Abstract
Here, we report that the untreated rabbit reticulocyte lysate contains over 300 different endogenous microRNAs together with the major components of the RNA-induced silencing complex and thus can be used as a model in vitro system to study the effects of microRNAs on gene expression. By using this system, we were able to show that microRNA hybridization to its target resulted in a very rapid and strong inhibition of expression that was exerted exclusively at the level of translation initiation with no involvement of transcript degradation or deadenylation. Moreover, we demonstrate that the magnitude of microRNA-induced repression can only be recapitulated in the context of a competitive translating environment. By using a wide spectrum of competitor cellular and viral RNAs, we could further show that competition was not exerted at the level of general components of the translational machinery, but relied exclusively on the presence of the poly(A) tail with virtually no involvement of the cap structure.
INTRODUCTION
microRNAs (miRNAs) are small non-coding RNAs (18–25 nt long) that are encoded by the cell genome. Once associated with the RNAi-induced silencing complex (RISC), they can regulate gene expression by interacting, in most cases, with the 3′ untranslated region (3′-UTR) of the messenger RNA (mRNA) to affect its translation and/or stability. miRNAs have been found in plants, animals and viruses, some of which are very well conserved during evolution, thus suggesting an important role (1,2). Interestingly, miRNAs were shown to be implicated in most of the biological processes studied so far (i.e. development, cell growth, cell division, etc.) (3,4). This is also reflected by the fact that about 60% of human coding genes possess conserved target-sites for miRNAs (5,6) showing the extent of miRNA-dependent regulation of gene expression.
Interaction between miRNAs and target mRNAs generally involves a full-match base pairing at the seed region (nucleotides 2–8 at the miRNA 5′-end), followed by a bulge region (a few nucleotides long) and partial complementarity to the 3′-end of the miRNA (7–9). Interestingly, full pairing between the miRNA and an mRNA leads to degradation of the latter by an small interfering RNA (siRNA) response that first cleaves the target transcript at the site of interaction and then provokes the complete degradation by the cell (10–12). Nevertheless, very few cases of natural full matching interactions have been reported in animals (12,13). In contrast, for the predominant bulged target-sites, repression of protein synthesis mediated by miRNAs depends on the RISC complex, which essentially consists of Argonaute, and GW182 proteins (common to the siRNA pathway) (14,15). However, the actual mechanisms by which miRNAs regulate gene expression are not yet fully understood. Several proposed mechanisms involve translational repression at the initiation (16–21) or post-initiation steps (22–24), and also mRNA deadenylation and mRNA target degradation (25–28). Furthermore, even though the RISC machinery is required for repression, it is not fully clear whether it plays a direct role or if it allows the recruitment of other cellular factors that could account for this repression (29–34).
Cell-free extracts have been instrumental in understanding the molecular mechanism of translation, and thus it would be of great interest to develop an in vitro system that would be able to recapitulate translational repression mediated by miRNAs. Most existing in vitro systems that allow an miRNA response rely on ‘home-made’ cell-free extracts that are technically difficult to produce and yield a low-level of translational activity (17,19,26,27). Recently, an in vitro system based on the rabbit reticulocyte lysate (RRL) has been proposed (20,21), but it relies exclusively on exogenous artificial miRNAs that need to be pre-annealed to the target mRNA before translation and more importantly it was developed in the nuclease-treated RRL, a system which does not recapitulate the cap/poly(A) dependence (35–37).
This is a drawback as the cap and poly(A) tail of mRNAs were recently shown to be critical players in miRNA-dependent translational repression (16–19), thus their synergy must be recapitulated in vitro, in order to reproduce all aspects of miRNA regulation of translation. Here, we have exploited the properties of the untreated RRL that was previously described to be both cap and poly-A dependent for translation (38), to faithfully reproduce translational repression driven by endogenous miRNAs. Biochemical analysis showed that the RISC machinery, as well as high amount of endogenous miRNAs, is present in RRL. Moreover, functional assays using an exogenous mRNA bearing target sites for endogenous miRNAs showed that endogenous RISC components were able to recapitulate all major aspects of translational repression observed in vivo with no evident deadenylation or degradation of target transcripts. Finally yet importantly, we also show that no miRNA response can be observed in the nuclease-treated RRL despite the fact that the latter also contains endogenous miRNAs in similar quantities. However, addition of competitor mRNAs to the nuclease-treated RRL restored a potent miRNA response. Interestingly, only polyadenylated competitor mRNAs were able to restore an miRNA response in the nuclease-treated RRL independently of the presence of a cap in the 5′ end. This was further investigated by showing that addition of free poly(A) was sufficient to restore a potent miRNA response in trans. Taken together, our results suggest a role for poly(A)-binding protein (PABP) in translational repression independent of its role in deadenylation which has recently been demonstrated (39–41). Finally, we propose the use of the untreated RRL as a standard in vitro system, available to any user, that recapitulates many previously described features of the miRNA response: pre-miRNA processing, miRNA hybridization to their target site and their effects on translation (in the case of bulged target sites) and mRNA cleavage (in the case of a full match pairing between the miRNA and the target mRNA).
MATERIALS AND METHODS
DNA constructs and in vitro transcription
Plasmids containing target sites for miR451 (Luc-451X6, Luc PMX4 and Luc-451MutX6) and let7 (Luc-let7X6) were derived from the pGlobin-Renilla, pEMCV-Renilla and pHCV-Renilla vectors recently described (38). Target sites were constructed by hybridizing two synthetic oligodeoxyribonucleotides (Eurogentec) that contained the target motifs separated by the natural let-7a spacer from the lin41 gene and cloned into the 3′UTR of the digested (HindIII) vector. These target sites were amplified by polymerase chain reaction (PCR) and sequentially cloned in the EcoRV and XbaI restriction sites to produce a construct containing six target sites in the 3′UTR of the renilla luciferase gene. Plasmids were linearized at the EcoRI site to produce polyadenylated RNAs, and at the XbaI site for producing non-polyadenylated RNAs. The firefly coding plasmid was constructed by cloning the firefly luciferase coding region into the pGlobin-Renilla vector digested by BamHI and EcoRV (thus releasing the renilla luciferase coding region). pHCV-NS, pCrPV-LacZ vectors were linearized respectively at the MluI and SspI, as described in ref. 42. Uncapped RNAs were transcribed following the protocol described in ref. 38, and treated with RQ1 DNAse (Promega Co., Madison, WI, USA). In vitro synthesized transcripts were capped using the ScriptCap kit (Epicentre), which allows full capping of RNAs in the 5′–5′ orientation.
Radiolabeled RNAs were transcribed as described above but UTP and GTP were replaced by [α-32P]UTP (800 Ci/mmol, 10 mCi/ml) and [α-32P]GTP (800 Ci/mmol, 10 mCi/ml). Prior to translation, mRNAs were heat denatured at 65°C for 5 min and then immediately placed on ice.
Polyadenylation of mRNAs by the poly(A) polymerase was performed using the Poly(A) Tailing Kit, following the manufacturer’s protocol (Ambion).
Western blotting of RISC components
RRL volumes of 1, 2 and 3 µl, or HeLa cell S10 and S100 lysates (prepared as described in (43) were resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Proteins were then transferred to a nitrocellulose membrane by electroblotting and incubated with antibodies specific for Dicer, Ago2 and PABP. Western blotting against TNRC6 was performed following the protocol described in ref. (44). For this, 100 µl of untreated RRL were used to immunoprecipitate TNRC6 using 5 µl of anti-TNRC6 antibodies (Santacruz Technology) and 15 µl of protein A magnetic beads (Millipore). Beads together with antibodies were incubated with untreated RRL for 2 h, then washed three times with radioimmunoprecipitation assay (RIPA) buffer and loaded on 7.5% SDS–PAGE. Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane by electroblotting and incubated with antibodies specific for TNRC6 described in ref. (45).
Microarray assay
Microarray assay was performed by LC Science Company. Five micrograms of total RNA sample isolated from untreated RRL (Promega) were size-fractionated using a YM-100 Microcon centrifugal filter (Millipore) and the small RNAs (<300 nt) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for subsequent fluorescent dye staining. Hybridization was performed overnight on a µParaflo microfluidic chip using a micro-circulation pump (Atactic Technologies). On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide coding segment complementary to target microRNA (from miRBase, http://microrna.sanger.ac.uk/sequences/) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The detection probes were made by in situ synthesis using photogenerated reagent (PGR) chemistry. The hybridization melting temperatures were balanced by chemical modifications of the detection probes. Hybridization used 100 µl 6 × SSPE buffer [0.90 M NaCl, 60 mM Na2HPO4, 6 mM ethylenediaminetetraacetic acid (EDTA), pH 6.8] containing 25% formamide at 34°C. After hybridization, detection was carried out by fluorescence labeling using tag-specific Cy3 and Cy5 dyes. Hybridization images were collected using a laser scanner (GenePix 4000B, Molecular Device) and digitized using Array-Pro image analysis software (Media Cybernetics). Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter10 (Locally-weighted Regression) and P-values of the t-test were calculated; differentially detected signals were those with <0.01 P-values.
