Abstract
With the availability of a large amount of genomic data it is expected that the influence of single nucleotide variations (SNVs) in many biological phenomena will be elucidated. Here, we approached the problem of how SNVs affect alternative splicing. First, we observed that SNVs and exonic splicing regulators (ESRs) independently show a biased distribution in alternative exons. More importantly, SNVs map more frequently in ESRs located in alternative exons than in ESRs located in constitutive exons. By looking at SNVs associated with alternative exon/intron borders (by their common presence in the same cDNA molecule), we observed that a specific type of ESR, the exonic splicing silencers (ESSs), are more frequently modified by SNVs. Our results establish a clear association between genetic diversity and alternative splicing involving ESSs.
INTRODUCTION
The large amount of data on the human transcriptome has allowed several studies that, without exception, show a high prevalence of alternative splicing in the human transcriptome (1–3). The fact that most human genes undergo alternative splicing has raised doubts about the biological significance of most of the variants. One possibility is that a significant fraction of all variants are spurious products of the splicing machinery, without any functional relevance. Indeed, there are significant differences (e.g. the preservation of codon reading frame) between splicing variants that are conserved between human and mouse, and therefore deemed as functional, and those that are not, suggesting that a fraction of the splicing variants are spurious products (4). Some authors have even suggested that these products would have functional implications by down-regulating the expression of functional variants (5). On the other hand, some have argued that most of the splicing variants are products of a regulated process. For instance, Wang,E. et al. (6) observed that most of the splicing variants of human genes show differential expression among different tissues whereas variation between individuals was ∼2 to 3-fold less common. These results corroborate the hypothesis that alternative transcripts could have tissue specific functionalities. It has also been shown by our group that intron retention events are not randomly distributed regarding several parameters, again suggesting the notion that the majority of these splicing variants are not spurious and their expression is somehow regulated (7,8). Based on the current evidence it is reasonable to speculate that at least one third of all splicing variants are products of regulated expression.
In addition to the importance of 5′/3′-splicing sites, branch point and polypyrimidine tracts in the control of splicing, there are several known cis-regulatory splicing elements that contribute to the splicing process and are located in intronic or exonic regions [for a review see (9,10)]. Many lines of evidence suggest that these elements can act by stimulating (enhancing) or inhibiting (silencing) the inclusion of the respective exon, or the neighbor exon, in the mature RNA transcript. These features are taken into account for the nomenclature of splicing regulatory elements. Those present in exons and with the capacity of enhancing splicing are called exonic splicing enhancers (ESE) and those with the capacity of inhibiting the splicing are the exonic splicing silencers (ESS). Generally, these classes of elements are called exonic splicing regulators (ESRs).
Several studies suggest that ESS have a significant role in the control of alternative splicing. For example, (i) using a set of paralogous exons, where one copy showed constitutive splicing and the other alternative splicing, Zhang,Z. et al. (11) found that the alternative copy had significantly lower ESE and higher ESS densities than the constitutive copy; (ii) using designed exons constructed by random ligation of ESEs, ESSs and neutral sequences, Zhang,X. et al. (12) showed that negative correlation between ESS density and inclusion rate was stronger than the positive correlation between ESE density and inclusion rate; (iii) The set of motifs which bind the tissue-specific splicing factors Nova1 and Nova2 can act as ESEs or ESS depending on their position in the primary transcript. When located in alternative exons they mainly act as silencers (13).
In the present study we used single nucleotide variation (SNV) and cDNA data to compare the genetic diversity of ESRs located in constitutive and alternative exons. By establishing an association between the SNV alleles and distinct borders of alternative exons our results show that variations in ESSs, and not in ESEs, are more commonly associated with alternative splicing.
MATERIALS AND METHODS
Public data
We obtained genomic (build 36.1) and cDNA (mRNAs and ESTs) sequence data from UCSC Genome Browser (http://genome.ucsc.edu/, files: mrna.fa.gz and est.fa.gz). Additional sequences were obtained from NCBI Reference Sequence Project (http://www.ncbi.nlm.nih.gov/RefSeq, release 22). We also downloaded EST libraries annotation from eVOC (http://www.evocontology.org/).
