Abstract
We have characterized the mitochondrial transcription factor (Mtf1) and RNA polymerase (Rpo41) of Schizosaccharomyces pombe. Deletion mutants show Mtf1 or Rpo41 to be essential for cell growth, cell morphology and mitochondrial membrane potential. Overexpression of Mtf1 and Rpo41 can induce mitochondrial transcription. Mtf1 and Rpo41 can bind and transcribe mitochondrial promoters in vitro and the initiating nucleotides were the same in vivo and in vitro. Mtf1 is required for efficient transcription. We discuss the functional differences between Mtf1 and Rpo41 of S. pombe with Saccharomyces cerevisiae and higher organisms. In contrast to S. cerevisiae, the established model for mitochondrial transcription, S. pombe, a petite-negative yeast, resembles higher organisms that cannot tolerate the loss of mitochondrial function. The S. pombe and human mitochondrial genomes are similar in size and much smaller than that of S. cerevisiae. This is an important first step in the development of S. pombe as an alternative and complementary model system for molecular genetic and biochemical studies of mitochondrial transcription and mitochondrial–nuclear interactions. This is the first systematic study of the cellular function and biochemistry of Rpo41 and Mtf1 in S. pombe.
INTRODUCTION
Mitochondria have their own circular genome. Mitochondrial transcription is accomplished by the mitochondrial RNA polymerase and mitochondrial transcription factors [reviewed in ref. (1–3)]. The mitochondrial RNA polymerase is a single subunit RNA polymerase that is similar to the T3 and T7 bacteriophage RNA polymerases (4). Both the budding yeast Saccharomyces cerevisiae Rpo41 and human (POLRMT) RNA polymerases display high sequence similarity to the C-terminal domain of the phage RNA polymerases (5–7). In contrast to the autonomous bacteriophage polymerases, the mitochondrial RNA polymerases require accessory factors.
In S. cerevisiae, basic mitochondrial transcription is mediated by the mitochondrial RNA polymerase Rpo41 (3,4,8) and the mitochondrial transcription factor Mtf1 (9–11). Disruption of the Mtf1 gene causes a petite phenotype, reduced mitochondrial transcription and mitochondrial DNA copy number (5,10,12). Deletion of Rpo41 also causes a petite phenotype (13). Mtf1 facilitates specific binding of the mitochondrial RNA polymerase to the promoter sequences, like the sigma factors in bacteria; combining with the core RNA polymerase to produce a functional ‘holoenzyme’, to ensure correct initiation of transcription in the mitochondrial genome in S. cerevisiae (11,14–17). Rpo41 can initiate transcription in the absence of Mtf1 under certain conditions (17–19). The crystal structure of Mtf1 showed that it consists of two domains, a large α/β N-terminal domain and a smaller tetra-α helical C-terminal domain separated by a cleft (20). Structurally the N-terminal domain was closely related to a series of rRNA methyltransferases (21–24).
In higher organisms, the mitochondrial RNA polymerase was originally proposed to have three distinct transcription factors: transcription factor A (TFAM) and two protein paralogs of sc-mtTFB; mtTFB1and mtTFB2 (1,2,25–29). TFAM contains two tandem high-mobility group (HMG) boxes (30,31). Human TFAM, mtTFB2 and POLRMT have been shown to initiate transcription from promoter sequences in vitro, and mtTFB2 and POLRMT are sufficient to initiate transcription from promoter sequences alone (32–36). Mouse and human mtTFB1 have been shown to have rRNA methyltransferase activity and appear to play an important role in mitochondrial ribosomal subunit assembly (37–39). RNAi knock-down of Drosophila mtTFB2 in cell culture reduces the abundance of specific mitochondrial transcript RNA and RNAi knock-down of Drosophila mtTFB1 reduces mitochondrial protein synthesis (28,29).
Although the mitochondrial transcription machinery in S. cerevisiae and higher organisms have been extensively studied [reviewed in ref. (1–3)], the mitochondrial RNA polymerase and transcription factors in the fission yeast Schizosaccharomyces pombe have not been identified or characterized. Schizosaccharomyces pombe has been widely used for studies of cell cycle control and differentiation. The S. pombe mitochondrial genome is 19 431 base pairs (bp), in terms of size, it compares favorably with the human mitochondrial genome of 16 569 bp, in contrast to that of S. cerevisiae at 85 709 bp. It provides an alternative and complementary model to budding yeast for the investigation of the molecular mechanisms of mitochondrial transcription (40). Normal mitochondrial function relies on an orchestrated cross-talk between the nucleus and the mitochondrial genes. Identification of the mitochondrial RNA polymerase and transcription factors in S. pombe will also provide a new system for studying mitochondrial-nuclear interactions. We have recently shown that the predicted mitochondrial transcription factor (Mtf1) in S. pombe has a non-mitochondrial function as a nuclear transcription factor that regulates transcription of srk1, a kinase involved in the stress response and cell cycle progression (41). To fully understand this novel nuclear function of a putative mitochondrial transcription factor (Mtf1), it is therefore important to confirm and characterize the mitochondrial transcription factor activity of Mtf1 in S. pombe.
The mitochondrial genome of S. pombe includes genes that encode all of the RNA components of mitochondrial translation, eight protein-coding genes of the respiratory chain, a small ribosomal subunit protein, and three intron encoded DNA binding endonucleases (42). Mitochondrial transcription in S. pombe has been mapped to three promoters: Pma the major promoter where transcription of the mitochondrial chromosome is initiated at the large (21S) ribosomal RNA, an additional minimal promoter Pmi where transcription of the cox3 gene is initiated and Pin which is located downstream of the cob group II intron 5′-splice junction (43).
In this article, we describe the cloning of the S. pombe mitochondrial RNA polymerase, Rpo41 and the mitochondrial transcription factor, Mtf1. We analyzed the phenotypes of Mtf1 or Rpo41 deletion mutant cells and the effect of Mtf1 or Rpo41 deletions on mitochondrial membrane potential. Deletion of Mtf1 or Rpo41 leads to reduced transcription of the mitochondrial genes. Overexpression of Mtf1 or Rpo41 can increase transcription of the mitochondrial genes. We show biochemically, that purified Mtf1 and Rpo41 together can bind to the S. pombe mitochondrial promoters and can support transcription from the S. pombe mitochondrial promoters in vitro. We have used 5′-RACE to map mitochondrial transcription start sites in vivo and in vitro.
