Abstract
The programming of cellular networks to achieve new biological functions depends on the development of genetic tools that link the presence of a molecular signal to gene-regulatory activity. Recently, a set of engineered RNA controllers was described that enabled predictable tuning of gene expression in the yeast Saccharomyces cerevisiae through directed cleavage of transcripts by an RNase III enzyme, Rnt1p. Here, we describe a strategy for building a new class of RNA sensing-actuation devices based on direct integration of RNA aptamers into a region of the Rnt1p hairpin that modulates Rnt1p cleavage rates. We demonstrate that ligand binding to the integrated aptamer domain is associated with a structural change sufficient to inhibit Rnt1p processing. Three tuning strategies based on the incorporation of different functional modules into the Rnt1p switch platform were demonstrated to optimize switch dynamics and ligand responsiveness. We further demonstrated that these tuning modules can be implemented combinatorially in a predictable manner to further improve the regulatory response properties of the switch. The modularity and tunability of the Rnt1p switch platform will allow for rapid optimization and tailoring of this gene control device, thus providing a useful tool for the design of complex genetic networks in yeast.
INTRODUCTION
The field of synthetic biology encompasses the engineering of biological systems that exhibit new functions through the design of synthetic gene networks. The proper functioning of genetic networks encoding complex behaviors depends on the coordinated regulation of genetic responses, enzymatic activities and protein levels (1–4). As such, the genetic programming of biological systems depends on our ability to design genetic devices that can detect molecular signals and link these detection events to new types of genetic control and thus biological function.
Motivated by the versatility of sensing and actuation functions that RNA can exhibit and the relative ease with which RNA structures can be modeled and designed, researchers have engineered RNA-based devices that detect diverse molecular signals and link this information to the regulation of gene expression events (5). RNA devices generally couple RNA components that exhibit sensing, actuation and information transmission activities. By varying the regulatory RNA encoded within the actuator component, RNA devices have been developed that function in different organisms through a variety of gene-regulatory mechanisms, including translation (6–10), transcript degradation (11,12), transcriptional activation (13) and splicing (14–16). Recently, several examples of RNA devices that act through the modulation of RNase III processing activities in mammalian cells have been described (17–19). However, such regulatory strategies have not yet been extended to RNase III enzymes in microorganisms.
We previously developed a genetic control system based on directing transcript cleavage through the RNase III enzyme in the yeast Saccharomyces cerevisiae (Rnt1p), where Rnt1p hairpin substrates are placed in the 3′ untranslated region (UTR) of the targeted gene (20). We utilized cell-based screening strategies to develop libraries of synthetic Rnt1p hairpins that exhibit predictable gene-regulatory activity based on modules that are inserted into two key regions of the hairpin—the cleavage efficiency box (CEB) and the binding stability box (BSB). These studies indicated that the CEB and BSB modules function independently and can be combinatorially integrated into a single Rnt1p hairpin to achieve a wide range of gene-regulatory activities (20; A.H. Babiskin and C.D. Smolke, submitted for publication).
Here, we describe the design of a new class of RNA devices based on the synthetic library of Rnt1p regulatory elements, which we call an Rnt1p switch. We have designed a strategy for coupling RNA aptamers to Rnt1p hairpins to build RNA control systems in yeast that exhibit integrated sensing and actuation functions. Specifically, theophylline aptamers were integrated into the CEB region of the Rnt1p hairpin, which plays a key role in modulating Rnt1p cleavage rates, where binding of the small molecule to the aptamer sequence resulted in inhibition of cleavage activity. We demonstrated three different strategies for tuning the quantitative properties of the device response curve based on the integration of different modules into the Rnt1p switch platform. Aptamer/CEB, BSB and multiple switch modules were used to modulate the EC50 value and switching activity of the device. Finally, we demonstrated that these modules can be implemented combinatorially in a predictable manner to further improve the regulatory response properties of the switch. The application of these tuning strategies resulted in an increase of the fold-change from 1.93 to 2.47 in a single module switch and an increase from 1.93 to 5.57 when three copies of the single module switch were implemented.
MATERIALS AND METHODS
Plasmid construction
Standard molecular biology techniques were utilized to construct all plasmids (21). DNA synthesis was performed by Integrated DNA Technologies (Coralville, IA, USA) or the Protein and Nucleic Acid Facility (Stanford, CA, USA). All enzymes, including restriction enzymes and ligases, were obtained through New England Biolabs (Ipswich, MA, USA) unless otherwise noted. Pfu polymerase was obtained through Stratagene. Ligation products were electroporated with a GenePulser XCell (Bio-Rad, Hercules, CA, USA) into Escherichia coli DH10B (Invitrogen, Carlsbad, CA), where cells harboring cloned plasmids were maintained in Luria-Bertani media containing 50 mg/ml ampicillin (EMD Chemicals). Clones were initially verified through colony PCR and restriction mapping. All cloned constructs were sequence verified by Laragen (Los Angeles, CA, USA) or Elim Biopharmaceuticals (Hayward, CA, USA).
The construction of the Rnt1p characterization plasmid, pCS321, and the Rnt1p expression plasmid, pRNT1, have been previously described (20). The plasmid map for pCS321 is available in Supplementary Figure S1. Insertion of engineered Rnt1p substrates and appropriate controls into the 3′-UTR of a gene encoding a yeast-enhanced green fluorescent protein (yEGFP3) in pCS321 was performed through either digestion with appropriate restriction endonucleases and ligation-mediated cloning or homologous recombination-mediated gap-repair during transformation into S. cerevisiae strain W303 (MATa, his3-11, 15 trp1-1 leu2-3 ura3-1 ade2-1) through standard lithium acetate procedures (22). The single module Rnt1p switch (RS), its mutant tetraloop control, the BSB variants and the theophylline aptamer variant RS-theo3 with its own BSB variants were amplified for insertion with both techniques using the forward and reverse primers RS_fwd (5′ ATGGTATGGATGAATTGTACAAATAAAGAGCCTAGGAAACAAACAAACTTGATGCCCTTGG) and RS_rev (5′ AAATTCGCTTATTTAGAAGTGGCGCGCCCTCTCGAGTTTTTATTTTTCTTTTTACGATGCTGGTATC), respectively. Two of the theophylline aptamer variants required individually designed primers: RSnt_fwd (5′ ATGGTATGGATGAATTGTACAAATAAAGAGCCTAGGAAACAAACAAACTTGATGCCATTGG) and RS_rev for RSnt; and RS-theo2_fwd (5′ ATGGTATGGATGAATTGTACAAATAAAGAGCCTAGGAAACAAACAAACTTGGCCCTTGGCA) and RS-theo2_rev (5′ AAATTCGCTTATTTAGAAGTGGCGCGCCCTCTCGAGTTTTTATTTTTCTTTTTACGGCTGGTATCCA) for RS-theo2. A second switch module was amplified for insertion using the forward and reverse primers RSx2_fwd (5′ GTGCTCGAGAAACAAACAAACTTGATGCCCTTGGCA) and RSx2_rev (5′ CAGCTCGAGTTTTTATTTTTCTTTTTACGATGCTGGTATCCAGATG). A third switch module was amplified for insertion using the forward and reverse primers RSx3_fwd (5 GTGCCTAGGAAACAAACAAACTTGATGCCCTTGGCA) and RSx3_rev (5′ CAGCCTAGGTTTTTATTTTTCTTTTTACGATGCTGGTATCCAGATG). In the case of digestion and ligation, the PCR products were digested with the unique restriction sites AvrII and/or XhoI, which are located 3 nts downstream of the yEGFP3 stop codon and upstream of the ADH1 terminator. Following construction and sequence verification of the desired vectors, 100–500 ng of each plasmid was transformed into strain W303. In the case of gap-repair, 250–500 ng of the PCR product and 100 ng of plasmid digested with AvrII and XhoI were transformed into the yeast strain. All yeast strains harboring cloned plasmids were maintained on synthetic complete (SC) media with an uracil dropout solution and 2% dextrose at 30°C. A table of primers and template sequences used for each switch module is provided in Supplementary Table S1.