Splinted ligation assays of selected miRNAs
Total reticulocyte lysate RNA was prepared using the mirVana kit (Ambion), and specific miRNAs assayed by the splinted ligation method (46). In all cases, the ligation oligodeoxynucleotide was 5′-CGCTTATGACATTC/reversed-dC/-3′, and was 5′-end-labeled using T4 polynucleotide kinase (NEB) according to the supplier’s recommendation.
Bridge oligodeoxynucleotides (Eurofins-MWG-Operon) had three-carbon spacers at each end, and the following sequences:
for let7a, 5′-GAATGTCATAAGCGAACTATACAACCTACTACCTCA-3′;
miR-451, 5′-GAATGTCATAAGCGAACTCAGTAATGGTAACGGTTT-3′;
and miR-221, 5′-GAATGTCATAAGCGGAAACCCAGCAGACAATGTAGCT-3′.
The splinted ligation was performed in a 15-µl volume as described in ref. 46: 100 fmol bridge oligonucleotide, 100 fmol 32P-labeled ligation oligonucleotide and 1, 2 or 4 µg reticulocyte RNA were denatured at 95°C for 1 min in 75 mM KCl and 20 mM Tris–HCl, pH 8.0. After annealing at 65°C for 2 min and then at 37°C for 10 min, 400 U T4 DNA ligase (NEB) and ligase buffer [to a final of 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 10 mM DTT, 1 mM adenosine triphosphate (ATP)] was added to the reaction mixture, which was incubated at 30°C for 1 h. Reaction was terminated by heat inactivation at 75°C for 15 min followed by addition of 10 U of calf intestinal alkaline phosphatase (NEB) and incubation at 37°C for 15 min. Then 15 µl formamide dye was added and the material was fractionated on a 15% urea–polyacrylamide gel, which was dried and the bands quantified using a PhosphorImager.
Preparation of untreated RRL and in vitro translation assays
Untreated RRL was prepared essentially as previously described (38,47). Briefly, 1 ml of untreated RRL (Promega Co., Madison, WI, USA) was supplemented, before thawing, with 25 µM haemin (Fluka) and 25 µg/ml creatine phosphokinase (Rabbit Skeletal Muscle, Calbiochem). After thawing, RRL was further supplemented with 5 mg/ml creatine phosphate (Disodium Salt, Calbiochem), 50 µg/ml bovine liver tRNAs (Sigma–Aldrich) and 3 mM D-glucose (Sigma–Aldrich).
Translation reactions were performed in a final volume of 30 µl consisting of 20 µl of untreated RRL, 0.46 fmol of heat-denatured mRNAs, in the presence of KCl (100 mM), MgCl2 (0.5 mM) and amino acids mixture (20 µM each). When indicated, 2′-O-Me oligonucleotides complementary to miR-451 or let-7a were added to a final concentration of 35 nM. RRL under full translational condition was incubated together with the heat denatured mRNA for 1 h at 10°C, followed by 2 min at 20°C, 2 min at 25°C and 30 or 60 min at 30°C. The reaction was then stopped by the addition of 50 µl of luciferase lysis buffer to 10 µl of the translation reaction. When indicated, 5 pmol of competitor RNAs, or 1.2 pmol of free poly(A) RNA (400-nt average length, GE Healthcare), or 27 µM (final concentration) of free cap-analog (New England Bioloabs), were added to the extracts before translation.
Translation in wheat germ extract (Promega Co., Madison, WI, USA) was carried out using 0.46 fmol of mRNA following the manufacturer’s protocol. Translation reactions were stopped by the addition of 50 µl of lysis buffer.
Renilla luciferase activity was measured in a Veritas™ luminometer (Turner Biosystems), using the Renilla Luciferase Assay System (Promega Co., Madison, WI, USA).
mRNA integrity assay
Radiolabeled mRNAs (0.46 fmol) were translated as described above. At the end of translation, RNAs were extracted using TRIzol (Invitrogen) and loaded on a 4% polyacrylamide 7 M urea gel. The gel was dried and submitted to autoradiography using X-ray films (Fuji).
Quantitative PCR assays
miRNA quantification was carried out using the Ncode miRNA kit (Invitrogen) according to the manufacturer’s protocol using 1 µg of total RNA extracted from untreated RRL.
mRNA stability after translation was performed by extracting total RNA from 20 µl of the translation reaction using TRIzol (Invitrogen). Reverse transcription of 500 ng of total RNA was performed using qScript kit (Quanta). Quantitative PCR was then performed as described (48) using endogenous glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA as an internal control.
Poly(A) tail length assay
Monitoring of the poly(A) tail length was carried out using the Ncode miRNA kit (Invitrogen) with the following modifications: Total RNA extracted from RRL (after translation of the target mRNA) and the capped and polyadenylated Luc or Luc-451X6 mRNAs (after transcription) were polyadenylated following the manufacturer’s protocol (using 1 µg of total RNA or 0.1 ng of pure target mRNA). Reverse transcription of the polyadenylated RNAs was carried out using the universal RT primer provided with the kit, that anneals at the 3′end of the mRNA poly(A) tail. The resulting cDNAs were then used as template for PCR using a universal antisense primer (complementary of the specific sequence of the RT oligo used to reverse transcribe the target mRNA) and a specific sense primer complementary to the 3′ end of the renilla coding region. PCR products were then resolved on a 2% agarose gel.
Processing of miRNA precursors
Two-hundred femtomoles of 5′ end-labeled pre-miR-122 (Dharmacon) were incubated in 70% RRL (with 10 mM creatine phosphate, 100 mM KCl, 0.5 mM MgCl2, 0.1 mM each amino acid) at 30°C. At 10-min intervals, 10-µl samples were taken up to 1 h, then a final sample was taken at 90 min. Samples were denatured in 10 µl formamide dye and then separated on a 15 % urea–polyacrylamide gel. The intensity of the bands was quantified by phosphorimager analysis.
Statistical analysis
Data were tested for normality using the one-sample Kolmogorov–Smirnov test (N = 45 for each group of samples). Having verified the normal distribution (P-value of 0.25), statistical significances were calculated on the normally distributed data sets using a paired Student’s t-test.
RESULTS
A functional RISC machinery as well as endogenous miRNAs are present in the untreated RRL
The RRL has been used for some time to study the control of protein synthesis as it presents a high metabolic activity and contains all cytoplasmic components that are needed for efficient translation. Most workers use the nuclease-treated RRL in which endogenous mRNAs have been destroyed by the use of the calcium-dependent micrococcal nuclease (47). Although this system has been widely used over the last 30 years, it has also been criticized, as it does not faithfully reproduce physiological conditions pertaining in the cytoplasm of cells (35–37). Thus, we have recently designed an in vitro system based on the untreated reticulocyte lysate (which contains endogenous mRNAs, mainly globin and lipoxygenase) that faithfully recapitulates the cap and poly-A synergy (38).