MATERIALS AND METHODS
Fission yeast strains, media and techniques
The strains used in this study: 972h− and h−leu1-32 (gift from Jürg Bähler), strain (mtf1Δ1 and mtf1Δ2) mtf1::kan ade6-210/ade6-216 (generated from this study) and rpo41 Δ strain rpo41::kanMX4 ura4-D18 leu1-32 (Bioneer). Plasmid pREP3-Mtf1 and pREP3-Rpo41 were used to transform S. pombe strains. Plasmids pGEX-4T-Mtf1 and pET28a-Rpo41 were generated for the purification of Mtf1 and Rpo41 in E. coli BL21 strain.
PCR primer sequences and oligonucleotide sequences used in this study
Real-time PCR or PCR primer sequences for cloning and sequences of synthetic DNA promoter regions are listed in Supplementary Table S1.
Plasmid construction
For plasmid transformation into S. pombe h−leu1-32 strain, pREP3-Mtf1 was created by inserting PCR fragments of the Mtf1 coding sequence into the SmalI site in pREP3 (44). pREP3-Rpo41 was generated by insertion of a PCR fragment of Rpo41 into the SmalI site of pREP3.
For overexpression and purification of Mtf1 and Rpo41 in E. coli, the Mtf1 coding sequence was ligated into pGEX-4T-1 using the XhoI and EcoRI sites to construct the plasmid pGEX-4T-1-Mtf1 (N-terminal GST tagged Mtf1). The Rpo41 coding sequence was ligated into vector pET28a using the EcoRI and XhoI sites to create pET28a-Rpo41 (N terminal His tagged Rpo41). Clones were confirmed by DNA sequencing.
Disruption of mtf1+
Mtf1 was deleted in diploid cells by using a PCR-based approach using plasmid pFA6a-kanMX6 as the PCR template to generate strain mtf1Δ1 or mtf1Δ2 mtf1::kan ade6-210/ade6-216 [described in ref. (45)]. The primer sequence can be found in Supplementary Table S1. The disruption was confirmed by real-time PCR of the mRNA expression level of the Mtf1 gene beyond the insertion point and by PCR of the genomic DNA using carefully designed primer pairs. The diploid cells were mated and sporulated to generate haploid cells for phenotype analysis.
Cell staining and confocal microscopy
Cells were stained and visualized as described previously (41). Briefly, cells were grown to an OD 600 of 0.5, fixed and stained with 4′,6′-diamidino-2-phenylindole (DAPI) or calcofluor. Unfixed cells were stained directly with DiOC6 (3). Cells were stained with 50 μg/ml of calcofluor in buffer containing 50% glycerol and 0.3 mg/ml p-phenylenediamine. Cells were visualized by confocal microscopy with a Leika TCS-SP5 microscope.
Real-time PCR analysis
Real-time PCR was performed as described previously (41). In brief, reverse transcripts were generated from total cellular RNA, followed by quantitative real-time PCR using an iQ5 Continuous Fluorescence Detector System (Bio-Rad). At least three independent biological repeats and four technical repeats were done for all experiments. Error bars were generated from the standard deviation (calculated by the instrument software). Transcripts were detected for the mitochondrial large (21S) and small (15S) ribosomal RNAs (rns and rnl, respectively) cytochrome oxidase subunits 1, 2 and 3 (cox1, cox2 and cox3), apocytochrome b (cob1), ATP-synthase subunits 6 and 9 (atp6 and atp9), the mitochondrial small subunit ribosomal protein 3 (rps3) and the group I (SPMIT.03) and group II (SPMIT.06) introns that encode DNA binding endonucleases. The housekeeping gene β actin was used as a reference, and α tubulin as an internal control. N.B. Two of the mitochondrial genes (the group I intron encoded SPMIT.02 and atp8) were not detected by real-time PCR presumably because transcripts were too short to detect.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) experiments were carried out as described previously (41), in each reaction, pure GST tagged Mtf1 protein and/or His tagged Rpo41 was incubated with 5′ biotinylated oligonucleotide probes in the presence of competitor DNA (polydI/dC) in binding buffer, and then electrophoresed through a 6% polyacrylamide native gel at 4°C. DNA/protein complexes were transferred to nylon membrane (Hybond-N+) (GE) (46) and detected by chemiluminescence.
Expression and purification of recombinant Mtf1 and Rpo41
GST-tagged Mtf1 was expressed in E. coli BL21 cells from pGEX4T-1-Mtf1. Cells were grown to an OD of 0.5 and induced with 0.1 mM IPTG at 30°C for 6 h. Cell pellets were lysed in PBS buffer (pH 8.0), GST-tagged Mtf1 was purified using Glutathione–Sepharose 4B (GE 17-1969-01). His-tagged Rpo41 was expressed in E. coli BL21 cells from pET28a-Rpo41. Cells were grown to an OD of 0.5 and induced with 0.1 mM IPTG at 16°C for 20 h. His-tagged Rpo41 cell pellets were lysed in binding buffer (20 mM imidazole, 20 mM Na Phosphate, 0.5 M NaCl, pH 7.4, 1 mM phenylmethylsulfonyl fluoride) and purified using Ni2+-NTA–agarose and, after washing with binding buffer with added 20 mM imidazole, was eluted with 500 mM imidazole, 20 mM Na Phosphate, 0.5 M NaCl, pH 7.4. Both Mtf1 and Rpo41 were further purified using a cation exchange (SP) column. The eluate from the GST-column or nickel column was buffer-exchanged on a gel filtration column (HiTrap, GE) with SP binding buffer (20 mM Na Phosphate, 0.1 M NaCl, 1mM DTT, pH 7.4), and then loaded onto the SP column. After a 10 column volume wash with SP-binding buffer, the protein peak was eluted with 20 mM Na Phosphate, 0.5 M NaCl, 1 mM DTT, pH 7.4. The eluate was concentrated by centricon (Millipore), and then desalted by dialysis against SP-binding buffer.
5′-RACE to determine the mitochondrial transcription start sites in vivo
The wild-type cells were grown to an OD of 0.5, 10 ml of culture was used for extraction of total RNA (41). DNA was removed from the RNA sample by treatment with DNase I (Fermentas). Reverse transcription and PCR reactions were carried out using a SMARTTM RACE cDNA Amplification Kit (Clontech). The sequence of PCR primers can be found in Supplementary Table S1. The transcription start site was determined from the DNA sequence.
