Abstract
Sam68 plays an essential role in mouse spermatogenesis and male fertility. Herein, we report an interaction between Sam68 and the phosphorylated forms of the RNA polymerase II (RNAPII) in meiotic spermatocytes. RNase treatment decreased but did not abolish the interaction, consistently with in vitro binding of RNAPII to the Sam68 carboxyl-terminal region. Sam68 retention in the spermatocyte nucleus was dependent on the integrity of cellular RNAs, suggesting that the protein is recruited to transcriptionally active chromatin. Mouse knockout models characterized by stage-specific arrest of spermatogenesis and staining with the phosphorylated form of RNAPII documented that Sam68 expression is confined to the transcriptionally active stages of spermatogenesis. Furthermore, Sam68 associates with splicing regulators in germ cells and we report that alternative splicing of Sgce exon 8 is regulated in a Sam68-dependent manner during spermatogenesis. RNA and chromatin crosslink immunoprecipitation experiments showed that Sam68 binds in vivo to sequences surrounding the intron 7/exon 8 boundary, thereby affecting the recruitment of the phosphorylated RNAPII and of the general splicing factor U2AF65. These results suggest that Sam68 regulates alternative splicing at transcriptionally active sites in differentiating germ cells and provide new insights into the regulation of Sam68 expression during spermatogenesis.
INTRODUCTION
Transcriptional and post-transcriptional regulation of gene expression need to be finely tuned during mammalian spermatogenesis because synthesis and translation of mRNAs are temporally uncoupled at two steps of this differentiation program (1–3). During the first meiotic prophase, chromatin becomes unavailable for transcription due to DNA repair after homologous recombination (4,5). It follows a wave of intense transcription at the pachytene stage until the onset of chromatin condensation that precedes the first division (4). Later, when round spermatids differentiate into spermatozoa, extensive nuclear remodelling and compaction of the chromatin, which is favoured by the replacement of histones with the highly basic protamines, represses transcription (6). As a consequence of these processes, mRNAs are accumulated in the transcriptionally active stages of spermatogenesis and they are stored and protected by a profusion of ribonucleoproteins, to preserve them until translation occurs (3,7).
Several RNA binding proteins (RBPs) are highly expressed in germ cells and their essential function has been highlighted by the spermatogenetic defects arising in mouse knockout models for the corresponding genes (3). Remarkably, RBPs involved in almost all steps of mRNA processing are essential for the production of a fertile spermatozoon (3). For example, knockout of the gene encoding MSY2 leads to mRNA instability and spermatogenic arrest (8), whereas disruption of the Dazl gene leads to reduced translation of selected mRNAs and loss of germ cells (9,10). Other examples are provided by the infertility of knockout mice for RBPs involved either in splicing, such as hnRNP G/T (11), or in small non-coding RNAs metabolism, like the PIWI proteins (12–14).
Another RBP required for male fertility is the Signal transduction and activation of RNA (STAR) protein Sam68 (KHDRBS1) (15). The RNA-binding domain of STAR proteins, named GSG (GRP33/Sam68/GLD-1 homology), consists of a large hnRNP K Homology (KH) domain flanked by conserved regions required for homodimerization and RNA binding specificity (16,17). The STAR protein GLD-1 in Caenorhabditis elegans is required for meiotic differentiation of germ cells and for accumulation of target mRNAs during oogenesis (18,19). Mammalian STAR members are the Quaking proteins (QKs), involved in myelination in the nervous system (20) and the Sam68 subfamily, composed of Sam68 and the highly homologous SLM-1 and SLM-2 (16,17). Sam68 interacts with signalling proteins through its proline-rich and tyrosine-rich regions of binding to SH2 and SH3 domains and it was originally described as a scaffold protein in signal transduction pathways (16). Furthermore, Sam68 takes part in various aspects of RNA metabolism, from alternative splicing (21–25) to cytoplasmic utilization of mRNAs (15,26–28).
Knockout of the Sam68 gene in the mouse affected bone metabolism, neurological functions and fertility (15,29–31). The specific functions of Sam68 responsible for these defects have been only partially elucidated. In particular, it was shown that Sam68 translocates to the cytoplasm and associates with the polysomes during meiosis in spermatocytes (27), thereby regulating translation of a subset of mRNAs necessary for sperm differentiation (15). Notably, ablation of Sam68 also impaired meiotic progression and cell survival in pachytene spermatocytes (15), in which this RBP is exclusively localized in the nucleus (27).
Given the importance of Sam68 for spermatogenesis, here we have investigated further its function in male germ cells. Our results document that Sam68 interacts with the phosphorylated form of the RNA polymerase II (RNAPII) and binds to transcriptionally active chromatin in pachytene spermatocytes. Moreover, Sam68 interacts with splicing factors and its expression is required for skipping of exon 8 in Sgce mRNA. Our results strongly suggest that Sam68 function is intimately connected with nuclear RNA processing during germ cell differentiation.
MATERIALS AND METHODS
Cell isolation, culture and treatments
Testes from 17- to 60-day-old CD1 mice (Charles River, Italy) or C57/B6 wild-type or Sam68 knockout mice (15,29) were used to obtain pachytene spermatocytes and round spermatids by elutriation technique as described previously (32). Purified germ cells were collected, washed with phosphate-buffered saline (PBS) and used for experiments.
Immunoprecipitation experiments
Nuclear extracts from germ cells were prepared as previously described (22,25). Briefly, cells were re-suspended in hypotonic buffer [10 mM Tris/HCl pH 7.4, 10 mM NaCl, 2.5mM MgCl2, 1 mM DTT, protease inhibitor cocktail (Sigma-Aldrich), 30 U/ml RNase inhibitor (Invitrogen), 10 mM β-glycerophosphate, 0.5 mM NaVO4]. After incubation on ice for 7 min, samples were centrifuged at 700g for 7 min. Pelleted nuclei were re-suspended in hypotonic buffer supplemented with 90 mM NaCl and 0.5% Triton, sonicated and stratified on 30% sucrose. After centrifugation at 5000g for 15 min, soluble nuclear extracts were pre-cleared for 1 h on Protein A-Sepharose beads (Sigma-Aldrich) and for an additional 1 h on the Protein A-Sepharose beads in the presence of 5 µg of rabbit IgGs and 0.05% BSA. After centrifugation for 1 min at 1000g, supernatants were incubated with 5 µg of anti-Sam68 or 5 µg of rabbit IgGs for 3 h at 4°C under rotation. For co-immunoprecipitation experiments with RNAPII, nuclear extracts were immunoprecipitated with 2 µg of mouse H5 or H14 anti-phospho RNAPII antibodies. Where indicated, nuclear extracts were treated with 100 µg/ml RNase A (Sigma-Aldrich) in the absence of RNase inhibitor.
