Abstract
The RNA Pol II transcription complex pauses just downstream of the promoter in a significant fraction of human genes. The local features of genomic structure that contribute to pausing have not been defined. Here, we show that genes that pause are more G-rich within the region flanking the transcription start site (TSS) than RefSeq genes or non-paused genes. We show that enrichment of binding motifs for common transcription factors, such as SP1, may account for G-richness upstream but not downstream of the TSS. We further show that pausing correlates with the presence of a GrIn1 element, an element bearing one or more G4 motifs at the 5′-end of the first intron, on the non-template DNA strand. These results suggest potential roles for dynamic G4 DNA and G4 RNA structures in cis-regulation of pausing, and thus genome-wide regulation of gene expression, in human cells.
INTRODUCTION
Genome-wide studies have shown that Pol II transcription complexes pause just downstream of the transcription start site (TSS) at many human genes (1–3). Pausing may poise a polymerase for rapid induction of transcription upon receipt of the appropriate signal, or provide a checkpoint at which the transcription complex ensures that all factors are present for productive elongation. Pausing occurs only at a fraction of genes, so one or more features of genomic sequence or structure must contribute to pausing at human genes. Those features have not yet been defined. Identification of the local features of DNA architecture that contribute to DNA pausing has important implications for understanding mechanisms of genomic instability and the response of cells to chemotherapeutics.
G-rich intron 1 (GrIn1) elements are a recently identified feature of genomic structure (4). These conserved elements are present in almost one-half of all human genes and map to the 5′-end of the first intron and the non-template strand. They bear the signature sequence motif characteristic of regions with potential to form G4 structures, G≥3NxG≥3NxG≥3NxG≥3 (5–8). Their G-richness cannot be accounted for by sequences that would make them targets of well-defined regulatory mechanisms, such as CpG dinucleotides that undergo methylation, or motifs recognized by transcription factors or RNA processing factors. GrIn1 elements occupy a privileged genomic position, as they are located on average 200 nt downstream of the TSS, within 100 bp of the 5′-end of the first intron and on the non-template strand. An element at this intronic position may regulate transcription or RNA processing without conferring selective pressure on protein sequence.
The position, conservation and abundance of GrIn1 elements suggest that these elements might function in regulation of gene expression. The G-richness of the GrIn1 element confers the potential to form a dynamic structure upon transcription of a genomic region. This structure, called a G-loop, carries a co-transcriptional RNA/DNA hybrid on the template strand, and G4 DNA interspersed with single-stranded regions on the G-rich non-template (coding) strand (9–12). Persistent co-transcriptional RNA/DNA hybrids like those that characterize G-loops can contribute to genomic instability (11,13–16). They also prolong the denaturation of the DNA strands that normally accompanies transcription, enhancing the potential of DNA to form G4 structures that may function as regulatory targets.
Here, we address the possibility that GrIn1 elements correlate with transcriptional pausing. We show that genes that can be classified as paused are more G-rich in the region flanking the TSS than RefSeq genes or non-paused genes, and we demonstrate that there is a strong correlation between transcriptional pausing and the presence of a GrIn1 element. These results suggest that formation of G4 structures on the non-template strand of the DNA or at the 5′-end of the nascent mRNA may promote promoter proximal pausing. GrIn1 elements may thereby contribute to genome-wide regulation of gene expression of specific classes of genes and they may also influence cellular sensitivity to drugs that perturb the normal dynamics of formation of DNA structure during transcription, including topoisomerase poisons and compounds designed to target G4 structures.
MATERIALS AND METHODS
Sequence data, regulatory motif masking and statistical analysis
Sequence data for the 18 187 human RefSeq genes (NCBI 36 assembly) were downloaded from the Ensembl database 54 using BioMart (17,18). As previously, we defined G-richness as the frequency within each set of genes of 100 nt sequence that contains a G4 DNA signature motif, G≥3NxG≥3NxG≥3NxG≥3 (5). Intron sequence derivation and calculations of G-richness were performed as described (4). For genes that express alternative transcripts with different first introns, the 5′-most first intron was included in the analysis. Masking of regulatory motifs was performed as described (4). The χ2-test was performed with the statistics program R version 2.7.1.
