Abstract
DNA is not the static entity that structural pictures suggest. It has been longly known that it ‘breathes’ and fluctuates by local opening of the bases. Here we show that the effect of structural fluctuations, exhibited by AT-rich low stability regions present in some common transcription initiation regions, influences the properties of DNA in a distant range of at least 10 bp. This observation is confirmed by experiments on genuine gene promoter regions of DNA. The spatial correlations revealed by these experiments throw a new light on the physics of DNA and could have biological implications, for instance by contributing to the cooperative effects needed to assemble the molecular machinery that forms the transcription complex.
INTRODUCTION
The dynamical opening of intermittent flexible single stranded domains along the double stranded DNA molecule is an intriguing phenomenon involved in many biological processes: as shown by key structural studies (1,2), unwinding the double helix and local specific bubble formation are prerequisites for both DNA replication and gene transcription.
Moreover, many DNA–molecule interactions are also affected by the dynamics of the base pairs. For instance, NMR studies reveal that antitumor DNA binder drugs like nogalamycin require a dynamical transient opening of the base pairs to allow their docking and binding (3). The kinetics of base-pair opening dynamics has been well described by following the exchange of protons from imino groups with water (4), showing that particular regions of the helix open by the action of thermal fluctuations, and suggesting the importance of sequence effects in the opening of particular tracks (5). Experimental observations prove that DNA ‘breathing’ is particularly strong in AT-rich regions that exhibit premelting phenomena starting at physiological temperatures (6–9).
While the importance of DNA fluctuational openings is well recognized, it is generally assumed that its effects are local, i.e. that they concern only the base pairs involved in the opening or next to it. Our results definitely change this view, showing that the stability and structural evolution of regions situated at some distance along a DNA molecule are also affected.
As shown schematically in Figure 1, in this work we answer the following question: ‘does an AT-rich domain, prone to opening, influence the properties of DNA in neighboring regions, and, if so, how far along the helix?’ The study has been conducted both on sequences specially designed to allow a quantitative answer, and in natural gene promoter fragments to show the potential relevance in biology.
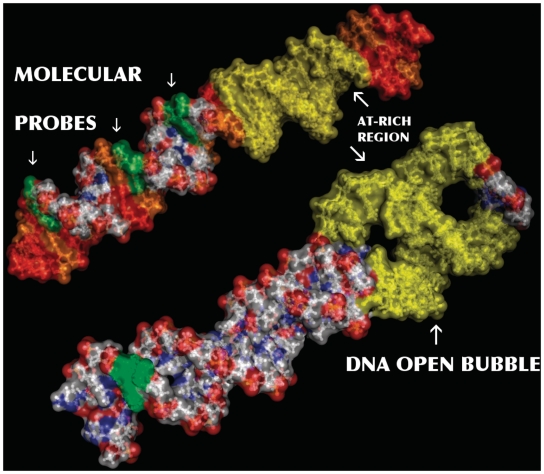
Figure 1.
Imaginary molecular view (PyMol Molecular Viewer; http://pymol.org) of the DNA molecule (S1) illustrating the primary goal of our experiment. The analytical chemical method, here reported, treats all the Guanines present in a DNA molecule as effective molecular probes (phosphorous green surface shaded). Therefore, any studied DNA fragment is populated by ad-hoc check points able to detect variations on the stacking helicoidal conformation. In this work we use such information to evaluate the influence of an intermediate structural conformation, a thermal induced bubble (yellow surface shaded), over distant regions along the molecule.
MATERIALS AND METHODS
Principle of the UV laser biphotonic photolysis method to detect local structural stability of DNA
This original method takes advantage of the specific oxidation chemistry of the guanine bases to turn each guanine site of a DNA sequence into a local probe of the helicoidal stacking in its vicinity. The method proceeds in two main steps: a DNA solution at the temperature of interest is first irradiated by a single high intensity UV laser pulse; then the sample is analyzed with standard biochemical methods to determine the outcome of the irradiation at each guanine site.
Guanines are the primary target for the one-electron oxidation of DNA, which is achieved by a high intensity 266 nm UV laser pulse, through a bi-photonic absorption process (10,11). This generates nucleobases radical cations, which evolve by hole transport processes dependent on the helicoidal stacking. Holes are trapped by guanines possessing the lowest oxidative potential (10,12–14). As shown in the Supplementary Figure S1, there are two dominant pathways of G+ transformation, leading to 8-oxo-7,8-dihydro-2-oxoguanine (8-oxodG) or oxazolone.
Moreover, the irradiation may also lead to inter-strand cross-linking of the GC pair, which provides an independent assessment of the local fluctuational opening of DNA that is discussed in the next section.
While oxazolone is the unique product resulting from one-electron oxidation of the free 2′-deoxyguanosine, 8-oxodG appears as soon as the nucleoside is incorporated in a helical structure. Hence, the measurement of the relative yield of these photoproducts provides an information on the local conformation and fluctuations of DNA. It is based on the cleavage of the DNA molecule by either formamidopyrimidine DNA glycosylase (Fpg protein) that acts preferentially at 8-oxodG sites or piperidine that acts preferentially at oxazolone sites.