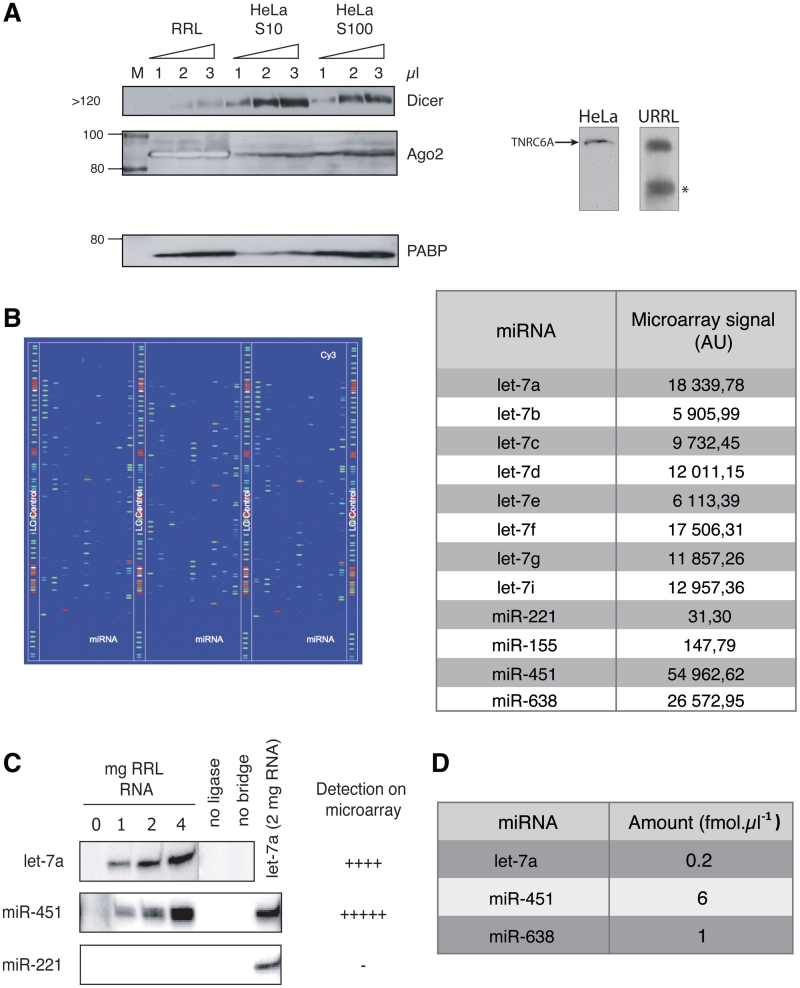
As this lysate has not been treated with nucleases, it was of interest to test for the presence of endogenous miRNAs and RISC components. For this, western blotting of RRL proteins was carried out using antibodies specific for Dicer, Argonaute2 (Ago2) and TNRC6A (the mammalian homolog of GW-182), as they all play a role in the maturation of miRNA precursors (Dicer, Ago2) or directly in the repression of targeted mRNAs (Ago2 and TNRC6). As a control, western blotting was also carried out on HeLa cell S10 and S100 lysates that were previously shown to contain RISC proteins (14) (Figure 1A). As shown, RRL contains endogenous Dicer in detectable amounts, although its expression level appears to be lower than that observed in S10 and S100 HeLa extracts (Figure 1A). TNRC6A was also detected both in HeLa and RRL (Figure 1A, right). However, Ago2 was much more abundant in the RRL than in S10 and S100 HeLa extracts, which is consistent with previous reports (49,50) (it should be noted that the bands corresponding to Ago2 in the RRL appear white as its concentration is saturating). The presence of most of the components of the RISC machinery in the RRL prompted us to test whether processing of a precursor miRNA (pre-miRNA) could take place. For this, a radiolabeled (5′-end label) miR-122 precursor was chosen, as it is not expressed in the rabbit reticulocyte lysate; this precursor was incubated in the RRL for different periods of times and the results are presented (Supplementary Figure S1). Interestingly, the pre-miR-122 was rapidly processed into two different intermediates molecules (Supplementary Figure S1, see intermed 1 and 2) and mature miR-122 molecules were observed as early as 10 min after the beginning of the incubation (Supplementary Figure 1, see miR-122) to reach 15% of the input pre-miR-122 after 90 min. It is noteworthy that the appearance of two putative processing intermediates is almost immediate (Supplementary Figure S1) and then seems to decrease with time.
Figure 1.
Rabbit reticulocyte lysate contains RISC components as well as endogenous miRNAs. (A) RISC proteins are present in the RRL. 1, 2 or 3 µl of RRL, HeLa cell S10 and S100 lysates were analyzed by western blotting with antibodies specific for Dicer, Ago2 and PABP as a loading control. TNRC6 was immunoprecipitated and analyzed by western blotting using specific antibodies. An asterisk corresponds to a non-specific band observed for TNRC6 in untreated RRL. (B) Endogenous miRNAs can be detected in untreated RRL. Total RNAs extracted from untreated RRL were hybridized to a microarray containing probes for most known mammalian miRNAs (LC Sciences). Each colored spot corresponds to an miRNA that is present in RRL; a total of more than 300 different miRNAs were detected. Microarray raw data signal is presented for let7, miR-221, miR-155, miR-451 and miR-638 (full data can be found in Supplementary Table S1). (C) Splinted ligation assays against let-7, miR-451 and miR-221 were carried out using increasing amounts of total RNA to validate microarray results. miRNAs not identified by microarray, such as miR-221, were not detected by splinted ligation. (D) Quantification of miRNAs present in untreated RRL. Quantitative PCR was carried out using specific primers against let-7, miR-451 and miR-638.
The next step was to look for the presence of endogenous miRNAs. For this, total RNA was purified from untreated RRL and hybridized on a microarray (LC Sciences) with probes against most known mammalian miRNAs (Figure 1B). Surprisingly, more than 300 different miRNAs could be detected in the untreated RRL (Figure 1B and Supplementary Table S1). Among them, some could be found at very high concentration and were those corresponding to miRNAs that are known to be upregulated during erythroid differentiation such as miR-451 (51,52) (which literally saturated the reading). On the other hand, miRNAs that were shown to be downregulated during erythroid cell differentiation process such as miR-155 and miR-221 (51,52) could hardly be detected on the microarray and were expressed about much less than miR-451 (Figure 1B, see table). Interestingly, ubiquitous members of the let-7 family of miRNAs (comprising let-7a to let-7i) were present at high concentration with the let-7a member being the most abundant.
In order to confirm these results, a splinted ligation assay (46) was performed against both miR-451 and let-7a (highly expressed) and miR-221 (virtually absent from the RRL) (Figure 1C, additional miRNAs were also tested and the results are presented in Supplementary Figure S2). As observed, both let-7a and especially miR-451 gave a strong band in a yield proportional to the amount of RRL RNA analyzed, whereas no endogenous miR-221 could be detected (Figure 1C).
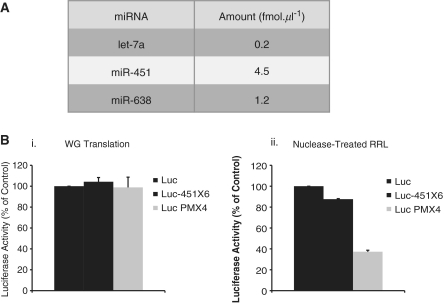
Finally, by using quantitative PCR, the amounts of endogenous miR-451, let-7a and miR-638 were quantified and the results are summarized in Figure 1D. This gave an estimated concentration of 0.2, 6 and 1 fmol per microliter of lysate for let-7a, miR-451 and miR-638, respectively. Interestingly, these concentrations of endogenous miRNAs remained essentially similar between different batches of lysate obtained from different rabbits (data not shown).
Taken together, these results show that the untreated RRL contains all the components required to recapitulate a miRNA response.
Endogenous miRNAs are functional and can trigger both a mi- and si- RNA response on target mRNAs
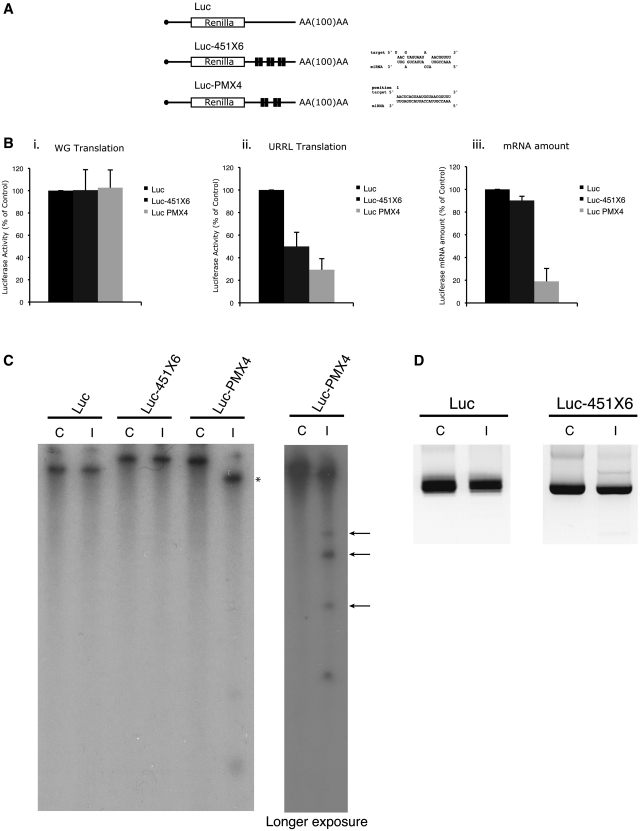
In order to test the effects of endogenous miRNAs on gene expression, different reporter constructs containing the beta globin 5′UTR driving translation of the Renilla luciferase open reading frame (ORF) and followed by four to six (depending on the construct) target sites for miR-451 in their 3′UTR were designed (Figure 2A). Two types of target site were created: (i) the first corresponds to a miRNA target with full complementarity to the seed in the 5′ end of the miRNA followed by a bulge and partial complementarity to the target RNA in the 3′end (named Luc-451X6 in Figure 2A) and (ii) the second is an siRNA target with full complementarity between the miRNA and the target RNA (named Luc PMX4 in Figure 2A). As a control, a luciferase construct containing no target site was used (Luc) (Figure 2A). The ability of miR-451 to bind to the target sites was first verified by annealing mature miR-451 to each of the three RNAs described above; a band shift can be observed when both the miR-451 sites together with the miR-451 oligos were added (Supplementary Figure S3). Capped and poly-adenylated RNAs were produced by in vitro transcription and then translated in the untreated RRL. Prior to translation, RNAs were heat-denatured at 65°C for 5 min. This was followed by a 1-h incubation at 10°C in the RRL to allow annealing of endogenous miR-451 and finally the reactions were incubated at 30°C for 30 min so that translation could occur. As a control, the RNAs were translated under the same experimental conditions in wheat germ extract as this system does not contain endogenous miR-451 (53). As shown in Figure 2B (panel i), translation efficiency in the wheat germ extract was virtually the same for the three mRNAs, indicating that the presence of target sites on the 3′ UTR does not affect translation in the absence of complementary endogenous miR-451. However, protein synthesis in untreated rabbit reticulocyte lysate (panel ii) resulted in a 2-fold inhibition of luciferase expression from the Luc-451X6 RNA and a 3-fold inhibition for Luc PMX4 compared to the control RNA containing no target sites (Luc). To avoid any non-specific effect the binding of the endogenous miRNAs to target mRNAs could have on overall translation, we also tested the levels of phosphorylated-eIF2α after translation of both Luc and Luc-451X6 mRNAs. It is important to note that we did not detect any difference of phosphorylated-eIF2α levels between Luc and Luc-451X6 samples after translation (data not shown), strongly suggesting that translational inhibition of Luc-451X6 did not depend on activation of the PKR pathway. Interestingly, when RNA levels were measured by quantitative-PCR after translation, we could not detect any significant difference between Luc and Luc-451X6 (Figure 2B, panel iii), whereas the amount of Luc PMX4 RNA (containing the siRNA target sites) was significantly lower, showing that RNA degradation had taken place. These data suggest that both an siRNA- and miRNA-mediated response can be observed in the RRL from endogenous miRNAs. Furthermore, similar results were also obtained using mRNAs containing target sites for let-7a (Supplementary Figure S4), thus indicating that multiple endogenous miRNAs are functional in the untreated RRL.