Microarray analysis of NCI-60 lines
Affymetrix GeneChip Human Exon 1.0 ST (GH Exon 1.0 ST) microarray analysis of NCI-60 cancer cell lines was carried out as described previously (8). In brief, microarrays were hybridized, usually in triplicate, following manufacturer’s instructions at GeneLogic (Gaithersburg, MD), and results normalized by robust multi-array analysis (19) using Partek Genomics Suite version 6.3. The GH Exon 1.0 ST microarray analysis of the NCI-60 lines characterized expression of 16 959 annotated genes, and probes were mapped to transcripts using exon designations assigned by SpliceCenter (20). Classification of genes as paused or non-paused was based on the difference in average probe set intensity level of expression and average standard deviation between the first exon and the other exons across all cell lines. Probe intensity criteria were first developed empirically for the topoisomerase 1 (TOP1) gene (8), and those criteria were applied to define paused genes from the larger database. At TOP1, it was noted that exon 1 was expressed both at higher level and in a manner less variable than the other exons. The increase in expression level of exon 1 was 1.24, and the reduction in standard deviation was 0.244. For subsequent analyses, paused genes were defined as exhibiting a difference in average intensities of the exons 2 through N as compared to exon 1 less than −1.24, and an increase in the standard deviations of the exons 2 through N as compared with exon 1 of greater than 0.244. Genes were classified as non-paused if they exhibited a difference in average intensities of the exons 2 through N as compared to exon 1 ≥0, and a decrease in average standard deviation of less than zero. Using these criteria, 3165 (19%) genes were classified as paused and 1401 (8%) as non-paused. The remaining genes did not fall into either category.
RESULTS
Inverse correlation between Pol II binding and G-richness
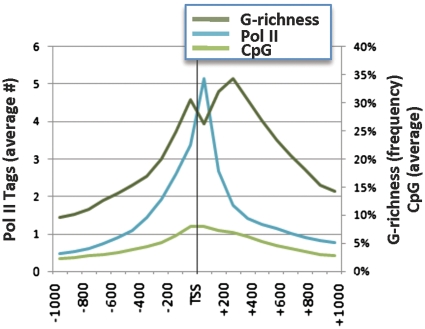
In human genes, two peaks of G-richness flank the TSS, centred on the region −100 to +1 and +200 to +300 (4). To ask if these peaks of G-richness correlate with binding by RNA Pol II, we graphed the frequency of G-richness and of Pol II binding sites as determined by Chromatin Immunoprecipitation-Sequencing (ChIP-Seq) for human T cells (1) in the 2 kb region flanking the TSS. The peak of Pol II binding, near +100, corresponded to a local minimum of G-richness, 200 bp downstream from the peak, near +300 (Figure 1). This peak represents the average of all Pol II molecules, regardless of pausing status. That the peak of Pol II binding coincides with a local minimum of G-richness is consistent with the A/T richness of most promoters.
Figure 1.
Inverse correlation between Pol II binding and G-richness at the TSS. Graph of the frequency Pol II binding sites (1), CpG dinucleotides, and the frequency of G-richness in the interval −1000 to +1000 around the TSS. G-richness was defined as the frequency within each set of genes of 100 nt sequences containing the G4 DNA signature motif, G≥3NxG≥3NxG≥3NxG≥3 (4).
CpG dinucleotides, which are sites for regulatory methylation, can contribute to local G-richness. We graphed the distribution of CpG dinucleotides in the region flanking the TSS, and showed that this comprised a relatively broad peak, which is not coincident with the peaks of G-richness and lies somewhat upstream of the peak of Pol II binding (Figure 1).
Promoters of paused genes are enriched in G4 motifs
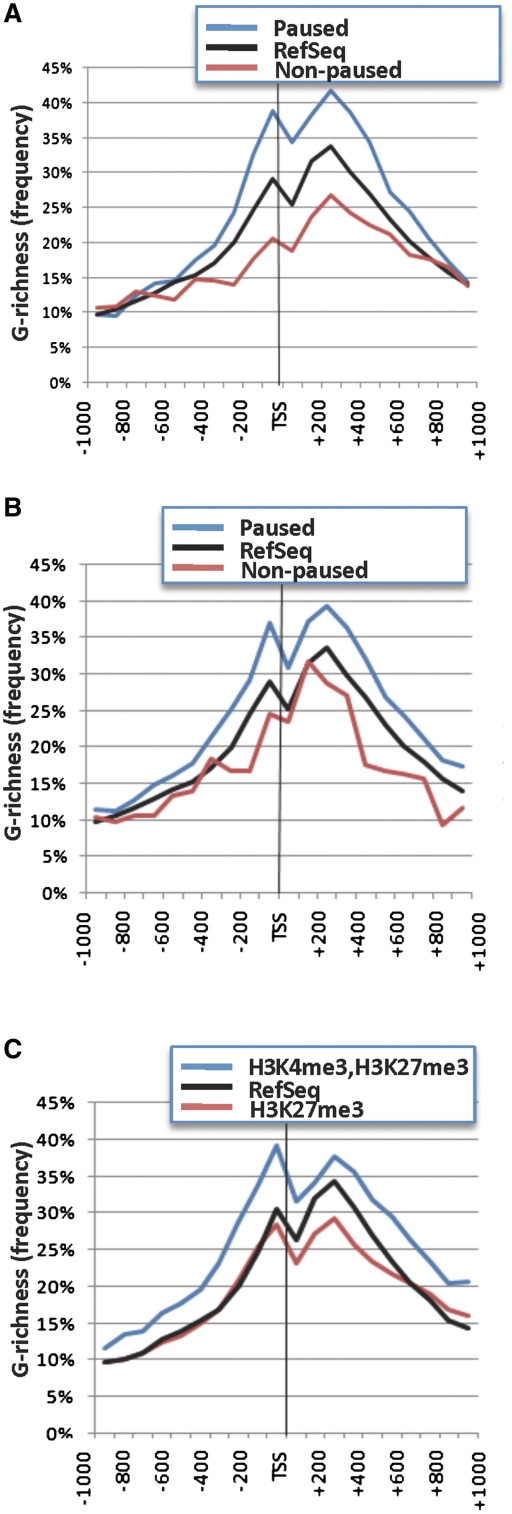