In vitro transcription and 5′-RACE analysis of RNA transcripts
DNA templates containing the promoters [Pma (738bp), Pmi (597bp) and Pin (431bp)] for in vitro transcription reactions were generated by PCR. A fourth predicted promoter (43) within rps3 was not present in the genome database at GeneDB (http://genedb.org/genedb/pombe/). His-tagged Rpo41 and GST-tagged Mtf1 proteins were expressed and purified in E. coli for in vitro transcription. Transcription reactions were performed using 150 nM of DNA template and 450 nM of Rpo41 in a reaction buffer containing 40 mM Tris (pH 7.9), 20 mM MgCl2, 10 mM DTT, 1 mM nucleotide triphosphates, 2 mM spermidine, 0.01% Triton X-100 and 40 U of RNase inhibitor in a total volume of 50 µl, 450 nM Mtf1 was added to reactions that included Mtf1. Reactions were incubated at 37°C for 3 h, and transcription was terminated by heating at 70°C for 10 min. The mixtures were extracted with phenol/chloroform followed by ethanol precipitation. The DNA template was digested with DNase I at 37°C for 1 h, DNase I was then inactivated at 70°C for 10 min followed by a further phenol/chloroform extraction. Reverse transcription and PCR reactions were carried out as described in the previous section. The sequences of PCR primers are shown in Supplementary Table S1. The transcription start sites were determined from the DNA sequence. This experiment was repeated three times with three different batches of purified Mtf1 and Rpo41.
In vitro transcription and semi-quantitative PCR
In vitro transcriptions were performed as described in the preceding section, in the presence and absence of Mtf1. Control RNA was added to 2 µg of each RNA transcript from in vitro transcription. The mixture of control and transcribed RNA was then reverse transcribed and then subjected to 28 cycles of PCR amplification. The sequence of the control RNA and primer sequences for the Pma, Pmi and Pin promoters can be found in Supplementary Table S1. The PCR products for each lane of the gel were then quantified relative to the control RNA which allowed the relative abundance of transcripts to be compared between transcription reactions and compensates for possible PCR or loading artifacts.
RESULTS
Cloning of mtf1 and rpo41 in fission yeast
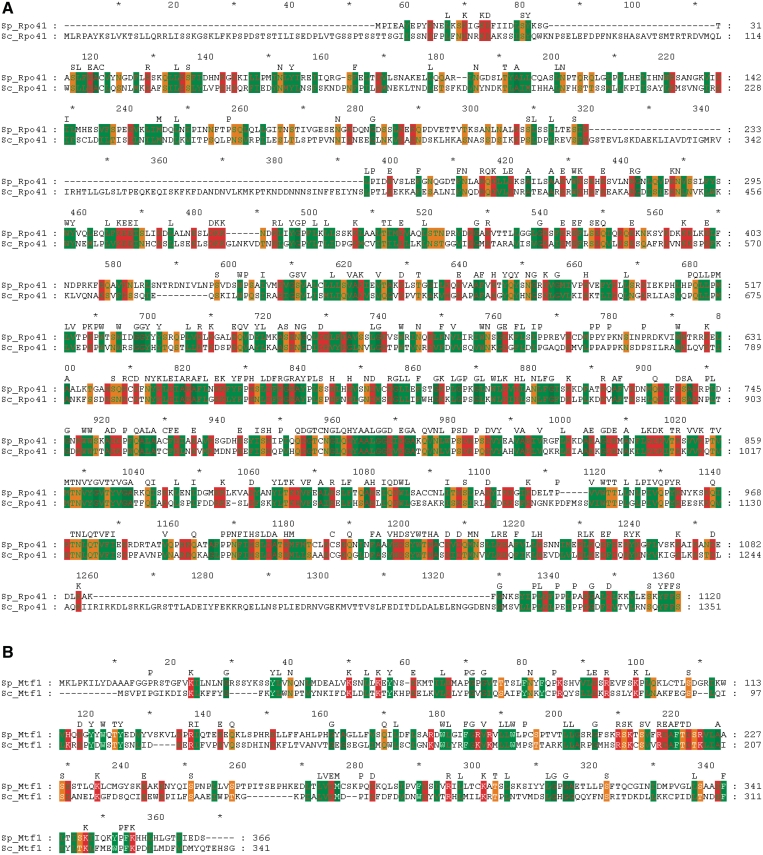
A search of the Welcome Trust Sanger Institute genome database GeneDB (http://genedb.org/genedb/pombe/) identified two putative proteins SPAC26H5.12 and SPAC1002.08c that are predicted by homology to be the mitochondrial RNA polymerase (Rpo41) and the mitochondrial transcription factor (Mtf1) in fission yeast (47). In a genome-wide protein localization study, Mtf1 was reported to localize to the mitochondrion, Rpo41 however, was localized as cytoplasmic dots (48). Mtf1 and rpo41 are encoded on chromosome I of the fission yeast genome. Mtf1 has ∼23% amino acid identity and ∼61% sequence similarity to the Mtf1 of S. cerevisiae and Rpo41 was found to have ∼36% amino acid identity and ∼74% sequence similarity to the Rpo41 of S. cerevisiae (Figure 1A and B) (49). We next cloned mtf1 and rpo41 into protein expression vectors to create plasmid pGEX-4T-Mtf1 (N-terminal GST tagged Mtf1) and pET28a-Rpo41 (N-terminal His tagged Rpo41), respectively (50).
Figure 1.
Alignment of S. pombe Mtf1 and Rpo41 with their S. cerevisiae homologs. Schizosaccharomyces pombe Mtf1 and Rpo41 were aligned with their S. cerevisiae homologs using Clustal W and Clustal X (49). Conserved residues are displayed above the alignment. Similar residues in S. pombe Mtf1 or Rpo41 are highlighted by the amino acid properties such that 1: green on red (DEHKR), 2: green on orange (NQST), 3: red on green (LIVMFYW) and 4: red on dark green (AG). ClustalW alignments were refined using GeneDoc (www.psc.edu/biomed/genedoc).