GST pull-down experiments
pGEX-4T1-Sam68 fusion protein plasmids were expressed in the Escherichia coli BL21 strain, grown at 30°C in LB medium to an OD600nm = 0.6 and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Gibco-BRL) for 3 h. The GST fusion proteins were purified from bacterial lysates using Sepharose-glutathione beads. Beads were pre-adsorbed as described above for the immunoprecipitation experiments and incubated with germ cell nuclear extracts ± 100 µg/ml RNase A. After washes in PBS-0.5% Triton, bound proteins were eluted in SDS-sample buffer.
Western blot analysis and antibodies
Western blot analyses were performed as previously described (22,25) using the following primary antibodies (1:1000 dilution): rabbit anti-Sam68 (sc-333), anti-Erk2, anti-RNAPII (N20) and anti-U2AF65 (Santa Cruz Biotechnology); mouse ASF/SF2 (1:100; US Biological), mouse anti-hnRNPA2 and anti-tubulin (Sigma-Aldrich); anti polymerase II H5 and H14 (Abcam).
Immunofluorescence analysis
Mouse spermatocytes or testis slices were fixed in 4% paraformaldheyde (PFA) and washed with PBS. Samples were permeabilized with 0.1% Triton X-100 for 7 min and incubated for 1 h in 0.5% BSA. Samples were then washed with PBS and incubated for 2 h at room temperature (RT) with antibodies against Sam68 (1:1000), RNAPII H5 (1:200), SCP3 (1:1000; gift of Paula Cohen), H1t (1:200, gift of Peter Moens), followed by 1 h of incubation with cy3-conjugated anti-mouse IgGs (Alexa) or FITC-conjugated anti-rabbit IgGs (Alexa) and analysed as previously reported (22,25).
Chromosome spreads
Chromosome spreads were performed as described (32). Briefly, germ cells were re-suspended in 200 µl of hypotonic solution and left on ice for 5 min. Cell suspension was spotted on 6 mm-well glass slides and nuclei were allowed to deposit for 10 min at RT. Nuclei were then fixed in 2% PFA for 6 min and rinsed in washing buffer (0.4% Photo-Flo, Kodak) for 3 min. Slides were then dried at RT for 10 min and processed for immunostaining or stored at −80°C for up to 2 months. Primary antibodies, anti-H1t (1:200), anti-SCP3 (1:1000), anti-Sam68 (1:200), diluted in ADB buffer (10% goat serum, 3% BSA, 0.05% Triton X-100, in PBS) were incubated over night at 37°C. Secondary antibodies (Alexa) were diluted 1:200 in ADB buffer and incubated for 1h at 37°C. Slides were mounted (ProLong Gold® antifade -Molecular Probes) and analysed by confocal microscopy as previously described (15,22,25).
RT–PCR analysis
Total RNA from wild-type and Sam68 knockout testis or isolated germ cells were prepared using TRIZOL (Invitrogen) and 1 µg RNA was used for RT–PCR with the M-MLV reverse transcriptase (Invitrogen) according to manufacturer’s instructions. A total of 10% of the RT reaction was used as template. Oligonucleotides used as PCR primers are listed in the Supplementary Table 1.
Cross-linked in vivo immunoprecipitation
Sam68-cross-linked in vivo immunoprecipitation (CLIP) was performed as previously described (33), with minor modifications. Cross-linked germ cells were re-suspended in PXL buffer (1 × PBS, 0.1% SDS, 0.5% deoxycholate, 0.5% NP-40), sonicated (3 × 30″ with BioRuptor) and incubated with Turbo DNase and Cocktail RNase (Ambion) for 15 min at 37°C. Length of digested RNA was ∼200–250 nucleotides. Extracts were centrifugated at 10 000g for 10 min, clarified with ProteinA-sepharose beads in the presence of BSA 0.05% for 2 h at 4°C under rotation and then immunoprecipitated with ProteinA-sepharose beads pre-adsorbed with 2 µg of rabbit IgGs or anti-Sam68 antibodies for 3 h at 4°C under rotation. Beads were washed with PXL buffer and High-salt Wash Buffer (5 × PBS, 0.1% SDS, 0.5% deoxycholate, 0.5% NP-40) and incubated with proteinase K. RNA was extracted and used for further analysis.
Chromatin immunoprecipitation
Germ cells were incubated with 1% formaldehyde in medium for the last 10 min of culture at RT, washed in cold PBS and harvested. Nuclei were isolated by lysing cells in hypotonic buffer (5 mM Pipes pH 8.0, 85 mM KCl and NP-40 0.5%). Nuclei were then re-suspended, lysed in a buffer containing 1% SDS, 10 mM EDTA and 50 mM Tris/HCl pH 8.1 and sonicated with 8 pulses (1−, 90% Amplitude) to yield chromatin size of 250–400 bp, clarified on ProteinA/agarose/salmon sperm DNA (Millipore) and used (100 µg of DNA/sample) for immunoprecipitation with 2 µg of H5, H14 antibodies or control rabbit IgGs (Sigma-Aldrich). Co-precipitated DNA was analysed by Quantitative real time PCRs as described (15). PCR primers are listed in the Supplementary Table 1.
RNA affinity chromatography
RNA affinity chromatography was performed as previously described (24,25). Sgce wild-type and mutated sequences (from −32 to +63) were amplified using Taq Plus Precision (Stratagene) and primers containing the T7 promoter. Amplified DNA was retro-transcribed using Biotinylated NTP Mix (Roche) and purified. The 23 dpp testis nuclear extract (100 µg) were pre-cleared on Streptavidine agarose beads (Sigma-Aldrich) in the presence of 1 µg of purified GST or GST-Sam681–277, 0.01% BSA and yeast tRNA for 90 min at 4°C under rotation and then incubated for 30 min at 30°C with Streptavidine beads pre-adsorbed with 500 ng of biotinylated RNA, 0.01% BSA and yeast tRNA. After stringent washes, beads were eluted in sample buffer for western blot analysis.
Electrophoretic mobility shift assay
The radio-labelled wild-type and mutated probes were generated by in vitro annealing and transcription of T7-Sgce wt T7-Sgce mutated using the MAXIScript RNA synthesis kit (Ambion) in the presence or absence of 32Pα-UTP. RNA probes (20 000 c.p.m.) were added to the reaction mixture in the presence or absence of GST-Sam681–277, pre-equilibrated in binding buffer (10 mM HEPES (pH 7.4); 10 mM KCl; 2 mM MgCl2; 100 mM NaCl; 200 ng/μl yeast tRNA; 1 mM DTT) for 5 min at 30°C, and incubated for 30 min on ice. For the competition assay, excess of cold probes were added to the reaction. Samples were loaded onto pre-run native 5% polyacrylamide gel in 0.5% TBE and resolved at 100 V for 3 h.