Figure 2.
Endogenous miRNAs are fully functional for si- and miRNA regulation of gene expression. (A) Schematic representation of the reporter RNAs used in this study. The renilla luciferase reporter gene (designated Luc) is driven by the human β-globin 5′UTR in which six miRNA bulged target sites for miR-451 (Luc-451X6) or four perfectly complementary target sites for miR-451 (Luc PMX4) were designed in the 3′UTR of the reporter gene as indicated in the figure. Target mRNA/miRNA interaction schemes were obtained using the RNA hybrid software (72). (B) Endogenous miRNAs are able to downregulate translation and stability of targeted mRNAs. Translation of 0.46 fmol of Luc (black bars), Luc-451X6 (dark gray bars) and Luc PMX4 (light gray bars) was carried out for 30 min in wheat germ extract (panel i) and untreated RRL (panel ii) showing a downregulation of Luc-451X6 and Luc PMX4 expression only in untreated RRL in which miR-451 is present. Stability of mRNAs in untreated RRL was monitored after translation by quantitative PCR (panel iii). Error bars correspond to the standard deviation calculated from three independent experiments. (C) Integrity of targeted mRNAs after translation in untreated RRL. Radiolabeled Luc, Luc-451X6 and Luc PMX4 mRNAs were analyzed on a denaturing polyacrylamide gel (left) before (Control lane: C) or after translation (Incubated lane: I). The right panel corresponds to the same samples from the left panel, ran for a shorter time and exposed for a longer period; asterisks indicate the 5′ cleavage product of the Luc PMX4 mRNA; arrows indicate potential Luc PMX4 target site fragments obtained after RISC-mediated cleavage of the mRNA. (D) Determination of the variations in the length of the poly(A) tail for Luc and Luc-451X6 constructs before and after translation by using a PCR-based approach (see ‘Materials and Methods’ section).
Recent data have indicated that miRNA hybridization could cause specific deadenylation of the transcript and this could be partly responsible for translation repression (28). Thus, we have monitored the integrity of target mRNAs after translation in the RRL. For this, radiolabeled Luc, Luc-451X6 and Luc PMX4 RNAs were extracted at the end of the translation reaction and subjected to PAGE on a 4% UREA-denaturing gel. It is important to note that such an approach allowed us to differentiate between the polyadenylated and non-polyadenylated forms of the target mRNAs as presented in Supplementary Figure S5. As shown on Figure 2C, no size difference could be observed for Luc and Luc-451X6 mRNAs before, and after translation (Figure 2C, Luc-451X6 lanes C and I), suggesting that no deadenylation had occurred during the period of incubation. This result was further confirmed by using a higher resolution PCR-based approach (see ‘Materials and Methods’ section) to specifically analyze the length of the poly(A) tail. Once again, this showed no difference whatsoever in poly(A) tail length before and after translation for Luc and Luc-451X6 mRNAs (Figure 2D). Interestingly, this was clearly not the case for Luc PMX4 RNA (containing siRNA target sites) which migrated at a lower molecular size following incubation in the RRL, suggesting that RNA degradation had taken place (Figure 2C, Luc PMX4 lanes C and I). It is noteworthy that hybridization of the miR-451 to the target PMX4 yielded a discrete band rather than a smear, suggesting that the RNA was cleaved at the sites of interaction with the miRNA (Figure 2C, see asterisks). This was confirmed by running the samples for a shorter time and exposing the film for longer to reveal the presence of three radiolabeled RNA fragments of smaller size (Figure 2C, see longer exposure; see arrows) that might correspond to the 3′ end of the target RNA cleaved at each of the four miR-451 target sites.
Taken together, these data indicate that endogenous rabbit reticulocyte miRNAs are functional and can control gene expression at the level of translation (miRNA response) or RNA degradation (siRNA response). Moreover, under our experimental conditions, translation inhibition observed upon miRNA association is not due to the deadenylation of the RNA target transcript.
Translation inhibition occurs rapidly and can be inhibited by addition of anti miR-451 oligonucleotides
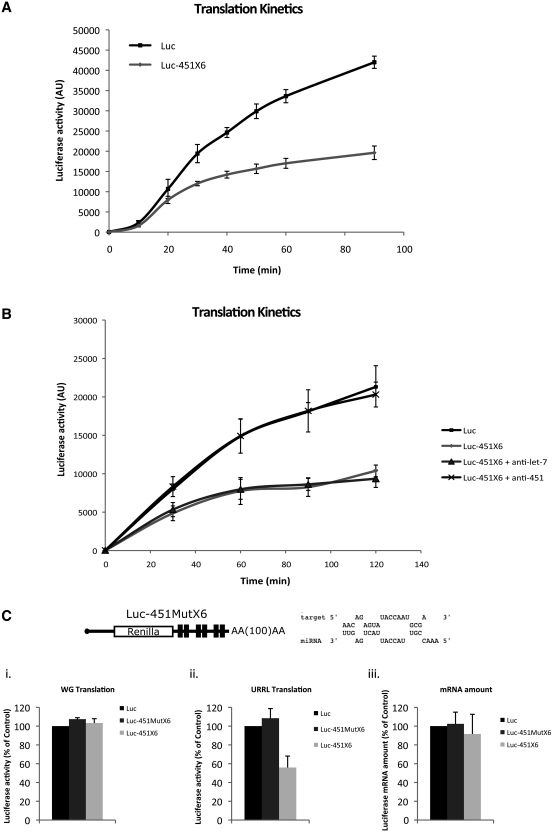
Kinetic studies of miRNA-mediated repression were then carried out in the RRL to monitor the translation rates at different time points from 0 to 90 min. As shown in Figure 3A, a 30% decrease in translational efficiency was observed as early as 20 min after the start of incubation. Interestingly, translation repression increased with time to reach a maximum of 55% after 90 min of incubation with no significant degradation of Luc-451X6 transcripts (data not shown). This indicates that assembly of the RISC complex and the resulting miRNA-mediated translation inhibition takes place rapidly in the RRL system.
Figure 3.
Translational repression mediated by endogenous miRNAs occurs rapidly and is specific to both the target site and the miRNA. (A) Translation kinetics of Luc and Luc-451X6 mRNAs was performed at different time points: 0, 10, 20, 30, 40, 50, 60 and 90 min after the beginning of translation in the untreated RRL. Error bars correspond to the standard deviation calculated from three independent experiments. (B) Translational repression mediated by miR-451 was counteracted by the addition of a specific antisense oligonucleotide. Translation of Luc and Luc-451X6 mRNAs was programmed in the untreated RRL after addition of oligonucleotides complementary to either let-7 or miR-451. Luciferase activity was measured at different time points: 0, 30, 60, 90 and 120 min after the beginning of translation. Error bars correspond to the standard deviation calculated from three independent experiments. (C) Translational repression mediated by miR-451 is specific for the target site. Six target sites for miR-451 with a mutated seed region were inserted in the 3′UTR of the Luc construct (Luc-451X6Mut, dark gray bars) and translation of this mRNA was compared to that of Luc (black bars) and Luc-451X6 (light gray bars) in wheat germ extract (panel i) and untreated RRL (panel ii). The relative stability of mRNAs in untreated RRL was monitored after translation by quantitative PCR and are represented as ‘mRNA amount' (panel iii). Error bars correspond to the standard deviation calculated from three independent experiments.