It is noteworthy that the relative yield ratio RFpg/Rpip is high in stable regions of DNA and decreases in regions where the helical structure is destabilized. As a result, the variation of RFpg/Rpip at each guanine site as a function of temperature has been found to follow the decay of the degree of helicoidal status and pairing probability of the bases as the melting of the duplex proceeds (15), but, instead of a global melting curve of a given sequence, it provides a measure of the local stability of the GC pairs. The value of the ratio RFpg/Rpip given by one particular guanine depends on the sequence of the neighboring sites (14). We paid a particular attention to this point in the selection of the sequences. Probes that carry the same label, such as G1, G2, correspond to guanines surrounded by the same adjacent base pairs. However, due to the sensitivity of the radical cation transformation pathway to the local sequence, comparisons of this ratio at different sites can only be used for a qualitative comparison between these sites. On the contrary, variations at a particular site, caused for instance by a temperature change, are quantitatively significant, with an accuracy that can be assessed by the dispersion of the data when the experience is reproduced several times in the same conditions.
Finally note that both temperature and sequence effects are not negligible in the electronic transfer mechanism underlying DNA oxidation (16). However, this does not affect the value of the chemical reactivity ratio RFpg/Rpip, as we are in single-hit ionization events per DNA fragment conditions and the radical cation transformation pathway is independent on how it is generated (14). Moreover, we are comparing the ionization in particular guanine probes surrounded by exactly the same bases. By probe we do not designate simply a guanine but also its environment. Our specially designed sequences conserve the following motif around the detection probe atacGat (sequences S1 to S3). And we managed to have found a consensus probe acGat in the real/biological sequences studied.
DNA–DNA crosslinking
The chemical basis of this method allows an independent cross-check. In addition to the two oxidative lesions that we detect, ionized Guanines also give rise to inter-strands adducts (cross links) with the complementary cytosine, which are extremely sensitive to the GC mutual position [A similar variation of this method has already been exploited in studying the TRF2-assisted strand invasion with telomeric DNA sequences in (17)]. Measuring the yield of cross linking at individual guanine sites versus temperature provides a complementary measurement of the local closing probability. We have exploited this technique to confirm the main results reported here (see more technical notes in the Supplementary Data).
Experimental details
Oligonucleotides
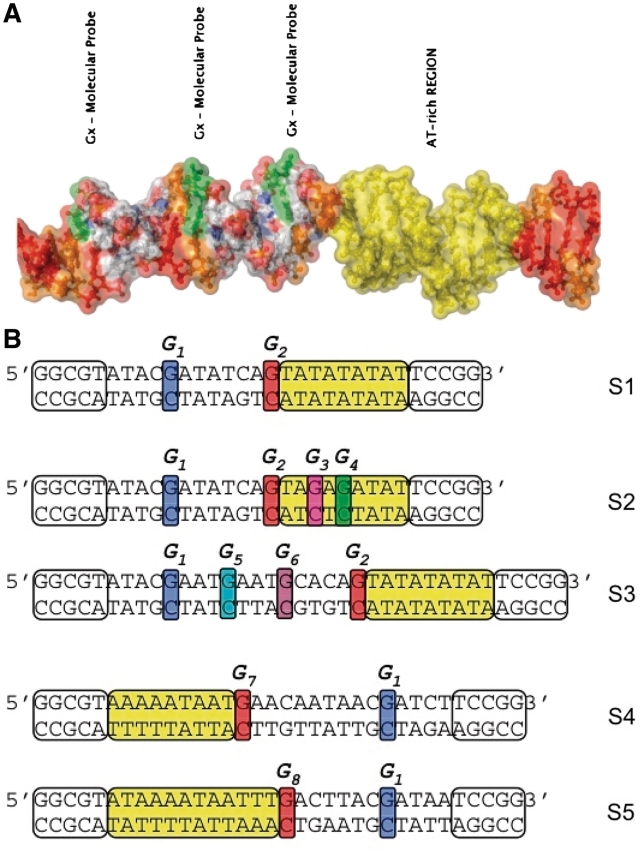
HPLC purified commercial oligonucleotides were purchased from Eurogentec. The sequences have been specifically designed (S1 to S3) or taken from promoter fragments in Escherichia coli and Yersinia pestis (sequences S4, S5) and then completed by GC-rich terminal regions (white boxes in Figure 2) to stabilize the double helix. Oligonucleotides (10 pmol) (Figure 2, upper strands) were 5′-labeled by [γ−32P]ATP in the presence of polynucleotide kinase. The labeled oligonucleotide was then annealed with an excess (2×) of its complementary strand, treated by Fpg to cleave background oxidative lesions, and gel purified using denaturing polyacrylamide (15% acrylamide and 8M urea) gel electrophoresis (PAGE). Gel-purified oligonucleotides were annealed by heating to 80°C and slow cooling, and checked by 15% native PAGE.
Figure 2.
Sequences of the DNA fragments investigated in this study. All the sequences contain a low-stability AT-rich region (yellow highlighted) able to nucleate temporal intermittent partial opening (DNA bubble). Guanines along the 5′-3′ strand are used as molecular probes. S1, S2, S3 are artificial sequences containing a large TATA box motif (modified in sequence S2) and S4 and S5 are natural sequences from DNA promoter regions of genes in E. coli 536 (bases −28 to −5 in gene ECP-0995) and Y. pestis CO92 (bases −42 to −20 in gene YPO2592, respectively. All fragments have been completed by GC-rich terminal domains (marked as white boxes) to stabilize these short DNA helices.
