Abstract
Post-translational poly(ADP-ribosyl)ation has diverse essential functions in the cellular response to DNA damage as it contributes to avid DNA damage detection and assembly of the cellular repair machinery but extensive modification eventually also induces cell death. While there are 17 human poly(ADP-ribose) polymerase (PARP) genes, there is only one poly(ADP-ribose) glycohydrolase (PARG) gene encoding several PARG isoforms located in different subcellular compartments. To investigate the recruitment of PARG isoforms to DNA repair sites we locally introduced DNA damage by laser microirradiation. All PARG isoforms were recruited to DNA damage sites except for a mitochondrial localized PARG fragment. Using PARP knock out cells and PARP inhibitors, we showed that PARG recruitment was only partially dependent on PARP-1 and PAR synthesis, indicating a second, PAR-independent recruitment mechanism. We found that PARG interacts with PCNA, mapped a PCNA binding site and showed that binding to PCNA contributes to PARG recruitment to DNA damage sites. This dual recruitment mode of the only nuclear PARG via the versatile loading platform PCNA and by a PAR dependent mechanism likely contributes to the dynamic regulation of this posttranslational modification and ensures the tight control of the switch between efficient DNA repair and cell death.
INTRODUCTION
Poly(ADP-ribosyl)ation is a post-translational modification of proteins involved in cellular processes as diverse as DNA repair, transcription, cell division or cell death (1). Poly(ADP-ribose) (PAR) is rapidly produced at DNA breaks by the founding member of the PARP family PARP-1, triggering local chromatin relaxation (2) and recruitment of repair factors which have strong affinity for PAR, such as the base excision repair/single-strand break repair factor XRCC1 (3–6). The amount of PAR produced, which reflects the severity of the DNA insult, directly contributes to the cell decision to initiate either survival- or death- programmes (7,8).
The PAR degrading enzyme poly(ADP-ribose) glycohydrolase (PARG) is encoded by a single gene but is present as multiple isoforms localized to different cellular compartments: full-length PARG111 is nuclear, PARG102 and PARG99 are cytoplasmic, whereas shorter isoforms (PARG60, PARG55) are targeted to the mitochondria (9–12). In mice, depleting all PARG isoforms is embryonically lethal, whereas a hypomorphic mutant is viable, but the mice are sensitive towards ionizing radiation and alkylating agents (13,14). Using shRNA to prevent the expression of all PARG isoforms in cells, we and others have shown that PARG is required for efficient repair of single- and double-strand breaks and oxidized bases (15–17).
Using a combination of live-cell microscopy and laser microirradiation, we and others could demonstrate the rapid recruitment of PARP-1 and XRCC1 to DNA damage sites and the role of PARP-1 activity in these protein relocalizations (4,18,19). Here, we show that all PARG isoforms except for the mitochondrial localized PARG fragment could be efficiently recruited to DNA damage sites. The recruitment of PARG was only partially dependent on PARP-1 and PAR synthesis. We identified a functional PCNA-binding motif within the PARG sequence and showed that binding to PCNA contributes to PARG recruitment at sites of DNA damage. Our results reveal that PARG is recruited to DNA damage sites via at least two pathways, one is PAR-dependent and the other is PCNA-dependent.
MATERIAL AND METHODS
Cell culture and transfection
Wild type, PARP-1−/− MEFs, PARP-2−/− MEFs, HeLa, HEK293T and U2OS (clone 2-6-3) cells were cultured in DMEM supplemented with 10% FCS and 50 µg/ml gentamicin. Cells grown on µ-slides (ibidi) or on gridded coverslips were cotransfected with polyethylenimine (Sigma). For microirradiation experiments cells were sensitized by incubation in medium containing BrdU (10 µg/ml) for 24–48 h. NU1025 (Sigma) was added to the cells 1 h before microirradiation at a concentration of 200 µM. For GFP-pulldown analyses, cells grown on Petri dishes were transfected with JetPEI (Polyplus).
Expression plasmids
The cDNA fragments encoding wild type and mutant human PARG were generated by PCR and subcloned into the EcoRI/SmaI sites of pEGFP-N1 (Clontech). Sequences of oligonuclotides and plasmids are available upon request. GFP-XRCC1, GFP-PARP-1, RFP-PCNA and PCNA-LacI-RFP expression construct were previously described (4,20).
Live-cell microscopy, microirradiation and photobleaching experiments
Live cell imaging, microirradiation and photobleaching experiments were carried out with a Leica TCS SP2/AOBS or Leica SP5/AOBS confocal laser scanning microscope, each equipped with a UV-transmitting HCX PL 63×/1.4 oil objective. GFP and RFP were excited with a 488 nm Ar laser line and a 561 nm DPSS laser line, respectively. The microscopes were equipped with a heated environmental chamber set to 37°C. Confocal image series were typically recorded with a frame size of 256 × 256 pixels and a pixel size of 90 nm.
Microirradiation was carried out as previously described (21). In brief, a preselected spot of ∼1 µm in diameter within the nucleus was microirradiated for 1 s with a 405 nm diode laser set to 50–80 µW. The laser power was measured after passing through the objective lens with a laser power meter (Coherent). Before and after microirradiation confocal image series of one mid z-section were recorded at 2 s time interval (typically six pre-irradiation and 150 post-irradiation frames). For evaluation of the recruitment kinetics, fluorescence intensities at the irradiated region were corrected for background and for total nuclear loss of fluorescence over the time course and normalized to the pre-irradiation value.