We next asked whether G-richness correlates with pausing, using three different operational definitions to classify genes as paused. One of these definitions distinguishes paused and non-paused genes based on relative expression of exon 1 and downstream exons, as determined by microarray analysis. The NCI-60 panel of cell lines includes 60 cell lines representing multiple tumor types for which drug sensitivity and transcriptome activity have been extensively studied and correlated (8,21–23). We calculated the frequency of G-richness in the region −1000 to +1000 for the genes classified as paused (19%) or not paused (8%) across all cell lines in the NCI-60 panel database, and for all RefSeq genes. Paused genes were more G-rich than RefSeq genes or than non-paused genes (Figure 2A).
Figure 2.
G4 motifs are enriched near promoters of paused genes. (A) Graph of the frequency of G-richness in genes defined as paused from the NCI-60 database in the interval −1000 to +1000 around the TSS. (B) Graph of the frequency of G-richness in genes in which Pol II is stably associated with the TSS in the absence of gene expression in the interval −1000 to +1000 around the TSS. This data set derives from analysis of primary resting human CD4+ T cells (1), and corresponds to the same data set for which genome-wide analysis Pol II position is presented in Figure 1. (C) Graph of the frequency of G-richness in the interval −1000 to +1000 around the TSS in genes carrying bivalent chromatin marks H3K4me3 and H3K27me3, as determined by analysis of primary resting human CD4+ T cells (1).
A second operational definition identifies paused genes as those at which Pol II is stably associated with the TSS in the absence of gene expression. Approximately one-third of genes in primary resting human CD4+ T cells were classified as paused by this criterion (1). We calculated the frequency of G-richness of the region flanking the TSS of those genes relative to RefSeq genes and non-paused genes in that data set. This analysis showed that paused genes were more G-rich in the region flanking the TSS than other genes (Figure 2B).
Chromatin marks can also be used to distinguish paused and non-paused genes. Histone modifications correlate with gene expression, with H3K4me3 characterizing active genes and H3K27me3 characterizing repressed genes. Some genes carry bivalent chromatin marks, with H3K4me3 near the promoter and H3K27me3 distributed more broadly along the gene and such bivalent marks can be used to distinguish paused genes from other genes (1,24). Calculation of the frequency of G-richness in the region from −1000 to +1000 showed that genes with bivalent H3K4me3 and H3K27me3 marks were more G-rich than RefSeq genes or inactive genes with monovalent H3K27me3 marks (Figure 2C).
The above analyses show that paused genes as defined by any of the three above criteria are more G-rich than non-paused genes. The G-richness of paused genes extends throughout the 2 kb interval analyzed, and includes regions both upstream and downstream of the promoter. Sequences upstream of the promoter may contribute to pausing by serving as sites for transcription factors that promote pausing. In this regard, it is interesting that genes classified as non-paused based on relative expression of exon 1 and downstream exons were comparatively G-poor (Figure 2A). This raises the possibility that transcription factors with G/C rich binding motifs may contribute to pausing at some genes, or conversely that transcription factors with A/T rich binding motifs may prevent pausing at others.