UV irradiation
Irradiation was performed in siliconized 0.5 ml eppendorf tubes by exposing DNA samples to a single UV-laser pulse (λ = 266 nm τ = 4−5 ns, energy 0.1 J/cm2) provided by the fourth harmonic generation of a Q-switched Nd:YAG laser, as described in (15,20). This was carried out in 10 μl aliquots (c = 1 nM) in TE, 25 mM NaCl buffer. The temperature was adjusted by storing the tubes in a PCR machine, programmed for stepwise temperature raising. After irradiation the samples were run on a sequencing gel that allows us to collect data for crosslinking, purified and analyzed by sequencing gel electrophoresis after Fpg or piperidine treatments.
Sequencing gel electrophoresis analysis
After irradiation the DNA samples were run on sequencing gel electrophoresis. The wet gel was exposed for phosphorimagery and the digital image was used for quantification of the cross-linked DNA. Note that on denaturing gels the cross-linked DNA strands migrate slower than the single stranded oligonucleotide, with a migration speed that depends on the position of the cross-linked GC base pair. This, and the partial piperidine lability of the inter-strand adducts, were used for the assignment of the slow-migrating bands. The same gel was also used for purification of the irradiated full-length oligonucleotides before cleavage. This purification step was necessary to exclude the partially piperidine-labile inter-strand adducts and the background level of the laser-induced direct single-strand breaks from the RFpg/Rpip measurement.
Irradiated gel-separated full-length oligonucleotides were eluted, passed through Sephadex G50 column, ethanol precipitated, resuspended in 20 μl TE with 30 mM NaCl buffer, 1 mM DDT and 100 μg/ml BSA, annealed and split in two 10 μl aliquots. One aliquot was incubated at 37°C with 4 ng of Fpg and the other with 1M piperidine at 90°C for 30 mn. The samples were lyophilized, resuspended in a formamide loading buffer and run on 15% sequencing acrylamide-8M urea gel. The dried gels were exposed to a phosphorimager screen and the images (as shown in Figure 3) were read out and quantified by a Fuji phosphorimager to determine the ratio RFpg/Rpip for each band, i.e. at each cleavage site. The quantification of RFpg/Rpip at each guanine site is done by the measurement of the cleaved fragments of DNA molecules radioactively labeled at one end with standard biochemical methods.
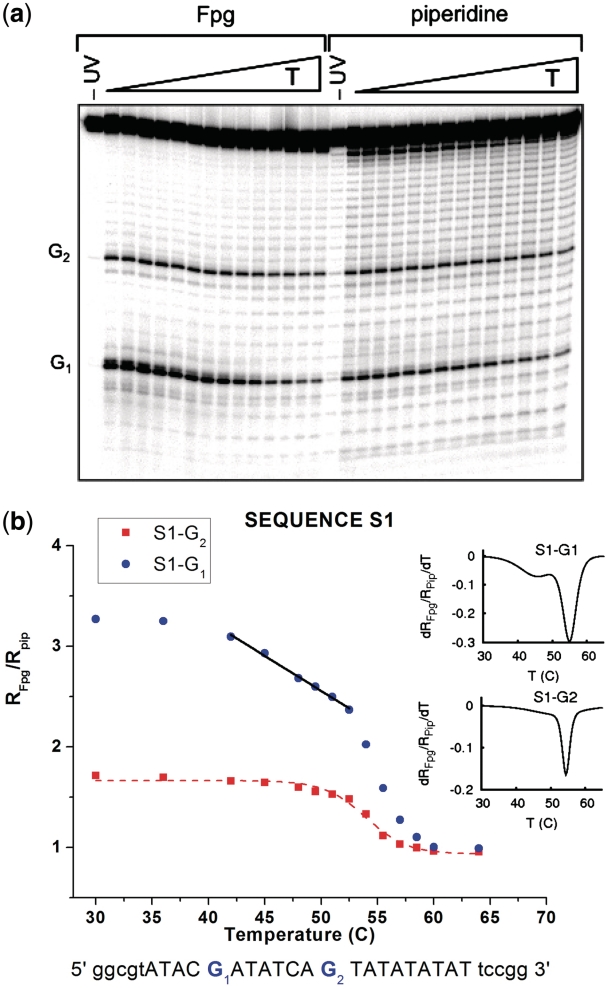
Figure 3.
Temperature dependence of the ratios RFpg/Rpip for the guanine probes G and G2 in sequence S1. The top panel (a) is an example of our experimental sequencing gel images. Each vertical stripe shows the results at a given temperature (increasing from left to right, as indicated on top of the image). A mark along a stripe corresponds to the value of RFpg, Rpip at a particular position along the DNA sequence. Characteristic points are indicated by a label on the left of the panel. The bottom panel (b) shows quantitative data extracted from the gel images analyzed in our experiment. The experimental errors are within the size of the data points. Data symbol markers are colored following the same convention as the Guanine probes highlighted in Figure 2. The straight lines, in the temperature domain corresponding to premelting, are linear fits, pointing out a two step melting transition in this domain. Dotted lines are non-linear curve fits following a sigmoidal one step melting transition typically found in common DNA melting curves.
The different steps of the experiment are illustrated schematically on Supplementary Figure S2.