RESULTS
Sam68 interacts with phosphorylated RNAPII
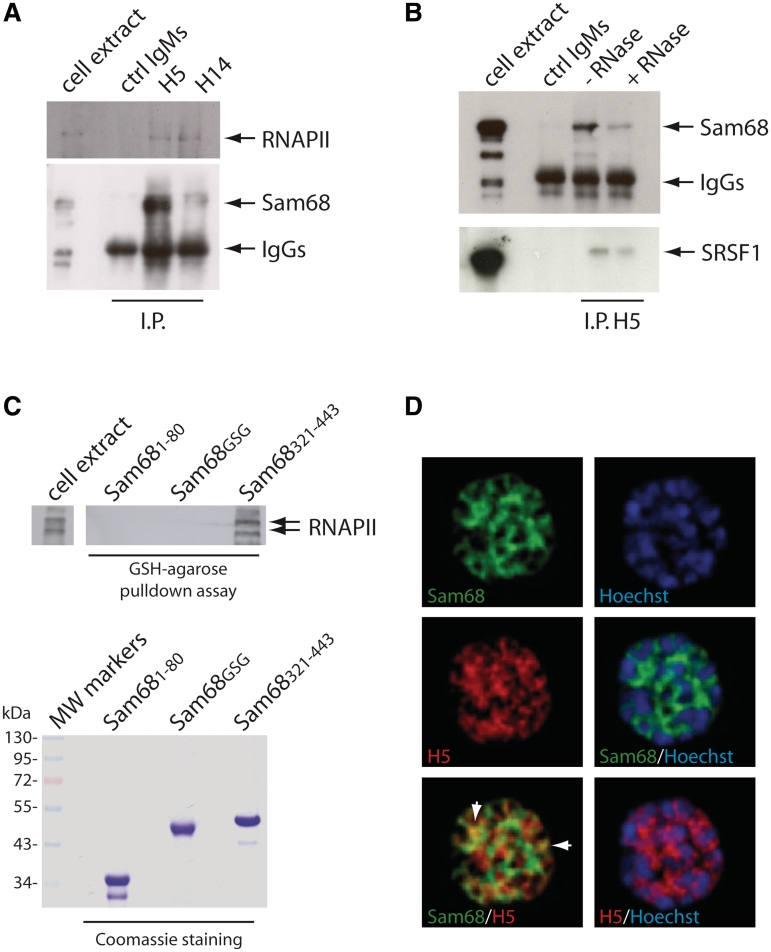
Sam68 regulates signalling events and transcription in a RNA binding-independent fashion, whereas its binding to RNA is required for modulation of splicing and translation (16,17). Thus, we asked whether its function in germ cells was related to RNA processing events. Many splicing regulators are recruited to nascent pre-mRNAs by their interaction with the phosphorylated carboxyl-terminal domain (CTD) of RNAPII (34). To investigate whether Sam68 interacts with RNAPII in pachytene spermatocytes, we immunoprecipitated the polymerase from spermatocyte nuclear extracts with antibodies specific for its phosphorylated CTD (34). The H5 antibody recognizes RNAPII phosphorylated on serine 2, whereas the H14 antibody recognizes the CTD phosphorylated on serine 5. We found that Sam68 was co-immunoprecipitated specifically with both antibodies (Figure 1A). RNase A treatment of the extracts partially impaired the interaction between Sam68 and serine 2-phosphorylated RNAPII (Figure 1B). However, the treatment also affected interaction of the polymerase with SRSF1 (Figure 1B), a splicing factor that interacts directly with the CTD (35). To test whether Sam68 could directly interact with RNAPII, we performed pull-down experiments in vitro. GST-fusion proteins containing the N-terminal region (amino acids 1–80), the RNA binding GSG domain (amino acids 90–276) or the C-terminal region (amino acids 321–443) of Sam68 were incubated with spermatocyte nuclear extracts. RNAPII specifically interacted with the C-terminal region of Sam68, but not with the GSG domain or the N-terminal region (Figure 1C). Since the C-terminal region of Sam68 does not bind RNA, this experiment shows that RNAPII and Sam68 can physically interact in vitro.
Figure 1.
Sam68 associates with phosphorylated RNAPII. (A) Co-immunoprecipitation of RNAPII and Sam68. Nuclear extracts from wild-type pachytene spermatocytes (500 µg) were immunoprecipitated with 1 µg of control IgMs, H5 antibody or H14 antibody. Immunoprecipitated proteins were analysed in western blot using anti-RNAPII and anti-Sam68 antibodies as indicated. (B) The co-immunoprecipitation experiment described in (A) was performed using either control IgMs or the H5 antibody and spermatocyte nuclear extracts in the presence or absence of RNase A in the immunoprecipitation buffer. Immunoprecipitated samples were analysed by western blot with anti-Sam68 and anti-SRSF1 antibodies. (C) The western blot analysis of RNAPII in pull-down assay using GST fusion proteins containing the N-terminus (amino acids 1–80), GSG domain or C-terminus (amino acids 321–443) of Sam68 and spermatocyte nuclear extracts. Coomassie staining of the purified GST-fusion proteins used in the assay is shown below. (D) Deconvolution images of a spermatocyte nucleus stained with antibodies for Sam68 (green), serine 2-phosphorylated RNAPII (H5, red). DNA was visualized by Hoechst staining. Merged images are shown as indicated. White arrows indicate overlapping signal of Sam68 and RNAPII in euchromatic regions of the nucleus.
Sam68 binds to nucleic acids in pachytene spermatocytes
We showed previously that Sam68 is localized in the nucleus of pachytene spermatocytes (15,27). Deconvolution microscopy documented that both Sam68 and serine 2-phosphorylated RNAPII (H5 staining) were excluded from the heterochromatic chromatin, whereas their localization showed partial overlap in euchromatic domains of spermatocyte nuclei (Figure 1D). These results suggested that Sam68 is recruited to nascent transcripts by the RNAPII. To investigate whether nuclear localization of Sam68 required its association with nucleic acids, we treated spermatocyte nuclei with DNase or RNase after permeabilization (36). Immunofluorescence analysis indicated that Sam68 remains associated with chromatin after Triton X-100 solubilization in control spermatocytes (Figure 2A and B). By contrast, treatments with DNase or RNase resulted in partial or complete lost of Sam68 staining (Figure 2A and B). Notably, analysis of RNAPII revealed a different behaviour for the serine 2- and 5-phosphorylated protein. Phosphorylation of serine 5 correlates with the positioning of RNAPII in pre-initiation complexes on the DNA and with stalled or delayed activity at sites of alternative exons (34). Serine 2- phosphorylation, instead, correlates with the elongating activity of RNAPII during transcription (34). H5 staining showed that RNAPII engaged in active transcription behaved like Sam68 and its retention in the permeabilized nucleus required RNA integrity (Figure 2A). In contrast, serine 5-phosphorylated RNAPII (H14 staining) was partially retained in the nuclei after the same treatment (Figure 2B), likely due to the pool of polymerase bound to pre-initiation complexes.