Identification of splicing events
A catalog of all splicing variants reported by the alignment of cDNAs on the human genome was obtained as previously described (7,8,14,15). Briefly, the coordinates of exon/intron borders for all cDNAs mapped onto the human genome were compared against each other to identify all splicing variants for all human genes. We used the software SIM4 (http://globin.cse.psu.edu/html/docs/sim4.html) for a more refined definition of the exon/intron borders. To increase the reliability of splicing events identified, we have chosen for further analysis only those events supported by at least two ESTs from two distinct libraries.
Exon classification
The definition of exonic and intronic regions was based on the genomic coordinates of cDNA sequences classified as ‘mRNA’ in GenBank. Regarding alternative splicing, four groups of exons were defined for the current analyses: exons reporting different donor or acceptor sites formed the Cryptic group (alternative splice site, 35 391 exons), exons missing in two or more transcripts formed the Skipping group (46 586 exons) and exons reporting an intron retention formed the Retention group (8310 exons). These three groups represent the major forms of alternative splicing. A fourth group, named Alternative (60 383 exons), was formed by the union of the three groups of alternative exons mentioned above excluding redundant exons among these groups. Finally, the Constitutive group (70 801 exons) was composed by exons for which no alternative exon/intron borders were detected.
Mapping the ESRs in exons
Eight different data sets of putative regulatory elements (six ESEs and two ESSs) were obtained from the literature (16–21). Four (SF2_IgM, SRP40, SRP55 and SC35) out of six ESE data sets were discovered in vitro by using the SELEX methodology while the other two were discovered in silico. Regarding the SELEX-ESEs, only those oligomers with a score equal or higher than the threshold scores defined by the original study were considered as ESEs. For the remaining data sets of ESEs (RESCUE and PESE), a list of ESE motifs was obtained from the supplementary material associated with the articles of Fairbrother et al. (18) and Zhang and Chasin (19). The PESS data set of silencers was also obtained from Zhang and Chasin (19) and the data set of ESS reported by Wang,Z. et al. (20) will be called ESS herein.
To identify the ESR motifs in the exons of our in-house database we perform a pairwise alignment between each set of ESRs and the exon sequences. The ESR counts were calculated independently for each group of exons analyzed in this study (Constitutive, Alternative, Cryptic, Skipping and Retention).
Mapping SNVs in exons
To make sure the SNVs were correctly indexed in our exon database, we mapped all 17 804 036 SNVs available in the dbSNP (release 130) in the genomic sequences used for our analysis. Relative positions of SNVs regarding exonic, intronic and intergenic regions were defined by comparing SNV cDNA coordinates.
Mapping exonic SNVs in published ESRs motifs
Sequence tags comprising each SNV were generated by extracting from the reference human genome the corresponding variant nucleotide plus ten nucleotides flanking the SNVs (totaling a 21 nucleotide tag, which we will be called SNP-tag herein). We only extracted tags from the same strand orientation of the Refseq gene which contains the exon where the SNV was mapped. The alignment of these tags with the published ESR motifs defined whether or not the SNV was mapped into a known ESR.
Finding isoform-associated SNVs and defining putative ESR motifs
Alignments between all human mRNAs and the genome were searched for the presence of mismatches. The position of the mismatches was then compared to the genomic position of SNVs. These analyses resulted in 106 271 mismatches that that co-occur with SNVs. For 96 756 of these mismatches, the discordant nucleotide reported by the mRNA corresponded to one of the alleles reported in the dbSNP for the respective SNV. Since the mRNA sequences are supposedly of high quality, this last number strongly suggests that the great majority of mismatches reported in the alignments are due to SNVs. Among these mismatches we selected 3533 SNVs where each allele was completely associated with alternative exon/intron borders (we refer to these as isoform-associated SNVs). Considering all the alleles of these SNVs, we obtained 7087 sequence tags (17 tri-allelic and 2 quadri-allelic SNVs). SNVs that presented the same allele in cDNAs reporting different exon/intron borders were not included in the category of isoform-associated SNVs.