In order to test the specificity of endogenous miR-451 to mediate translational repression, 2′-O-methyl modified antisense oligoribonucleotides were used (Figure 3B). These oligonucleotides can specifically interact with endogenous miR-451 to preclude it from being incorporated into the RISC complex and/or to interact with its mRNA target sequence (54). The modified oligonucleotides were added before the pre-incubation of the mRNAs in the RRL and translation was monitored at 30, 60, 90 and 120 min. As a control, an antisense oligonucleotide complementary to let-7a was also used in a parallel incubation. As observed in Figure 3B, translation of the RNA construct containing the miR451 sites (Luc-451X6) was inhibited compared to the control Luc mRNA. However, addition of the antisense oligonucleotide directed against endogenous miR-451 abolished this inhibition and restored Luc-451X6 translation to levels similar to that of the control RNA. This was observed at early and later times, even when translation was strongly repressed in the absence of the antisense oligonucleotide (Figure 3B, see 90 and 120 min). The specificity of this effect was demonstrated by the fact that an antisense oligonucleotide not specific to miR451 (directed against let-7a) could not relieve translation repression of Luc-451X6 mRNA (Figure 3B).
Finally, in order to test for the specificity of the interaction between miR-451 and its target site, a new target motif was designed (Luc-451MutX6) which has several mutations in the seed region (Figure 3C). The corresponding in vitro synthesized capped and polyadenylated Luc, Luc-451X6 and Luc-451MutX6 RNAs were translated in wheat germ extract and the RRL. As shown previously (Figure 2), no difference could be observed in the translation efficiency of these three constructs in the wheat germ extract (Figure 3C, panel i). However, the RNA containing mutations in the seed region was no longer repressed in the RRL system (Figure 3C, panel ii, compare Luc-451MutX6 with Luc-451X6) confirming the importance of the seed match in the miRNA response. Analysis of the RNA level by quantitative real time (RT)-PCR did not show any significant difference between the three constructs indicating that the effect was not due to RNA degradation (Figure 3C, panel iii).
Translational repression shows a target site additive effect and can be saturated by high amounts of RNA
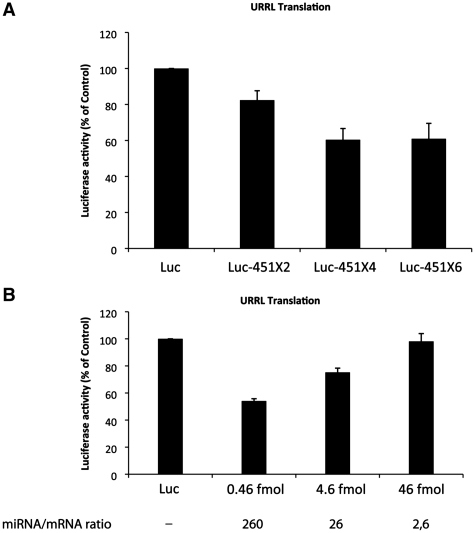
Target site cooperation was shown to improve miRNA translational repression in various cultured cell lines (55–57). Therefore, luciferase RNAs containing two, four or six target sites for miR-451 in their 3′UTR were designed, transcribed and translated in the untreated RRL. As seen in Figure 4A, translation of an RNA bearing only two target sites for miR-451 was reduced by only 20% compared to the control RNA. However, translation of the same RNA in which four target motifs have been inserted was inhibited by 40% compared to the control RNA, but the addition of two additional miR-451 sites did not further repress translation. These data show that instead of cooperation, increasing the number of target sites exerts an additive repressive effect on the target mRNA up to a certain level after which the miRNA response cannot be improved.
Figure 4.
Translational repression mediated by miR-451 can be saturated by high amounts of mRNAs and shows target site additive repression. (A) Target site additive effect improves translational repression. Untreated RRL was programmed with Luc and Luc-451 bearing 2, 4 or 6 target sites mRNAs (designated, Luc-451X2, X4 and X6) and luciferase activity was measured 30 min after the beginning of translation. Error bars correspond to the standard deviation calculated from three independent experiments. (B) Untreated RRL was programmed with 0.46, 4.6 or 46 fmol of Luc and Luc-451X6 mRNAs and luciferase activity with Luc-451X6 was measured as a percentage of that observed with the same concentration of Luc mRNA after 30 min of translation. The molar ratio of miR-451 to the target mRNA is indicated on the bottom of the figure. Error bars correspond to the standard deviation calculated from three independent experiments.
To further investigate this issue, we have varied the concentration of exogenous mRNA added to the RRL. The standard experimental conditions that have been tested so far correspond to a ratio of 260 molecules of endogenous miRNA per one molecule of luciferase Luc-451X6 mRNA (corresponding to 0.46 fmol of target mRNA); thus the final ratio is 43 miRNA molecules per target site. This was made variable by changing the concentration of exogenous target Luc-451X6 mRNA (Figure 4B). At a 30 to 1 ratio (corresponding to 4.6 fmol of target mRNA), which corresponds to five miRNA molecules per target site, translational repression dropped to 30% compared to the control RNA with no target sites (Figure 4B, see 4.6 fmol). Interestingly, at a ratio of <1 miRNA per target site (corresponding to 46 fmol of target mRNA), no repression at all could be observed (Figure 4B, see 46 fmol). This result suggests that the RISC machinery can be saturated by high amounts of target mRNA.
Internal ribosome entry site-mediated translation is insensitive to miRNAs
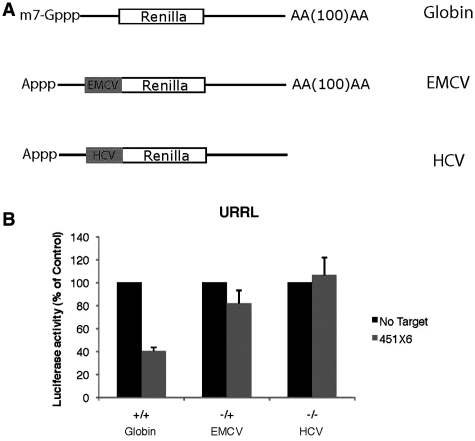
Translation initiation mediated by some internal ribosome entry sites (IRES) has been reported to be refractory to miRNA regulation (17,18). Therefore, we tested whether IRES-driven translation could be affected by miRNAs in our system. For this, target sites for miR-451 were inserted in the 3′UTR of constructs coding for the renilla luciferase driven by the encephalomyocarditis (EMCV) and hepatitis C virus (HCV) IRESes. EMCV and HCV IRES were chosen because of their different requirement for initiation factors; EMCV requires the entire set of initiation factors with the exception of eIF4E (58), whereas HCV can bypass the need for all eIF4 initiation factors (59). These constructs were used to produce uncapped and polyadenylated EMCV RNAs (–/+), and uncapped non-polyadenylated HCV RNAs (–/–) as found in native virions. Untreated RRL was then programmed with these constructs together with a capped and polyadenylated β-Globin RNA control (Figure 5A).
Figure 5.
Translation mediated by internal ribosome entry sites is refractory to miRNA-mediated regulation. (A) Schematic representation of target RNAs used in this experiment: ‘Globin’ corresponds to a capped and polyadenylated luciferase coding RNA driven by the human β-Globin 5′UTR, ‘EMCV’ corresponds to an uncapped (A-capped) and polyadenylated RNA driven by the EMCV IRES, ‘HCV’ corresponds to an uncapped and non-polyadenylated RNA driven by the HCV IRES. (B) Translation of 0.46 fmol of Globin, EMCV and HCV RNAs bearing no target sites (No Target, black bars) or 6 target-sites for miR-451 (451X6, gray bars), was carried out in untreated RRL for 60 min. Error bars correspond to the standard deviation calculated from three independent experiments.
As observed (Figure 5B), translation driven by the HCV IRESes in the untreated RRL was not inhibited at all by the binding of miR-451. Interestingly, translation of the EMCV RNA showed only a slight inhibition (18% of the control) (Figure 5B).
This suggests that translation initiation mediated by IRES elements is resistant to miRNA repression and, above all, strongly argues that translational repression mediated by miRNAs occurs at the level of translation initiation.