Strand biased G-richness downstream of the TSS at paused genes
The results above (Figure 2) establish that paused genes are more G-rich than other genes. How might G-richness contribute to the mechanism of pausing? Pausing could in principle be caused by formation of G4 structures in either the DNA or the nascent transcript. If G-loop formation contributes to the mechanism of pausing, then G-richness of paused genes is predicted to exhibit a strand bias, with G-rich regions downstream of the TSS concentrated in the non-template strand (9,10,12). We therefore compared the frequency of non-template and template strand G-richness in the 2 kb region spanning the TSS for genes classified as paused and non-paused based on relative expression of exon 1 and downstream exons in the NCI-60 database, and for all RefSeq genes. For all three groups of genes, there was clear strand asymmetry in G-richness downstream of the TSS, with greater G-richness on the non-template strand. Notably, paused genes were more G-rich than RefSeq genes, which were more G-rich than non-paused genes (Figure 3).
Figure 3.
Strand-biased G-richness downstream of the TSS at paused genes. Graph of the frequency of G-richness of the non-template (dark lines) and template (pale lines) strands in the interval −1000 to +1000 around the TSS for genes in the NCI-60 database classified as paused (left) or non-paused (center), and RefSeq genes (right).
G-richness of the genes analyzed, exhibited a characteristic distribution. For all three groups, more genes were G-rich on the non-template strand than on the template strand.
For all three groups of genes, upstream of the TSS and on the non-template strand, the maximum frequency of G-richness fell within the region from −100 to −1, where 40% of paused genes were G-rich, compared with 22% of non-paused genes and 30% of all RefSeq genes. Downstream of the TSS and on the non-template strand, maximum frequency of G-richness fell within the region from +200 to +300, where 42% of the paused genes were G-rich, compared with 28% of the non-paused genes and 35% of the RefSeq genes. Downstream of the TSS and on the non-template strand, a peak in the frequency of G-richness was also evident among paused and RefSeq genes, but not non-paused genes.
Transcriptional regulatory motifs account for some but not all G-richness near the TSS of paused genes
G-richness can reflect the presence of DNA sequence elements with well-characterized functions, including CpG dinucleotides that are targets of methylation as well as motifs for some common transcription factors that recognize G-rich sites in duplex DNA, including SP1 (RGGCGKR), KLF (GGGGTGGGG), EKLF (AGGGTGKGG), MAZ (GGGAGGG), EGR-1 (GCGTGGGCG) and AP-2 (CGCCNGSGGG). To eliminate contributions from these elements, we analyzed the distribution of G-richness with these sites masked. The frequency of G-richness may be greatly underestimated following masking, because masking is carried out based on DNA sequence alone, independent of information on whether a motif actually serves as a binding site for its cognate factor. Moreover, in the absence of knowledge regarding whether a specific motif contributes to pausing, masking may even eliminate from the tally genes bearing a motif that promotes pausing. Nonetheless, masking provides a convenient view of how canonical motifs affect the genomic landscape.
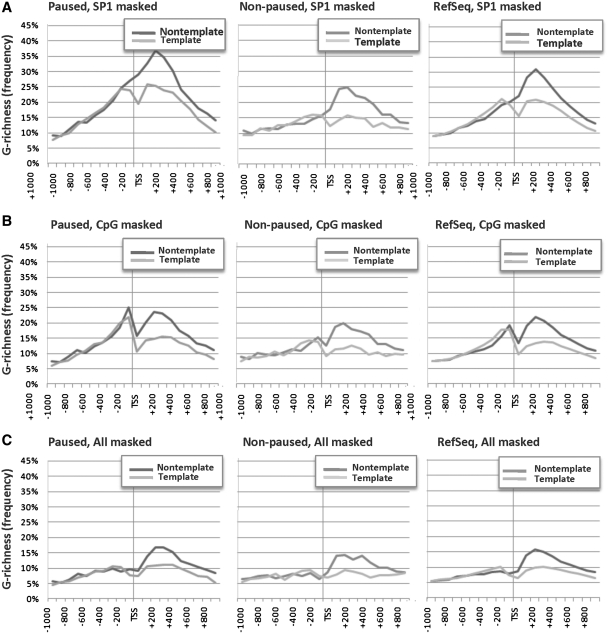
We first masked SP1 binding motifs, separately analyzing all RefSeq genes and the paused and non-paused genes identified in the NCI-60 database. Eliminating SP1 motifs primarily affected the region upstream of the TSS, eliminating the peak of G-richness upstream of the TSS in the non-template (but not template) strand of all three sets of genes (Figure 4A). Downstream of the TSS, G-richness of the non-template strand for paused genes was still greater (37%) than for non-paused genes (24%) or RefSeq genes (31%).
Figure 4.