Rnt1p substrate characterization assays
Saccharomyces cerevisiae cells harboring pCS321-based plasmids were grown on SC media with an uracil dropout solution and the appropriate sugars (2% raffinose, 1% sucrose) overnight at 30°C. The cells were back-diluted the following morning into a 4.3 ml combination of SC media and 25 mM theophylline dissolved directly into the same SC media to an optical density at 600 nm (OD600) of 0.1 (∼200 µl) and grown again at 30°C. The theophylline solution and SC media were mixed to achieve the appropriate concentration of theophylline (0–5 mM) in the test tube at 5 ml. After 1 h, the test tube volume was brought up the final volume of 5 ml by adding 0.5 ml of 20% galactose (2% final concentration) for induction or water (non-induced control) to the cell cultures. The cells were grown for another 4.5 h before measuring the fluorescence levels.
Fluorescence quantification
On the Quanta flow cytometer (Beckman Coulter, Fullerton, CA, USA), the distribution of GFP fluorescence was measured with the following settings: 488 nm laser line, 525 nm band-pass filter and photomultiplier tube setting of 5.83 (pCS321-based) or 4.50 (pCS1585-based). Data were collected under low flow rates until 5000 viable cell counts were collected. A non-induced cell population was used to set a gate to represent GFP-negative and GFP-positive populations. Final data values are reported as the average of the median GFP-positive fluorescence from three independently-grown samples and normalized to a construct baring no module inserts (‘no insert’) at the same theophylline concentration. Standard error is determined from the standard deviation of the triplicate samples.
Quantification of cellular transcript levels
Total RNA from S. cerevisiae was collected by a standard hot acid phenol extraction method (23) and followed by DNase I (New England Biolabs) treatment to remove residual plasmid DNA according to manufacturer’s instructions. cDNA was synthesized from 5 µg of total RNA with gene-specific primers for yEGFP3 and ACT1 (24) (rnt1p_rtpcr_rev2 and ACT1_rtpcr_rev, respectively) and SuperScript III Reverse Transcriptase (Invitrogen) according to manufacturer’s instructions. The forward and reverse primers for yEGFP3 quantification are rnt1p_rtpcr_fwd2 (5′ CGGTGAAGGTGAAGGTGATGCTACT) and rnt1p_rtpcr_rev2 (5′ GCTCTGGTCTTGTAGTTACCGTCATCTTTG), respectively. The forward and reverse primers for ACT1 quantification are ACT1_rtpcr_fwd (5′ GGCATCATACCTTCTACAACGAAT) and ACT1_rtpcr_rev (5′ GGAATCCAAAACAATACCAGTAGTTCTA), respectively. Relative transcript levels were quantified in triplicate from three identical reactions from the cDNA samples by using an appropriate primer set and iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) on an iCycler iQ quantitative real-time PCR (qRT-PCR) machine (Bio-Rad) according to the manufacturer’s instructions. For each run, a standard curve was generated for yEGFP3 and a house-keeping gene, ACT1, using a dilution series for a control representing no insertion of an Rnt1p substrate. Relative yEGFP3 and ACT1 levels were first individually determined for each sample and then the yEGFP3 values were normalized by their corresponding ACT1 values.
In vitro transcription of Rnt1p substrates
Rnt1p switches were PCR-amplified to include an upstream T7 promoter site using forward and reverse primers Rnt1p_col-T7_fwd_prmr (5′ TTCTAATACGACTCACTATAGGATGGTATGGATGAATTGTACAAATAAAGCCTA) and Rnt1p_col-T7_rev_prmr (5′ AAATTCGCTTATTTAGAAGTGGCGC), respectively. About 500 ng of PCR product was transcribed with T7 RNA Polymerase (New England Biolabs) in the presence and absence of α-P32-GTP. The 25 µl reaction consisted of the following components: 1× RNA Pol Reaction Buffer (New England Biolabs), 3 mM rATP, 3 mM rCTP, 3 mM rUTP, 0.3 mM rGFP, 1 µl RNaseOUT (Invitrogen), 10 mM MgCl2, 2 mM DTT, 1 µl T7 Polymerase and 0.5 µCi α-P32-GTP. Unincorporated nucleotides were removed from the reactions by running the samples through NucAway Spin Columns (Ambion, Austin, TX, USA) according to the manufacturer’s instructions.
Rnt1p expression and purification
The pRNT1 plasmid was transformed into E. coli strain BL21 using the Z-competent E. coli Transformation Kit and Buffer Set (Zymo Research, Orange, CA, USA) according to manufacturer’s instructions. Rnt1p was collected as a protein extract as previously described (25). Briefly, an overnight culture of BL21 cells harboring pRNT1 was back-diluted to an OD600 of 0.5. Once the culture reached an OD600 of 1.1–1.4, it was induced with 1 mM IPTG and grown for an additional 3 h. The cells were centrifuged at 2500g for 12 min at 4°C and the resulting cell pellet was frozen in a −80°C freezer. After weighing the frozen cell pellet, the cells were resuspended in 4 ml Ni2+ buffer [25% (v/v) glycerol, 1 M NaCl, 30 mM Tris pH 8.0] per gram of harvested cells. The resuspension was sonicated (Heat Systems-Ultrasonics, Inc.) twice with the following settings: 2 × 30 s, output control 5 and 50% duty cycle. Cellular debris was removed by centrifugation at 20 000g for 30 min at 4°C and the supernatant was filtered through a 0.2 µm pore size Acrodisc 25 mm syringe filter (Pall Life Sciences, Ann Arbor, MI, USA).
Rnt1p was purified from the resulting supernatant with one 1 ml HisTrap HP column (GE Healthcare) on an AKTA fast protein liquid chromatography (FPLC) machine (GE Healthcare). Elution of the protein was performed with an imidazole concentration of 150 mM in Ni2+ buffer and the protein was collected in six-ml fractions. Protein purification was confirmed by analyzing an aliquot of each fraction on a SDS–PAGE gel (NuPAGE 4–12% Bis–Tris Gel, Invitrogen) and protein function was confirmed by incubating an aliquot of each fraction with a control Rnt1p substrate and analyzing the resulting cleavage products on an 8% denaturing polyacrylamide gel. Positive fractions were pooled and concentrated to <3 ml volume using a Centricon Centrifugal Filter Device (10 000 MWCO; Millipore) according to the manufacturer’s instructions. The concentrated protein was then injected into a Slide-A-Lyzer Dialysis Cassette (10 000 MWCO; Pierce Biotechnology) and buffer-exchanged twice with Rnt1p Storage Buffer [50% (v/v) glycerol, 0.5 M KCl, 30 mM Tris pH 8.0, 0.1 M DTT, 0.1 M EDTA] at 4°C. The first buffer exchange took place for 4 h and the second buffer exchange occurred overnight. The purified Rnt1p was stored in aliquots at −20°C.
In vitro Rnt1p substrate cleavage assay
Cleavage assays were performed on Rnt1p substrates as previously described (25,26). Briefly, a 10 µl mixture of RNA and Rnt1p were incubated at 30°C for 30 min in Rnt1p reaction buffer [30 mM Tris (pH 7.5), 150 mM KCl, 5 mM spermidine, 20 mM MgCl2, 0.1 mM DTT and 0.1 mM EDTA (pH 7.5)] in the presence or absence of 10 mM theophylline. The RNA concentration was 0.05 µM and the Rnt1p concentration was 20.7 µM. The cleavage reaction products were separated on an 8% denaturing polyacrylamide gel run at 35 W for 30 min. Gels were transferred to filter paper and analyzed for relative substrate and product levels through phosphorimaging analysis on a FX Molecular Imager (Bio-Rad).
