli. Mol. Microbiol. 2008;68:1018–1028. doi: 10.1111/j.1365-2958.2008.06212.x. [DOI] [PubMed] [Google Scholar]
- 8.Biller SJ, Burkholder WF. The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning. Mol. Microbiol. 2009;74:790–809. doi: 10.1111/j.1365-2958.2009.06893.x. [DOI] [PubMed] [Google Scholar]
- 9.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 11.Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 12.Graham JE, Sherratt DJ, Szczelkun MD. Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA. Proc. Natl Acad. Sci. USA. 2010;107:20263–20268. doi: 10.1073/pnas.1007518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham JE, Sivanathan V, Sherratt DJ, Arciszewska LK. FtsK translocation on DNA stops at XerCD-dif. Nucleic Acids Res. 2010;38:72–81. doi: 10.1093/nar/gkp843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol. Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, Bustamante C, Cozzarelli NR. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 16.Saleh OA, Perals C, Barre FX, Allemand JF. Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004;23:2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivanathan V, Allen MD, de Bekker C, Baker R, Arciszewska LK, Freund SM, Bycroft M, Lowe J, Sherratt DJ. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat. Struct. Mol. Biol. 2006;13:965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivanathan V, Emerson JE, Pages C, Cornet F, Sherratt DJ, Arciszewska LK. KOPS-guided DNA translocation by FtsK safeguards Escherichia coli chromosome segregation. Mol. Microbiol. 2009;71:1031–1042. doi: 10.1111/j.1365-2958.2008.06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bigot S, Saleh OA, Cornet F, Allemand JF, Barre FX. Oriented loading of FtsK on KOPS. Nat. Struct. Mol. Biol. 2006;13:1026–1028. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- 20.Yates J, Zhekov I, Baker R, Eklund B, Sherratt DJ, Arciszewska LK. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol. Microbiol. 2006;59:1754–1766. doi: 10.1111/j.1365-2958.2005.05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nolivos S, Pages C, Rousseau P, Le Bourgeois P, Cornet F. Are two better than one? Analysis of an FtsK/Xer recombination system that uses a single recombinase. Nucleic Acids Res. 2010;38:6477–6489. doi: 10.1093/nar/gkq507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arciszewska LK, Baker RA, Hallet B, Sherratt DJ. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J. Mol. Biol. 2000;299:391–403. doi: 10.1006/jmbi.2000.3762. [DOI] [PubMed] [Google Scholar]
- 23.Recchia GD, Aroyo M, Wolf D, Blakely G, Sherratt DJ. FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J. 1999;18:5724–5734. doi: 10.1093/emboj/18.20.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capiaux H, Lesterlin C, Perals K, Louarn JM, Cornet F. A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep. 2002;3:532–536. doi: 10.1093/embo-reports/kvf116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perals K, Cornet F, Merlet Y, Delon I, Louarn JM. Functional polarization of the Escherichia coli chromosome terminus: the dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarity. Mol. Microbiol. 2000;36:33–43. doi: 10.1046/j.1365-2958.2000.01847.x. [DOI] [PubMed] [Google Scholar]
- 26.Massey TH, Aussel L, Barre FX, Sherratt DJ. Asymmetric activation of Xer site-specific recombination by FtsK. EMBO Rep. 2004;5:399–404. doi: 10.1038/sj.embor.7400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ip SC, Bregu M, Barre FX, Sherratt DJ. Decatenation of DNA circles by FtsK-dependent Xer site-specific recombination. EMBO J. 2003;22:6399–6407. doi: 10.1093/emboj/cdg589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 30.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 31.Crisona NJ, Weinberg RL, Peter BJ, Sumners DW, Cozzarelli NR. The topological mechanism of phage lambda integrase. J. Mol. Biol. 1999;289:747–775. doi: 10.1006/jmbi.1999.2771. [DOI] [PubMed] [Google Scholar]
- 32.Steiner WW, Kuempel PL. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J. Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallet B, Arciszewska LK, Sherratt DJ. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol. Cell. 1999;4:949–959. doi: 10.1016/s1097-2765(00)80224-5. [DOI] [PubMed] [Google Scholar]
- 34.Gunderson CW, Boldt JL, Authement RN, Segall AM. Peptide wrwycr inhibits the excision of several prophages and traps holliday junctions inside bacteria. J. Bacteriol. 2009;191:2169–2176. doi: 10.1128/JB.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barre FX, Aroyo M, Colloms SD, Helfrich A, Cornet F, Sherratt DJ. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blakely GW, Davidson AO, Sherratt DJ. Binding and cleavage of nicked substrates by site-specific recombinases XerC and XerD. J. Mol. Biol. 1997;265:30–39. doi: 10.1006/jmbi.1996.0709. [DOI] [PubMed] [Google Scholar]