Transcriptional regulatory motifs do not account for G-richness near TSS of paused genes. Graph of the frequency of G-richness of non-template (dark lines) and template (pale lines) strands in the interval −1000 to +1000 around the TSS for paused genes (left), non-paused genes (centre) and all human RefSeq genes (right), with the following motifs masked: (A) SP1 motifs. (B) CpG motifs. (C) CpG, SP1, MAZ, KLF, EKLF, EGR-1 and AP-2 motifs.
Masking of CpG dinucleotides reduced the frequency of G-richness both upstream and downstream of the TSS in all three sets of genes (Figure 4B). Even after masking, there was clear strand asymmetry in G-richness downstream of the TSS for all groups of genes. In addition, G-richness of paused genes remained greater at both upstream and downstream peaks (25 and 24%, respectively) than G-richness of non-paused genes (15 and 20%) or RefSeq genes (19 and 22%). Thus, although CpG content corresponded with pausing in both upstream and downstream regions, it did not account for all of the G-richness surrounding the TSS. We note that a peak of G-richness upstream of the TSS that was eliminated by masking SP1 motifs (Figure 4A) persisted after masking CpG motifs (Figure 4B), suggesting that SP1 motifs make their primary contribution to G-richness upstream and not downstream of the TSS.
Finally, we maximally depleted common G-rich motifs by masking binding motifs for six common transcription factors that bind G-rich sites, including SP1, KLF, EKLF MAZ, EGR-1 and AP-2, as well as CpG motifs. To maximize depletion of these canonical motifs, they were masked before eliminating CpG motifs. This stringent masking diminished the peaks of G-richness upstream and downstream of the TSS in all three classes of genes, but affected the upstream peak most profoundly (Figure 4C). Following stringent masking, the strand asymmetry in G-richness downstream of the TSS persisted, although only small differences were evident in non-template strand G-richness at both upstream and downstream peaks of paused genes (11 and 17%, respectively) relative to non-paused genes (7 and 14%, respectively). The very high stringency of masking is likely to be responsible for this considerable decrease in frequency of G-richness, and these small differences are unlikely to be significant.
GrIn1 elements correlate with pausing
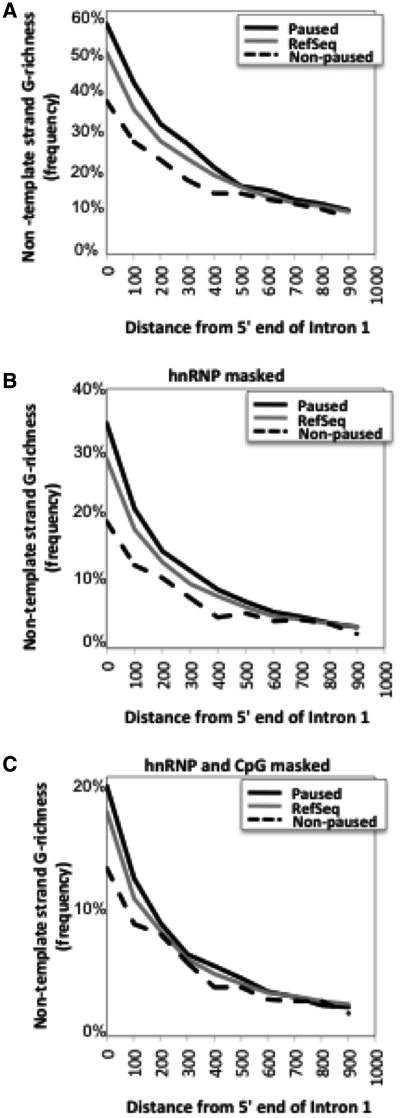
We previously found that almost one-half of all human genes contain G-rich elements on the non-template DNA strand at the 5′-end of the first intron, referred to as GrIn1 elements (4). To ask if a difference in GrIn1 element frequency characterizes paused and non-paused genes, we calculated G-richness for 1000 bp of the first introns for RefSeq genes and genes classified as paused or non-paused in the NCI-60 database. (This analysis was restricted to introns at least 1000 bp in length in order to include a constant number of genes along the length distribution. We previously showed (4) that setting the lower limit of intron size to either 100 bp or 1000 bp generates an essentially identical distribution of G-richness.) A fraction of genes in all three groups exhibited a peak of G-richness at the very 5′-end of the first intron, consistent with the presence of a GrIn1 element (Figure 5A). GrIn1 elements were present in 57% of paused genes, 38% of non-paused genes and 50% of RefSeq genes. The difference between the fraction of paused and non-paused genes containing GrIn1 elements was highly significant (χ2 = 82; P < 10−10).
Figure 5.