RESULTS
Design of a ligand-responsive RNA switch based on Rnt1p processing
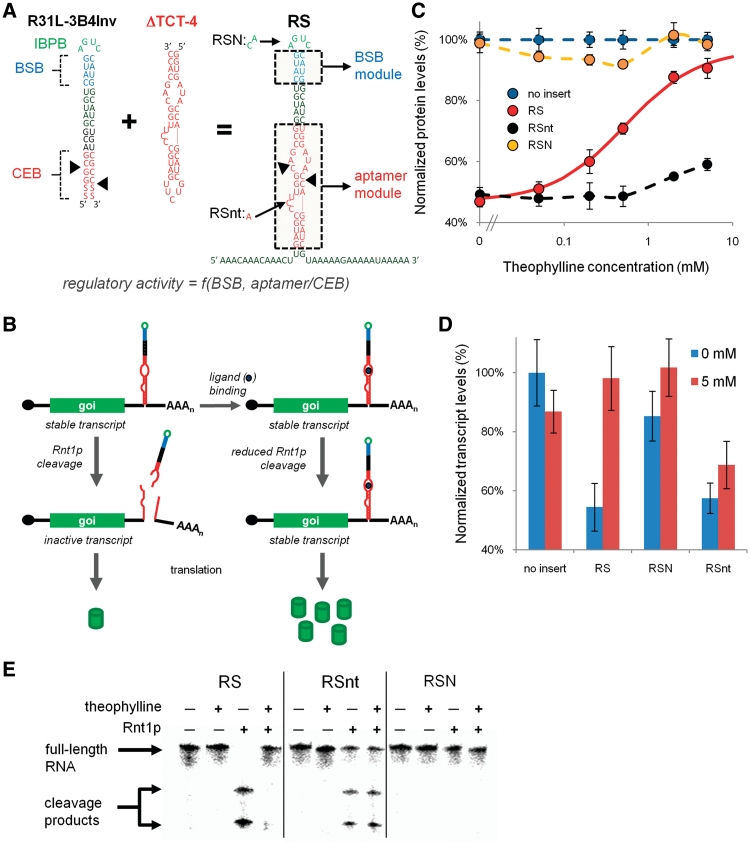
We set out to utilize the synthetic Rnt1p genetic control elements developed in early work from our laboratory in the design of a new class of RNA devices based on the modulation of Rnt1p processing, called an Rnt1p switch. The first step in building an integrated RNA device platform is identifying a physical coupling strategy between the sensor (i.e., aptamer) and actuator (i.e., Rnt1p hairpin) components that maintains the activity of each component, but allows ligand binding to the sensor to modify the activity of the actuator. Rnt1p substrates adopt hairpin structures that contain three critical regions for enzyme binding and processing: the initial binding and positioning box (IBPB), comprising an AGNN tetraloop; the BSB, comprising the base-paired region immediately below the tetraloop; and the CEB, comprising the nucleotides adjacent to the two cleavage sites (27) (Figure 1A). Rnt1p initially binds to the tetraloop (the IBPB) and the interaction is stabilized by the BSB. The enzyme then cleaves the hairpin at two locations within the CEB: between the 14th and 15th nts upstream of the tetraloop and the 16th and 17th nts downstream of the tetraloop. While the CEB has no structural or sequence requirements, the BSB and IBPB have more rigid sequence and structural requirements (28). Therefore, we identified the CEB as a possible region of the Rnt1p hairpin where the integration of aptamer sequences may not disrupt enzyme binding and processing.
Figure 1.
Design and implementation of Rnt1p switches as post-transcriptional genetic control elements. (A) The design of an Rnt1p switch (RS) by the integration of a sensor component (ΔTCT-4 aptamer) into the actuator component (R31L-3B4Inv Rnt1p hairpin). R31L-3B4Inv contains the consensus regions of a Rnt1p substrate: the CEB, BSB and IBPB. The nucleotide modifications associated with the Rnt1p and ligand-binding controls (RSN and RSnt, respectively) are indicated on RS. Color scheme is as follows: CEB and aptamer, red; BSB, blue; IBPB, green. Black triangles indicate locations of cleavage sites by Rnt1p. The regulatory activity of the synthetic Rnt1p switches is a function of the modular elements in the BSB and CEB/aptamer, which are indicated by the dashed boxes on RS. (B) The synthetic Rnt1p switches control the destabilization of target transcripts by controlling Rnt1p cleavage in the 3′-UTR of transcripts encoding a gene of interest (goi). Barrels represent protein molecules and dark blue circles represent the ligand molecule. In the absence of ligand binding to the aptamer module, the transcript is inactivated by Rnt1p cleavage and translation inhibited. Ligand binding to the aptamer module in the CEB inhibits Rnt1p cleavage activity and stabilizes the transcript, resulting in increased protein production. (C) The dose–response curves of RS and the Rnt1p and ligand-binding controls, RSN and RSnt, respectively, indicate that decreased gene expression is caused by Rnt1p cleavage and that cleavage is inhibited by theophylline. The following concentrations of theophylline are used for all response curves (in mM): 0, 0.05, 0.2, 0.5, 2, 5. Normalized protein levels were determined by measuring the median GFP levels from cells harboring constructs with the indicated switch through flow cytometry analysis, and values are reported relative to a construct lacking a hairpin module (set to 100%). Solid curves indicate the theoretically-determined model fit. The model parameters for the curve fit are provided in Table 1. Dashed curves are utilized for control constructs that are not fit to the model and are generated through Microsoft Excel’s smooth line option. (D) Analysis of transcript levels of the Rnt1p switch, RS and the controls, RSN and RSnt, supports the proposed mechanism of inhibition of Rnt1p processing due to ligand binding. Relative transcript levels are determined by measuring transcript levels of yEGFP3 and a house-keeping gene, ACT1, through qRT-PCR and normalizing the yEGFP3 levels with their corresponding ACT1 levels. Normalized transcript levels for each construct are reported relative to that from an identical construct lacking a hairpin module. (E) Cleavage reaction assays and analyzes by denaturing polyacrylamide gel electrophoresis on the Rnt1p switch RS and the controls, RSnt and RSN, support the proposed mechanism of inhibition of Rnt1p processing due to ligand binding. The top band corresponds to uncleaved full-length RNA; the bottom bands correspond to the three cleavage products expected from Rnt1p processing. Two of the expected cleavage products differ in size by 2 nt and cannot be resolved into individual bands under the conditions used for this assay. RNA was added to the final concentration of 0.05 µM in each reaction. When present, Rnt1p was added to the final concentration of 20.7 µM and theophylline was added to the final concentration of 10 mM.
We designed an Rnt1p switch platform based on direct replacement of the CEB with an aptamer sequence (Figure 1A). Ligand binding in the CEB can inhibit Rnt1p cleavage by inducing secondary or tertiary structural changes in the region that Rnt1p is unable to process effectively or by sequestering the CEB nucleotides. The Rnt1p switch is located in the 3′-UTR of a target transcript, where it acts to modulate the stability of that transcript (Figure 1B). Specifically, in the absence of ligand the hairpin is processed by Rnt1p, resulting in transcript destabilization and a decrease in gene expression. However, ligand binding at the integrated aptamer component will result in inhibition of Rnt1p processing of the hairpin structure, resulting in transcript stabilization and an increase in expression. Thus, the Rnt1p switch acts as a gene expression ‘ON’ switch, where gene expression is expected to increase with increasing ligand concentration. In addition, A-rich spacer sequences were placed around the device to more readily enable the extension of the system to the integration of multiple devices in the 3′-UTR (Figure 1A).