For FRAP analysis, a region of interest was selected and photobleached for 300 ms with all laser lines of the Ar-laser and the 561 nm DPSS laser set to maximum power at 100% transmission. Before and after bleaching, confocal image series were recorded at 150 ms time intervals (typically 10 pre-bleach and 200 post-bleach frames). Mean fluorescence intensities of the bleached region were corrected for background and for total nuclear loss of fluorescence over the time course and normalized to the mean of the last four prebleach values.
For the quantitative evaluation of microirradiation and photobleaching experiments, data of at least nine nuclei were averaged and the mean curve and the standard error of the mean calculated and displayed using Microsoft Excel software.
Indirect immunofluorescence microscopy
Cells grown on glass coverslips were treated as described in the legends of the figures, and fixed with ice cold methanol/acetone for 15 min (for PAR) or in ice-cold methanol for 5 min (for PCNA). Cells were permeabilized with PBS, 0.1% tween and incubated overnight at 4°C with primary antibodies diluted in PBS, 0.1% tween, 1 mg/ml BSA: mouse monoclonals anti-PAR 10H (IgG3k, 1:1000), anti-GFP (IgG1, 1:500, Roche) or anti-PCNA (IgG2a, 1:200, Dianova). Secondary antibodies used were Alexa Fluor goat anti-mouse IgG3 (568) or IgG1 (488 or 568) (1:2000, Molecular Probes, Invitrogen). Cells were counterstained with DAPI, mounted using Mowiol (Roche) and observed on a Leica DMRA2 equipped with an Orca-ER CCD camera (Hammamatsu) and the capture software OpenLab 4.1 (Improvision). Alternatively images were taken with a PerkinElmer UltraView Vox spinning disk microscope equipped with a Plan-Apochromat 63× /1.4 oil objective using Volocity software 5.0 for image capturing. DAPI, GFP and RFP were excited with a 405, 488 and a 561 nm DPSS laser line, respectively.
Zymogram
PARG activity gel assay was performed as described by Amé et al. (15).
PAR-blot
32P-labelled PAR was synthetised and purified as described by Amé et al. (22). PCNA (ENZO Life), histone H1 (ENZO Life) and BSA (Sigma) purified recombinant proteins were spotted onto nitrocellulose sheet as indicated in the figure, and the membrane was incubated with 32P-labelled PAR for 1.5 h at 25°C. After six washes of 10 min in PBS, the membrane was autoradiographied. Similar amounts of purified proteins were analysed by SDS–PAGE gel electrophoresis followed by Coomassie staining to verify the integrity and quantities of proteins used in the assay.
GFP-pull down assays
HEK393T cells (1.5 × 107) were transfected with the plasmids encoding GFP-tagged PARG as described in figure legend. Thirty-six hours post-transfection, cells were lysed in 20 mM Tris–HCl pH 7.5, 120 mM NaCl, 0.1% NP40, 0.5 mM Pefabloc and miniCOmplete protease inhibitor cocktail (Roche Diagnostic) for 20 min on ice, centrifuged at 132 000 rpm for 20 min at +4°C and supernatant was recovered. For the experiments including H2O2 treatment, cells were treated for 10 min with 1 mM H2O2 in complete medium at 37°C, washed twice with 1× PBS, pelleted and resuspended in 300 µl of 20 mM Tris–HCl pH 7.5, 400 mM NaCl, 5 mM DTT, 0.1% NP40, 20% glycerol, 0.5 mM Pefabloc and protease inhibitor. After four cycles of freeze/thaw and centrifugation at 132 000 rpm for 20 min at +4°C, supernatant was dilute to reach a final concentrention of NaCl of 120 mM. Protein extracts were incubated 2.5 h at +4°C with GFP-binder (Chromotek) on a rotating wheel. After four washes with 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% NP40, 0.5 mM Pefabloc, the immunoprecipitated proteins were solubilized in Laemmli buffer by boiling 4 min at 95°C and analysed by SDS–PAGE electrophoresis followed by western blotting. The membranes were probed with anti PCNA (PC10, DAKO, 1/1000) then with anti-GFP (Roche, 1/1000) monoclonal antibodies.