GrIn1 elements correlate with pausing. (A) Graph of the frequency of non-template strand G-richness within 1 kb downstream of the TSS for paused, non-paused genes and RefSeq genes (top). (B) Graph of the frequency of non-template strand G-richness as in Panel A, but with hnRNP A (UAGGGU/A) and hnRNP H (GGGA) motifs masked. (C) Graph of the frequency of non-template strand G-richness as in Panel B, with CpG motifs, hnRNP A (UAGGGU/A) and hnRNP H (GGGA) motifs masked.
Motifs for hnRNP proteins and CpG dinucleotides contribute to but do not account for GrIn1 elements
Two hnRNP proteins involved in RNA processing recognize motifs containing runs of three or more guanines in single-stranded DNA or RNA, hnRNP A (UAGGGU/A) and hnRNP H (GGGA) (25,26). These motifs contribute to but are not sufficient for binding, so the tally of motifs will overestimate their functional contribution of to G-richness of the intron. After masking these motifs, 34% of paused genes, 19% of non-paused genes and 28% of RefSeq genes retained a peak of G-richness, differences comparable with those observed upon analyzing the unmasked genes (Figure 5B). Masking CpG motifs in addition to hnRNP A and H binding motifs reduced the frequency of G-richness at the 5′-end of intron 1, so that a peak of G-richness was evident in 19% of paused genes, 13% of non-paused genes and 17% of RefSeq genes (Figure 5C). Thus, even with all these motifs masked, there was a greater frequency of GrIn1 elements in paused genes than in other gene classes.
DISCUSSION
We have identified a correlation between G-richness near the TSS and pausing in human genes. This correlation emerged from a genome-wide analysis, which examined genes classified as paused in the NCI-60 panel of cell lines or in primary resting T cells. The analysis defined pausing by three different operational criteria: relative levels of transcripts from exon 1 and downstream exons; association of Pol II with the TSS in the absence of transcription; and bivalent histone marks. Downstream but not upstream of the TSS, G-richness of paused genes was biased to the non-template DNA strand. G-rich consensus recognition motifs for sequence-specific DNA or RNA binding proteins, or of CpG dinucleotides, accounted for some but not all G-richness of paused genes. We emphasize that while the correlation between G-richness and pausing was strong, it did not apply to all genes. Additional mechanisms undoubtedly contribute to pausing and G-richness is likely to be only one of the many factors that modulate pausing at any given gene.
The correlation between pausing and G-richness was particularly apparent at the 5′-end of the first intron, where paused genes proved significantly more likely to carry GrIn1 elements, defined as at least one G4 motif within the first 100 bp of the first intron, on the non-template DNA strand (4). GrIn1 elements characterized 57% of paused genes and only 38% of non-paused genes. The genomic position of GrIn1 elements is consistent with a possible role in promoter-proximal pausing. GrIn1 elements lie at the very 5′-end of the first intron, or ∼ 200–300 bp downstream of the TSS, as the median distance from the TSS to the 5′-end of the first intron is 198 bp for human genes and GrIn1 elements are about 100 bp in length. Promoter-proximal pausing occurs in the region +20 to +50 relative to the TSS (2). A regulatory element 200–300 bp downstream from the TSS could readily communicate with Pol II or other components of the transcription apparatus, to cause Pol II to pause.
G4 motifs and the mechanism of pausing
The correlation between G4 motifs and pausing suggests that dynamic structures formed upon transcription of a region bearing G4 motifs may contribute to regulation of pausing in cis. Figure 6 illustrates those structures, which may promote pausing by distinctive mechanisms: (i) A G4 DNA structure formed behind the advancing polymerase may be recognized by factors that regulate pausing, which in turn cause polymerase to pause. A compelling precedent for a cis-regulatory role for G4 DNA has recently been provided by evidence that G4 DNA formation controls pilin gene antigenic variation in Neisseria gonorrhea (27). In addition, the human TOP1 gene has recently been found regulated by pausing in the first intron at conserved G4 DNA elements (8). Alternatively, a G4 structure in the DNA might serve as a roadblock to an advancing polymerase, suggested by in vitro analysis of transcription on G-rich templates (28), as well as evidence that G4 motifs can block progression of DNA polymerase or even the translation machinery (29–31). In that case, pausing would not occur during the first round of transcription, but after a ‘pioneering’ round of transcription that enabled a G4 DNA structure to form. (ii) A G4 RNA structure in the 5′-end of the nascent transcript may communicate a pause to the transcription apparatus. This mechanism of pausing has been extensively documented in prokaryotes, where RNA hairpins interact with the polymerase complex to promote pausing at specific sites (32). In human cells, the Trans-Activating Response (TAR) element of the HIV-1 retrovirus has been shown to form a stem–loop structure recognized by Trans-Activator of Transcription (TAT) and associated factors to promote transcription (33). (iii) A stable co-transcriptional RNA/DNA hybrid may communicate a signal for pausing via the RNA processing apparatus or the transcription apparatus. Single molecule imaging has provided dramatic evidence of how co-transcriptional RNA/DNA hybrids can contribute to ‘pile-ups’ of Pol I actively transcribing the G-rich rDNA in budding yeast (34).