The theophylline aptamer ΔTCT-4 (29) was selected for the initial development of the Rnt1p switch platform due to this sequence having the highest reported affinity for the small molecule. However, the placement of this aptamer in the previously characterized Rnt1p substrate R31-27 (28), which is the same base hairpin that was used in the development of the Rnt1p cleavage library (20), resulted in the disruption of effective Rnt1p cleavage of the hairpin in the absence of ligand (data not shown). To lower the baseline of the ‘OFF’ state and thus provide a greater potential switching range, we utilized a different Rnt1p base hairpin in the switch design that had been previously reported to have the highest rate of Rnt1p processing in vitro (R31L-3B4Inv) (27). For all experiments, the highest external theophylline concentration applied to the yeast cells was 5 mM. At concentrations >10 mM, theophylline begins to become toxic to yeast and the reliability of the data decreases. All ligand titration data were fit to a simple binding model with Prism 5 (GraphPad) as:
where Y represents the normalized gene expression data for a given theophylline concentration (t), b represents the amount of gene expression in the absence of theophylline and M is the theoretical maximal ‘ON’ state determined by the model. The value of b was determined experimentally and not fit in the non-linear regression. The regulatory range of the switch is characterized by two measures, the fold-change, determined as the ratio of the ‘ON’ state to the ‘OFF’ state, and the dynamic range, determined as the difference between the ‘ON’ and the ‘OFF’ states, where the ‘OFF’ and ‘ON’ states are defined as the value of Y (t) at theophylline concentrations of 0 and 5 mM, respectively. The responsiveness of the switch was determined as the EC50 value of the dose-response curve (reported in terms of extracellular theophylline concentration). M gives a measure of the regulatory range of the switch in the case where the ligand is not cytotoxic at concentrations >5 mM. The theoretical fold-change and dynamic range based on the measured values of b and the model-calculated parameter M are reported for all switches in Supplementary Table S2.
We placed the theophylline aptamer in several locations within the CEB to screen for the most effective aptamer integration site (Supplementary Figure S2). One integration site location resulted in a functioning RNA switch (RS-theo1-B00), which retained the ability to be effectively cleaved by Rnt1p in the ‘OFF’ state and exhibited theophylline dependent increases in target gene expression levels (Figure 1C). In the switch naming system, the ‘theo1’ indicates the identity of the integrated aptamer (ΔTCT-4 aptamer) and ‘B00’ indicates the identity of the BSB module contained in R31L-3B4Inv, which is the same as the BSB module used in the development of the cleavage library (20). We refer to the parent Rnt1p switch RS-theo1-B00 as RS, and for other switches we will only indicate modules different than the parent switch for simplicity. The RS module switches from 47% to 91% as theophylline concentrations increase from 0 to 5 mM, resulting in a fold-change of 1.93 and a dynamic range of 44% (Table 1). The EC50 value for RS was determined to be 0.54 mM, where previous studies have indicated a substantial drop in theophylline across the cell membrane ranging from ∼300- to 1500-fold (18,30) (Liang, J.C., Michener, J.K. and Smolke, C.D., unpublished results, 2008). To verify that the reduced gene expression levels from RS were due to Rnt1p cleavage and that the theophylline-dependent response was due to the integrated aptamer sequence, we characterized two control hairpins: RSN, containing a mutant CAUC tetraloop that had been previously shown to impede Rnt1p activity in vivo (20), and RSnt, containing a single nucleotide mutation in the theophylline aptamer that obstructs the associated ligand-binding activity. The dose–response curve of RSN demonstrates gene expression levels at or around 100% for all theophylline concentrations, whereas the dose–response curve of RSnt demonstrates a consistent level of reduced gene expression independent of theophylline (Figure 1C).
Table 1.
Relevant parameters for all RS-based switches and the Rnt1p and ligand-binding controls, RSN and RSnt
| Switch | EC50 (mM) | b = Y (0 mM) % | Y (5 mM) % | Fold-change | Dynamic range % | M % |
|---|---|---|---|---|---|---|
| RS | 0.54 ± 0.05 | 47 ± 2 | 91 ± 3 | 1.93 ± 0.10 | 44 ± 4 | 97 ± 1 |
| RSN | 98 ± 3 | 101 ± 5 | 1.03 ± 0.06 | 3 ± 6 | ||
| RSnt | 49 ± 2 | 59 ± 2 | 1.20 ± 0.07 | 10 ± 3 | ||
| RS-theo2 | 1.47 ± 0.30 | 52 ± 2 | 84 ± 2 | 1.60 ± 0.06 | 32 ± 3 | 94 ± 3 |
| RS-theo3 | 1.17 ± 0.09 | 41 ± 4 | 101 ± 4 | 2.47 ± 0.26 | 60 ± 6 | 116 ± 2 |
| RS-B03 | 0.48 ± 0.08 | 79 ± 4 | 102 ± 4 | 1.29 ± 0.08 | 23 ± 5 | 105 ± 1 |
| RS-B05 | 0.56 ± 0.07 | 51 ± 1 | 97 ± 4 | 1.91 ± 0.10 | 46 ± 4 | 104 ± 2 |
| RS-B06 | 0.59 ± 0.11 | 56 ± 5 | 101 ± 6 | 1.82 ± 0.19 | 46 ± 7 | 108 ± 3 |
| RS-B07 | 0.84 ± 0.04 | 44 ± 2 | 100 ± 4 | 2.30 ± 0.13 | 57 ± 5 | 110 ± 1 |
| RS-B12 | 0.47 ± 0.08 | 44 ± 2 | 94 ± 4 | 2.11 ± 0.12 | 50 ± 4 | 99 ± 2 |
| RS×2 | 1.07 ± 0.02 | 20 ± 1 | 79 ± 3 | 3.91 ± 0.19 | 59 ± 3 | 92 ± 0 |
| RS×3 | 1.81 ± 0.06 | 10 ± 0 | 57 ± 1 | 5.57 ± 0.26 | 47 ± 1 | 74 ± 1 |
| RS-B07×2 | 1.81 ± 0.11 | 16 ± 1 | 69 ± 4 | 4.24 ± 0.36 | 53 ± 4 | 89 ± 2 |
| RS-B12×2 | 0.89 ± 0.07 | 22 ± 0 | 73 ± 1 | 3.30 ± 0.08 | 51 ± 1 | 81 ± 2 |
| RS-theo3-B07 | 1.13 ± 0.10 | 37 ± 2 | 91 ± 2 | 2.47 ± 0.14 | 54 ± 3 | 103 ± 2 |
| RS-theo3-B12 | 1.14 ± 0.13 | 36 ± 1 | 84 ± 1 | 2.31 ± 0.09 | 48 ± 2 | 95 ± 2 |
b and Y (5 mM) are experimentally determined values corresponding to the normalized protein levels at 0 mM and 5 mM theophylline, respectively. The EC50 and theoretical maximal output (M) are parameters determined by fitting the dose-response data to the binding model. The fold-change is the ratio of Y (5 mM) to b and the dynamic range is the difference between these two values. The theoretical fold-change and dynamic range determined by M instead of Y (5 mM) are reported in Supplementary Table S2.
Rnt1p processing is expected to destabilize the transcript and thus reduce transcript levels. We monitored relative steady-state transcript levels of RS and its controls, RSN and RSnt, in the presence and absence of ligand by qRT-PCR and confirmed that decreased transcript levels are associated with decreased protein expression (Figure 1D). In order to further verify the competition between ligand-binding and Rnt1p cleavage, we performed in vitro cleavage assays with purified Rnt1p and radiolabeled Rnt1p substrates in the presence and absence of theophylline (Figure 1E). We observed no Rnt1p activity on the mutated tetraloop control (RSN) and the inability of theophylline to impact Rnt1p activity with the inactive aptamer control (RSnt). In contrast, Rnt1p was able to cleave the switch (RS) in the absence of theophylline, but that activity was inhibited when theophylline was added. We also performed identical reactions without Rnt1p to examine any non-specific effects of theophylline on the Rnt1p substrates, which confirmed that theophylline has no effect on how the RNA runs in the gel (Figure 1E). Taken together, the results indicate that the knock down in expression levels and the theophylline response exhibited by RS are due to Rnt1p cleavage and ligand binding to the integrated aptamer, respectively.