Experiments with sequences S1, S4, S5 were triplicated and with S2 and S3 duplicated. The reproducibility of the absolute values of room temperature ratio in fully independent experiments was 5–10%. Within a given experiment the relative error on the ratio was below 5% for ratios above 1.5 and slightly above for smaller ratios.
Differential scanning calorimetry experiments
Differential scanning calorimetry (DSC) measurements were performed using a Nano-Differential Scanning Calorimeter III Model CSC 6300 at a rate of 1°C/min. DNA samples were HPLC purified commercial oligonucleotides bought from Eurogentec. All samples were prepared in 100 mM NaCl, 10 mM TRIS and 50 μM EDTA buffer. The excess specific heat shown in Supplementary Figure S3 was computed by removing pre- and post-transitional baselines determined by fitting straight lines to the curve before the first transition and after the main peak.
Selection of sequences based on biological fragments
While sequences S1, S2, S3 were specifically designed for this study, sequences S4 and S5 belong to fragments found in different genomes. We performed a massive database search using both BLAST (http://blast.ncbi.nlm.nih.gov/) and RSAT (http://rsat.ulb.ac.be/rsat/) (18). We found, with a high degree of identity, the pattern (w)m(N)pwwACGA, with 9 ≤ m ≤ 12, 4 ≤ p ≤ 6 in multitude parts of genomes belonging to either eukaryotic and prokaryotic organisms. Note that this pattern keeps the main idea explored in this work: having a large AT-rich domain separated by a buffer region from the main probe labeled G1 used along all our experiments.
The exact pattern selected in S4, has been found in different portions of 36 independent genes, belonging to the same or to a different organism. Similarly, the exact pattern of S5 was found identically in 12 genes of the bacteria Y. pestis and Y. pseudotuberculosis, (see Supplementary Figure S4).
Most of the regions were these patterns are located, have relevant biological importance. In order to reduce the number of candidates, we decided to restrict our selection to portions of promoters, −80 to +20 around the transcription-starting site (+1). This is a crucial region for gene expression and for the attachment of all the molecular machinery involved in transcription.
Therefore, we constructed sequence S4 with a portion of the gene ECP-0995, from E. coli 536 (bases 1053574 to 1053595 in the genome). This region is part of a promoter (−28 to −5) related to the expression of trimethylamine-N-oxide reductase 1 precursor (19). On the other hand, Sequence S5 contains the portion of the gene YPO2592 from Y. pestis, CO92 (bases 2914936–2914959 in the genome). This region is part of a promoter −42 to −20 possibly related to the expression of a hypothetical protein similar to Streptococcus pneumoniae transmembrane protein CAP33FM TR:O86896 (EMBL:AJ006986) and to internal region of Campylobacter jejuni probable enterochelin uptake permease CeuC TR:Q9PMU6 (EMBL:AL139078).
RESULTS
In order to answer the question raised in the introduction, we use a novel physicochemical methodology able to provide a mapping of the structural fluctuations along a DNA molecule.
Although this is generally achieved with the help of special molecular constructs involving a dye or a fluorophore (20), only specific sites can be observed and DNA may be locally perturbed by the probe. Our method does not need any external additive or structural modification to the DNA under study (see ‘Materials and Methods’ section for the rest of details). As we have discussed, the value of the relative yield of the two types of biochemical reactions at each guanine site that we show along this section, RFpg/Rpip, provides a measure of the degree of helicoidal stacking at the probe site. In addition, its decay versus temperature has been found to follow the decay of the local helicoidal structure as the melting of the duplex proceeds (15). As a result, we get a collection of snapshots of the structural states around all the guanine sites on a radio-labeled strand.
In summary, every guanine acts in this method as an intrinsic molecular probe for the local structural stability of the DNA helix. Instead of the melting curve for a full DNA segment we record the local structural evolution resolved in space.
The set of DNA fragments selected for our study are described in Figure 2. In a first step, let us focus on three of them, S1 to S3, which were specially designed for this study. As a common peculiarity they contain a TATA box segment included to investigate its particular structural effect and its range of influence. The relevance of this particular motif in genetics, which was the first identified core promoter element and plays an important role in the case of TATA-dependent genes, is well known (21).
The first test sequence (S1) includes a large TATA box of 9 bp. We focused our attention on two Guanines that belong to the strand marked 5′−3′ in Figure 2. Probe G1 is separated from the TATA box by a buffer region made of 7 bp, including two strong GC pairs. Probe G2 is adjacent to the TATA box. Figure 3 shows the temperature dependence of the ratio RFpg/Rpip for the two probes. As discussed in ‘Materials and Methods’ section, values of RFpg/Rpip for different probes cannot be considered as quantitative measures to compare the closing probability of different base pairs because they also depend on the average configuration of DNA near the guanine probes. Conversely, for a given probe, the variation of the ratio RFpg/Rpip provides a quantitative measure for the evolution a base pair in the helix stacking conformation.
Nevertheless, the much larger value of RFpg/Rpip for probe G1 far from the TATA box than for probe G2 next to it suggests that, at room temperature, the probability for a GC pair to remain closed in an helicoidal conformation is significantly reduced when it is next to the TATA box on the 5′ side. This is consistent with an earlier theoretical analysis that showed that a GC pair adjacent to a TATA box on the 5′ side has an opening probability that is strongly enhanced (22).