Mtf1 or Rpo41 are essential in fission yeast
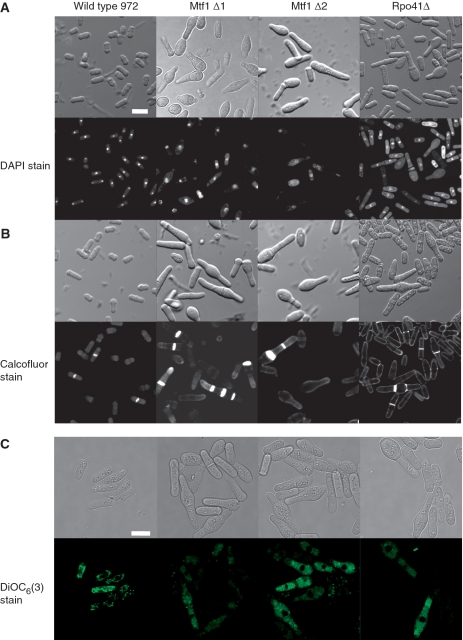
To investigate Mtf1 deletion phenotypes, we generated two mtf1 deletion mutants (mtf1Δ1 and mtf1Δ2) by insertion of a Kanamycin resistance gene into the C- or the N-terminal domain of the Mtf1 coding sequence on the chromosome in a diploid strain. An Rpo41 diploid deletion mutant was supplied by Bioneer. Mtf1 or Rpo41 diploid deletion cells were mated and sporulated, and haploid segregates were visualized by microscopy after germination. S. pombe Mtf1 and Rpo41 deletion mutants appear to share the same phenotypes. Cells lacking Mtf1 (mtf1Δ1 and mtf1Δ2) or Rpo41 (rpo41Δ) can not form colonies or grow in liquid medium and only undergo a few cell divisions after germination on plates. Mtf1Δ1 and mtf1Δ2 or rpo41Δ cells have abnormal cell morphology (Figure 2); they became elongated or enlarged and ‘egg’ or ‘bottle’ shaped compared to wild-type cells. DAPI staining showed some visible nuclear defects in mtf1Δ1 and mtf1Δ2 or rpo41Δ cells (Figure 2A). Calcofluor staining revealed Mtf1 or Rpo41 deletion cells to have abnormal septation including multiple septa (Figure 2B). These phenotypes together suggest that Mtf1 and Rpo41 are essential for cell growth and cell morphology in fission yeast. The mtf1Δ1 and mtf1Δ2 or rpo41Δ deletion phenotypes could be rescued by transformation of plasmids that overexpress Mtf1 or Rpo41 (plasmid pREP3-Mtf1 or pREP3-Rpo41, described in the following section) into the diploid strain. After germination, the haploid cells with Mtf1 or Rpo41 chromosomal deletions were rescued on G418 plates by plasmids that overexpress Mtf1 or Rpo41 (data not shown).
Figure 2.
The phenotype of mtf1 or rpo41 deletions. (A) Wild-type 972 cells and mtf1Δ1 and mtf1Δ2 or rpo41Δ cells grown on YES/YES + G418 plates and cells were fixed and stained with DAPI. The top row shows phase contrast images of unstained cells, the bottom row shows DAPI stained cells viewed by confocal microscope. Bar is 10 μm. (B) Wild-type 972 cells and mtf1Δ1 and mtf1Δ2 or rpo41Δ cells were grown on plates and cells were fixed and stained with Calcofluor. The top row shows phase contrast images of unstained cells, the bottom row shows Calcofluor stained cells viewed by confocal microscope. (C) Wild-type 972 cells, mtf1Δ1, mtf1Δ2 and rpo41Δ cells were stained with the green fluorescent mitochondrial stain DiOC6(3) and cells were observed by confocal microscope. Bar is 10 μm.
To examine the effect of Mtf1 or Rpo41 deletion on the mitochondrion, we performed mitochondrial staining with a fluorescent dye DiOC6 (3) which has been used to detect mitochondrial membrane potential in live cells (51). This dye is a green fluorescent lipophilic cation that can pass through the plasma membrane by the virtue of its lipophilicity and then accumulates in the mitochondrion through the attraction of the positive charge on the molecule to the high negative mitochondrial membrane potential. Mitochondrial staining with DiOC6 (3) showed that wild-type cells have intense green staining with long filamentous structures but the mtf1Δ1, mtf1Δ2 or rpo41Δ cells stain dimly showing dispersed green signals with no structure, suggesting that Mtf1 or Rpo41 deletion resulted in a low mitochondrial membrane potential (Figure 2C).
Mtf1 and Rpo41 are both associated with the transcription of mitochondrial genes in fission yeast
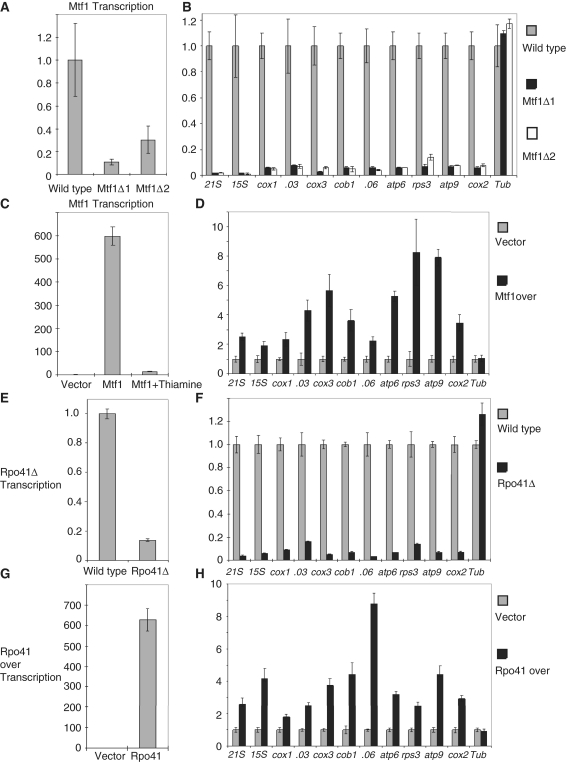
To investigate the role of Mtf1 in the transcription of mitochondrial genes, we performed real-time PCR experiments in the two types of Mtf1 deletion cells to analyze the transcription of nine of the mitochondrial genes (that encode 21S and 12S rRNAs, cox1, cox3, cob1, atp6, rps3, atp9 and cox2) and two of the genes encoded by the group I (SPMIT.03) and group II (SPMIT.06) introns that encode DNA binding endonucleases. We first confirmed that transcript abundance of the mtf1 gene in mtf1Δ1 and mtf1Δ2 cells was reduced to background levels compared to the wild-type cells (Figure 3A). We next showed that transcription of each of the mitochondrial genes was nearly abolished in the mtf1Δ1 and mtf1Δ2 cells compared to wild-type cells, in comparison, transcription of α tubulin is relatively unchanged in wild type and both mtf1 deletion strains (Figure 3B).