Figure 2.
Nuclear localization of Sam68 requires the integrity of nucleic acids. Purified pachytene spermatocytes were permeabilized on microscope slides in a buffer containing 0.1% Triton X-100 and incubated for 15 min with medium alone (Control) or DNase or Rnase as indicated. At the end of the incubation, cells were washed three times with PBS and fixed for immunofluorescence analysis with the anti-Sam68 and H5 (A) or H14 (B) antibodies. DNA was stained by Hoechst dye. (C) The western blot analysis of Sam68 and β-tubulin in control or treated (DNase or RNase) pachytene spermatocytes after sequential extractions with the indicated buffers.
The requirement of RNA integrity for Sam68 nuclear localization was also tested biochemically. Sequential extraction experiments showed that only a small fraction of Sam68 was recovered in the cytosol of spermatocytes after hypotonic lysis. Treatment of the nuclear pellets with 0.1% Triton X-100 was not sufficient to extract the protein, which was largely solubilized only in 1% SDS-buffer after sonication (Figure 2C). However, in the presence of DNase or RNase, Sam68 extraction in hypotonic and 0.1% Triton X-100 buffers was notably increased (Figure 2C). Tubulin was extracted with the cytosolic fraction by hypotonic buffer under all conditions. These results indicate that association with nucleic acids is required for the accumulation of Sam68 in the nucleus of pachytene spermatocytes and suggest a role for Sam68 in RNA metabolism in germ cells.
Stage-specific expression of Sam68 during meiotic prophase I
Genome integrity as well as transcriptional and post-transcriptional regulation of gene expression are highly dynamic processes during the first prophase of male meiosis (1–7). We previously observed that Sam68 expression levels oscillate during differentiation of spermatocytes (27). Mouse spermatogenesis is divided in XII stages, defined by specific cell associations in the seminiferous tubule (1,4,6,7). A closer examination revealed that Sam68 was barely detectable in pachytene spermatocytes from Stage II/III of the seminiferous tubule, whereas pachytene spermatocytes at more advanced stages of development (Stage XI) expressed high levels of the protein (Figure 3A). To investigate whether expression of Sam68 correlates with a specific physiological status of meiotic germ cells, we used several approaches. First, we co-stained testicular sections with antibodies for Sam68 and H1t, a testis-specific histone expressed in mid-pachytene spermatocytes starting from Stage IV tubules (37). As shown in Figure 3B, Sam68 and H1t expression overlapped in testicular sections, suggesting that Sam68 is also expressed from the mid-pachytene stage of meiosis. In addition, we precisely staged meiotic progression by staining nuclear spreads with antibodies for Sam68 and SCP3, the axial/lateral component of the synaptonemal complex that maintains the homologous chromosomes paired (38). In leptotene spermatocytes, before the pairing of homologous chromosomes, SCP3 forms short stretches and Sam68 is absent (Figure 3C, panels a–d). Sam68 is absent also at zygotene (Figure 3C, panels e–h), when chromosomes start to synapse and in early pachytene spermatocytes (Figure 3C, panels i–l), when homologues are fully synapsed but SCP3 staining is incompletely compacted. By contrast, Sam68 was detected in spermatocytes at mid-pachytene stage (Figure 3C, panels m–p), when fully compacted SCP3 marks the axis of the synapsis and in diplotene spermatocytes (Figure 3C, panels i–l).
Figure 3.
Stage-specific expression of Sam68 during meiotic prophase. (A) Immunohistochemical analysis of Sam68 expression in adult mouse testis. Mouse testicular sections representing the indicated stages of the cycle of the seminiferous tubule were stained with anti-Sam68 antibody and counterstained with hematoxilin to detect cell nuclei. Arrows indicate representative pachytene spermatocytes with different expression levels of the protein. The stage of the seminiferous tubule was assessed by staining an adjacent section with PAS and it is labelled in the lower left side of each panel. (B) Co-staining of testicular sections from adult wild-type mice with anti-Sam68 (red) and anti-H1t (green) antibodies. Nuclei were visualized by DAPI staining. Representative pachytene (Pc) spermatocytes are indicated by arrows. (C) Co-staining of nuclear spreads obtained from mouse spermatocytes with antibodies against SCP-3 (red) and Sam68 (green). DAPI was used for the DNA staining (blue). Sam68 is absent in leptotene (a–d), zygotene (e–h) and early pachytene spermatocytes (i–l, early Pc indicated by the arrow), but it is expressed in mid pachytene (m–p) and diplotene (i–l) spermatocytes.
To confirm expression of Sam68 from the mid-pachytene stage of prophase I, we analysed knockout mouse models with well established meiotic phenotypes. Atm−/− mice display a meiotic arrest at the zygotene stage, as a result of abnormal chromosome synapsis and subsequent chromosome fragmentation (39). In Atm−/− testis, Sam68 is strongly expressed by the somatic Sertoli cells at the basis of the tubule, but it is absent in the meiotic spermatocytes accumulating at the luminal pole of the tubule (Figure 4A, panels a–d). Similarly, in the H2ax−/−testis, where meiosis is arrested at the zygotene/early pachytene stage (40), Sam68 is expressed only by Sertoli cells (Figure 4A, panels e–h). However, in these animals, some germ cells proceed further in prophase I (40) and we found Sam68-positive spermatocytes in some adjacent seminiferous tubules (data not shown). In Mlh1−/− mice, spermatogenesis is interrupted at metaphase of the first meiotic division, due to lack of chiasmata formation (41). Hence, spermatocytes progress normally through most of prophase I and Sam68 was highly expressed in the Mlh1−/− meiotic cells (Figure 4A, panels i–l).
Figure 4.
Sam68 expression in knockout models of meiotic arrest. (A) Sam68 staining (red) in Atm−/− (a–d), H2AX−/− (e–h) and Mlh−/− testis (i–l). Zg, zygotene; Pc, pachytene; St, Sertoli cells. Sam68 is strongly expressed by the somatic Sertoli cells at the basis of the tubules in the Atm−/− and H2ax−/− testis, where meiosis is arrested at the zygotene/early pachytene stage. In Mlh1−/− mice, spermatocytes progress through prophase I and Sam68 was highly expressed in the meiotic cells (i–l). Panels d, h and l are enlarged images of c, g and k. (B) Sam68 staining (red) in Atm−/−Spo11+/− (a–c) and Spo11−/− testis (d–f). Sam68 is expressed in Pc spermatocyes of Atm−/−Spo11+/− mice and in Pc-like spermatocytes of Spo11−/− mice, respectively.