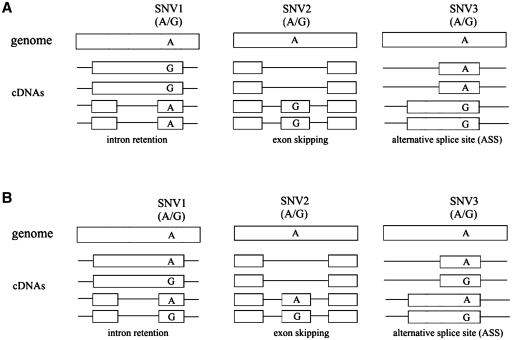
To make statistical inferences about a possible enrichment of known regulatory elements in the isoform-associated SNV data set, we randomly created 1000 control data sets from a pool of 46 336 SNVs also mapped in alternative exons but without alleles in complete association with alternative exon/intron borders. A schematic view of our approach is shown in Figure 1. Distinct control data sets were created for the three main groups of alternative exons (Cryptic, Retention and Skipping). Each control data set is of exactly the same size as the isoform-associated SNV data set (1385 SNVs for Cryptic, 1780 SNVs for Retention and 958 SNVs for Skipping). Similarly, we create control data sets, with 3533 SNVs each, to use in the comparison with the main group of alternative exons (which includes all three forms of alternative exons). Next, we used the same strategy described above to generate tags around the SNVs of the control data sets and search for known ESR motifs. For each replicate data set we counted the number of ESRs that were part of an SNP-tag, and used the distribution of these values to test the null hypothesis that our set of isoform-associated SNVs are not associated with ESRs. The P-value was defined as the fraction of the ranked values observed in the control data sets which were greater than the observed value in the case set.
Figure 1.
Schematic representation of the computational approach used in this study to identify splicing regulatory elements. (a) Isoform-associated SNVs. Schematic view of association between genetic and splicing variation. A sine qua non condition is the presence of both the SNV and the alternative exon/intron border in the same cDNA molecule allowing the direct association between the alternative exon/intron border and the presence of the SNV. (b) Control. Schematic view of SNVs not associated with splicing variation, which defined the control set used in the simulation.
Defining the ancestral and derived alleles of SNVs
In order to establish the polarity of the ESR modification imposed by SNVs and so define events as gains, losses or maintenance/alteration of ESRs, we compared the alleles of human SNVs to the orthologous alleles of the chimpanzee (Pan troglodytes) genome. To perform this analysis we used data from the table ‘snp130OrthoPt2Pa2Rm2’ available at the UCSC Genome Browser (http://genome.ucsc.edu/), which contains the othologs of 11 797 184 human SNVs in four species of primates, including the chimpanzee.
Statistical test
For all comparisons of proportions presented in this article we used the chi-square distribution to evaluate the statistical significance of the difference between the expected and observed values. We used the chi-square test implemented in the function prop.test of R statistical software (http://www.r-project.org/).
RESULTS AND DISCUSSION
Alternative exons are enriched in SNVs when compared to constitutive exons
We first compared the density of SNVs between constitutive and alternative exons. Alternative exons, when taken as an unique group, have ∼10% more SNVs than constitutive exons (5.09 and 4.52 SNVs per 1000 nucleotides for alternative and constitutive exons, respectively, P-value = 2.53−102, chi-square test).
Comparisons among three sub-groups of alternative exons reveals that the Skipping group shows a significantly lower SNV density than the other two groups. (Skipping = 5.01 versus Cryptic = 5.19 and Retention = 5.17 SNVs per 1000 nucleotides, P-value <3.81−9 for both comparisons). This reduced genetic diversity of skipped exons in relation to other forms of alternative exons may reflect a stronger selective constraint. This result is in accordance with the findings from Wang,E. et al. (6), who showed that skipped exons are more conserved among four mammalian genomes and seem to be most important in tissue-specific alternative splicing.