For both probes G1,2 we notice a sharp drop when the temperature is raised above about 55°C. It corresponds to the thermal denaturation (melting), i.e. the complete separation of the strands, in the domain monitored by the probes. There is a second important result to notice on Figure 3b). The signal for probe G1, which stays roughly constant from room temperature to 38°C starts to decay at this temperature with a slope much smaller than the slope near the melting temperature, but nevertheless significant. This peculiarity appears clearly as a small side peak if we compute the derivative versus temperature of a smooth curve passing by all the points of the signal of probe G1, as shown in the inset of Figure 3b). The same phenomenon is not detected for probe G2, adjacent to the TATA box, since it strongly feels the destabilizing effect of the TATA box even at room temperature.
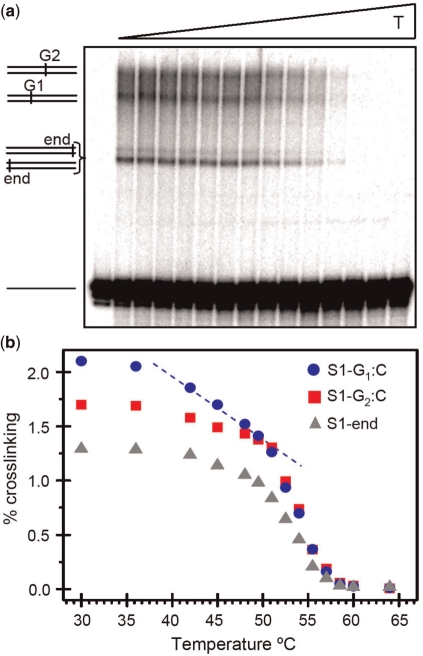
This observation suggests that, in the temperature range 38 − 55°C, the premelting effect in the TATA box (6,9) extends its influence as far as probe G1, i.e. at least 7 bp away. Strikingly, the same two-phase decay (premelting and melting) is observed in the cross-linking yield of probe G1 (Figure 4), but not for the GC pairs at the ends of the sequence, which provides an independent confirmation of this effect. (More details of this cross-check in Supplementary Data)
Figure 4.
Temperature dependence of the cross-linking yield for guanine probes G1 and G2 of sequence S1. The cross linking results also show the data for the two GC ends of the sequence (triangles marked S1-end), which are not distinguished by this method, and give rise to a strong band in the gel image. (a) shows an example of the sequencing gel image. Each vertical stripe shows the results at a given temperature (increasing from left to right, as indicated on top of the image). A mark along a stripe corresponds to the cross-linking yield at a particular position along the DNA sequence. Characteristic signatures in the gel are indicated by a label on the left of the panel. (b) quantitative data extracted from the gel images. The experimental error bars are within the size of the data points. The pre-melting effect at probe G1 is clearly visible, in good agreement with the results obtained from the RFpg/Rpip ratio (Figure 3).
The molecular sketch of Figure 1, based on sequence S1, points out the significance of this result, showing how far the influence of an AT-rich region may extend in the molecular structure of DNA.
Further tests on specific sequences were done in order to test and evaluate the range of this effect. The first one was to modify the TATA box to reduce the premelting (sequence S2 in Figure 2). The second one was to move the probe farther to test the range of the influence of the AT-rich region (sequence S3).
For the first check the TATA box has been modified by introducing two stronger GC pairs in this weak AT region. These isolated mutations, acting as ‘impurities’ in the track, are expected to be unable to fully prevent the tendency of the TATA box to open below the melting temperature, but constrain its fluctuations. This point has been checked by differential scanning calorimetry (DSC) of DNA in solution, a particular thermoanalytical technique able to determine the specific heat of the sample versus temperature, allowing to detect fine energy variations associated to conformational changes with great sensitivity.
DSC results for sequences S1 and S2 are shown in the Supplementary Data. In summary, they point out the existence of a main peak in the specific heat, directly associated to the melting of the double helix, preceded at lower temperature by an additional smaller peak that can be understood as a signature of an increase of the energy fluctuations prior to melting, attesting to the existence of pre-melting effects in both sequences. However, for sequence S2, in which the TATA box is constrained by GC pairs, the data show that the precursor peak appears at a temperature closer to melting, which confirms that premelting is less pronounced for sequence S2 as expected from its design.
On the other hand, sequence S3, leaves the TATA box intact but extends the buffer region that separates it from probe G1. In addition, we made it less prone to fluctuational opening by including GC pairs within the buffer. A common bonus in these new sequences is that they introduce additional Guanines which can be used to probe the state of the DNA molecule at new positions.