These observations strongly suggested that expression of Sam68 in male meiotic prophase I requires progression beyond the early pachytene stage. This hypothesis was confirmed by generating Atm−/−Spo11+/− mice. Haploinsufficiency of Spo11, which encodes for a topoisomerase-like protein required for double-strand breaks (DSBs) formation in meiotic cells (42), partially rescues the phenotype of Atm knockout and allows progression of meiosis up to metaphase I (43). In line with the hypothesis, Sam68 expression was readily detected in the more differentiated spermatocytes at the luminal pole of the seminiferous tubules in these mice (Figure 4B, panels a–c).
Thus, the analyses reported here suggest that accumulation of Sam68 requires progression of spermatocytes beyond the early pachytene stage of the first meiotic prophase.
Sam68 expression correlates with RNAPII activity in mouse spermatocyte
Homologous recombination causes DNA DSBs and RNA transcription is inactivated in leptotene and zygotene spermatocytes (4,5). To test whether repression of Sam68 expression follows formation of DSBs, we analysed testicular sections from Spo11−/− mice. Lack of SPO11 impedes DSBs and meiosis arrests at the zygotene stage, due to lack of homologous synapses between chromosomes (42). We found that Sam68 was expressed in Spo11−/− spermatocytes (Figure 4B, panels d–f), suggesting that formation of DSBs is required for its repression in meiosis.
The Spo11−/−spermatocytes display features of more mature meiotic cells (pachytene-like), such as active transcription and expression of H1t (44). Thus, we asked whether Sam68 expression correlated with the transcriptional activity of wild-type meiotic germ cells. Testicular sections were co-stained with an anti-Sam68 antibody and the H5 antibody, to detect serine 2-phosphorylated RNAPII engaged in transcript elongation (34). We found that Sam68 and phosphorylated RNAPII were co-expressed in pachytene spermatocytes and round spermatids (Figure 5A). In contrast, both signals were absent in zygotene spermatocytes (Figure 5A) and in elongating spermatids (data not shown), when transcription is repressed (6,7). To confirm these observations, wild-type meiotic germ cells were purified by elutriation and analysed by immunofluorescence. As shown in Figure 5B, RNAPII is unphosphorylated in zygotene spermatocytes when Sam68 is not detected (Figure 5B, panels a–d). In contrast, both signals were detected in pachytene spermatocyte (Figure 5B, panels a–d). Notably, RNAPII was not phosphorylated in all the pachytene spermatocytes analysed; however, its phosphorylation on serine 2 perfectly correlated with the expression of Sam68 (Figure 5B, e–h).
Figure 5.
Sam68 is co-expressed with phosphorylated RNAPII in male germ cells. (A) Testis section from a 25-day-old wild-type mouse was stained with anti-Sam68 (a) and H5 (b; anti-pser2 RNAPII) antibodies and co-stained with Hoechst (c) to detect nuclei. Arrows indicate the pachytene (Pc) and zygotene (Zg) spermatocytes. (B) Wild-type male germ cells were isolated by elutriation and analysed by immunofluorescence using the anti-Sam68 (a,e) and H5 (b,f; anti-p-ser2 RNAPII) antibodies as indicated. Nuclei were stained with Hoechst (c,g) to identify cell stages by nuclear morphology. Arrows in panels d and h indicate the zygotene (Zg) and pachytene (Pc) spermatocytes in the population examined. (C) Immunofluorescence analysis of isolated germ cells with the anti-Sam68 (a) and anti-γH2AX (b) antibodies. Nuclei were stained with Hoechst (c). In leptotene spermatocytes (Lp; arrows in panel d), γH2AX is localized in nuclear foci and Sam68 is absent; in pachytene spermatocytes (Pc; arrows in panel d), γH2AX is localized in the sex body whereas Sam68 is excluded from it.
We then probed whether Sam68 co-localizes with transcriptionally active chromatin in late pachytene spermatocytes. During the meiotic prophase, the X and Y chromosomes condense to form a transcriptionally repressed domain named sex body (45). This macro-chromatin domain is a site of accumulation of the phosphorylated form of H2AX (γH2AX) (44). Co-staining of pachytene spermatocytes with a γH2AX antibody revealed that Sam68 is excluded from the transcriptionally repressed sex body (Figure 5C). These results strongly indicate that Sam68 expression in germ cells correlates temporally and physically with the transcriptionally active stages of spermatogenesis.
Sam68 regulates the alternative splicing of Sgce in male germ cells
Sam68 modulates alternative splicing of selected targets in somatic cells (21–25), in some cases through interaction with other splicing factors or transcriptional regulators (22,25,46,47). To investigate whether Sam68 plays a role in alternative splicing also in male germ cells, we first analysed its potential interaction with components of the splicing machinery. Fractionation experiments of germ cell lysates to yield cytosolic (S), nuclear soluble (NS) and nuclear matrix-attached insoluble (NI) fractions showed that Sam68 and most splicing factors are retained in the NI fraction (Supplementary Figure 1A), similarly to what described in somatic cells (48). Moreover, Sam68 co-immunoprecipitated with SRSF1 and hnRNP A2 in both meiotic and post-meiotic male germ cells (Supplementary Figure 1B). In vitro pull-down assays showed that both splicing factors bind to the C-terminus of Sam68 in an RNase-independent manner (Supplementary Figure 1C), but not with the GSG RNA binding domain, suggesting a direct protein–protein interaction.
Next, we asked whether Sam68 is involved in alternative splicing in male germ cells. The best characterized Sam68-dependent splicing event in mouse cells is exon 8 inclusion in the epsilon sarcoglycan (Sgce) mRNA (23). In mouse neuronal progenitor cells, Sam68 represses inclusion of exon 8, possibly by binding to elements located in the flanking introns (23). We found that skipping of Sgce exon 8 occurs also in testis at various stages of development (Supplementary Figure 1D). Importantly, ablation of Sam68 strongly suppressed this splicing event in developing testis (Figure 6A). The effect was ascribable to a role of Sam68 in germ cells, because skipping of exon 8 was observed in purified wild-type pachytene spermatocytes and round spermatids, whereas it was almost abolished in knockout germ cells (Figure 6B).
Figure 6.