Alternative exons are enriched in ESRs when compared to constitutive exons
Next, the density of ESRs was compared between constitutive and alternative exons. Table 1 shows that ESR motifs are enriched in the group of alternative exons. RESCUE-ESEs and PESEs were the exceptions, presenting an opposite trend. These exceptions are, in fact, expected since RESCUE-ESEs are identified from a set of constitutive exons and PESEs are identified from a set of exons with high inclusion levels (18,19).
Table 1.
Comparison of ESR density between constitutive and alternative exons
| ESR | Constitutive (10 650 372) | Alternative (22 622 280) | Percent change# | P-value* |
|---|---|---|---|---|
| RESCUE | 0.11305 | 0.08943 | −20.8 | 0 |
| SF2_IgM | 0.05466 | 0.06135 | 12.2 | 0 |
| SC35 | 0.04156 | 0.04534 | 9.0 | 0 |
| SRP40 | 0.04275 | 0.04400 | 2.8 | 6.97−61 |
| SRP55 | 0.02514 | 0.02530 | 0.06 | 0.01 |
| PESE | 0.07030 | 0.06398 | −8.9 | 0 |
| PESS | 0.01430 | 0.01916 | 33.9 | 0 |
| ESS | 0.00006 | 0.00013 | 116.6 | 2.76−59 |
*P-value for two-tailed chi-square test. Between brackets is the total number of nucleotides analyzed in each exon group. #Approximate percent change of Alternative compared with Constitutive exons. Positive and negative values represents excess and depletion, respectively.
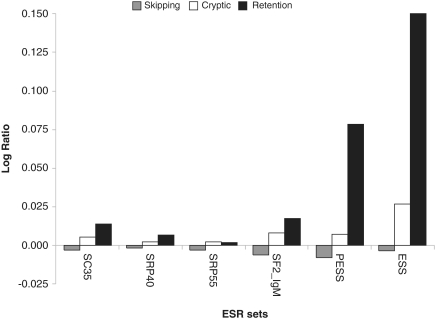
Exons belonging to the Skipping group showed a significant depletion in the density of SELEX-ESEs and ESSs when compared to the other groups of alternative exons (Figure 2). These results corroborate the results of Kurmangaliyev and Gelfand (22), who observed similar results in a comparison between skipped exons and exons with alternative splicing sites with mutations in their splice sites. However, they contradict the findings from Wang,J. et al. (23), who performed a similar analysis and found a significantly lower density of SELEX-ESEs in skipped exons when compared to constitutive exons. We believe that the discrepancy between our result and those of Wang,J. et al. (23) may be due to differences in the group of constitutive exons used in both studies. Currently, the coverage of the human transcriptome is deeper when compared to 2005, and probably a significant proportion of exons defined as constitutive in their work is currently defined as alternative.
Figure 2.
Comparison of ESR density between the three main types of alternative exons. Values represent the log10 of the ratio between each individual group of alternative exons and the Alternative group (pool of the three types).
Alternative exons show higher proportion of ESRs modified by a SNV than constitutive exons
We next compared the proportion of ESRs modified by a SNV in both constitutive and alternative exons. Generally, ESRs in alternative exons are proportionally more modified by a SNV than those ESRs in constitutive exons (P-value <1.26−16, chi-square test, highest significant P-value from Table 2). The only exception was for ESS. Despite the fact that PESE and RESCUE sets are enriched in constitutive exons, we also observed a higher proportion of these motifs mapped in SNVs of alternative exons than in SNVs of constitutive exons. Moreover, these two sets of ESRs showed the most significant differences in comparisons between constitutive and alternative exons (Table 2). Consistent with our previous observations, the Skipping group showed the lowest proportion of ESRs affected by SNVs among the groups of alternative exons (Supplementary Table S5).
Table 2.