RESULTS AND DISCUSSION
Except the mitochondrial PARG, all PARG isoforms are recruited to laser-induced DNA damage sites
To follow the redistribution of GFP-tagged PARG to sites of DNA damage in living cells, we generated DNA lesions at preselected subnuclear sites in HeLa cells using laser microirradiation as previously described (4,21). We found that PARG111-GFP (Figure 1A) was recruited to DNA damage sites (Figure 1B and C), but more slowly (maximum accumulation reached in more than 4 min) than two other central repair proteins GFP-PARP-1 and GFP-XRCC1 [Figure 1D and E and Mortusewicz et al. (4)]. Comparable recruitment kinetics of PARG111-GFP could be observed in immortalized mouse embryonic fibroblasts (MEF, Figure 1F). The cytoplasmic isoforms PARG102 and PARG99 were shown previously to shuttle between the cytoplasm and the nucleus, and to accumulate in the nucleus following γ-irradiation (23). Here, we could show that both isoforms were recruited with similar kinetics (Figure 1G and H) but accumulated less than PARG111-GFP (maximal intensity reached, MAX Int, reflecting the amount of protein accumulated, was 1.47 ± 0.07 for PARG102-GFP and 1.38 ± 0.04 for PARG99-GFP compared to 2.22 ± 0.11 for PARG111-GFP, Supplementary Figure S1C and D). In contrast, PARG461-976-GFP, which corresponds to the previously described hPARG59 containing the regulatory segment and the mitochondrial localization signal (11,12,24), and thus localized to the mitochondria, was never recruited (Figure 1I). To summarize, PARG111 and to a lesser extend PARG102 and PARG99 but not mitochondrial PARG461–976 accumulate at DNA damage sites in vivo.
Figure 1.
Recruitment of PARG isoforms to laser-induced DNA damage sites. (A) Schematic representation of PARG and fusion proteins used. (B) Live cell imaging of a microirradiated HeLa cell expressing PARG111-GFP. Comparision of PARG111-GFP (C), GFP-PARP-1 (D) and GFP-XRCC1 (E) recruitment to DNA damage sites in HeLa cells and quantitative evaluation of recruitment kinetics showing mean curves. Recruitment of the PARG isoforms PARG111-GFP (F), PARG102-GFP (G) and PARG99-GFP (H) to laser-induced DNA damage sites in mouse embryonic fibroblasts (MEFs). (I) The mitochondrial isoform PARG461-976-GFP shows no accumulation at microirradiated sites. Where no recruitment can be observed, sites of irradiation are indicated by arrows. Error bars represent the standard error of the mean. Scale bar: 5 µm.
Recruitment of PARG to DNA damage sites depends partly on PAR synthesis
We next examined whether the recruitment of PARG was dependent on PAR synthesized at DNA damage sites. Treatment of MEFs with the PARP inhibitor NU1025 or KU0058948 (data not shown) slowed down (time required for 50% of the maximum accumulation, tMAX1/2, of 72.51 ± 4.25 s, compared to 54.11 ± 3.03 s in absence of NU1025) and decreased the amount of PARG111-GFP recruited to DNA damage sites (MAX Int of 1.81 ± 0.09, compared to 2.11 ± 0.11 in absence of NU1025, Figure 2B and C and Supplementary Figure S1C and D). This indicates that the recruitment of PARG111-GFP depends partly on PAR synthesized at the damage site. However, it is apparently independent of PARP-1 or PARP-2, since PARG111-GFP recruitment is not affected in MEFs either lacking PARP-1 (Parp-1−/−, Figure 2B and C) or PARP-2 (Parp-2−/−, Supplementary Figure S1A and B). By examining the mobility of PARG molecules accumulated at DNA damage sites by FRAP analyses, we found a minor increase of PARG111-GFP mobility in the presence of the PARP inhibitor, indicating a reduced binding of PARG111-GFP to DNA damage sites, which could account to some extent for the different recruitment kinetics in the presence or absence of the PARP inhibitor (Figure 2D). Altogether, these results indicate that the recruitment of PARG only partially depends on PAR synthesis, and point to the existence of a PAR-independent recruitment mechanism.
Figure 2.
PAR-dependent recruitment of PARG to DNA damage sites. (A) Schematic representation of PARG and fusion proteins used. (B) Live cell imaging of PARG111-GFP recruitment in wt MEFs (upper row), Parp-1−/− MEFs (middle row) and in wt MEFs treated with the PARP inhibitor NU1025 (lower row). (C) Comparison of PARG111-GFP recruitment kinetics in wt MEFs (green curve), Parp-1−/− MEFs (red curve) and wt MEFs treated with NU1025 (blue curve). (D) Quantitative evaluation of FRAP data showing mean curves. To analyse the mobility of PARG111-GFP at DNA damage sites in wt MEFs (green curve), Parp-1−/− MEFs (red curve) and wt MEFs treated with NU1025 (blue curve) the microirradiated region was bleached 5 min after microirradiation and the fluorescence recovery was measured. (E) Accumulation of the N-terminal domain of PARG (PARG1–469-GFP) at laser-induced DNA damage sites. (F) Quantitative evaluation of PARG1–469-GFP recruitment kinetics in wt MEFs (green curve), Parp-1−/− MEFs (red curve) and wt MEFs treated with NU1025 (blue curve) showing mean curves. (G) The C-terminal domain of PARG (PARG478–976-GFP) shows no accumulation at DNA damage sites. Where no recruitment can be observed, sites of irradiation are indicated by arrows. Error bars represent the standard error of the mean. Scale bar: 5 µm.
We next aimed to unravel these PAR-dependent and PAR-independent mechanisms for PARG recruitment to DNA damage sites by analysing the recruitment of different PARG domains. Whereas the C-terminal half of PARG (PARG478–976-GFP) was not recruited (Figure 2G), the N-terminal half of PARG (PARG1-469-GFP) considered to act as a putative regulatory domain (Figure 2A) was efficiently recruited to DNA lesions, but its recruitment was independent of PARP-1 and PAR (Figure 2E and F). This suggests that a sequence motif or a domain present in the N-terminal part mediates the PAR-independent recruitment, whereas the presence of the C-terminal part of PARG containing the catalytic domain probably confers PAR-dependent recruitment.