Figure 6.
Regulation of transcriptional pausing at G4 motifs. Model of dynamic nucleic acid structures that may contribute to pausing upon transcription of a G-rich region. Mechanisms that contribute to pausing may include: (i) G4 DNA formed behind an advancing polymerase may be recognized by factors that promote pausing, (ii) G4 DNA structure formed in a ‘pioneering’ round of transcription may serve as a roadblock during the next round of transcription, (iii) a G4 RNA structure in the nascent transcript may communicate a pause to the transcription complex, as occurs in prokaryotes and (iv) a stable co-transcriptional RNA/DNA hybrid may promote pausing, via signals transmitted through the RNA processing apparatus.
Polymerase pausing is transient (35) and specific regulatory mechanisms may enable a polymerase to exit the paused state. A polymerase that pauses upon encountering a G4 structure could resume transcription upon elimination of that structure, e.g. by a G4 helicase; or if the polymerase/G4 interaction was interrupted by another factor. In this regard, it is interesting that the hnRNP proteins which interact with RNA in the nucleus contain structural domains (RRM/RBD domains or RGG domains) that recognize and may destabilize G4 structures (36), raising the possibility that they may compete with components of the transcription apparatus for binding to G4 structures.
No single mechanism is likely to account for pausing at every gene. Moreover, the genome-wide analysis that we carried out does not show that all genes that pause carry GrIn1 elements; or that GrIn1 elements are simple identifiers of genes that pause. Nonetheless, the model in Figure 6 should provide a useful starting point for future experiments that elucidate the mechanism of pausing at individual genes and classes of genes.
G-richness and genomic instability in AID-expressing tumors
We have previously shown that G-rich regions are targets of translocations in B cell lymphomas that express the DNA deaminase, AID, although not in T cell leukemias, which do not express AID (11). AID associates with a pausing factor, Spt5 (37). The connection we have established between G-richness and pausing suggests that Spt5 may recruit AID to G-rich paused regions to initiate instability. High levels of AID expression characterize ovarian, breast and prostate malignancies (38) as well as B cell lymphomas. Our results suggest that G-rich sites of pausing may also be targeted for instability in those tumor types.
G4 motifs and drug sensitivity
A role for G4 structures in polymerase pausing has implications for improved understanding of the mechanisms of several classes of drugs, including G4-binding small molecule ligands, G4 aptamers and topoisomerase I poisons. Small molecules that target G4 structures are currently in active development, with telomeres and rDNA as specifically prominent targets (39–42). Our results suggest that interactions with transcription-induced structures may contribute to both the effects and side effects of these drugs. G4 aptamers have also shown promise in treatment of cancer, but their mechanism of action is complex (43). Our results raise the possibility that transcription-induced G4 structures may compete with aptamers for binding key factors, thereby causing unanticipated off-target effects. This could, for example, explain cell type specificity of some aptamers, as binding competition would be determined by the genes expressed in a given cell type.
Camptothecin, a topoisomerase I poison, is the prototype for an important class of cancer chemotherapeutics (44). Treatment of cells with camptothecin has been shown to diminish Pol II pausing (45), an observation which can be explained in terms of the model shown in Figure 6. Formation of co-transcriptional RNA/DNA hybrids is very sensitive to local superhelicity (16,34,46,47). Camptothecin treatment prolongs the half-life of the covalent topoisomerase I/DNA intermediate on the DNA, and may thereby diminish not only local superhelicity but also stability of the local structure containing a co-transcriptional hybrid that promotes pausing. This will contribute to reducing pausing at a subset of genes in camptothecin-treated cells. In this regard, it is interesting that the TOP1 gene, which encodes topoisomerase I, carries a GrIn1 element and is itself regulated by transcriptional pausing (8), which may render TOP1 expression sensitive to local superhelicity, and to camptothecin. The effect of camptothecin on transcript levels is likely to differ from gene to gene, depending on details of local regulation of gene expression and DNA architecture.