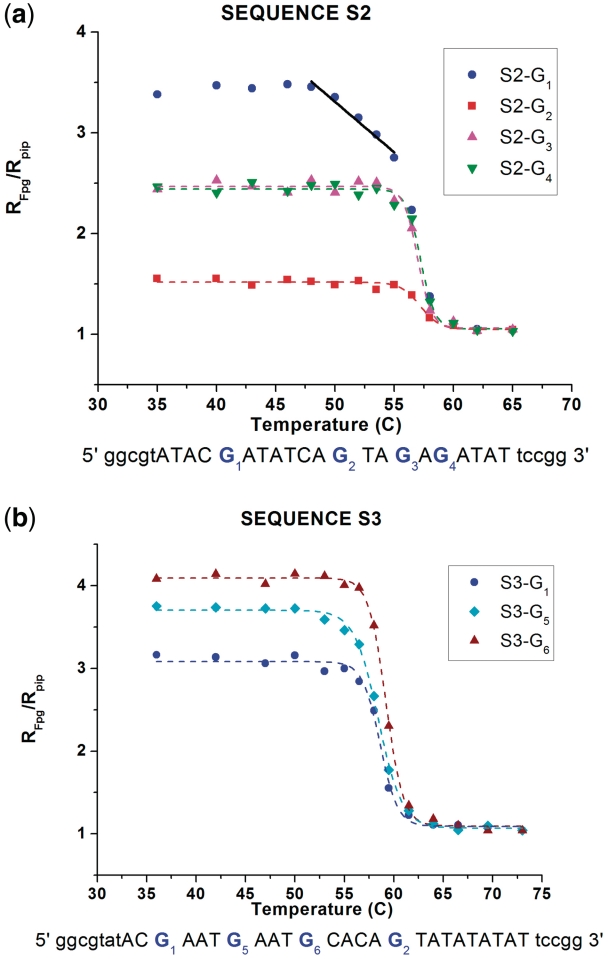
UV irradiation results for sequences S2 and S3 are shown in Figure 5. When the TATA box is stabilized by two GC pairs (sequence S2), the melting transition is slightly raised and the precursor effect, leading to a linear decay of the closing probability of probe G1 below the melting temperature, is still observed. However, it starts at 49°C instead of 38°C. We must emphasize that this result is quantitatively meaningful because the probe is exactly the same as in sequence S1, including the neighboring bases since only 2 bp inside the TATA box have been modified.
Figure 5.
Temperature dependence of the ratios RFpg/Rpip for the guanine probes G1 to G6 in sequences S2 (a) and S3 (b).
Therefore, such a significant change demonstrates that a mutation in the TATA box has a noticeable effect 10 bp away from the first mutation in the sequence. Probes G3 and G4 inside the modified TATA box show moderate room temperature values of RFpg/Rpip, what means a possible medium stable helical conformation. Since the neighboring bases to both probes are not exactly the same as in G1 or G2, these values should only be considered as a qualitative indication rather than a quantitative measure due to the sequence effects.
The experiment with sequence S3, which has a longer buffer region and a higher GC content between the unmodified TATA box and probe G1, shows that the influence of the TATA box does not extend as far as 14 bp. Probe G5 that is only 10 bp away from the TATA box shows a very small precursor effect, much weaker than probe G1 in sequence S1. This is not surprising since probe G5 is 2 bp further away from the TATA box than probe G1, and moreover the additional GC pair (probe G6) that separates probe G5 from the TATA box tends to insulate it from the fluctuations of the TATA box. Probe G6, which is only 5 bp away from the TATA box does not show precursor effects, pointing out the role of the local environment, for instance by comparison with probe G5 that is surrounded by two AT pairs while probe G6 is flanked by a GC pair on the side of the TATA box.
To summarize our results so far, all the observations obtained with artificially designed sequences converge to a consistent perspective: premelting effects in an AT-rich region such as the TATA box influence the fluctuations and conformation of DNA to some distance, about 10 bp away, inducing a non canonical conformational state prior to the full melting. Such an effect is strongly attenuated upon Guanine–Cytosine mutation in particular positions.
Our theoretical study (see Supplementary Data) also confirms a relationship between the size and stability of the bubble and buffer segments with respect to the effect exerted in distant regions.
DISCUSSION
In order to assert the possible biological significance of our results, we must prove that this phenomena is not a specificity of the artificial sequences that we designed. It should also occur in natural DNA sequences containing typical AT motifs like the ones that appear in many genome regions.
In particular, AT rich fragments are a common feature within promoter regions, having crucial functions in the process of transcription. For instance, in the case of TATA dependent genes (21,23), this well known core promoter element plays an important role promoting the transcription either by itself or in cooperation with other core promoters elements (24,25). In bacterial promoters, the UP element, located immediately 5′ to the −35 element, (−40 and −60) has also a recognizable pattern of AT-rich sequences. It takes part in the promoter recognition mechanism (26) and enhances RNA polymerase binding by complexing with α- subunits and stimulates the intrinsic transcription observed without an UP element (27,28). A particular study also has found in the lac promoter that a specific AT-rich 10-bp sequence around the −15 position seems to enhance both RNA polymerase binding and open complex formation (29).
Our goal here was to identify in genomes similar AT-rich patterns to our artificial ones, and test their influence over a distant region. Therefore we performed an exhaustive study and database search (18) in genomes from different eukaryotic and prokaryotic organisms, (details are given both in ‘Materials and Methods’ section and in Supplementary Data). The search pattern (w)m(N)pwwACGA, where w stands for weak, i.e. A or T, and N for any nucleotide, with 9 ≤ m ≤ 12, 4 ≤ p ≤ 6, was chosen in order to have a large AT-rich domain separated by a buffer region from the same probe G1 used in the previous experiments, including the base pairs on each side. It is interesting to note that this pattern was found in a wide variety of genes in different organisms. We finally restricted our selection to promoter portions between −45 to +20 around the transcription-starting site (+1), due to the previously mentioned importance of AT-rich motifs in such regions.