Proportion of ESRs affected by SNVs in both Constitutive and Alternative exons
| ESR | Constitutive | Alternative | Percent change# | P-value* |
|---|---|---|---|---|
| RESCUE | 0.03900 | 0.04614 | 18.3 | 1.72−203 |
| SF2_IgM | 0.07658 | 0.08064 | 5.3 | 7.82−22 |
| SC35 | 0.07341 | 0.08319 | 13.3 | 3.95−89 |
| SRP40 | 0.06326 | 0.06889 | 8.9 | 2.69−36 |
| SRP55 | 0.06240 | 0.06721 | 7.7 | 1.26−16 |
| PESE | 0.05819 | 0.06800 | 16.8 | 6.82−173 |
| PESS | 0.05858 | 0.06724 | 14.7 | 4.87−32 |
| ESS | 0.12006 | 0.11569 | −3.6 | 0.8 |
Proportions were obtained by dividing the number of ESRs affected by a SNV by the total of ESRs, within each group.
*P-value for two-tailed chi-square test. #Approximate fold change of Alternative compared with Constitutive exons. Positive and negative values represents excess and depletion, respectively.
The observations that the alternative exons show a higher density of SNVs, ESRs and also a higher proportion of ESRs modified by SNVs, suggest that this genetic variation could to some extent be one of the causal factors distinguishing alternative and constitutive splicing. In fact several studies analyzed the impact of single nucleotide polymorphism in the regulation of transcript isoform expression in tissue-specific and non-specific manners (24–26) and validated some causative SNPs occurring in splicing regulators (27).
ESS associated with alternative splicing are more modified by SNVs
We decided to further explore this putative association between the SNVs and alternative splicing by examining those cDNAs that reported both an alternative exon/intron border and known SNV. Two categories of SNVs were used. The first category contains SNVs with alleles in complete association with different exon/intron borders (isoform-associated SNVs). The second category, the control set, contains the remaining SNVs mapped to alternative exons, i.e. those SNVs without a complete association with alternative exon/intron borders (Figure 1).
Among the set of sequence tags derived from the isoform-associated SNVs we found that ∼86% contained at least one ESR already described. Is there an enrichment of any particular type of ESR in this set of sequence tags in comparison to tags derived from SNVs not associated with alternative borders? To answer this question, re-sampling was performed comparing the isoform-associated SNV data set to 1000 control data sets, each comprised of the same number of SNVs but not associated with alternative exon/intron borders. The analysis was performed independently for each set of published ESRs using alternative exons either as an unique group or divided into the three categories previously discussed.
Table 3 shows that isoform-associated tags are enriched in ESSs in comparison with the control data set when the alternative exons are analyzed as an unique group. This is true for both sets of ESS analyzed (PESS and ESS). Moreover, the isoform-associated SNVs is significantly depleted in ESEs when compared to the replicate data sets (Table 3). Interestingly, Zhang,X. et al. (12), showed that the absolute number of ESS correlates significantly (R2 = 0.78, P-value < 5−47) with the non-inclusion rate (negative correlation with inclusion rate) of exons, when other splicing signals are constant. Moreover, they found a significant positive correlation (R2 = 0.53, P-value < 3e−6) between inclusion level and the ratio ESE/ESE + ESS. Consistent with this, we found this ratio to be significantly lower in the SNP-tags of our experimental data set when compared to the SNP-tags of control data sets (data not shown). Together, these results suggest that the influence of SNVs on some types of alternative splicing occur predominantly through their effects on ESSs.
Table 3.
Difference in ESR densities between experimental and control sets of SNVs
| ESR | Exon group | P-value |
|---|---|---|
| Enhancers | ||
| RESCUE | Skipping | 0.74 |
| Retention | 0.87 | |
| Cryptic | 0.98a | |
| Alternative | 1a | |
| SC35 | Skipping | 0.95a |
| Retention | 1a | |
| Cryptic | 0.85 | |
| Alternative | 1a | |
| SRP40 | Skipping | 0.86 |
| Retention | 0.99a | |
| Cryptic | 0.87 | |
| Alternative | 0.9 | |
| SRP55 | Skipping | 0.19 |
| Retention | 0.86 | |
| Cryptic | 0.44 | |
| Alternative | 0.46 | |
| PESE | Skipping | 0.11 |
| Retention | 1a | |
| Cryptic | 1a | |
| Alternative | 1a | |
| SF2_IgM | Skipping | 0.09 |
| Retention | 1a | |
| Cryptic | 0.93 | |
| Alternative | 1a | |
| Silencers | ||
| PESS | Skipping | 0.92 |
| Retention | 0b | |
| Cryptic | 0b | |
| Alternative | 0b | |
| ESS | Skipping | 0.1 |
| Retention | 0.09 | |
| Cryptic | 0.01b | |
| Alternative | 0b |
aSignificantly lower than control.
bSignificantly higher than control.