Sam68 regulates alternative splicing of Sgce exon 8 in male germ cells. RT–PCR analysis of Sgce exon 8 inclusion in total RNA extracted from Sam68 wild-type or knockout testes at 8, 16 and 30 days post partum (dpp) (A) or from isolated wild-type or knockout spermatocytes and spermatids. (B) Bands corresponding to mRNAs containing or not, exon 8 are indicated on the right of the panels. (C and D) Chromatin immunoprecipitation (ChIP) analysis of Sam68 wild-type and knockout germ cells. Sonicated chromatin (100 µg of DNA/sample) was immunoprecipitated with 2µg of H5, H14 antibodies or control rabbit IgGs and co-precipitated DNA was analysed by real time PCR with primers (black arrows in the scheme) spanning the Sgce transcription unit as indicated. (E) CLIP analysis of the binding of Sam68 to Sgce pre-mRNA. After UV crosslink, Sam68 wild-type and knockout germ cell extracts were sonicated and treated with DNase and RNase to yield RNA fragments of ∼200 to 250 nt. Wild-type and knockout germ cell extracts were immunoprecipitated with 2 µg of rabbit IgGs or anti-Sam68 antibodies and co-precipitated RNA was analysed by real time PCR with primers (black arrows in the scheme) spanning the Sgce transcription unit as indicated. (F) Electrophoretic mobility shift assays (EMSAs) of the binding of purified GST-Sam681–277 to a labelled Sgce probe containing the sequences encoded at the intron 7/exon 8 boundary. The position of the free probe and the probe complexed with GST-Sam681–277 are shown on the left side. Competition with the wild-type (middle panel) or mutated (right panel) cold probes is shown. The scheme above the gels shows the wild-type and mutated sequence used in the EMSAs. The mutated bases that interfere with the Sam68 consensus are shown in violet. (G) The western blot analysis of RNA pulldown assay of U2AF65 binding to Sgce exon 8. Biotinylated RNAs encoding the Sgce wild-type and mutated exon 8 sequences (from −32 to +63) were bound to Streptavidine agarose beads and incubated with testicular nuclear extract (100 µg) in the presence of 1 µg of purified GST or GST-Sam681–277, as indicated.
Changes in the phosphorylation status of RNAPII have been shown to affect alternative splicing (34,46). To characterize further the role played by Sam68 in Sgce splicing, we performed chromatin immunoprecipitation (ChIP) analyses in wild-type and Sam68−/− germ cells using antibodies for the phosphorylated forms of RNAPII. We found that RNAPII occupancy of the Sgce transcription unit was affected by ablation of Sam68. Decreased occupancy of the elongating polymerase (H5) was observed at the exon 5/intron 5 and exon 8/intron 8 boundaries, with minor changes in the rest of the gene (Figure 6C). The profile of the pausing polymerase (H14) in Sam68−/− germ cells was affected at the intron 6/exon 7 and exon 8/intron 8 boundaries, as well as within intron 7 (Figure 6D). Notably, the region in intron 7 overlapped with some of the binding sites for Sam68 proposed by in vitro experiments (23). UV CLIP experiments to map the in vivo binding sites of Sam68 in the Sgce pre-mRNA showed a peak of Sam68 binding at the intron 7/exon 8 boundary, nearby the 3′splice site (Figure 6E).
To investigate the ability of Sam68 to bind this region, we performed mobility shift assays using a probe covering 30 nucleotides upstream the regulated 3′splice site, which includes two putative consensus sequences for Sam68 binding (Figure 6F). Sam68 bound this sequence in a concentration dependant fashion. Importantly, binding was completed out by the addition of the cold probe in the assay, but not by the addition of a probe mutated in the consensus sequences (Figure 6F).
Exon inclusion is favoured by definition of the 3′splice site by binding of the general splicing factor U2AF65 and assembly of the U2 snRNP (49). To determine whether Sam68 interfered with U2AF65 binding at the 3′splice site, we performed RNA affinity chromatography experiments with biotinylated oligos, using probes covering from −32 to +63 of Sgce exon 8. As shown in Figure 6G, U2AF65 from testis nuclear extracts is recruited to exon 8 splice sites and addition of recombinant GST-Sam68 to the reaction reduced this binding. Moreover, mutation of the putative consensus sites decreased the ability of GST-Sam68 to compete for binding (Figure 6G). These results suggest that Sam68 modulates Sgce alternative splicing by affecting recruitment of phosphorylated RNAPII and U2AF65 at the 3′splice site of exon 8.
DISCUSSION
Post-transcriptional regulation of gene expression is essential for spermatogenesis. A large number of mRNAs need to be accumulated during the transcriptionally active stages of germ cell differentiation to account for the lack of transcription that occurs during homologous recombination and sperm differentiation (1–3). In addition, testis expresses a high number of tissue-specific splicing variants (7,50). We previously reported that Sam68 knockout male mice are sterile (15), highlighting the crucial role of this RBP in germ cells. Herein, we demonstrate that expression of Sam68 temporally correlates with the transcriptionally active stages of spermatogenesis. Sam68 is predominantly nuclear in germ cells and RNA integrity is required for its retention in the nucleus. In this cellular compartment, Sam68 associates with the phosphorylated forms of RNAPII and with splicing regulators, thereby modulating alternative splicing of mRNA targets, such as Sgce, in male germ cells. These results point to Sam68 as a new marker of the transcriptionally competent stages of spermatogenesis, which plays a role in testis-specific gene expression.
In male germ cells, transcription is repressed at two distinct developmental stages: during the process of homologous recombination, which occurs in the first meiotic prophase and during the terminal differentiation of the haploid spermatids (3–7). We have used the H5 antibody to check the status of RNAPII activity and found a perfect correlation between phosphorylation of the CTD on serine 2 and the reported activity of RNAPII (4,5). The H5 antibody failed to stain germ cells at the zygotene stage of meiotic prophase and elongated spermatids, in which RNA transcription is known to be silenced (4–7). Our results also show that only a subgroup of pachytene spermatocytes are positive to H5 staining. Remarkably, we observed that H5 staining perfectly overlapped with Sam68 expression in these cells. Spermatocytes increase in size during the pachytene stage through the accumulation of mRNAs and proteins required to sustain two subsequent cell divisions without an intervening interphase. Histological staging of meiosis and genetic analyses of mice with specific arrest of meiotic progression have allowed us to restrict Sam68 expression from the mid-pachytene to the meiotic divisions (Stage IV–XII of the seminiferous cycle). Germ cells from Atm−/− and H2ax−/− mice did not express Sam68. However, this RBP was still expressed when early meiotic arrest occurred in the absence of DSBs and transcriptional repression, such as in the Spo11−/− pachytene-like cells (42–44). Together, with the results of H5 staining, our observations suggest that mouse spermatocytes re-activate their transcriptional program and accumulate Sam68 protein from the mid-pachytene stage of prophase I.