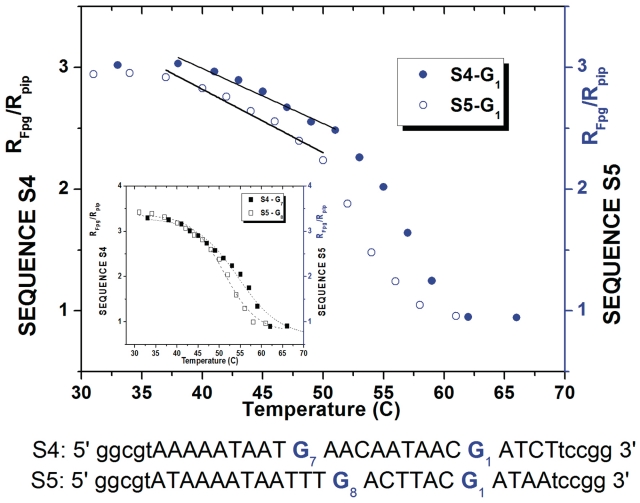
The two examples chosen for our study, are segments existing in genes from bacteria E. coli 536 (ECP-0995) and Y. pestis CO92 (YPO2592). They correspond to sequences S4 and S5 shown in Figure 2. The fragments contain the exact ACGAT pattern of probe G1 to allow us to make quantitative comparisons with previous experiments. They also include another guanine adjacent to the AT region, but with an environment slightly different from probe G2 in sequences S1 to S3. Therefore a quantitative comparison between the results for these probes G7 and G8 and the previous results for probe G2 should be made with caution. A major difference with the artificial sequences is that the reference probe G1 is on the 3′ side of the AT-rich region. The temperature variation ratio RFpg/Rpip for the guanine probes of sequences S4 and S5 is plotted in Figure 6.
Figure 6.
Ratio RFpg/Rpip for the guanine probes of sequences S4 and S5 extracted from gene promoters ECP-0995 and YPO2592, versus temperature. The structural intermediate in the premelting regime (stressed by the straight lines) is clearly detected by probes G1 in regions far from the AT-rich fluctuating zone. It appears as a double step structural transition for both probes (G1) in the two bacterial DNA fragments. The inset represents the evolution of Guanines G7,8 located next to the bubble region.
The first noticeable result is that the 3′ side and the 5′ side of an AT-track have very different properties. This is attested by the measures of RFpg/Rpip for probes adjacent to the AT motif. While probe G2 on the 5′ side in sequences S1, S2, S3 showed a very large departure from the native helix configuration, probes G7 and G8, on the 3′ side in sequences S4 and S5 show instead a high helicoidal character. This asymmetry has already been observed in a theoretical calculation of the opening probabilities of GC pairs adjacent to a TATA box (22).
More importantly, our measurements of RFpg/Rpip for probe G1 in the natural promoter sequences S4 and S5, plotted in Figure 6, show that the same signature of precursors effects to melting, induced by the influence of a distant AT-rich region, is observed, as for probe G1 in our reference sequence S1.
Further studies of mutations in the AT-rich motif are needed to clarify the limit of the effect in gene promoter regions and the possible biological consequences.
CONCLUSION
In conclusion, we throw a new light on the biophysics of DNA providing a unique measure of the correlation length of the conformational fluctuations of DNA. We have confirmed that large premelting fluctuations in AT-rich regions are able to influence the helicoidal structure and stability of the molecule over a significant distance. Our findings suggest that the effect appears to be particularly strong 10 bp away from the AT-rich region, i.e. after a full turn of the double helix. This suggests that geometrical effects play a role in our observations. A possible important consequence derived from this fact concerns current sequence analysis protocols, suggesting that they might have to consider this kind of ‘long-range’ effect to be meaningful.
It is important to notice that the existence of precursor effects and their influence far away from the AT-rich motifs is not a specificity of our artificial reference sequence. They do exist in natural promoter sequences with relevant biomolecular implications, and may extend as far as 11 bp away from the AT-rich region (sequence S4). A transcription factor binding site profile (30) shows that the studied sequences contain possible binding sites for 5–7 recognized transcription factors, which implies that the precursor effects detected may cover several protein-binding domains along the promoter sites.
Since precursor effects were found in a sequence pattern that exists in the promoter regions of genes in a large variety of organisms, the spatial correlations revealed by these experiments could have biological implications by contributing to the cooperative effects needed to assemble the molecular machinery that forms the transcription complex. Our results suggest future research on the possible role of structural fluctuations of a neighboring AT-rich region in protein binding to DNA or DNA–drug interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Public Funding (laboratoire de Physique of Ecole Normale Supérieure de Lyon). Funding for open access charge: Public Funding (department of Physics).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This paper is dedicated to Jean Cadet (CEA-CENG, France) on the occasion of his 70th anniversary.