Specific siRNA activity, but not translational repression, occurs in the nuclease-treated RRL
RRL is generally treated with micrococcal nuclease in order to degrade endogenous mRNAs and to allow the study of translation of a single exogenous mRNA species. Since endogenous miRNAs are thought to be incorporated into RISC, they should be protected against the micrococcal nuclease treatment. To verify this, we performed quantitative PCR against different miRNAs previously described in the untreated RRL. Interestingly, the amounts of endogenous miRNAs were almost identical between untreated and nuclease-treated RRL (compare Figure 6A with Figure 1D), indicating that either mature miRNAs are too small to be degraded by the micrococcal nuclease or that they are protected by the RISC proteins. As nuclease-treated RRL contains endogenous miRNAs at levels similar to those of the untreated RRL (Figure 6A), it was of interest to compare their activity. For this, nuclease-treated RRL as well as wheat germ extract were programmed with 0.46 fmol of Luc, Luc-451X6 and Luc PMX4 mRNAs (Figure 6B). As expected, translation in the wheat germ extract yielded no significant differences between translation rates of each mRNA (Figure 6B, left). Surprisingly, in the nuclease-treated RRL, which does not contain any endogenous mRNA, we only detected a weak inhibition (15% compared to Control) of Luc-451X6 mRNA (Figure 6B, right). However, Luc PMX4 expression was still strongly impaired to a level of inhibition similar to that previously observed in the untreated RRL (70% compared to Control, Figure 6B, right). These results indicate that endogenous miRNAs contained in the nuclease treated RRL can still recapitulate an siRNA response but are unable to mediate inhibition of translation by an miRNA mechanism.
Figure 6.
Specific siRNA activity, but not translational repression, occurs in the nuclease-treated RRL. (A) Quantification of miRNAs present in nuclease-treated RRL. Quantitative PCR was carried out using specific primers against let-7, miR-451 and miR-638. (B) Translation of 0.46 fmol of Luc (black bars), Luc-451X6 (dark gray bars) and Luc PMX4 (light gray bars) was carried out for 30 min in wheat germ extract and nuclease-treated RRL. Error bars correspond to the standard deviation calculated from three independent experiments.
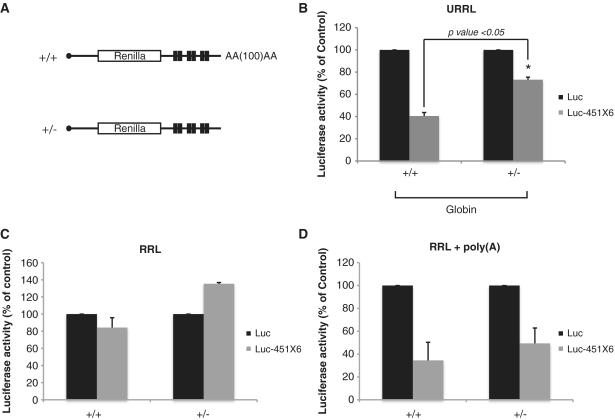
The miRNA response in nuclease-treated RRL can be restored by poly(A) tail containing RNAs
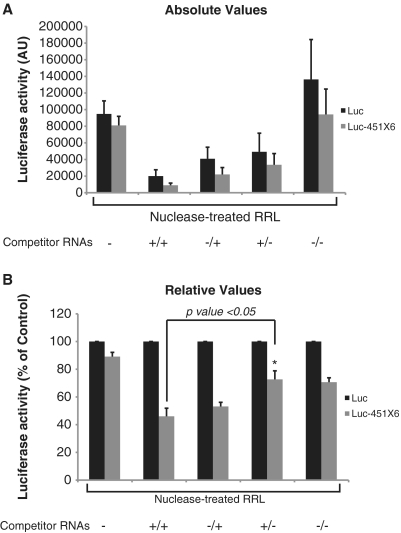
Nuclease-treated RRL has been shown to be poorly cap and poly(A) dependent for translation as opposed to crude RRL (38). One of the reasons for this lack of cap/poly(A) synergy could be the lack of competition with endogenous mRNAs. This could also be responsible for the absence of an miRNA response in the nuclease-treated RRL. To test this hypothesis, we have monitored translation of Luc and Luc-451X6 mRNAs in the presence of a 104-fold excess of competitor mRNAs. For this, 5 pmol of in vitro transcribed capped and polyadenylated competitor (+/+), capped/non-polyadenylated (+/–), uncapped/polyadenylated (–/+) and uncapped/non-polyadenylated (–/–), mRNAs (containing the β-globin 5′UTR followed by the firefly luciferase coding sequence) were added to each translation reaction before the addition of Luc or Luc-451X6 mRNAs. As shown in Figure 7, addition of these competitor RNAs led to an inhibition of both Luc and Luc-451X6 translation, with the capped and polyadenylated (+/+) having the biggest effect (Figure 7, top panel see lane +/+). Interestingly, an efficient miRNA response could only be restored upon addition of competitor mRNAs that contained a poly(A) tail whereas addition of non-polyadenylated mRNAs only had a marginal effect on miRNA activity (Figure 7, compare +/– and –/– lanes with the non-competitor lane). Interestingly, a potent miRNA activity in nuclease-treated RRL could be restored by the addition of poly(A) containing transcripts independently of the presence of a cap structure at their 5′end (Figure 7, bottom panel compare +/+ and –/+ and +/– lanes with no competitor lane). This suggests that only the presence of the poly(A) tail is required to restore miRNA-mediated translational repression in the nuclease-treated lysate. However, it could be argued that the presence of the poly(A) tail could restore a miRNA activity because of the fact that poly(A) containing transcripts are better translated than non-poly(A) containing RNAs. Thus, the effects observed would only be the consequence of an increased competition for ribosomes.
Figure 7.
Polyadenylated competitor mRNAs restore a strong miRNA activity in the nuclease-treated RRL. Translation of 0.46 fmol of Luc (black bars), and Luc-451X6 (gray bars) was carried out for 60 min in untreated RRL (URRL) or nuclease-treated RRL alone (RRL) or in the presence of 5 pmol of capped/polyadenylated firefly luciferase competitor mRNAs (+/+), uncapped/polyadenylated (–/+), capped/non-polyadenylated (+/–) and uncapped/non-polyadenylated mRNAs (–/–). Total renilla luciferase activity (A) as well as luciferase activity normalized against Luc expression (B) are presented. Statistical significance [Luc-451X6 (+/+) versus Luc-451X6 (+/–)] was calculated using the paired Student’s t-test; *P < 0.05. Error bars corresponds to three independent experiments.
Free poly(A) RNA is sufficient to restore a microRNA response in nuclease-treated RRL
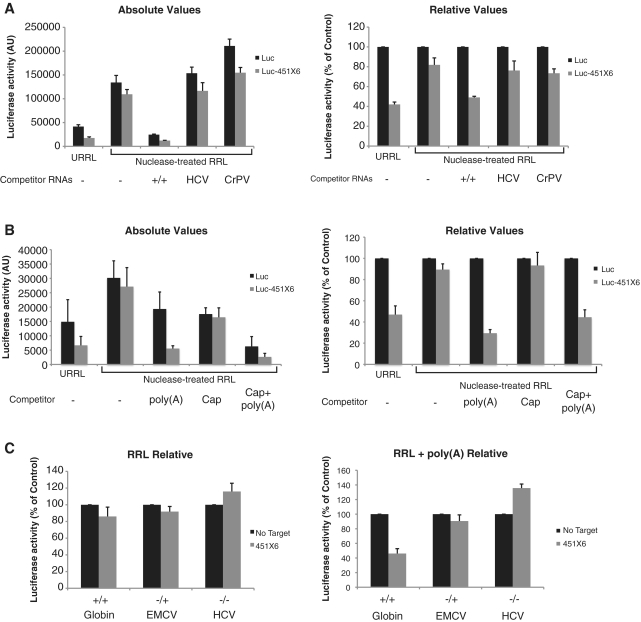
In order to investigate this effect further, we have added uncapped and non-polyadenylated mRNAs driven by the HCV and cricket paralysis virus (CrPV) IRES to nuclease-treated lysates prior to translation. Indeed, these two viral RNAs are able to recruit ribosomes very efficiently on their IRES sequence with no need for a cap structure and a poly(A) tail (see Supplementary Figure S6A for relative translational efficiencies of each competitor RNA tested). As observed, addition of 5 pmol of HCV and CrPV competing mRNAs failed to restore a potent miRNA activity (Figure 8A, compare RRL to HCV and CrPV lanes). These results indicate that the competition for ribosomes is not the major cause of inhibition mediated by miRNAs, therefore suggesting a specific role for the poly(A) tail.
Figure 8.
Free poly(A) RNA is sufficient to restore a microRNA response in nuclease-treated RRL. (A) Translation of 0.46 fmol of Luc (black bars), and Luc-451X6 (gray bars) was carried out for 60 min in untreated RRL (URRL) or nuclease-treated RRL alone (RRL) or in the presence of 5 pmol of capped/polyadenylated Firefly-coding competitor mRNAs under control of the β-globin 5′UTR (+/+) or uncapped/non-polyadenylated mRNAs under the control of the HCV and CrPV IRESes. (B) Translation of 0.46 fmol of Luc (black bars), and Luc-451X6 (gray bars) was carried out for 1 h in untreated RRL (URRL) or nuclease-treated RRL alone (RRL) or in the presence of 1.2 pmol of free poly(A) [poly(A)] (GE Healthcare), 27 µM of free cap analog (Cap analog) (New England Biolabs) or 1.2 pmol of free poly(A) together with 27 µM of cap analog [Cap analog + poly(A)]. Total luciferase activity (left panel) as well as luciferase activity normalized against Luc expression (right panel) are presented. (C) Translation of 0.46 fmol of Globin, EMCV and HCV RNAs bearing no target sites (No Target, black bars) or 6 target-sites for miR-451 (451X6, gray bars), was carried out in RRL (left panel) and RRL in the presence of free poly(A) RNA (right panel) for 60 min. Error bars correspond to three independent experiments.