PARG catalytic activity regulates PARG localization to DNA damage sites
To assess whether PARG activity could influence its relocalization to DNA damage sites, we followed the recruitment of an inactive PARG, obtained by mutating glutamic acids 755 and 756 to alanines (Figure 3A), according to Patel et al. (25). Inactivity of this mutant was verified by zymogram (Figure 3B). PARGE755,756A-GFP showed a remarkable faster recruitment to DNA damage sites than its wild-type counterpart (Figure 3C and 3D, compare the green and black curves and see Supplementary Figure S1C and D). Interestingly, its recruitment was affected in Parp-1−/− cells, but even more dramatically in the presence of a PARP inhibitor (Figure 3C and D, compare the green, red and blue curves and Supplementary Figure S1C and D). The dramatic decrease of PARGE755,756A-GFP recruitment in presence of a PARP inhibitor can be partially attributed to an increase of mobility, as shown by FRAP analyses (Figure 3E), which indicates a weaker binding of PARGE755,756A-GFP to DNA damage sites in the absence of PAR. These results reveal the involvement of PARP-1 and PAR in the recruitment of PARG111. The slower recruitment kinetics of PARGE755,756A-GFP in cells treated with a PARP inhibitor than in cells lacking PARP-1 could result from the presence of inactive PARP-1 molecules at the damage site that could hamper the recruitment of PARG to the site of lesion. This hypothesis is strengthened by the slightly more efficient recruitment of PARGE755,756A-GFP in Parp-1−/− MEFs treated with NU1025 than in wt MEFs treated with the same PARP inhibitor (Figure 3D and data not shown). Of note, the residual recruitment of PARGE755,756A-GFP in the presence of a PARP inhibitor was comparable to that of wild-type PARG111-GFP in similar conditions (compare Figures 3D and 2E), confirming the existence of a PAR-independent mechanism. Taken together, these results highlight the important role of PAR in the recruitment of PARG to DNA damage sites.
Figure 3.
Inactive PARG is recruited with higher efficiency to laser-induced DNA damage sites. (A) Schematic representation of PARG and PARGE755,756A-GFP. Mutated amino acid positions are indicated in red. (B) Zymogram showing the PAR-degrading capacity of the indicated GFP-tagged PARG proteins. (C) Live cell imaging of PARGE755,756A-GFP recruitment in wt MEFs (upper row), Parp-1−/− MEFs (middle row) and in wt MEFs treated with the PARP inhibitor NU1025 (lower row). (D) Recruitment kinetics of PARGE755,756A-GFP in wt MEFs (green curve), Parp-1−/− MEFs (red curve) and wt MEFs treated (blue curve) or not (black curves) with NU1025. (E) Mobility of PARGE755,756A-GFP at DNA damage sites in wt MEFs (green curve), Parp-1−/− MEFs (red curve) and wt MEFs treated with NU1025 (blue curve). (F) Immunodetection of PAR in PARGKD cells expressing shRNA-insensitive PARGE755,756A-GFP or PARG111-GFP, at the time indicated after 10 min of treatment with 0.5 mM H2O2. Error bars represent the standard error of the mean. Scale bar: 5 µm.
The dramatic differences in the recruitment of PARGE755,756A-GFP and PARG111-GFP suggest that the catalytic activity of PARG111-GFP could affect its accumulation at DNA damage sites, by efficiently and promptly removing PAR produced in response to DNA damage. We tested this hypothesis by analysing the amount of PAR produced in H2O2-treated PARGKD cells which lack endogenous PARG (15), transfected with plasmids encoding shRNA-insensitive PARGE755,756A-GFP or PARG111-GFP. PAR could be detected in H2O2-treated and untreated PARGKD cells as previously shown (15) and also in cells expressing PARGE755,756A-GFP, whereas in cells expressing PARG111-GFP PAR could never be detected (Figure 3F). Therefore, the fast PAR degradation at DNA damage sites observed in presence of an overexpressed active PARG masks the contribution of PARP-1/PAR in its recruitment, that can only be clearly revealed with the catalytically inactive PARGE755,756A-GFP. Taken together, our data suggest that there are several modes of PARG recruitment to DNA damage sites: one PARP-1/PAR dependent (represented by the shift of the green to the red curve in Figure 3D), one PAR-dependent and possibly PARP-1 independent (red to blue curve in Figure 3D), and one PAR-independent (blue curve to no recruitment in Figure 3D) that we next aimed to analyse in more detail.