A recent study by Woolfe et al. (28), analyzed a small set of well curated SNPs (a total of 87) associated to exon skipping, which they compared to a large set of HapMap SNPs which were putatively neutral with respect to splicing. Using an approach different from ours, they also found that alterations of ESSs were significantly overrepresented when compared to alterations which are putatively neutral with respect to splicing. Moreover, they also found that the degree of ESS alterations was even greater for events of alternative splice site than that for exon skipping. The concordance between these two studies, which used different approaches to define the association between SNVs and splicing variants, corroborates the important role played by ESSs in the splicing regulation.
Analyzing the polarity of the ESRs changes imposed by SNVs
Can we further discriminate the effect of SNVs in ESSs? If we assume that the derived allele increases transcriptome variability by allowing the use of alternative exon/intron borders, we can try to better understand the effect of SNVs on ESS by defining a pattern of ESS gain or loss with the emergence of a derived allele. To this end we defined the polarity of change by assuming the reference chimpanzee genome as the ancestral allele, as done by others (29).
Results in Table 4 confirm that exon skipping does not seem to be primarily regulated by SNVs that create ESSs. When we independently analyzed the events of ESS gain from non-ESR motifs, the difference between case and control sets does not exist (11 SNVs in the isoform-associated set against 12 SNVs in the control set, P-value = 0.56). The difference is restricted to those events of ESS gain from an ancestral ESE (29 SNVs in the case set against 44 SNVs in the control set, P-value = 0.99). This suggests that the significant depletion of SNVs involved in ESS gain observed for this type of alternative splicing reflects depletion in the number of SNVs that affect ESEs.
Table 4.
SNP counts of ESS loss, gain and maintenance found in isoform-associated SNV data set and control data set
| Isoform-associated SNV set | Control# | P-value | |
|---|---|---|---|
| Skipping | |||
| ESS loss | 38 | 23–63 | 0.67 |
| ESS gain | 40 | 40–57 | 0.99a |
| ESS maintenance | 22 | 10–45 | 0.6 |
| Cryptic | |||
| ESS loss | 81 | 39–87 | 0.005b |
| ESS gain | 68 | 51–115 | 0.97a |
| ESS maintenance | 54 | 19–60 | 0.005b |
| Intron retention | |||
| ESS loss | 111 | 63–115 | 0.01b |
| ESS gain | 108 | 75–140 | 0.53 |
| ESS maintenance | 90 | 43–93 | 0b |
#Range for 1000 replicate data sets.
aSignificantly lower than the control.
bSignificantly higher than the control.
These findings differ from those of Woolfe et al. (28), and show that the mechanism of splicing regulation among the skipped exons is more complex than just an increase in the proportion of ESS gains. We note, however, that these authors compared SNVs alleles associated to skipped exons to a group of SNVs alleles belonging to a heterogeneous set of exons. This differs from our approach, in which the isoform-associated and control SNVs alleles were all from skipped exons, and may explain the differences between the studies.
For cryptic and intron retention, the predominant pattern involves loss and alteration/maintenance of ESS. Based on the significant frequency of ESS loss, we predict that the derived allele could be generating a decrease in ESS strength in those cases where ESS is maintained.
Final remarks
The results reported here support the view that ESRs have a higher genetic diversity in alternative exons when compared to constitutive exons. We believe that this genetic variation could to some extent be one of the major features distinguishing alternative from constitutive splicing. Furthermore, we provide evidence that this effect is mainly due through SNVs acting on ESS.