Many pre-mRNA processing events occur during gene transcription through the recruitment of RNA processing factors by the CTD of RNAPII (34). For instance, 5′-capping enzymes bind directly to the CTD phosphorylated on serine 5. In addition, several splicing factors are recruited to splice sites by the CTD (34), indicating that mRNA splicing and transcription are temporally and physically coupled. Thus, the CTD acts as a scaffold to recruit many of the players required for pre-mRNA processing. The phosphorylation status of the CTD is also indicative of RNAPII activity. The non-phosphorylated protein is usually found in the pre-initiation transcriptional complex at the promoter region. Phosphorylation of serine 5 correlates with transition to transcriptional initiation, whereas phosphorylation of serine 2 correlates with the elongating activity of RNAPII (34). We report evidence that Sam68 is involved in co-transcriptional events in mouse germ cells. First, we found that Sam68 is tightly co-expressed with the phosphorylated form of RNAPII. Furthermore, endogenous Sam68 co-immunoprecipitate with the phosphorylated polymerase in spermatocytes. RNAPII interacted in vitro with the C-terminus of Sam68, which does not bind RNA. However, treatment of germ cell extracts with RNase decreased the binding of Sam68 to RNAPII. Although we cannot formally exclude that the interaction with RNAPII is mediated by another protein bound to nascent RNA, it is likely that Sam68 can directly interact with the CTD. Nevertheless, in germ cells this interaction seems to be stabilized by concomitant binding to mRNAs, possibly because a fraction of Sam68 is deposited on pre-mRNAs as RNAPII proceeds in transcript elongation.
Although testis is known to have a peculiar pattern of splicing events and expresses a large number of splicing factors (50–52), not much is known on the regulatory pathways generating transcript diversity. Few testis-specific splicing regulators, possibly contributing to tissue specific control of gene expression, have been identified (52). Two such examples are RBMY and hnRNPG-T, which were shown to regulate splicing of the testis-specific TLE4-T exon in cooperation with the SR-like protein TRA2β (53). On the other hand, RBMY antagonized the function of TRA2β and the SR protein 9G8 in the regulation of a testis-enriched exon from the Acinus gene (54). Notably, haploinsufficiency of Hnrnpg-t strongly impairs spermatogenesis (11), suggesting that its role in splicing is essential. Thus, the RNA context and the cell-specific expression of splicing factors might finely tune expression of crucial testis-specific splicing events in male germ cells.
Another testis-specific splicing regulator is SLM-2 (55), a close homologue of Sam68 (16,17). Our results suggest that Sam68 might also contribute to splicing regulation in germ cells undergoing differentiation. Although Sam68 is usually described as an ubiquitous protein (16,29), its expression is finely tuned during spermatogenesis, with stage-specific expression during meiosis and spermiogenesis. Sam68 is a multifunctional RBP, with roles in transcription, splicing, RNA export and translation (15–17,24–28). In addition, Sam68 interacts with signalling proteins and participates as scaffold in the coordination of pathways triggered by membrane receptors in a RNA-independent manner (16). Our work suggests that Sam68 function in germ cells is tightly linked to nuclear RNA processing events. Sam68 is predominantly nuclear in germ cells and this localization requires RNA integrity. Furthermore, Sam68 expression correlates with the transcriptional activity of germ cells and we found that it forms complexes with transcriptionally active RNAPII and with splicing regulators. Together with the evidence that it is required for Sgce exon 8 splicing in meiotic and post-meiotic germ cells, these results suggest that Sam68 is an important regulator of co- and post-transcriptional events during spermatogenesis. In line with this notion, we found that ablation of Sam68 expression in germ cells affected the profile of RNAPII in the Sgce gene. In particular, recruitment of serine 5-phosphorylated RNAPII in intron 7 required Sam68. Since RNAPII pausing was shown to correlate with regulation of alternative exons during transcription (34,46), it is possible that Sam68-dependent deposition of serine 5-phosphorylated RNAPII plays a role in exon 8 splicing. Importantly, we also found that Sam68 directly binds to sequences located at the intron 7/exon 8 boundary and that it affects the interaction of U2AF65 with the 3′ splice site of exon 8 in vitro. Thus, Sam68 might modulate Sgce exon 8 splicing by multiple mechanisms. It is likely that Sam68 regulates also other splicing events in spermatocytes and spermatids. Genome-wide analyses of the changes in alternative splicing caused by ablation of Sam68 in germ cells will now be required to determine the general contribution of this RBP to proteome diversity in the testis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Telethon (GGP09154 to C.S.); Associazione Italiana Ricerca sul Cancro (AIRC to C.S. and M.B.); Association for International Cancer Research (AICR); Lance Armstrong Foundation (LAF to C.S.); Italian Ministry of Education (PRIN 2008 to R.G.); Long-Term Fellowship grant from the Human Frontier Science Program (HFSP to M.P.P.). Funding for open access charge: Associazione Italiana Ricerca sul Cancro and Italian Ministry of Education (PRIN 2008).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Drs André Nussenzweig and Manuela Pellegrini for the H2ax knockout testicular sections, Drs Maria Jasin and Scott Keeney for the Spo11, Atm and Mlh1 knockout mice, Drs Paula Cohen and Peter Moens for the generous gift of antibodies. We are also indebted to Dr Flavia Botti for assistance with histochemical analysis and to Dr Simona Pedrotti for help with germ cell purification and analyses.