Replacement of the aptamer sequence modulates ligand responsiveness and Rnt1p processing
The response curve of the Rnt1p switch can be modulated by three parameters associated with the genetic device: the strength of the ligand–aptamer interaction, the binding affinity of Rnt1p for the hairpin and the cleavage rate of Rnt1p on the hairpin. The ligand–aptamer interaction will primarily affect the EC50 of the dose–response curve, where aptamers with stronger affinities (lower KD values) will result in decreased EC50 values. However, modifications that result in stronger interactions between the hairpin and Rnt1p will result in increased EC50 values. Finally, modifications to the cleavage rate of Rnt1p on the hairpin will result in changes to the dynamic range or fold-change of the switch based on Michaelis–Menten enzyme kinetics, where increased Rnt1p activity corresponds to an increase in Vmax resulting in a lowering of the baseline in the absence of theophylline. The two regions of an Rnt1p hairpin that can be experimentally altered to achieve these affects are the CEB (or integrated aptamer sequence) and the Rnt1p binding regions (BSB and IBPB). We first examined the ability to modulate the EC50 value associated with the Rnt1p switch by incorporating aptamers with different ligand affinities. However, because the aptamer sequence is located within the CEB there may be unpredicted effects of integrating alternative aptamer sequences on the Rnt1p processing rates.
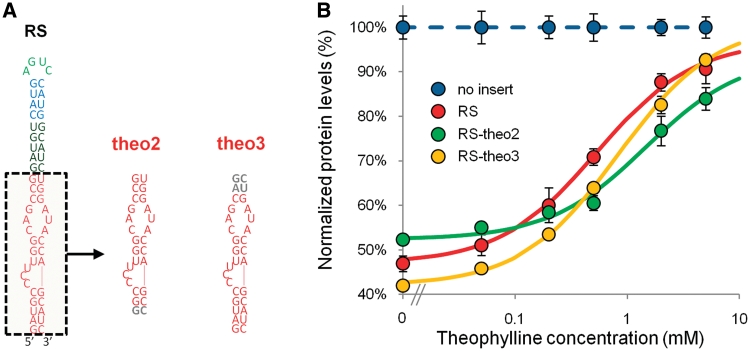
We replaced the aptamer component in RS with two alternative theophylline aptamer sequences: Δ-33 (RS-theo2) and a ΔTCT-4 variant (RS-theo3) (29) (Figure 2A). The Δ-33 aptamer (KD = 0.32 µM) exhibits a slightly decreased affinity relative to the ΔTCT-4 aptamer (KD = 0.29 µM) (29). However, the effect of the varied aptamer sequences on the switch-response curve was unknown, as aptamer integration required removal of terminal loops and different numbers of base-pairs at the base of the switch, which have been shown to affect the KD of the aptamer (29), and required nucleotide modifications of the hairpin stem, which have been shown to affect both Rnt1p binding and cleavage (27). The EC50 values of RS-theo2 (1.47 mM) and RS-theo3 (0.83 mM) were greater than that exhibited by RS (0.54 mM), and both switches exhibited changes in the baseline expression levels of the dose-response curves (Figure 2B and Table 1). The increased EC50 values for both switches indicate that the aptamer modifications decreased ligand affinity. In terms of Rnt1p cleavage, RS-theo2 exhibited decreased Rnt1p processing and RS-theo3 exhibited increased processing as determined from the baseline levels in the absence of ligand (RS: 47%; RS-theo2: 52%; RS-theo3: 42%). Therefore, RS-theo3 exhibited a larger dynamic range (DR) and fold-change (fold) than RS (RS-theo3: 51% DR, 2.21 fold; RS: 44% DR, 1.93 fold), although its EC50 value was ∼50% greater than that of RS, whereas RS-theo2 exhibited a decreased dynamic range and fold-change (RS-theo2: 32% DR, 1.60 fold). These results indicate that the sequence and structure of aptamer components, in addition to their binding properties, can impact the regulatory range exhibited by Rnt1p switches.
Figure 2.
Tuning the response curve of the Rnt1p switch through the integration of different theophylline aptamers. (A) Switch and aptamer module sequences illustrating the replacement of the aptamer sequence in RS with different theophylline aptamers: theo2 and theo3. Gray lettering is used to indicate the nucleotides in theo2 and theo3 that differ from theo1. (B) The dose–response curves of RS, RS-theo2 and RS-theo3 indicate a shift in EC50 values and variations in the baseline levels. Data are reported as indicated in Figure 1C. The model parameters for the curve fit are provided in Table 1.
Incorporation of synthetic BSBs modulates ligand responsiveness and processing of the Rnt1p switch
Modifications to the binding region of the Rnt1p hairpin can also be used to tune the switch response. A set of synthetic BSB modules was previously described (A.H. Babiskin and C.D. Smolke, submitted for publication), which can be used to modulate Rnt1p binding affinities. However, the synthetic BSB modules were shown to affect Rnt1p binding and processing (A.H. Babiskin and C.D. Smolke, submitted for publication), such that changes in both the EC50 value and baseline expression level of the dose-response curve are expected from their integration into the Rnt1p switch.
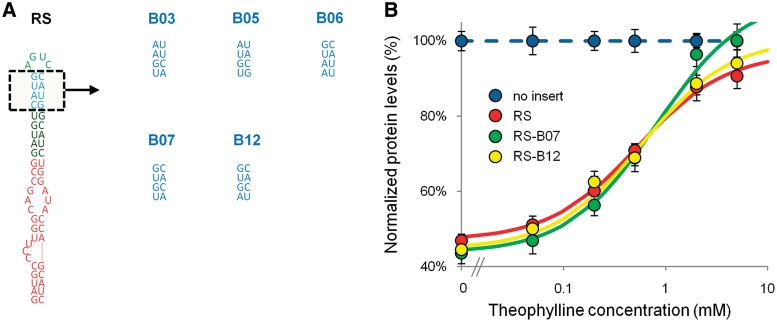
We incorporated several synthetic BSB modules (B03, B05, B06, B07, B12) into the BSB of RS (Figure 3A, Supplementary Table S3). We observed a shift in the baseline expression levels for the dose-response curves of all BSB-modified Rnt1p switches, where expression levels at 5 mM theophylline were restored to ∼90–100% normalized protein levels (Figure 3B, Table 1, Supplementary Figure S3). The primary effect of the incorporated BSBs was observed with shifts in the baseline levels, which were likely a result of altered processing rates by Rnt1p. Two of the switches, RS-B07 and RS-B12, exhibited decreases in the baseline expression levels (44%) relative to RS (47%), thereby resulting in an increased dynamic range and fold-change (RS-B07: 57% DR, 2.30 fold; RS-B12: 50% DR, 2.11 fold; RS: 44% DR, 1.93 fold). The remainder of the switches exhibited increases in the baseline levels, resulting in slightly reduced or similar dynamic range and fold-change (Table 1). The majority of the modified BSB Rnt1p switches exhibited slight shifts in the EC50 values (from 0.48 to 0.59 mM). However, RS-B07 exhibited a substantially higher EC50 value of 0.81 mM. The improvement in switch activity for RS-B07 and RS-B12 as determined by the fold-change was due to the increased processing by Rnt1p. However, RS-B12 exhibited greater sensitivity to lower concentrations of theophylline relative to RS-B07 as determined by the EC50 values for these switches (RS-B12: 0.58 mM; RS-B07: 0.81 mM). These results indicate that the set of synthetic BSB modules can be employed to tune the baseline expression exhibited by Rnt1p switches while maintaining restoration of the ‘ON’ state.