Figure 3.
The transcription of mitochondrial genes is associated with Mtf1 and Rpo41. (A) Real-time PCR of mtf1 mRNA abundance in wild-type cells and mtf1Δ1 and mtf1Δ2 cells, showing that mtf1 mRNA abundance is reduced in mtf1Δ1 and mtf1Δ2 cells. (B) Real-time PCR of transcript abundance of the mitochondrial genes in wild-type cells and mtf1Δ1 and mtf1Δ2 cells. NB the intronic genes SPMIT.03 and SPMIT.06 are labeled .03 and .06 for convenience. (C) Real-time PCR of mtf1 mRNA abundance in cells transformed with vector only and cells transformed with pREP3-Mtf1 overexpressing Mtf1 without thiamine and with 60 μM thiamine. (D) Real-time PCR of transcription abundance of the mitochondrial genes in cells transformed with vector only and cells transformed with pREP3- Mtf1 overexpressing Mtf1, showing that transcription of all of the mitochondrial genes is elevated when Mtf1 is overexpressed. (E) Real-time PCR of rpo41 mRNA abundance in wild-type cells and rpo41Δcells. We note that rpo41 mRNA abundance is reduced to background level in rpo41Δcells. (F) Real-time PCR of transcription abundance of the mitochondrial genes in wild-type cells and rpo41Δcells, showing that transcription is reduced in rpo41Δcells. (G) Real-time PCR of rpo41 mRNA abundance in cells transformed with vector only and cells transformed with pREP3-Rpo41 overexpressing Rpo41. (H) Real-time PCR of transcription abundance of the mitochondrial genes in cells transformed with vector only and cells transformed with pREP3-Rpo41 that over expresses Rpo41, showing that transcription of the mitochondrial genes is elevated. Act1 was used as a reference for all of the Real-time PCR experiments and each experiment was biologically repeated at least three times. Error bars were generated from the standard deviation (calculated by the software on the Bio-Rad iQ5 Continuous Fluorescence Detector System) of four technical repeats.
We next tested the effects of Mtf1 over expression on the transcript abundance of mitochondrial genes in S. pombe by real-time PCR. The Mtf1 coding sequence was cloned into the pREP3 vector under the control of the thiamine repressible nmt1 promoter to generate plasmid pREP3-Mtf1. This plasmid was transformed into the h−leu1-32 strain (44) and cells were grown in the absence or in the presence of 60 μM thiamine. Initially we confirmed that mtf1 mRNA abundance in cells over expressing mtf1 was higher than for cells transformed with the pREP3 vector alone. The presence of 60 μM thiamine can repress transcription of mtf1 to minimal levels although this suppression is not complete (Figure 3C). Under these conditions, Mtf1 protein levels were shown to be induced in the absence of thiamine and repressed in the presence of thiamine as described previously (41). This was consistent with the measurement of mtf1 mRNA abundance in cells over expressing mtf1. We next showed that transcription of the mitochondrial genes was elevated in cells over expressing Mtf1 but remained unchanged in cells transformed with the vector only. In contrast, transcription of the nuclear gene α tubulin was unaffected in cells over expressing Mtf1 (Figure 3D). The effect of Mtf1 deletion and over expression on the transcription of mitochondrial genes suggests that Mtf1 does indeed associate with the transcription of mitochondrial genes.
To associate Rpo41 with the transcription of the mitochondrial genome, we performed real-time PCR to detect transcript abundance of the mitochondrial genes in rpo41Δ cells. We first showed that transcription of the rpo41 gene was reduced in the rpo41Δ cells (Figure 3E). Transcription of the mitochondrial genes was also reduced in the rpo41Δ cells compared to wild-type cells, but transcription of the nuclear gene α tubulin remained at similar levels in rpo41Δ and wild-type cells (Figure 3F). We next investigated the effect of Rpo41 over expression on the transcription of the mitochondrial genes. We constructed the plasmid pREP3-Rpo41 by cloning the Rpo41 coding sequence under the control of the thiamine repressible nmt1 promoter into the pREP3 vector, and transformed this plasmid into the h−leu1-32 strain. We tested the transcription of the mitochondrial genes by real-time PCR in cells in which Rpo41 is over expressed in the absence of thiamine. We first confirmed that transcription of rpo41 in cells over expressing rpo41 was higher than for cells transformed with the pREP3 vector alone in the absence of thiamine (Figure 3G). We next showed that transcription of the mitochondrial genes was induced when Rpo41 was over expressed, while the transcription of the nuclear gene α tubulin was not effected by over expression of Rpo41 (Figure 3H). To confirm that the transcriptional induction of the mitochondrial genes was due to the induction of Rpo41 expression we performed the analogous experiment under conditions where Rpo41 was not expressed (in the presence of 60 µM thiamine); we found that transcription of all of the mitochondrial genes remained unchanged (data not shown).
Mtf1 and Rpo41 together bind to the mitochondrial promoters in fission yeast
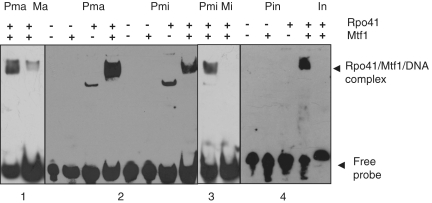
We have shown that Mtf1 and Rpo41 are associated with the transcription of the mitochondrial genes. To further explore the potential interactions between Mtf1, Rpo41 and the mitochondrial promoters biochemically, we performed EMSA using biotin and streptavidin-based detection (52). We over-expressed and purified Mtf1 and Rpo41 using plasmid pGEX-4T-Mtf1 (N-terminal GST tagged Mtf1) and pET28a-Rpo41 (N-terminal His tagged Rpo41) in E. coli BL21 cells (Supplementary Figure S1). Pma, Pmi and Pin promoter sequences (5′ biotinylated oligonucleotide DNA) and purified Mtf1 and Rpo41 were used in EMSA. Mtf1 alone did not cause a detectable shift of the Pma, Pmi and Pin promoter DNA but Rpo41 could generate a weak retarded species in the gel on its own with Pma, Pmi and Pin promoter DNA (for Pin the band shift is too weak to be seen). Mtf1 and Rpo41 together caused a strong super-shift of the probes (Figure 4). However, upon prolonged exposure we have also observed a weak retarded species for Pma, Pmi and Pin promoter DNAs with Mtf1 alone under different conditions (41). We noted that in the presence of the Pma, Pmi and Pin and Rpo41, addition of Mtf1 eliminated the band-shift by Rpo41 alone and caused a strong super-shift, suggesting that Mtf1 and Rpo41 together can bind to the promoters much more efficiently whereas Rpo41 or Mtf1 can only bind weakly to the promoters. To investigate the specificity of the binding that we observed, we generated mutated Pma, Pmi and Pin promoters that contained three-base mutations within the core sequence of the promoters, and performed EMSA with the mutated promoters. Mtf1 and Rpo41 with the mutant promoters exhibited a significantly reduced band shift compared to the wild-type promoters (Figure 4). This suggests that Mtf1 and Rpo41 bind to the promoters specifically and that the core sequences of the promoters are required for recognition by Mtf1 and Rpo41.