FUNDING
US National Cancer Institute (P01 CA77852 to N.M.); Basic and Cancer Immunology Training Grant (CA009537 to J.E.); US National Institutes of Health (R01 GM41712 and NIH R01 GM65988 to N.M.); Cancer Research Institute Tumor Immunology Pre-doctoral Training Grant (to J.E.); Intramural Research Program of the National Cancer Institute, Center for Cancer Research support (Z01 BC 006150-19LMP to Y.P. and W.C.R.). Funding for open access charge: P01 NCI CA77852.
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
We thank members of our laboratories for helpful discussions.
REFERENCES
- 1.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 3.Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- 4.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinhold WC, Mergny JL, Liu H, Ryan M, Pfister TD, Kinders R, Parchment R, Doroshow J, Weinstein JN, Pommier Y. Exon array analyses across the NCI-60 reveal potential regulation of TOP1 by transcription pausing at guanosine quartets in the first intron. Cancer Res. 2010;70:2191–2203. doi: 10.1158/0008-5472.CAN-09-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 10.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duquette ML, Huber MD, Maizels N. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 2007;67:2586–2594. doi: 10.1158/0008-5472.CAN-06-2419. [DOI] [PubMed] [Google Scholar]
- 12.Vallur AC, Maizels N. Activities of human exonuclease 1 that promote cleavage of transcribed immunoglobulin switch regions. Proc. Natl Acad. Sci. USA. 2008;105:16508–16512. doi: 10.1073/pnas.0805327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilera A. mRNA processing and genomic instability. Nat. Struct. Mol. Biol. 2005;12:737–738. doi: 10.1038/nsmb0905-737. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 15.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl Acad. Sci. USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, Conti C, Nakamura AJ, Das BB, Nicolas E, et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009;10:887–893. doi: 10.1038/embor.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard T, Andrews D, Caccamo M, Cameron G, Chen Y, Clamp M, Clarke L, Coates G, Cox T, Cunningham F, et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 19.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix geneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan MC, Zeeberg BR, Caplen NJ, Cleland JA, Kahn AB, Liu H, Weinstein JN. SpliceCenter: a suite of web-based bioinformatic applications for evaluating the impact of alternative splicing on RT-PCR, RNAi, microarray, and peptide-based studies. BMC Bioinformatics. 2008;9:313. doi: 10.1186/1471-2105-9-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, et al. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 22.Weinstein JN, Pommier Y. Transcriptomic analysis of the NCI-60 cancer cell lines. C. R. Biol. 2003;326:909–920. doi: 10.1016/j.crvi.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 25.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994b;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 27.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodford KJ, Howell RM, Usdin K. A novel K(+)-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J. Biol. Chem. 1994;269:27029–27035. [PubMed] [Google Scholar]
- 30.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Stevens M, De Clercq E, Balzarini J. The regulation of HIV-1 transcription: molecular targets for chemotherapeutic intervention. Med. Res. Rev. 2006;26:595–625. doi: 10.1002/med.20081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maizels NM. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc. Natl Acad. Sci. USA. 1973;70:3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 37.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, San-Martin BR, Barreto V, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pauklin S, Sernandez IV, Bachmann G, Ramiro AR, Petersen-Mahrt SK. Estrogen directly activates AID transcription and function. J. Exp. Med. 2009;206:99–111. doi: 10.1084/jem.20080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brassart B, Gomez D, De Cian A, Paterski R, Montagnac A, Qui KH, Temime-Smaali N, Trentesaux C, Mergny JL, Gueritte F, et al. A new steroid derivative stabilizes g-quadruplexes and induces telomere uncapping in human tumor cells. Mol. Pharmacol. 2007;72:631–640. doi: 10.1124/mol.107.036574. [DOI] [PubMed] [Google Scholar]
- 40.De Cian A, Mergny JL. Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res. 2007;35:2483–2493. doi: 10.1093/nar/gkm098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drygin D, Siddiqui-Jain A, O'Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, et al. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 42.Sparapani S, Haider SM, Doria F, Gunaratnam M, Neidle S. Rational design of acridine-based ligands with selectivity for human telomeric quadruplexes. J. Am. Chem. Soc. 2010;132:12263–12272. doi: 10.1021/ja1003944. [DOI] [PubMed] [Google Scholar]
- 43.Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khobta A, Ferri F, Lotito L, Montecucco A, Rossi R, Capranico G. Early effects of topoisomerase I inhibition on RNA polymerase II along transcribed genes in human cells. J. Mol. Biol. 2006;357:127–138. doi: 10.1016/j.jmb.2005.12.069. [DOI] [PubMed] [Google Scholar]
- 46.Hraiky C, Raymond MA, Drolet M. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J. Biol. Chem. 2000;275:11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- 47.Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, Baaklini I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front. Biosci. 2003;8:d210–d221. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]