Figure 3.
Tuning the response curve of the Rnt1p switch through the integration of different synthetic BSB modules. (A) Switch and BSB module sequences illustrating the replacement of the BSB sequence with synthetic BSB modules (B03, B05, B06, B07 and B12). The gene-regulatory activities of the synthetic BSB modules as previously determined in the context of an Rnt1p hairpin control element are provided in Supplementary Figure S3. (B) The dose–response curves of RS, RS-B07 and RS-B12 indicate that the two synthetic BSB modules decrease baseline levels and exhibit minor effects on the EC50 of the response curve. The dose–response curves of the other BSB-incorporated switches are presented in Supplementary Figure S3. Data are reported as indicated in Figure 1C. The model parameters for the curve fit are provided in Table 1.
The application of multiple switch modules decreases theophylline responsiveness and increases fold-change
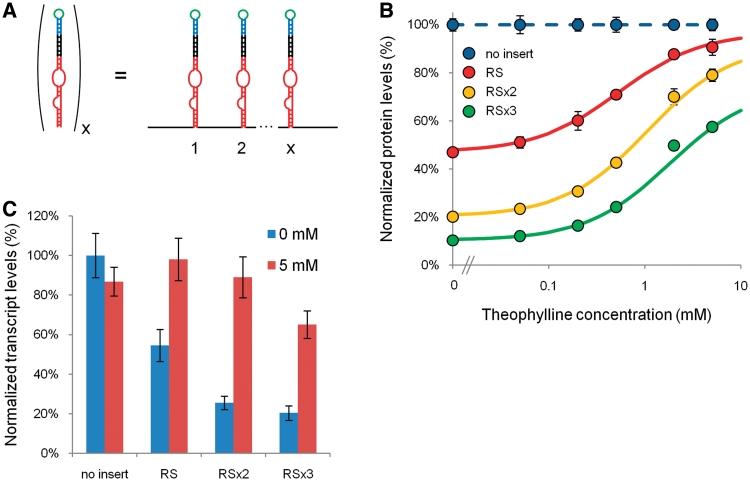
We examined a third tuning strategy for the Rnt1p switch regulatory response based on integrating multiple copies of a switch in the 3′-UTR of a target transcript. The integration of multiple copies of cis-acting RNA switches in the 3′-UTR of target transcripts has previously been demonstrated to modulate the regulatory response by decreasing baseline expression levels (11,31). However, it is also expected that this tuning strategy will result in decreased sensitivity to the ligand or an increased EC50 value for a given regulatory system. Based on a mechanism of competition inhibition, we anticipated that the RS switch design would demonstrate close to full restoration of gene activity under maximal ligand concentrations, resulting in significant increases in the fold-change and dynamic range of the system.
We built two-copy (RS×2) and three-copy (RS×3) RS switch constructs (Figure 4A). We observed an increase in the EC50 value and a decrease in baseline expression levels with increasing switch copy number (Figure 4B and Table 1). The effect of multiple Rnt1p switches on the EC50 value of the system was determined to be nearly additive. However, the effect of multiple switches on the baseline level (b) was determined to be multiplicative:
Where x is the number of modules. At the highest concentration of theophylline added to the system (5 mM), the gene expression levels decreased with increasing switch number. We expected the ON state expression levels to be multiplicative with the theoretical maximal state (M) (Supplemental Table S2); while this relationship was observed for RS×2, it was not observed for RS×3. With three copies of RS, the full switch dynamic range cannot be observed when limited to 5 mM theophylline, as evidenced by the lack of saturation of the dose-response curve. The fold-change of the system increased with increasing switch number from 1.93 (RS) to 3.91 (RS×2) to 5.57 (RS×3). We verified that the observed trends in protein expression were consistent with transcript levels, supporting increased Rnt1p processing of the multiple switch modules (Figure 4C). Based on the observed trends, we expect that we would likely observe a decrease in regulatory activity when over three copies of the Rnt1p switch are implemented, as further decreases in baseline levels would be limited and the EC50 value would be well above 2 mM theophylline, thus substantially limiting the ON state expression levels that could be accessed. These results indicate that the integration of multiple switch copies can be used to predictably tune the dynamic range and EC50 value exhibited by a given Rnt1p switch.
Figure 4.
Predictive tuning of the Rnt1p switch-response curve through the integration of multiple copies of the switch module. (A) Schematic representing the integration of multiple switch modules. Each Rnt1p switch is insulated with spacer sequences as indicated in Figure 1A. Two and three copies of the original RS switch were examined for their effects on the regulatory response. (B) The dose–response curves of RS, RS×2 and RS×3 indicate increasing dependence on theophylline concentration and decreasing baseline levels with each added switch module. Data are reported as indicated in Figure 1C. The model parameters for the curve fit are provided in Table 1. (C) The transcript levels of RS, RS×2 and RS×3 support increased Rnt1p processing of the multiple switch modules. Data are reported as indicated in Figure 1D.
Combined tuning strategies support the rational design of Rnt1p switch control systems with enhanced regulatory properties
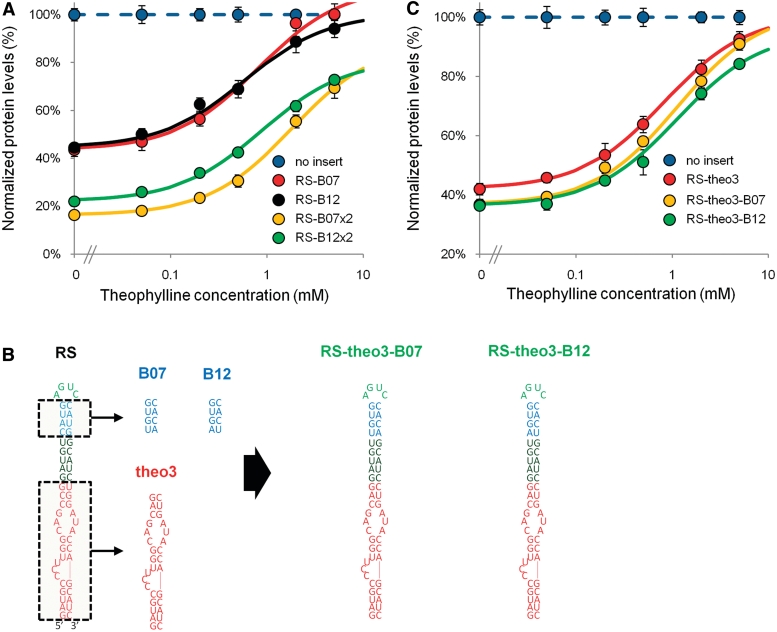
Our studies identified three different strategies for tuning the regulatory response of an Rnt1p switch based on altering the aptamer/CEB, BSB and number of switch modules. We propose that these tuning modules can be combined to rationally design Rnt1p switches with enhanced regulatory properties. We first examined the combined implementation of synthetic BSB modules with multiple copies of the modified Rnt1p switches. We built genetic control systems that contained two copies of Rnt1p switches incorporating the BSB modules that resulted in the largest single-copy switch dynamic ranges (RS-B07, RS-B12). Both two-copy switch systems (RS-B07×2, RS-B12×2) exhibited decreased baseline expression levels that followed the previously described multiplicative trends (Figure 5A and Table 1). Although both switches exhibited the expected increased dependence on theophylline, the EC50 values were not additive as had been observed with RS. Limitations in theophylline concentration had a more substantial impact on the experimentally attainable dynamic range of RS-B07×2 compared with that of RS×2, as RS-B07×2 exhibits an EC50 value that is nearly twice that of RS×2. Even with this limitation, RS-B07×2 exhibits a larger fold-change than that of RS×2. However, the theoretical fold-change [based on the theoretical maximal output of the switch (M) from the model fit] of RS-B07×2 increases from 4.24 to 5.42, whereas that of RS×2 only increases to 4.54. RS-B12 experienced the lowest level of gene expression in its ON state (at 5 mM theophylline and M) when compared with the other switches incorporating synthetic BSB modules (Table 1, Supplementary Table S2). Therefore, due to the multiplicative effect associated with multiple switch modules, we expected the ON state for RS-B12×2 to be diminished greater than the other two module switches. Although the baseline was reduced multiplicatively for RS-B12×2 and the theoretical fold-change enhanced, the decrease to the ON state resulted in smaller improvements in the fold-change relative to RS×2 and RS-B07×2 from their respective single module switch.