Figure 4.
Promoter recognition by Mtf1 and Rpo41. Six percent of polyacrylamide native gels showing EMSA with the mitochondrial promoters Pmi, Pma and Pin in the present of purified Mtf1 or Rpo41 or both proteins together. Note that Mtf1 and Rpo41 together can bind to the promoters more efficiently. The wild-type Pma contains the core promoter sequence ATATATGT, which is mutated to ATCGCTGT (Ma). Similarly, the wild-type Pmi contains the core promoter sequence TTATATGT and Pin contains ATATATGTG, which is mutated ATCGCTGT (Mi or In). Ma, Mi and In all show a reduction in affinity compared the wild-type promoter sequence, indeed for Mi or In, binding is completely abolished.
In vivo mapping of mitochondrial promoters in fission yeast by 5′-RACE
The mitochondrial transcription initiation sites in fission yeast were previously mapped to Pma, Pmi by in vitro capping of RNA with guanylyl-transferase but Pin could not be detected by this method. Pin was detected by primer extension analysis (43). We performed 5′-RACE to examine the mitochondrial transcription initiation sites for Pma, Pmi and Pin in vivo. Total RNA was extracted from cells and treated with DNase I to remove DNA from the RNA sample. RNA was reverse transcribed and the cDNA amplified by 5′-RACE PCR. The transcription start sites were determined by DNA sequencing. Supplementary Figure S2A shows that the 5′-RACE products for Pma, Pmi and Pin are the correct size (153, 250 and 141 bases, respectively). Comparison of the in vivo 5′-RACE products shows that Pma generates the most intense product band, and that Pin generates the least intense product band, which is consistent with previous observations that Pma is the major mitochondrial promoter, Pmi a minor promoter and Pin a weak promoter (43). 5′-RACE products for Pmi, Pma and Pin were gel purified for DNA sequencing. Supplementary Figure S2B–D shows the DNA sequencing results that unambiguously indicate the transcription initiation sites for Pmi, Pma and Pin. The transcription initiation sites for Pmi and Pma are the same as those previously identified through in vitro capping of the RNA. However, for Pin the initial nucleotide of the RNA transcript is a G (Supplementary Figure S2D), and not U as previously described (43). The 5′-RACE technique appears to be more accurate than primer extension analysis for the transcription initiation site of Pin.
In vitro transcription of the mitochondrial promoters by Rpo41/Mtf1 and analysis of RNA transcripts by 5′-RACE
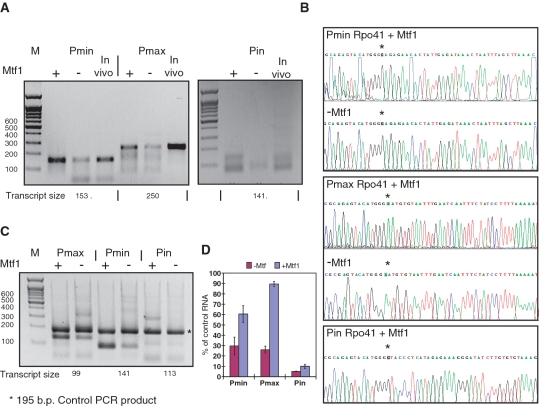
To examine the transcriptional activity of Mtf1 and Rpo41, we performed in vitro transcription with purified Rpo41 in the presence and absence of Mtf1 (Supplementary Figure S1) using the Pma, Pmi and Pin promoters. The DNA templates for Pma, Pmi and Pin promoters were generated by PCR to give template sizes of 738, 597 and 431 base pairs, which in turn gave rise to transcript sizes of 598, 426 and 377 nts, respectively. The RNA transcripts were then subjected to 5′-RACE analysis. The RNA transcripts were reverse transcribed and the cDNA was amplified by PCR using a forward linker primer and a sequence specific reverse primer. 5′-RACE products were then gel purified for DNA sequencing. Figure 5A shows the 5′-RACE products of RNA transcripts generated from the in vitro transcription with and without Mtf1 for Pma, Pmi and Pin promoters. The corresponding cellular RNAs, extracted from cells for 5′-RACE analysis of Pma, Pmi and Pin promoters were used as a control to indicate the size of the in vivo transcripts. The 5′-RACE products of the RNA transcripts synthesized by Rpo41 in vitro are the same size as the control in vivo transcripts in the presence or absence of added Mtf1. Figure 5B shows the DNA sequencing results and indicates that Rpo41/Mtf1 or Rpo41 alone can initiate transcription in vitro. We note that the in vitro start sites for all three promoters are exactly the same as the in vivo mapped start sites for Pma, Pmi and Pin, confirming the transcriptional activity of Mtf1 and Rpo41 in vitro (Figure 5B and Supplementary Figure 2B–D). The 5′-RACE products of Pin transcripts in the absence of Mtf1 were insufficient for DNA sequencing. Although the Rpo41 from both fission and budding yeast resembles the single polypeptide bacteriophage RNA polymerases, we note that both yeast enzymes can tolerate the inclusion of an N-terminal His tag, and remain active in in vitro transcription reactions, in contrast to T7 RNA polymerase (50,53). Compared to Pma and Pmi, in vitro, the Pin promoter is much weaker (Figure 5A), indeed the relative promoter strengths that are observed in vitro are remarkably similar to those that are observed in vivo (Supplementary Figure S2A and Figure 5A). Figure 5A shows that for all three promoter sequences, inclusion of Mtf1 in transcription reactions gave rise to significantly more 5′-RACE PCR product. Although in vitro Rpo41 alone can also initiate transcription in the absence of Mtf1, the presence of Mtf1 significantly improves the efficiency of transcription. To further investigate this, we performed semi quantitative PCR with in vitro RNA transcripts prepared with and without Mtf1. An equal amount of a control RNA was spiked into each RNA sample before the reverse transcription and PCR amplification was performed. The control RNA produces a PCR product of 195 nts. The PCR products corresponding to the RNA transcripts from Pma, Pmi and Pin are 99, 141, 113 nts, respectively (Figure 5C). Inclusion of the control RNA allows the intensity of the PCR products from Pma, Pmi and Pin to be normalized relative to the control RNA, and eliminates possible PCR and loading artifacts, so that relative promoter efficiencies can be compared. Figure 5D shows that Mtf1 increases transcription efficiency for all three promoters in vitro significantly.