REFERENCES
- 1.Braun RE. Post-transcriptional control of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 1998;9:483–489. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- 2.Elliott D. Pathways of post-transcriptional gene regulation in mammalian germ cell development. Cytogenet. Genome Res. 2003;103:210–216. doi: 10.1159/000076806. [DOI] [PubMed] [Google Scholar]
- 3.Paronetto MP, Sette C. Role of RNA-binding proteins in mammalian spermatogenesis. Int. J. Androl. 2010;33:2–12. doi: 10.1111/j.1365-2605.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- 4.Monesi V. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J. Cell Biol. 1964;22:521–532. doi: 10.1083/jcb.22.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 6.Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–561. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 7.Kleene KC. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech. Dev. 2001;106:3–23. doi: 10.1016/s0925-4773(01)00413-0. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Medvedev S, Yu J, Tang LC, Agno JE, Matzuk MM, Schultz RM, Hecht NB. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc. Natl Acad. Sci. USA. 2005;102:5755–5760. doi: 10.1073/pnas.0408718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, Dorin J, Cooke HJ. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds N, Collier B, Maratou K, Bingham V, Speed RM, Taggart M, Semple CA, Gray NK, Cooke HJ. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum. Mol. Genet. 2005;14:3899–3909. doi: 10.1093/hmg/ddi414. [DOI] [PubMed] [Google Scholar]
- 11.Ehrmann I, Dalgliesh C, Tsaousi A, Paronetto MP, Heinrich B, Kist R, Cairns P, Li W, Mueller C, Jackson M, et al. Haploinsufficiency of the germ cell-specific nuclear RNA binding protein hnRNP G-T prevents functional spermatogenesis in the mouse. Hum. Mol. Genet. 2008;17:2803–2818. doi: 10.1093/hmg/ddn179. [DOI] [PubMed] [Google Scholar]
- 12.Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 13.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 14.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, Palombi F, Stefanini M, Geremia R, Richard S, et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J. Cell Biol. 2009;185:235–249. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim. Biophys. Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Sette C, Messina V, Paronetto MP. Sam68: a new STAR in the male fertility firmament. J. Androl. 2010;31:66–74. doi: 10.2164/jandrol.109.008136. [DOI] [PubMed] [Google Scholar]
- 18.Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MH, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 2001;15:2408–2420. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larocque D, Richard S. QUAKING KH domain proteins as regulators of glial cell fate and myelination. RNA Biol. 2005;2:37–40. doi: 10.4161/rna.2.2.1603. [DOI] [PubMed] [Google Scholar]
- 21.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 22.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chawla G, Lin CH, Han A, Shiue L, Ares M, Jr, Black DL. Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol. Cell Biol. 2009;29:201–213. doi: 10.1128/MCB.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paronetto MP, Cappellari M, Busà R, Pedrotti S, Vitali R, Comstock C, Hyslop T, Knudsen KE, Sette C. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010;70:229–239. doi: 10.1158/0008-5472.CAN-09-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, Manley JL, Sette C. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J. 2010;29:1235–1247. doi: 10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coyle JH, Guzik BW, Bor YC, Jin L, Eisner-Smerage L, Taylor SJ, Rekosh D, Hammarskjöld ML. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol. Cell Biol. 2003;23:92–103. doi: 10.1128/MCB.23.1.92-103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paronetto MP, Zalfa F, Botti F, Geremia R, Bagni C, Sette C. The nuclear RNA-binding protein Sam68 translocates to the cytoplasm and associates with the polysomes in mouse spermatocytes. Mol. Biol. Cell. 2006;17:14–24. doi: 10.1091/mbc.E05-06-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grange J, Belly A, Dupas S, Trembleau A, Sadoul R, Goldberg Y. Specific interaction between Sam68 and neuronal mRNAs: implication for the activity-dependent biosynthesis of elongation factor eEF1A. J. Neurosci. Res. 2009;87:12–25. doi: 10.1002/jnr.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard S, Torabi N, Franco GV, Tremblay GA, Chen T, Vogel G, Morel M, Cléroux P, Forget-Richard A, Komarova S, et al. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 2005;1:e74. doi: 10.1371/journal.pgen.0010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukong KE, Richard S. Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav. Brain Res. 2008;189:357–363. doi: 10.1016/j.bbr.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Bianchi E, Barbagallo F, Valeri C, Geremia R, Salustri A, De Felici M, Sette C. Ablation of the Sam68 gene impairs female fertility and gonadotropin-dependent follicle development. Hum. Mol. Genet. 2010;19:4886–4894. doi: 10.1093/hmg/ddq422. [DOI] [PubMed] [Google Scholar]
- 32.Barchi M, Geremia R, Magliozzi R, Bianchi E. Isolation and analyses of enriched populations of male mouse germ cells by sedimentation velocity: the centrifugal elutriation. Methods Mol. Biol. 2009;558:299–321. doi: 10.1007/978-1-60761-103-5_18. [DOI] [PubMed] [Google Scholar]
- 33.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 35.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 36.Chiodi I, Biggiogera M, Denegri M, Corioni M, Weighardt F, Cobianchi F, Riva S, Biamonti G. Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J. Cell Sci. 2000;113:4043–4053. doi: 10.1242/jcs.113.22.4043. [DOI] [PubMed] [Google Scholar]
- 37.Drabent B, Bode C, Bramlage B, Doenecke D. Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem. Cell Biol. 1996;106:247–251. doi: 10.1007/BF02484408. [DOI] [PubMed] [Google Scholar]
- 38.Lammers JH, Offenberg HH, van Aalderen M, Vink AC, Dietrich AJ, Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, Brown K, Rottinghaus S, Jackson SP, Tagle D, Ried T, et al. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development. 1998;125:4007–4017. doi: 10.1242/dev.125.20.4007. [DOI] [PubMed] [Google Scholar]
- 40.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 42.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 43.Barchi M, Roig I, Di Giacomo M, de Rooij DG, Keeney S, Jasin M. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 2008;4:e1000076. doi: 10.1371/journal.pgen.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahadevaiah SK, Bourc'his D, de Rooij DG, Bestor TH, Turner JM, Burgoyne PS. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J. Cell Biol. 2008;182:263–276. doi: 10.1083/jcb.200710195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monesi V. Differential rate of ribonucleic acid synthesis in the autosomes and sex chromosomes during male meiosis in the mouse. Chromosoma. 1965;17:11–21. doi: 10.1007/BF00285153. [DOI] [PubMed] [Google Scholar]
- 46.Batsché E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 47.Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 49.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 50.de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010;38:2825–2838. doi: 10.1093/nar/gkq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008;36:4823–4832. doi: 10.1093/nar/gkn463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elliott DJ, Grellscheid SN. Alternative RNA splicing regulation in the testis. Reproduction. 2006;132:811–819. doi: 10.1530/REP-06-0147. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Bourgeois CF, Pang S, Kudla M, Dreumont N, Kister L, Sun YH, Stevenin J, Elliott DJ. The germ cell nuclear proteins hnRNP G-T and RBMY activate a testis-specific exon. PLoS Genet. 2009;5:e1000707. doi: 10.1371/journal.pgen.1000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dreumont N, Bourgeois CF, Lejeune F, Liu Y, Ehrmann IE, Elliott DJ, Stévenin J. Human RBMY regulates germline-specific splicing events by modulating the function of the serine/arginine-rich proteins 9G8 and Tra2-{beta} J. Cell Sci. 2010;123:40–50. doi: 10.1242/jcs.055889. [DOI] [PubMed] [Google Scholar]
- 55.Venables JP, Vernet C, Chew SL, Elliott DJ, Cowmeadow RB, Wu J, Cooke HJ, Artzt K, Eperon IC. T-STAR/ETOILE: a novel relative of SAM68 that interacts with an RNA-binding protein implicated in spermatogenesis. Hum. Mol. Genet. 1999;8:959–969. doi: 10.1093/hmg/8.6.959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.