PARG interacts with PCNA through a PIP box-containing PBD
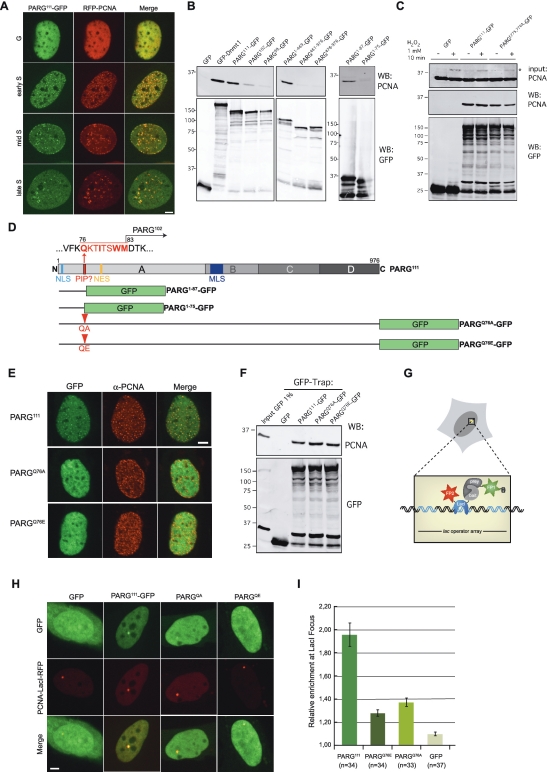
In the course of our study, we noticed that PARG111-GFP displayed a punctuate localization in a fraction of cells, resembling replication foci. We could show that PARG111-GFP colocalizes with co-expressed RFP-PCNA or endogenous PCNA at each stage of S-phase (Figure 4A and E), suggesting that PARG is present at the replication forks throughout S-phase. In contrast, PARG102-GFP and PARG99-GFP were never found to colocalize with PCNA at sites of replication (data not shown), suggesting that the first 1–83 residues of PARG are necessary for this localization to replication foci. In order to test whether PARG interacts with PCNA, we overexpressed GFP-tagged PARG isoforms in HEK293T cells and immunoprecipitated the protein complexes using anti-GFP-coupled beads. Copurifying endogenous PCNA was assessed by western blot (Figure 4B). PCNA was efficiently copurified with the positive control GFP-DNMT1 (26), but not with GFP alone. PCNA also interacted with PARG111-GFP, but the interaction was dramatically decreased, although not completely abolished, with PARG102-GFP and PARG99-GFP. This demonstrates that PARG interacts with PCNA and that the very N-terminal part is involved in this interaction. This is further confirmed by the observed interaction of PCNA with PARG1–469-GFP, but not with PARG461–976-GFP and PARG478–976-GFP.
Figure 4.
PARG interacts with PCNA and is present at replication foci. (A) Colocalization of PARG with PCNA throughout S-phase. Spinning disk microscopy images of wt MEFs coexpressing PARG111-GFP and RFP-PCNA in different S-phase stages. PARG111-GFP colocalizes with RFP-PCNA at sites of replication throughout S-phase. (B) Interaction of PARG with PCNA analysed by GFP-pull down assays. The indicated GFP-tagged proteins were overexpressed in HEK293T cells, recovered by GFP-pulldown and copurified PCNA was assessed by western blot with anti PCNA antibodies (upper panels). The presence of GFP-tagged protein is revealed with an anti GFP antibody (lower panels). (C) Effect of H2O2 treatment on the interaction between PARG and PCNA. PARG111-GFP and PARGE755,756A-GFP overexpressing HEK293T cells were treated with 1 mM H2O2 for 10 min before performing a GFP-pull down assay as described in (B). The asterisk indicates mono-ubiquitinated PCNA, visible in the input of H2O2-treated cells (1% input was loaded on the gel). (D) Schematic representation of PARG including the identified PIP box sequence and fusion proteins used. Mutated amino acid positions are indicated in red. (E) Spinning disk microscopy images of wt MEFs expressing PARG111-GFP, PARGQ76A-GFP or PARGQ76E-GFP and stained with a mouse-monoclonal antibody against PCNA to mark sites of replication. (F) Effect of the mutation of the PARG PIP domain on its interaction with PCNA. GFP-pull down assays with the indicated PARG constructs were performed as in (B). (G) Schematic outline of the F2H assay (see text for details). (H) U2OS.2-6-3 cells with a stably integrated lac operator array coexpressing PCNA-LacI-RFP (bait protein) and various GFP-tagged PARG fusions (prey proteins). Colocalization of the red PCNA-LacI-RFP signal with a green prey signal at the lac operator array indicates interaction. (I) Quantification of the relative enrichment of PARG111-GFP, PARGQ76A-GFP, PARGQ76E-GFP and GFP at the lac operator array. For quantification, the fluorescence signal at the lac operator array was measured and divided by the overall fluorescence measured in the nucleus. Error bars represent the standard error of the mean. Scale bar: 5 µm.
We then tested whether the interaction between PARG and PCNA was modified in response to DNA damage. PCNA was co-immunoprecipitated with PARG-111-GFP with comparable efficiency whether cells were treated or not with 1 mM H2O2 for 10 min before lysis (Figure 4C). However, since the overexpression of PARG-111-GFP alters the production of H2O2-triggered PAR synthesis as shown in Figure 3F, we performed the same experiment with the inactive PARGE755,756A-GFP mutant. Whereas the mutation of PARG catalytic domain had no significant effect on binding to PCNA, the H2O2 treatment slightly decreased the interaction between the two proteins, suggesting that PARG activity can regulate this interaction in response to DNA damage. However, we do not want to conclude from these experiments that normal endogenous PARG and PCNA interact less in response to DNA damage, since overexpressing the inactive PARG mutant could also have repercussions on the PAR level of the damaged cells, by a dominant-negative effect. The only way to really test the potential DNA-damage dependent change in PARG-PCNA interaction would be to look at complexes formed by endogenous proteins, but the lack of specific anti PARG antibodies precludes from such experiments.