In order to investigate this further, we performed similar experiments using free cap analog or free poly(A) as competitors (Figure 8B). As free poly(A) was shown to stimulate PABP activity both in plants and mammals (60,61), we tested whether free poly(A) could also modulate miRNA activity. For this, exogenous free poly(A) (400 nt in average length) was added to the translation reaction in the same molar amount to that of the previous experiments using competitor mRNAs. As observed, addition of free poly(A) to the nuclease-treated RRL restored miRNA activity (by 60%) to a level of magnitude close to that observed in crude RRL (Figure 8B compare the URRL and poly(A) lanes). To rule out any effect of the 5′ cap, we also added free cap-analog alone or in the presence of free poly(A) (Figure 8B, see Cap analog lane). As observed, addition of free cap analog to the nuclease-treated RRL, could not restore the miRNA activity in the nuclease-treated RRL even though the overall level of global translation efficiency was significantly affected (Figure 8B, see lane Cap analog). Furthermore, the addition of both free cap analog and free poly(A), although leading to a strong translational inhibition of both Luc and Luc-451X6 mRNAs, did not further increase miRNA activity compared to free poly(A) alone [Figure 8B, compare the poly(A) lane to Cap analog + poly(A)]. Interestingly, we could observe a dose-dependent effect of the addition of free poly(A) RNA in restoring a miRNA repression in nuclease-treated RRL (Supplementary Figure S6B). However, this dose-dependent effect was observed within a narrow range (<2-fold increase) of poly(A) RNA (Supplementary Figure S6B).
Because IRES-mediated translation appeared to be refractory to translational repression mediated by miRNAs in untreated RRL, we tested whether this was also the case in RRL upon addition of free poly(A) RNA. For this, capped and polyadenylated Globin RNAs, uncapped and polyadenylated EMCV RNAs (–/+), and uncapped non-polyadenylated HCV RNAs (–/–), containing miR-451 target-sites (451X6) or not (No target), were programmed in RRL (Figure 8C, left). As expected, translation of all these RNAs (including Globin) was not regulated by endogenous miR-451 (Figure 8C, left). However, upon addition of free poly(A) RNA, translation of Globin-451X6 RNA was downregulated by 2-fold compared to the Globin RNA bearing no miRNA target sites (Figure 8C, right). Interestingly, translation of EMCV and HCV RNAs was still insensitive to miRNA-mediated repression even in the presence of free poly(A) RNA. This result suggests that the effect of free-poly(A) in stimulating miRNA activity is only exerted on cap-dependent translation.
Free poly(A) restores translational repression on a non-polyadenylated target RNA in a trans fashion
Due to the effect of the poly(A) RNA addition for efficient miRNA repression, we next wondered whether poly(A) tail removal from target RNAs could also affect repression. For this, untreated RRL was programmed with capped and polyadenylated (+/+) and capped and non-polyadenylated (+/–) Luc and Luc-451X6 mRNAs (Figure 9A). As observed, removal of the poly(A) tail from the target mRNA resulted in an attenuated miRNA-mediated repression, which dropped to 25% of the control RNA (i.e. bearing no target sites) (Figure 9B). Interestingly, translation of non-polyadenylated Luc-451X6 RNAs in the nuclease treated RRL was not inhibited but rather moderately stimulated (40%) by miR-451 (Figure 9C, left). However, upon addition of free poly(A) RNA to nuclease-treated RRL, translational repression was restored both for capped and polyadenylated Luc-451X6 RNAs (65% compared to the control) and also for capped and non-polyadenylated Luc-451X6 RNAs (51% compared to the control) (Figure 9C, right).
Figure 9.
Free poly(A) restores translational repression of a non-polyadenylated target RNA in trans. (A) Schematic representation of the RNAs used in this experiment corresponding to the Luc and Luc-451X6 RNAs that are capped and polyadenylated (+/+) or capped and non-polyadenylated (+/–). (B) Translation of 0.46 fmol of capped/ polyadenylated and capped/non-polyadenylated Luc (black bars), and Luc-451X6 (gray bars) RNAs in untreated RRL for 60 min. Statistical significance (Luc-451X6 +/+ versus Luc-451X6 +/–) was calculated using the paired Student’s t-test; *P < 0.05. (C) Translation of the same RNAs was carried out in nuclease-treated RRL (left panel) and in nuclease-treated RRL in the presence of 1.2 pmol of free poly(A) RNA (right panel). Error bars correspond to three independent experiments.
Taken together, these results suggest that the poly(A) tail is required whether in cis or in trans to achieve efficient repression mediated by miRNAs.
DISCUSSION
Despite an extensive amount of work, the molecular mechanism by which miRNAs control translation remains elusive. Based on experiments and results obtained using different systems, some reports describe translational repression at the initiation and post-initiation steps, linked in some cases to the deadenylation of the target mRNA which contributes to translational repression, while others reports a rapid degradation of the target mRNAs or even co-translational proteolysis of the nascent polypeptide (9,16–34). In vitro translation systems have been instrumental for the study of translational regulation as they allow the control of many parameters that are important in protein synthesis such as mRNA amount, salt conditions, initiation factor concentrations, time of incubation and mRNA degradation. The most commonly used in vitro system is the RRL, which was conceived and developed about 30 years ago and has been widely used since (47). Recently, Novina and colleagues (20,21) have developed an interesting approach to study miRNAs in the nuclease-treated RRL. However a major drawback comes from the fact that the nuclease-treated RRL does not recapitulate the cap and poly(A) synergy (35–37), which seems to be a major determinant in the miRNA response. In addition, Novina and colleagues have been using exogenous artificial miRNAs and so this approach does not allow investigation of all aspects of miRNA repression such as pre-miRNA processing, miRNA binding and nucleation of the RISC complex.
Here, we report that the non-nuclease-treated RRL (38) is capable of faithfully recapitulating translational control of gene expression mediated by miRNAs. Importantly, our system exploits the presence of endogenous miRNAs that are found in the rabbit reticulocyte lysate, namely miR-451 that has been recently shown to be implicated in the maturation of erythroid cells (51,52,62,63).
We first showed that the RISC protein components are present in the RRL (Figure 1) and are fully active to process miRNA precursors (Supplementary Figure S1). By western blotting, we detected low levels of Dicer, while Ago2 was highly abundant. Interestingly, processing of pre-miR-451 has been recently shown to occur through a dicer-independent (but Ago2 dependent) pathway (64). This could possibly explain the relatively low abundance of dicer compared to Ago2 (Figure 1A). Finally, we also detected TNR6CA (also known as GW182), which is an important player of the miRNA repression pathway.
We could then show that translational repression of mRNAs targeted by endogenous miRNAs recapitulated all major aspects of miRNA-dependent regulation. Indeed, translation is repressed by about 2-fold (Figure 2) as has been observed not only for natural targets in cultured cells (1,65–69) but also in other in vitro cell-free extracts based on insect embryos (26). Importantly, this effect on gene expression is specific to the binding of the miRNA to its target and can be reversed by addition of complementary oligonucleotides and saturating amounts of target mRNAs (Figures 3 and 4). Moreover, regulation of translation was particularly sensitive to modifications impairing complementarity within the seed region of the miRNA, and showed target site cooperation (Figures 3 and 4). Most importantly, target mRNAs were neither deadenylated nor degraded by the action of the miRNA (Figure 2), and this lack of deadenylation could be explained by the fact that our in vitro system allowed us to measure translational activity after only 30 min of incubation. This is particularly interesting as it allows the effects that miRNAs can have on translation to be uncoupled from those on deadenylation and transcript degradation. However, transcript degradation can be reproduced by having full pairing between the miRNA and the mRNA target, which triggers a siRNA response (Figure 2). Interestingly, in a recent report by Fabian and coworkers (39), miRNA-dependent deadenylation was shown to occur 1 h after the beginning of the translation reaction. This could therefore explain our lack of visible deadenylation as we monitor mRNA integrity between 30 and 60 min after the start of translation. Finally, we also show that repression occurs at the level of translation initiation as RNAs under control of the EMCV and HCV IRESes were not repressed by the action of miR-451 (Figure 5).