REFERENCES
- 1.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 2.Murakami KS, Matsuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 3.Robinson H, Liaw Y, van der Marel GA, van Boom JH, Wang AH-J. NMR studies on the binding of antitumor drug nogalamycin to DNA hexamer d(CGTACG) Nucleic Acids Res. 1990;18:4851–4858. doi: 10.1093/nar/18.16.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gueron M, Kochoyan M, Leroy JL. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987;328:89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- 5.Dornberger U, Leijon M, Fritzsche H. High base pair opening rates in tracts of GC base pairs. J. Biol. Chem. 1999;274:6957–6962. doi: 10.1074/jbc.274.11.6957. [DOI] [PubMed] [Google Scholar]
- 6.Erfurth SC, Peticolas WL. Melting and premelting phenomenon in DNA by laser Raman scattering. Biopolymers. 1975;14:247–264. doi: 10.1002/bip.1975.360140202. [DOI] [PubMed] [Google Scholar]
- 7.Chan SS, Breslauer KJ, Hogan ME, Kessler DJ, Austin RH, Ojemann J, Passner JM, Wiles NC. Physical studies of DNA premelting equilibria in duplexes with and without homo dA.dT tracts: correlations with DNA bending. Biochemistry. 1990;29:6161–6171. doi: 10.1021/bi00478a008. [DOI] [PubMed] [Google Scholar]
- 8.Chan SS, Breslauer KJ, Austin RH, Hogan ME. Thermodynamics and premelting conformational changes of phased (dA)5 tracts. Biochemistry. 1993;32:11776–11784. doi: 10.1021/bi00095a005. [DOI] [PubMed] [Google Scholar]
- 9.Movileanu L, Benevides JM, Thomas GJ., Jr Determination of base and backbone contributions to the thermodynamics of premelting and melting transitions in B DNA. Nucleic Acids Res. 2002;30:3767–3777. doi: 10.1093/nar/gkf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douki T, Ravanat J-L, Angelov D, Wagner JR, Cadet J. Effects of duplex stability on charge transfer efficiency within DNA. Top Curr. Chem. 2004;236:1–25. [Google Scholar]
- 11.Angelov D, Berger M, Cadet J, Getoff N, Keskinova E, Solar W. Comparison of the effects of high-power UV-laser pulses and ionizing radiation on nucleic acids and related compounds. Radiat. Phys. Chem. 1991;37:717–727. [Google Scholar]
- 12.Spassky A, Angelov D. Influence of the local helical conformation on the guanine modifications generated from one-electron DNA oxidation. Biochemistry. 1997;36:6571–6576. doi: 10.1021/bi962761d. [DOI] [PubMed] [Google Scholar]
- 13.Douki T, Angelov D, Cadet J. UV laser photolysis of DNA: effect of duplex stability on charge-transfer efficiency. J. Am. Chem. Soc. 2001;123:11360–11366. doi: 10.1021/ja016426a. [DOI] [PubMed] [Google Scholar]
- 14.Angelov D, Beylot B, Spassky A. Origin of the heterogeneous distribution of the yield of guanyl radical in UV laser photolyzed DNA. Biophys. J. 2005;88:2766–2778. doi: 10.1529/biophysj.104.049015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spassky A, Angelov D. Temperature-dependence of UV laser one-electron oxidative guanine modifications as a probe of local stacking fluctuations and conformational transitions. J. Mol. Biol. 2002;323:9–15. doi: 10.1016/s0022-2836(02)00878-1. [DOI] [PubMed] [Google Scholar]
- 16.Mickley Conron SM, Thazhathveetil AK, Wasielewski MR, Burin AL, Lewis FD. Direct measurement of the dynamics of hole hopping in extended DNA G-Tracts. An unbiased random walk. J. Am. Chem. Soc. 2010;132:14388–14390. doi: 10.1021/ja106991f. [DOI] [PubMed] [Google Scholar]
- 17.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat. Strut. Mol. Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 18.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochhut B, Wilde C, Balling G, Middendorf B, Dobrindt U, Brzuszkiewicz E, Gottschalk G, Carniel E, Hacker J. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol. Microbiol. 2006;61:584–595. doi: 10.1111/j.1365-2958.2006.05255.x. [DOI] [PubMed] [Google Scholar]
- 20.Brauns EB, Murphy CJ, Berg MA. Complex local dynamics in DNA on the picosecond and nanosecond time scales. J. Am. Chem. Soc. 1998;120:2449–2456. doi: 10.1103/PhysRevLett.88.158101. [DOI] [PubMed] [Google Scholar]
- 21.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 22.Beger R, Prohofsky EW. Calculated enhancement of open base pair probability downstream of a (TATA)2 box. Biophys. J. 1992;61:58–62. doi: 10.1016/S0006-3495(92)81815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 2007;20:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- 24.Corden J, Wasylyk B, Buchwalder A, Sassone-Corsi P, Kedinger C, Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980;209:1405–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- 25.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nature Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 26.Hook-Barnard IG, Hinton DM. Transcription initiation by mix and match elements: Flexibility for polymerase binding to bacterial promoters. Gene Regul. Syst. Biol. 2007:275–293. [PMC free article] [PubMed] [Google Scholar]
- 27.Hook-Barnard IG, Hinton DM. The promoter spacer influences transcription initiation via σ70 region 1.1 of Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA. 2009;106:737–742. doi: 10.1073/pnas.0808133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Tolstorukov M, Zhurkin V, Garges S, Adhya S. A mutant spacer sequence between -35 and -10 elements makes the Plac promoter hyperactive and cAMP receptor protein-independent. Proc. Natl Acad. Sci. USA. 2004;101:6911–6916. doi: 10.1073/pnas.0401929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinemeyer T, Wingender E, Reuter I. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:364–370. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.