Figure 5.
Combinatorial implementation of multiple tuning modules results in predictive tuning of the Rnt1p switch regulatory response curve. (A) The dose–response curves of switches that incorporate synthetic BSB and multiple switch modules (RS-B07, RS-B12, RS-B07×2 and RS-B12×2) indicate increased dependence on theophylline concentration and decreased baseline levels based on the combined activity of the two incorporated tuning modules. The model parameters for the curve fit are provided in Table 1. (B) Optimization of single-copy switch designs by incorporating synthetic BSB modules (B07 and B12) with the aptamer/CEB module (theo3) that exhibited the most improved switch-response curves. (C) The dose–response curves of RS3-theo3, RS-theo3-B07 and RS-theo3-B12 indicate that integration of optimized BSB and aptamer/CEB modules results in Rnt1p switches exhibiting improved dynamic range and fold-change over that observed with the individual modules. Data are reported as indicated in Figure 1C. The model parameters for the curve fit are provided in Table 1.
We next examined the combined implementation of synthetic BSB modules with alternative aptamer modules to improve Rnt1p switch regulatory range. We identified two synthetic BSB modules (B07, B12) and one alternative aptamer module (theo3) that resulted in increases in dynamic range and fold-change when integrated individually into the Rnt1p switch platform. Therefore, we designed two new Rnt1p switches that combined these modules: RS-theo3-B07 and RS-theo3-B12 (Figure 5B). The combined module switches exhibited changes in EC50 values and dynamic ranges that were improved from the effects of the individual tuning modules (Figure 5C and Table 1). Specifically, RS-theo3-B07 and RS-theo3-B12 exhibited increased EC50 values (1.13 and 1.14 mM, respectively) from those exhibited by RS-theo3 (0.83 mM), RS-B07 (0.81 mM) and RS-B12 (0.58 mM). In addition, both RS-theo3-B07 and RS-theo3-B12 exhibit baseline levels (37% and 36%, respectively) that are lower than the switches harboring the individual tuning modules (RS-theo3: 42%; RS-B07 and RS-B12: 44%) (Figure 5C). The rationally tuned Rnt1p switches, RS-theo3-B07 and RS-theo3-B12, exhibit the greatest fold-change of the single-copy switches generated in this study (RS-theo3-B07: 2.47; RS-theo3-B12: 2.31). The results demonstrate that the rational combination of independent tuning modules can be used to improve switch regulatory ranges in a manner that is predictable from the individual module effects.
DISCUSSION
We have developed a new class of synthetic RNA devices based on ligand-responsive modulation of RNase III cleavage activity in S. cerevisiae. Our design strategy incorporated a sensor component, encoded within an aptamer sequence, directly within a region of Rnt1p substrates known to effect enzyme cleavage—the CEB. This direct integration design strategy differs from commonly implemented design strategies for synthetic riboswitches that couple the aptamer component to the regulatory component through a linker sequence (5,32,33). As such the Rnt1p switch does not undergo substantial secondary structure rearrangements between gene-regulatory active and inactive states as is observed with many RNA switch platforms, but instead functions through inhibiting Rnt1p processing as a result of the slight structural changes or sequestering of CEB nucleotides associated with ligand binding at the aptamer/CEB domain. A similar direct integration design strategy was recently described for a synthetic RNA switch platform based on modulating Drosha (an RNase III enzyme in the RNAi pathway) processing of its substrates by integration of aptamer sequences into the basal segment region of pri-miRNA hairpins (19). In addition, a direct coupling strategy was previously described for the design of an RNA switch that modulated Dicer (another RNase III enzyme in the RNAi pathway) processing of shRNA substrates by coupling aptamer sequences to the terminal loop of the shRNA hairpin (17). These examples indicate that the direct integration of sensor and actuator components may present an effective design strategy for riboswitches based on the modulation of RNase III processing activities.
We had previously demonstrated that the CEB region has no rigid structural or sequence requirements for efficient Rnt1p cleavage (20). Therefore, the described Rnt1p switch platform is likely amenable to diverse aptamer sequences, supporting the ability to tailor Rnt1p switches to diverse molecular effectors. However, our earlier work also demonstrated that different nucleotide sequences in the CEB resulted in altered processing rates by Rnt1p in vivo (20). Therefore, it is also expected that changing the aptamer sequence within the CEB will result in unpredictable changes to the baseline levels exhibited by the switch due to effects of the aptamer sequence on Rnt1p cleavage activity. This was observed when integrating different theophylline aptamer sequences into the Rnt1p switch platform (Figure 2B and Table 1). Ultimately, the activity of an Rnt1p switch will be determined by two factors: (i) the ability of the aptamer sequence and structure to be effectively processed by Rnt1p; and (ii) the ability of the ligand to compete with Rnt1p binding and cleavage or decrease Rnt1p’s ability to cleave the ligand-bound CEB. For example, in our initial studies we examined different placements of the theophylline aptamer in the Rnt1p hairpin stem and observed several designs that were processed efficiently by Rnt1p, but where Rnt1p activity was not inhibited under ligand addition (data not shown). It is likely that the sequences and structures of certain aptamers will inhibit Rnt1p cleavage activity when placed in the CEB. Therefore, the successful design of Rnt1p switches responsive to new effector molecules will likely require screening of potential aptamer sequences within the switch platform for those sequences and structures that can be effectively cleaved by Rnt1p. In addition, newer in vivo screening methods will likely be effective in identifying the best aptamer candidates from in vitro enriched aptamer pools (34).
Optimization of the Rnt1p switch regulatory response was significantly aided by earlier foundational work conducted in our laboratory on developing synthetic CEB and BSB modules that can be implemented individually or in combination to rationally build tailored Rnt1p control elements (20; A.H. Babiskin and C.D. Smolke, submitted for publication). We demonstrated that synthetic BSB modules from a set of 16 previously described elements (A.H. Babiskin, and C.D. Smolke, submitted for publication) can be integrated into an Rnt1p switch to tune switch dynamics by modulating the baseline expression. Using this approach we identified two synthetic BSB modules (B07 and B12) that increase the dynamic range and fold-change of the switch response. However, it was also observed that the rank-order activities of the BSB modules as previously characterized in the Rnt1p hairpin substrate were different than that observed in the Rnt1p switch hairpin (RS) as determined by the baseline level in the absence of theophylline. This difference is likely due to the fact that the intervening stem between the CEB and BSB modules are different in the original Rnt1p hairpin substrate and the hairpin used for the Rnt1p switch. In addition, the aptamer sequence integrated into the CEB region also extends beyond the CEB module, introducing additional changes to that stem region. Therefore, the data indicate that the relative activities of the BSB elements may be affected by the stem sequence between the CEB and BSB modules.