We have also shown that the nuclease-treated RRL contained the same concentration of endogenous miRNAs as the untreated RRL; yet, we were surprised to find out that these miRNAs were unable to repress translation of a reporter gene in the case of bulged target sites (Figure 6). However, in the case of a perfect match target site, the same endogenous miRNAs fully recapitulated an siRNA effect with the same magnitude as that observed with untreated RRL. In order to understand the reasons of such a discrepancy between the mi- and siRNA mechanisms, we tested several different conditions. Because nuclease-treated RRL lacks endogenous mRNAs, we first reasoned that addition of competitor mRNAs, in similar amount to that of crude RRL, could restore a functional miRNA activity (Figure 7). Interestingly, this showed that only capped/polyadenylated and uncapped/polyadenylated RNAs were able to restore an effective repression mediated by miRNAs in nuclease-treated RRL, with virtually no effect of addition of non-polyadenylated RNAs (Figure 7). This suggested either a role of the poly(A) tail in this effect, or the need for competing RNAs that are efficiently translated.
In order to discriminate between these two possibilities, we have used viral IRESes from HCV and CrPV that are either naturally uncapped and non-polyadenylated (in the case of HCV) or that can function with no need for initiation factors (nor initiator tRNA in the case of CrPV). Interestingly, addition of competitor RNAs containing either the HCV or the CrPV IRESes failed to restore miRNA repression in the nuclease treated system (Figure 8A), suggesting that competition for general components of the translational machinery was not critical for the efficiency of the miRNA response; however, it rather points out to a specific role for some initiation factors that bind either the cap or the poly(A) tail. Thus, we next designed an experiment in which the nuclease treated lysate was supplemented by addition of cap analogue and free poly(A) prior to translation (Figure 8B). Interestingly, overall translational efficiency of Luciferase was affected to a similar extent by the addition of either the cap analog or the free poly(A) with addition of both having an additive effect (Figure 8B, black bars). However, and to our surprise, addition of free poly(A) alone was both necessary and sufficient to restore an miRNA response in the nuclease treated lysate, whereas addition of free cap analog failed to do so (Figure 8B). Interestingly, addition of free poly(A) could also restore efficient repression on non-polyadenylated RNAs which were not effectively repressed in crude RRL (Figure 9C). On the contrary, translation driven by the EMCV and HCV IRESes could not be repressed by miRNAs even upon addition of free poly(A) RNA, suggesting that only cap-dependent translation would be sensitive to miRNA-mediated translational inhibition.
While this work was in progress, several reports have addressed the critical role of PABP in the miRNA response at the level of transcript deadenylation (39–41) and also on translation where GW182 was proposed to compete with PABP for eIF4G binding (41). Our results nicely confirm and extend these data by showing that the poly(A) tail can also play a role in trans for the efficient repression of target mRNAs. But how is the poly(A) able to modulate the miRNA response in trans? Interestingly, several studies carried out using wheat germ cell-free extracts have shown that addition of free poly(A) can improve PABP binding to the eIF4F complex and stimulate its activity (60,70). Moreover, free poly(A) has been shown to stimulate translation of non polyadenylated mRNAs in trans in the nuclease-treated RRL by improving the interaction between PABP and eIF4G (61). However, a mechanism where GW182 and PABP compete for eIF4G binding, although possible, is nevertheless unlikely in our system as translation driven by the EMCV [which under normal conditions depends on the PABP/eIF4G interaction (71)] is not repressed by miRNAs. However, it could be possible that miRNAs regulate the activity of the eIF4F complex. Thus, it would be conceivable that free poly(A) as well as polyadenylated competitor mRNAs could improve PABP binding to the eIF4F complex, thus allowing miRNAs to effectively regulate eIF4F activity and inhibit translation of the target mRNA. This would be also in agreement with the fact that the HCV IRES, which is independent of the eIF4F complex for translation remains unaffected by miRNAs.
Taken together, we describe here a new in vitro cell-free extract that faithfully reproduces the regulation of translation mediated by miRNAs, which is commercially available, easy to use and yielding highly reproducible results. Interestingly, our system allows translation repression to be studied at very early times when deadenylation has not yet taken place.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ANRS and ANR; E.P.R. was a recipient of “Ministère de la Recherche et de la Technologie” and “Fondation pour la Recherche Medicale (FRM)” fellowships. Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank W. Filipowicz for Ago2 and TNRC6 antibodies, D. Scadden for Tudor-SN antibody and S. Morley for PABP antibody. We thank B. Blanquier and IFR128 for qPCR facilities. We also thank Dr. Marie Sémon for statistical analysis of our data.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R, Su B. Small but influential: the role of microRNAs on gene regulatory network and 3′UTR evolution. J. Genet. Genomics. 2009;36:1–6. doi: 10.1016/S1673-8527(09)60001-1. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 9.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci. STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 10.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 11.Song J-J, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 12.Yekta S, Shih I-H, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 13.Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- 14.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Humphreys DT, Westman BJ, Martin DIK, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4 E/cap and poly(A) tail function. Proc. Natl Acad. Sci. USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 18.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 19.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol. Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 21.Wang B, Yanez A, Novina CD. MicroRNA-repressed mRNAs contain 40 S but not 60 S components. Proc. Natl Acad. Sci. USA. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 23.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 24.Petersen CP, Bordeleau M-E, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell. Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Rivas FV, Wohlschlegel J, Yates JR, III, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell. Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borman AM, Michel YM, Kean KM. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 2000;28:4068–4075. doi: 10.1093/nar/28.21.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel YM, Poncet D, Piron M, Kean KM, Borman AM. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- 37.Svitkin YV, Evdokimova VM, Brasey A, Pestova TV, Fantus D, Yanagiya A, Imataka H, Skabkin MA, Ovchinnikov LP, Merrick WC, et al. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J. 2009;28:58–68. doi: 10.1038/emboj.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soto Rifo R, Ricci EP, Décimo D, Moncorgé O, Ohlmann T. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007;35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates RISC-associated mRNA in human cells. Mol. Cell. Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricci EP, Herbreteau CH, Decimo D, Schaupp A, Datta SAK, Rein A, Darlix J-L, Ohlmann T. In vitro expression of the HIV-2 genomic RNA is controlled by three distinct internal ribosome entry segments that are regulated by the HIV protease and the Gag polyprotein. RNA. 2008;14:1443–1455. doi: 10.1261/rna.813608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunt SL, Jackson RJ. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moser JJ, Chan EK, Fritzler MJ. Optimization of immunoprecipitation-western blot analysis in detecting GW182-associated components of GW/P bodies. Nat. Protoc. 2009;4:674–685. doi: 10.1038/nprot.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maroney PA, Chamnongpol S, Souret F, Nilsen TW. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA. 2007;13:930–936. doi: 10.1261/rna.518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelham HR, Jackson RJ. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 1976;67:247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- 48.Ricci EP, Mure F, Gruffat H, Decimo D, Medina-Palazon C, Ohlmann T, Manet E. Nucleic Acids Res. 2009;37:4932–4943. doi: 10.1093/nar/gkp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakravarty I, Bagchi MK, Roy R, Banerjee AC, Gupta NK. Protein synthesis in rabbit reticulocytes. Purification and properties of an Mr 80,000 polypeptide (Co-eIF-2A80) with Co-eIF-2 A activity. J. Biol. Chem. 1985;260:6945–6949. [PubMed] [Google Scholar]
- 50.Zou C, Zhang Z, Wu S, Osterman JC. Molecular cloning and characterization of a rabbit eIF2C protein. Gene. 1998;211:187–194. doi: 10.1016/s0378-1119(98)00107-3. [DOI] [PubMed] [Google Scholar]
- 51.Masaki S, Ohtsuka R, Abe Y, Muta K, Umemura T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem. Biophys. Res. Commun. 2007;364:509–514. doi: 10.1016/j.bbrc.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 52.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur. J. Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 53.Heimberg AM, Sempere LF, Moy VN, Donoghue PCJ, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc. Natl Acad. Sci. USA. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007;8:R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bi X, Goss DJ. Wheat germ poly(A)-binding protein increases the ATPase and the RNA helicase activity of translation initiation factors eIF4A, eIF4B, and eIF-iso4F. J. Biol. Chem. 2000;275:17740–17746. doi: 10.1074/jbc.M909464199. [DOI] [PubMed] [Google Scholar]
- 61.Borman AM, Michel YM, Malnou CE, Kean KM. Free poly(A) stimulates capped mRNA translation in vitro through the eIF4G-poly(A)-binding protein interaction. J. Biol. Chem. 2002;277:36818–36824. doi: 10.1074/jbc.M205065200. [DOI] [PubMed] [Google Scholar]
- 62.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl Acad. Sci. USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 66.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 68.Seitz H. Redefining microRNA targets. Curr. Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 69.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 70.Khan MA, Goss DJ. Translation initiation factor (eIF) 4B affects the rates of binding of the mRNA m7G cap analogue to wheat germ eIFiso4F and eIFiso4F.PABP. Biochemistry. 2005;44:4510–4516. doi: 10.1021/bi047298g. [DOI] [PubMed] [Google Scholar]
- 71.Svitkin YV, Imataka H, Khaleghpour K, Kahvejian A, Liebig HD, Sonenberg N. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA. 2001;7:1743–1752. [PMC free article] [PubMed] [Google Scholar]
- 72.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.