Carefull examination of the PARG primary sequence led us to identify a motif at the N-terminus resembling a PIP (PCNA-interacting-peptide) box, 76-QKTITSW-82, found in various PCNA binding domains (PBD) of replication and repair associated proteins (27,28) (Figure 4D). An N-terminal PARG construct encompassing this motif (PARG1–87-GFP, Figure 4D) was able to bind PCNA, whereas a shorter construct ending just before this motif (PARG1–75-GFP) failed to bind PCNA, demonstrating the functionality of this sequence acting as a PIP box (Figure 4B). Since the Gln residue Q76 has been shown to be essential for PCNA interaction through the PBD of several proteins (28), we mutated it to either Ala or Glu in PARG111-GFP (Figure 4C). Both mutations impaired the localization of the proteins PARGQ76A-GFP and PARGQ76E-GFP at replication foci (Figure 4E). However, when these mutants were tested for their interaction with PCNA by co-immunoprecipitation in HEK293T overexpressing cells, they both unexpectedly still showed strong binding to PCNA (Figure 4F). This result does not fit with the lack of colocalization of these mutants with PCNA in vivo, as shown in Figure 4E. We therefore propose that the interaction observed in the immunoprecipitation assays might result from interactions with third proteins that could reinforce the link between PCNA and these mutants. Alternatively, we cannot exclude that another domain of PARG participates in the interaction with PCNA. This hypothesis is strengthened by the residual binding observed for PARG102-GFP to PCNA (Figure 4B). To further test the interaction of PARG and its PIP mutants with PCNA in vivo, we used a fluorescent two-hybrid assay developed for the direct visualization of protein–protein interactions in living cells (20). Briefly, the fluorescent bait, consisting of PCNA fused to LacI and RFP is tethered to a stably integrated chromosomal lac operator array in the nucleus and interaction is assayed for by looking for co-localization with GFP-tagged prey proteins at this defined spot (Figure 4G). Using this technique, we could show that PARG111-GFP interacts with PCNA-LacI-RFP in vivo, whereas GFP did not (Figure 4H). Introducing a point mutation at the conserved Gln residue within the PIP box strongly impaired the interaction of PARGQ76A-GFP or PARGQ76E-GFP with PCNA-LacI-RFP, without completely abolishing it, as shown by the quantitative evaluation of the enrichment of these proteins at the lac operator array (Figure 4H and I). Taken together, these results demonstrate that PARG interacts with PCNA and that the PIP domain is involved in this interaction in vivo.
PARG interaction with PCNA favours its recruitment to DNA damage sites
Besides its essential implication in DNA replication, PCNA is also an active player in various repair pathways, such as mismatch repair, nucleotide excision repair and base excision repair [see for review ref. (27,29)]. As such, PCNA is recruited to locally introduced DNA damage (5,18,19,30–32), although not as fast as the scaffold BER protein XRCC1 (4). We thus wondered whether the interaction of PARG with PCNA could regulate the dynamics of PARG at DNA damage sites.
We compared the recruitment of the PIP-bearing fusion protein PARG1-87-GFP with that of the PIP-lacking fusion protein PARG1–75-GFP to laser-induced DNA damage sites. In contrast to PARG1–75-GFP, PARG1–87-GFP was recruited to DNA damage sites, although with low efficiency (Figure 5A and B), indicating that PARG can also be recruited to DNA damage sites via its interaction with PCNA. We then examined the recruitment of full length PARG harbouring a mutation in the PIP box. The Q76A (or Q76E, data not shown) mutation diminished the recruitment of PARG at DNA lesions introduced in wild-type MEFs (Figure 5C and Supplementary Figure S1E and F, MAX Int of 1.77 ± 0.05 for PARGQ76A-GFP compared to 2.22 ± 0.11 s for PARG111-GFP). These results support the involvement of PCNA in the efficient recruitment of PARG to DNA damage sites. Moreover, recruitment of PARGQ76A-GFP was decreased upon PARP inhibition or in Parp-1−/− cells (Figure 5C), though not totally abolished, pointing to the existence of an additional mechanism for PARG recruitment to DNA damage sites that is PCNA- and PAR-independent. Of note, the accumulation of PARGQ76A-GFP (or PARGQ76E-GFP, data not shown) in the presence of a PARP inhibitor was only slightly more affected than that of wild-type PARG111-GFP in the same condition (Figure 5D and Supplementary Figure S1F, MAX Int of 1.67 ± 0.09 for PARGQ76A-GFP in the presence of NU1025 compared to 1.81 ± 0.09 for PARG111-GFP in the presence of NU1025). These results suggest that the PCNA-mediated recruitment of PARG could partly depend itself on PAR. This is further supported by the fact that PARG1–87-GFP accumulates more slowly in Parp-1−/− cells or in the presence of NU1025, whereas PARG1–75-GFP is not recruited at all in any condition (Figure 5A). Indeed, that PARP-1 inhibition could partially decrease PCNA relocalization to DNA damage sites was reported previously by Okano et al. (5) and Lan et al. (18), whereas Godon et al. (19) made the opposite observation of an increased accumulation of PCNA at microirradiated sites upon chemical inhibition of PARPs, but not in cells lacking PARP-1. These contradictory data prompted us to look at the recruitment of PCNA in our experimental system. We found that recruitment of Cherry-PCNA at laser-induced DNA damages was hindered by PARP inhibition, but not in Parp-1−/− cells (Figure 5E and F and Supplementary Figure SIE and F). This reminds what we observed for PARG111 and PARGE755,756A which were both recruited less efficiently upon PARP inhibition than in the absence of PARP-1 (Figures 2C and 3C). Moreover, the NU1025 PARP inhibitor impedes more severely the recruitment of PARGE755,756A-GFP in Parp-1+/+ cells than it did in Parp-1−/− cells, supporting the hypothesis that it is the presence of inactive PARP-1 molecules that likely affects the access of PCNA and PARG to DNA damage sites. In addition, we failed to show a direct interaction of recombinant PCNA with purified PAR on dot blot assays (Figure 5G), supporting the fact that PCNA recruitment to DNA damage sites unlikely depends on a direct binding to PAR.