We also demonstrated that the different tuning modules can be implemented combinatorially to predictably tailor the switch-response curve based on the individual activities of each module. For example, by integrating the BSB modules (B07 and B12) with the aptamer/CEB module (theo3) that most improved the switch-response curve into a single Rnt1p switch, we were able to build single-copy switches exhibiting a greater dynamic range, fold-change and lowered baseline expression level than any of the switches harboring one of these modules (Figure 5C and Table 1). However, as one drawback of this tuning strategy, modules that improved the switching activity of the response curve generally also resulted in increased EC50 values. This trade-off between switching activity and sensitivity highlights the requirement for developing higher affinity aptamers or aptamers with appropriate affinities for the intended application (35).
A third tuning strategy was examined to improve switching activity based on the implementation of multiple copies of Rnt1p switches in a single target transcript. Two-copy switch systems were examined for the original Rnt1p switch (RS) and two BSB-tuned switches that exhibited improved regulatory responses (RS-B07 and RS-B17). In general, the implementation of a second copy of a given switch resulted in decreased baseline levels, increased fold-change, and increased EC50 values (Figure 4B, Figure 5A and Table 1), where the observed effects on baseline levels were multiplicative. However, the observed dynamic range (difference between the ON and the OFF states) did not increase with increasing switch number, due to the response curves not saturating at 5 mM theophylline. In addition, the maximal ON state (M), determined by extrapolating the data with the binding model curve fit, indicated that the theoretical dynamic range only slightly increased for RS-B07×2 and RS-B17×2 (Supplementary Table S2). This limited improvement in theoretical dynamic range was due to substantial decreases in M with increasing switch number, potentially due to non-specific effects resulting from the additional structures such as interference with translation or transcript destabilization or due to increased residual cleavage by Rnt1p of the substrate in the ligand-bound form. We also built a three-copy version of RS (RS×3), which exhibited additional increases in the fold-change over the two-copy system. However, the implementation of three Rnt1p switch modules exhibited limiting returns in the decreased baseline levels and increased the EC50 of the response curve to a level approaching that which would not allow proper observation of switch activity due to limitations in theophylline concentrations that can be applied to the yeast culture.
This work extends the utility of Rnt1p control elements for cellular engineering applications by developing an integrated RNA platform for building ligand-responsive Rnt1p-based control devices in yeast. By taking a synthetic biology approach to the development of a tool for designing precise genetic control elements based on building Rnt1p controllers from synthetic modules (20; A.H. Babiskin and C.D. Smolke, submitted for publication), we were able to identify readily implementable tuning and optimization strategies for the more complex Rnt1p-based regulatory device. Specifically, we demonstrated that three tuning strategies, based on the implementation of synthetic aptamer/CEB, BSB and switch modules independently or in combination, can be used to optimize the regulatory response of the engineered control system. Importantly, the effects of the tuning modules when implemented combinatorially were predictable from their individual activities. An interesting aspect of this engineered riboswitch platform is that our single-copy switches generally exhibit fully restored gene expression activity at maximum ligand concentration compared to constructs lacking a switch, such that the observed differences between switch activities are largely due to changes in baseline levels.
An advantage of such RNA devices over traditional genetic control elements, such as those based on promoter systems, is the ability to tune regulatory activities to match application-specific requirements and to tailor the input processing and output gene-regulatory functions of these devices. In addition, because they act post-transcriptionally, RNA devices can be coupled with any promoter of interest, allowing researchers to build layered control schemes and thus more sophisticated genetic networks. While the Rnt1p switches exhibit relatively moderate gene-regulatory activities (in terms of fold-change), recent research has demonstrated the application of other engineered RNA controllers exhibiting similar activities to the regulation of diverse cellular phenotypes (11,12,34,36). Therefore, the integration of compatible aptamers into this switch platform that respond to broad classes of molecules of interest, including non-toxic exogenous chemicals, primary metabolites and chemicals of industrial interest, will advance the engineering of genetic circuits in yeast for diverse biotechnological applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Foundation (CAREER award CBET-0917705 to C.D.S.); Alfred P. Sloan Foundation, fellowship (to C.D.S.). Funding for open access charge: National Science Foundation CBET-0917705.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Hoff for assistance in the expression and purification of Rnt1p; and S. Bastian and F.H. Arnold for assistance in sonication and FPLC.
REFERENCES
- 1.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:2006.0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl Acad. Sci. USA. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 4.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 5.Win MN, Liang JC, Smolke CD. Frameworks for programming biological function through RNA parts and devices. Chem. Biol. 2009;16:298–310. doi: 10.1016/j.chembiol.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grate D, Wilson C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg. Med. Chem. 2001;9:2565–2570. doi: 10.1016/s0968-0896(01)00031-1. [DOI] [PubMed] [Google Scholar]
- 7.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 8.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 9.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma V, Nomura Y, Yokobayashi Y. Engineering complex riboswitch regulation by dual genetic selection. J. Am. Chem. Soc. 2008;130:16310–16315. doi: 10.1021/ja805203w. [DOI] [PubMed] [Google Scholar]
- 11.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl Acad. Sci. USA. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Thompson KM, Syrett HA, Knudsen SM, Ellington AD. Group I aptazymes as genetic regulatory switches. BMC Biotechnol. 2002;2:21. doi: 10.1186/1472-6750-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007;35:4179–4185. doi: 10.1093/nar/gkm425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DS, Gusti V, Dery KJ, Gaur RK. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol. Biol. 2008;9:23. doi: 10.1186/1471-2199-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An CI, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol. Syst. Biol. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beisel CL, Chen YY, Culler SJ, Hoff KG, Smolke CD. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq954. doi:10.1093/nar/gkq954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babiskin AH, Smolke CD. A synthetic library of RNA control modules for predictable tuning of gene expression in yeast. Mol. Syst. Biol. 2011 doi: 10.1038/msb.2011.4. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 22.Gietz R, Woods R. In: Guide to Yeast Genetics and Molecular and Cell Biology, Part B. Guthrie C, Fink G, editors. Vol. 350. San Diego, CA: Academic; 2002. pp. 87–96. [Google Scholar]
- 23.Caponigro G, Muhlrad D, Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng R, Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1980;77:3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamontagne B, Elela SA. Purification and characterization of Saccharomyces cerevisiae Rnt1p nuclease. Method. Enzymol. 2001;342:159–167. doi: 10.1016/s0076-6879(01)42543-2. [DOI] [PubMed] [Google Scholar]
- 26.Lamontagne B, Elela SA. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 2004;279:2231–2241. doi: 10.1074/jbc.M309324200. [DOI] [PubMed] [Google Scholar]
- 27.Lamontagne B, Ghazal G, Lebars I, Yoshizawa S, Fourmy D, Elela SA. Sequence dependence of substrate recognition and cleavage by yeast RNase III. J. Mol. Biol. 2003;327:985–1000. doi: 10.1016/s0022-2836(03)00231-6. [DOI] [PubMed] [Google Scholar]
- 28.Lamontagne B, Hannoush RN, Damha MJ, Abou Elela S. Molecular requirements for duplex recognition and cleavage by eukaryotic RNase III: discovery of an RNA-dependent DNA cleavage activity of yeast Rnt1p. J. Mol. Biol. 2004;338:401–418. doi: 10.1016/j.jmb.2004.02.059. [DOI] [PubMed] [Google Scholar]
- 29.Zimmermann GR, Wick CL, Shields TP, Jenison RD, Pardi A. Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA. 2000;6:659–667. doi: 10.1017/s1355838200000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch AL. The metabolism of methylpurines by Escherichia coli. I. Tracer studies. J. Biol. Chem. 1956;219:181–188. [PubMed] [Google Scholar]
- 31.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 33.Suess B, Weigand JE. Engineered riboswitches: overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 34.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Beisel CL, Smolke CD. Design principles for riboswitch function. PLoS Comput. Biol. 2009;5:e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.