Figure 5.
In vitro transcription from the mitochondrial promoters and 5′-RACE analysis of the transcripts. (A) In vitro transcription of Pmi, Pma and Pin promoters by purified Rpo41 was performed with and without Mtf1. RNA transcripts were then used for 5′-RACE analysis. Extracted cellular RNA was used as a control. The 5′-RACE products were run on a 2% agrose gel. The transcript sizes corresponding to the 5′RACE products are labeled at the bottom of the gel. (B) DNA sequence results of 5′-RACE products showing the in vitro transcription initiation site for Pmi, Pma and Pin promoters with and without Mtf1. The position corresponding to the initial nucleotide is marked with an asterisk. (C) Semi quantitative PCR of the RNA transcripts from in vitro transcription of Pmi, Pma and Pin promoters. The PCR products are run on 2% agarose gel. The size of the control PCR product and the transcript sizes for Pmi, Pma and Pin are labeled on the gel. (D) Quantitation of (C) showing that Mtf1 increases Rpo41 transcriptional efficiency.
DISCUSSION
In fission yeast the product of SPAC1002.08c is predicted to be Mtf1. Here we confirm the predicted mitochondrial transcription factor activity of this protein. We have recently shown that Mtf1 has an extra-mitochondrial function as a nuclear transcription factor. As a nuclear transcription factor Mtf1 regulates transcription of srk1, a kinase involved in the stress response and cell cycle progression. Mtf1 localizes to the nucleus, and interacts specifically with the srk1 promoter sequence in vivo and in vitro. Srk1 is induced in cells where Mtf1 is overexpressed and abolished in cells where Mtf1 is deleted. Over expression of Mtf1 causes cell elongation (41). Thus Mtf1 functions as a transcription factor in both the nucleus and the mitochondrion. It was therefore important to confirm the mitochondrial transcription factor activity of Mtf1 and not simply rely on predicted protein functions; and in this context it was also important to characterize its putative mitochondrial partner the RNA polymerase Rpo41.
In S. cerevisiae cells lacking Mtf1 or Rpo41 exhibit a slow-growing or petite-colony phenotype and cannot grow on non-fermentable carbon sources and are respiratory deficient (10,11). We found in this study that S. pombe cells lacking Mtf1 or Rpo41 can not form colonies or grow in liquid medium; they can only undergo several cell divisions on YES plates and display abnormal cell morphology and disturbed mitochondrial membrane potential (Figure 2). Thus the phenotypes that we observe for Mtf1 or Rpo41 deletion mutant cells in S. pombe are different from those reported for S. cerevisiae and reflect the contrasting tolerances for mitochondrial loss between the two organisms. Schizosaccharomyces pombe is a petite-negative yeast, and resembles higher organisms in that it cannot tolerate the loss of mitochondrial function. Disruption of the nuclear encoded mitochondrial DNA polymerase (pog1) is sufficient to deplete mtDNA in S. pombe and pog1Δ cells are defective in mitochondrial function (54), pog1Δ cells have a slow growing phenotype and show similar cell morphology as Mtf1Δ or Rpo41Δ cells. Deletion of Mtf1 or Rpo41 causes a reduction in mitochondrial membrane potential in S. pombe. Thus the phenotypes for Mtf1 and Rpo41 deletion mutants that we observe in this study are consistent with the idea that Mtf1 and Rpo41 have a role in S. pombe mitochondrial function. These data also suggest a link between the role of Mtf1 and Rpo41 in mitochondrial transcription and the aberrant phenotypes observed in the deletion cells. Deletion of either Mtf1 or Rpo41 caused a reduction in transcription of mitochondrial genes to background levels (Figure 3B and F), suggesting that both Mtf1 and Rpo41 are required for the transcription of mitochondrial genes. Significantly, cells in which Mtf1 has been deleted contain normal levels of Rpo41, but mitochondrial transcription remains low, indicating that in the absence of Mtf1, Rpo41 retains no residual RNA polymerase activity in vivo although we observed that Rpo41 alone caused a band shift to the mitochondrial promoters in EMSA and that Rpo41 alone can transcribe the Pma, Pmi and Pin promoters at low levels in vitro (Figure 5A and B). We also showed that over-expression of Mtf1 leads to an increase in mitochondrial transcription (Figure 3D) although in cells that over express Mtf1, Rpo41 levels were limiting. From this we infer that an increase in Mtf1 levels led to an increase in transcriptional efficiency by Rpo41. In S. cerevisiae Mtf1 and Rpo41 combine as a holoenzyme to initiate transcription at the promoter sequence, before Mtf1 is released from the transcription complex (55). In this model, Mtf1 is a component of the Mtf1/Rpo41 holoenzyme for transcriptional initiation and also exists in a free form. An excess of Mtf1 would be predicted to increase the concentration of the ‘holoenzyme’ complex and therefore increase transcriptional initiation at mitochondrial promoters. This was shown to be the case with Mtf1 homologs mtTFB2 in Drosophila (28) and human mtTFB1/mtTFB2 (26). When Mtf1 and Rpo41 are present together, the complex (55) melts the promoter from −4 to +2 without requiring initiating NTPs (19). Direct interactions between S. cerevisiae Mtf1 and its human homolog mtTFB2 and the non-template strand have recently been observed (56–58). The influence of Mtf1 deletion or over expression on mitochondrial transcription in cells is consistent with the notion that Mtf1 is a mitochondrial transcription factor. We next provide biochemical evidence through EMSA and in vitro transcription experiments to show that Mtf1 is a mitochondrial transcription factor and Rpo41 the mitochondrial RNA polymerase (Figures 4, 5A and B). The transcriptional start sites identified by 5′-RACE in vitro map to the same positions as the mitochondrial promoters in vivo, demonstrating the transcriptional activity of Mtf1 and Rpo41 (Supplementary Figure 2B–D and Figure 5B).