Figure 5.
PARG interaction with PCNA contributes to its accumulation at laser-induced DNA damage sites. (A) Accumulation and recruitment kinetics of PARG1-87-GFP to laser-induced DNA damage sites. The recruitment kinetics of PARG1-87-GFP in wt MEFs (green curve), Parp-1−/− MEFs (red curve) and wt MEFs treated with NU1025 (blue curve) as well as the recruitment kinetics for PARG111-GFP in wt MEFs (light green) are shown for comparison. (B) PARG1-75-GFP lacking the newly identified PIP box does not accumulate at DNA damage sites. (C) Mutation of the newly identified PIP box of PARG (PARGQ76A-GFP) decreases PARG accumulation to DNA damage sites. Recruitment kinetics of PARGQ76A-GFP in wt MEFs (green curve), Parp-1−/− MEFs (red curve) and wt MEFs treated with NU1025 (blue curve) as well as the recruitment kinetics for PARG111-GFP in wt MEFs (light green) are shown for comparison. (D) Comparison of PARGQ76A-GFP (dark blue curve) and PARG111-GFP (light blue curve) recruitment kinetics in wt MEFs treated with NU1025. Error bars represent the standard error of the mean. Scale bar: 5 µm. (E) Live cell imaging of cherry-PCNA recruitment in wt MEFs (upper row), Parp-1−/− MEFs (middle row) and in wt MEFs treated with the PARP inhibitor NU1025 (lower row). (F) Comparison of cherry-PCNA recruitment kinetics in wt MEFs (red curve), Parp-1−/− MEFs (blue curve) and wt MEFs treated with NU1025 (green curve). Error bars represent the standard error of the mean. Where no recruitment can be observed, sites of irradiation are indicated by arrows. Scale bar: 5 µm. (G) PCNA does not bind PAR on polymer blot assay. Recombinant purified BSA, histone H1 and PCNA were spotted on nitrocellulose membrane as indicated before incubation with 32P-labelled PAR (right panel). Same amounts of proteins were analysed by SDS–PAGE gel electrophoresis and Coomassie staining to control the quality and quantities of the proteins used in the assay.
In summary, in this study we identified two modes of recruitment for PARG to DNA damage sites. Given the importance of PARG in the cellular response to DNA damage through the regulation of PAR levels, it was tempting to speculate that PARG might be targeted to DNA damage sites by direct recognition of PARylated proteins, like histones or PARP-1. We found that both PAR and PARP-1 actually have an influence on PARG recruitment, probably either by enabling PARG access to the damage sites by the local opening of chromatin or by serving as a docking site for PARG recruitment. Besides, we could identify a second and unexpected mode of recruitment through a PBD-mediated interaction of PARG with PCNA. This newly identified interaction of PARG with PCNA could have evolved to enhance the recruitment efficiency of PARG to DNA damage sites. As PCNA shows a slow and constant accumulation at DNA damage sites and also remains associated with repair sites for a prolonged period of time, this interaction could also ensure that PAR levels are kept low at later steps of repair via the constant recruitment of PARG. Alternatively, and considering that (i) PCNA is involved in the long-patch BER pathway (33), (ii) PARP-1 participates in the decision between short- and long-patch BER (34), (iii) PARG interacts with PARP-1 and XRCC1 (35) and PCNA (this study) and (iv) PAR is proposed to be a source of energy favouring long-patch repair in conditions of ATP shortage (36), the PCNA-dependent accumulation of PARG at DNA damage sites identified here could also be regarded as an additional regulatory level for long-patch base BER.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique, Université de Strasbourg, Electricité de France and Ligue Contre le Cancer Comité du Haut-Rhin (to V.S.) and Nanosystems Initiative Munich (NIM) and grants from the Deutsche Forschungsgemeinschaft [SFB 646 and 684 (to H.L.)]; Fellowship from Association pour la Recherche sur le Cancer (E.F.). V.S. is an ‘Equipe Labellisée Ligue Contre le Cancer’. Funding for open access charge: Ligue Contre le Cancer.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank F. Dantzer for helpful discussions and D. L. Spector for providing U2OS.2-6-3 cells containing a lac operator array.