A possible caveat of our approach is that we cannot directly distinguish between causal and associated SNVs since an isoform-associated SNV may be in linkage disequilibrium with a different causal variant. However, our re-sampling analysis addresses this issue by examining if the isoform associated SNVs are associated to ESRs as frequently as non-isoform associated SNVs (used as a ‘control’). Using this approach we were able to show that ESS are significantly overrepresented among isoform-associated SNVs, supporting their functional role in splicing regulation.
The emergence of next generation sequencing is beginning to provide a huge amount of both genomic and expressed sequence data. We believe that the strategy used in this manuscript will be very useful in the next few years to further explore the role of SNVs in alternative splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (2007/55790-5 to S.J.S.); Ph.D. fellowship (2007/59721-8 to R.F.R.). Funding for open access charge: Ludwig Institute for Cancer Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Daniel Ohara for technical support.
REFERENCES
- 1.Mironov A, Fickett J, Gelfand M. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modrek B, Lee C. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 4.Resch A, Xing Y, Alekseyenko A, Modrek B, Lee C. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 2004;32:1261–1269. doi: 10.1093/nar/gkh284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis B, Green R, Brenner S. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang E, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore S, Schroth G, Burge C. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galante P, Sakabe N, Kirschbaum-Slager N, de Souza S. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakabe N, de Souza J, Galante P, de Oliveira P, Passetti F, Brentani H, Osório E, Zaiats A, Leerkes M, Kitajima J, et al. ORESTES are enriched in rare exon usage variants affecting the encoded proteins. C R Biol. 2003;326:979–985. doi: 10.1016/j.crvi.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 9.Pagani F, Baralle F. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Burge C. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Zhou L, Wang P, Liu Y, Chen X, Hu L, Kong X. Divergence of exonic splicing elements after gene duplication and the impact on gene structures. Genome Biol. 2009;10:R120. doi: 10.1186/gb-2009-10-11-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Arias M, Ke S, Chasin L. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA. 2009;15:367–376. doi: 10.1261/rna.1498509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe B, Darnell R. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 14.Kirschbaum-Slager N, Parmigiani R, Camargo A, de Souza S. Identification of human exons overexpressed in tumors through the use of genome and expressed sequence data. Physiol. Genomics. 2005;21:423–432. doi: 10.1152/physiolgenomics.00237.2004. [DOI] [PubMed] [Google Scholar]
- 15.Galante P, Vidal D, de Souza J, Camargo A, de Souza S. Sense-antisense pairs in mammals: functional and evolutionary considerations. Genome Biol. 2007;8:R40. doi: 10.1186/gb-2007-8-3-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Zhang M, Krainer A. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Chew S, Cartegni L, Zhang M, Krainer A. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairbrother W, Yeh R, Sharp P, Burge C. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Chasin L. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Rolish M, Yeo G, Tung V, Mawson M, Burge C. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Smith P, Zhang C, Wang J, Chew S, Zhang M, Krainer A. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 22.Kurmangaliyev Y, Gelfand M. Computational analysis of splicing errors and mutations in human transcripts. BMC Genomics. 2008;9:13. doi: 10.1186/1471-2164-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Smith P, Krainer A, Zhang M. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33:5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stranger B, Nica A, Forrest M, Dimas A, Bird C, Beazley C, Ingle C, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwan T, Grundberg E, Koka V, Ge B, Lam K, Dias C, Kindmark A, Mallmin H, Ljunggren O, Rivadeneira F, et al. Tissue effect on genetic control of transcript isoform variation. PLoS Genet. 2009;5:e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge B, Pokholok D, Kwan T, Grundberg E, Morcos L, Verlaan D, Le J, Koka V, Lam K, Gagné V, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 27.Coulombe-Huntington J, Lam K, Dias C, Majewski J. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet. 2009;5:e1000766. doi: 10.1371/journal.pgen.1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolfe A, Mullikin J, Elnitski L. Genomic features defining exonic variants that modulate splicing. Genome Biol. 2010;11:R20. doi: 10.1186/gb-2010-11-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fairbrother W, Holste D, Burge C, Sharp P. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.