REFERENCES
- 1.Hakmé A, Huber A, Dollé P, Schreiber V. The macroPARP genes Parp-9 and Parp-14 are developmentally and differentially regulated in mouse tissues. Dev. Dyn. 2008;237:209–215. doi: 10.1002/dvdy.21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mortusewicz O, Leonhardt H. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol. Biol. 2007;8:81. doi: 10.1186/1471-2199-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl Acad. Sci. USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp. Cell Res. 2007;313:2920–2936. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 11.Niere M, Kernstock S, Koch-Nolte F, Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol. Cell Biol. 2008;28:814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whatcott CJ, Meyer-Ficca ML, Meyer RG, Jacobson MK. A specific isoform of poly(ADP-ribose) glycohydrolase is targeted to the mitochondrial matrix by a N-terminal mitochondrial targeting sequence. Exp. Cell Res. 2009;315:3477–3485. doi: 10.1016/j.yexcr.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl Acad. Sci. USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amé JC, Fouquerel E, Gauthier LR, Biard D, Boussin FD, Dantzer F, de Murcia G, Schreiber V. Radiation-induced mitotic catastrophe in PARG-deficient cells. J. Cell Sci. 2009;122:1990–2002. doi: 10.1242/jcs.039115. [DOI] [PubMed] [Google Scholar]
- 16.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erdelyi K, Bai P, Kovacs I, Szabo E, Mocsar G, Kakuk A, Szabo C, Gergely P, Virag L. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. Faseb J. 2009;23:3553–3563. doi: 10.1096/fj.09-133264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, Favaudon V. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zolghadr K, Mortusewicz O, Rothbauer U, Kleinhans R, Goehler H, Wanker EE, Cardoso MC, Leonhardt H. A fluorescent two-hybrid assay for direct visualization of protein interactions in living cells. Mol. Cell Proteomics. 2008;7:2279–2287. doi: 10.1074/mcp.M700548-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Mortusewicz O, Roth W, Li N, Cardoso MC, Meisterernst M, Leonhardt H. Recruitment of RNA polymerase II cofactor PC4 to DNA damage sites. J. Cell Biol. 2008;183:769–776. doi: 10.1083/jcb.200808097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amé JC, Hakme A, Quenet D, Fouquerel E, Dantzer F, Schreiber V. Detection of the nuclear Poly(ADP-ribose)-metabolizing enzymes and activities in response to DNA damage. Methods Mol. Biol. 2009;464:267–283. doi: 10.1007/978-1-60327-461-6_15. [DOI] [PubMed] [Google Scholar]
- 23.Haince JF, Ouellet ME, McDonald D, Hendzel MJ, Poirier GG. Dynamic relocation of poly(ADP-ribose) glycohydrolase isoforms during radiation-induced DNA damage. Biochim. Biophys. Acta. 2006;1763:226–237. doi: 10.1016/j.bbamcr.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Botta D, Jacobson MK. Identification of a regulatory segment of poly(ADP-ribose) glycohydrolase. Biochemistry. 2010;49:7674–7682. doi: 10.1021/bi100973m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel CN, Koh DW, Jacobson MK, Oliveira MA. Identification of three critical acidic residues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARG catalytic domain. Biochem. J. 2005;388:493–500. doi: 10.1042/BJ20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iida T, Suetake I, Tajima S, Morioka H, Ohta S, Obuse C, Tsurimoto T. PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells. 2002;7:997–1007. doi: 10.1046/j.1365-2443.2002.00584.x. [DOI] [PubMed] [Google Scholar]
- 27.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 29.Stoimenov I, Helleday T. PCNA on the crossroad of cancer. Biochem. Soc. Trans. 2009;37:605–613. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- 30.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashiguchi K, Matsumoto Y, Yasui A. Recruitment of DNA repair synthesis machinery to sites of DNA damage/repair in living human cells. Nucleic Acids Res. 2007;35:2913–2923. doi: 10.1093/nar/gkm115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perucca P, Cazzalini O, Mortusewicz O, Necchi D, Savio M, Nardo T, Stivala LA, Leonhardt H, Cardoso MC, Prosperi E. Spatiotemporal dynamics of p21CDKN1A protein recruitment to DNA-damage sites and interaction with proliferating cell nuclear antigen. J. Cell Sci. 2006;119:1517–1527. doi: 10.1242/jcs.02868. [DOI] [PubMed] [Google Scholar]
- 33.Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- 34.Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 2001;276:32411–32414. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- 35.Keil C, Grobe T, Oei SL. MNNG-induced cell death is controlled by interactions between PARP-1, poly(ADP-ribose) glycohydrolase, and XRCC1. J. Biol. Chem. 2006;281:34394–34405. doi: 10.1074/jbc.M606470200. [DOI] [PubMed] [Google Scholar]
- 36.Petermann E, Ziegler M, Oei SL. ATP-dependent selection between single nucleotide and long patch base excision repair. DNA Repair. 2003;2:1101–1114. doi: 10.1016/s1568-7864(03)00117-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.