A band shift of the mitochondrial promoter DNA by S. pombe Mtf1 could only be detected upon prolonged exposure of the gel in the EMSA experiments. We note that direct interactions between promoter DNA and S. cerevisiae Mtf1 and human mtTFB2 have also been detected (9,19,56–58). Schizosaccharomyces pombe Rpo41 on its own however, can cause a band shift to Pma, Pmi and Pin (Figure 4), we observed that Rpo41 on its own can also initiate low levels of transcription from all three promoters in vitro (Figure 5A). This is in contrast to the situation in S. cerevisiae. Saccharomyces cerevisiae Rpo41 can not recognize and melt the mitochondrial promoter on its own, or initiate RNA synthesis unless the promoter is pre-melted around the transcription start site (18,19). We also note that both Mtf1 and Rpo41 together produce a more intense supershifted band, suggesting that the Mtf1/Rpo41 holoenzyme binds to all three promoter sequences more efficiently. This is also consistent with the observation that Mtf1 overexpression can induce mitochondrial transcription in vivo and in vitro (Figures 3D, 5A and B).
As the first systematic study of the cellular function and biochemistry of Rpo41 and Mtf1 in S. pombe, these studies represent a further significant step in the development of S. pombe as an alternative and complementary model system for molecular genetic and biochemical study of mitochondrial transcription. We anticipate that in the future the convenience and malleability of S. pombe will also establish it as a valuable model system for the study of mitochondrial-nuclear interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Project 985; Shanghai Pujiang Programme and Natural Science Foundation (grant No. 30970061 to D.C. and 31070666 to A.M.); National Key 973 grant (2010CB912602); City of Shanghai Key project grant (09DJ1400601). Funding for open access charge: Laboratory fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Judith Jaehning for critical reading of the manuscript and discussion. Yalin Huang, Xia Sun, Qian Huang, Dihan Shen, Min Shi for help with the confocal microscopy.
REFERENCES
- 1.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Jaehning JA. Mitochondrial transcription: is a pattern emerging? Mol. Microbiol. 1993;8:1–4. doi: 10.1111/j.1365-2958.1993.tb01197.x. [DOI] [PubMed] [Google Scholar]
- 4.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 5.Greenleaf AL, Kelly JL, Lehman IR. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc. Natl Acad. Sci. USA. 1986;83:3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly JL, Lehman IR. Yeast mitochondrial RNA polymerase. Purification and properties of the catalytic subunit. J. Biol. Chem. 1986;261:10340–10347. [PubMed] [Google Scholar]
- 7.Tiranti V, Savoia A, Forti F, D’Apolito MF, Centra M, Rocchi M, Zeviani M. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the expressed sequence tags database. Hum. Mol. Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 8.Shadel GS, Clayton DA. Mitochondrial transcription initiation. Variation and conservation. J. Biol. Chem. 1993;268:16083–16086. [PubMed] [Google Scholar]
- 9.Schinkel AH, Groot Koerkamp MJ, Tabak HF. Mitochondrial RNA polymerase of Saccharomyces cerevisiae: composition and mechanism of promoter recognition. EMBO J. 1988;7:3255–3262. doi: 10.1002/j.1460-2075.1988.tb03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lisowsky T, Michaelis G. Mutations in the genes for mitochondrial RNA polymerase and a second mitochondrial transcription factor of Saccharomyces cerevisiae. Mol. Gen. Genet. 1989;219:125–128. doi: 10.1007/BF00261167. [DOI] [PubMed] [Google Scholar]
- 11.Jang SH, Jaehning JA. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J. Biol. Chem. 1991;266:22671–22677. [PubMed] [Google Scholar]
- 12.Shadel GS, Clayton DA. A Saccharomyces cerevisiae mitochondrial transcription factor, sc-mtTFB, shares features with sigma factors but is functionally distinct. Mol. Cell. Biol. 1995;15:2101–2108. doi: 10.1128/mcb.15.4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Shadel GS. Stability of the mitochondrial genome requires an amino-terminal domain of yeast mitochondrial RNA polymerase. Proc. Natl Acad. Sci. USA. 1999;96:8046–8051. doi: 10.1073/pnas.96.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkley CS, Keller MJ, Jaehning JA. A multicomponent mitochondrial RNA polymerase from Saccharomyces cerevisiae. J. Biol. Chem. 1985;260:14214–14223. [PubMed] [Google Scholar]
- 15.Schinkel AH, Koerkamp MJ, Touw EP, Tabak HF. Specificity factor of yeast mitochondrial RNA polymerase. Purification and interaction with core RNA polymerase. J. Biol. Chem. 1987;262:12785–12791. [PubMed] [Google Scholar]
- 16.Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol. Cell. 2006;22:329–338. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 17.Amiott EA, Jaehning JA. Sensitivity of the yeast mitochondrial RNA polymerase to +1 and +2 initiating nucleotides. J. Biol. Chem. 2006;281:34982–34988. doi: 10.1074/jbc.M608638200. [DOI] [PubMed] [Google Scholar]
- 18.Matsunaga M, Jaehning JA. Intrinsic promoter recognition by a “core” RNA polymerase. J. Biol. Chem. 2004;279:44239–44242. doi: 10.1074/jbc.C400384200. [DOI] [PubMed] [Google Scholar]
- 19.Tang G, Paratkar S, Patel SS. Fluorescence mapping of the open complex of yeast mitochondrial RNA polymerase. J. Biol. Chem. 2009;284:5514–5522. doi: 10.1074/jbc.M807880200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubot FD, Chen CJ, Rose JP, Dailey TA, Dailey HA, Wang BC. Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription. Protein Sci. 2001;10:1980–1988. doi: 10.1110/ps.11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, O’Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 22.O’Farrell HC, Scarsdale JN, Rife JP. Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli. J. Mol. Biol. 2004;339:337–353. doi: 10.1016/j.jmb.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 23.O’Farrell HC, Pulicherla N, Desai PM, Rife JP. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA. 2006;12:725–733. doi: 10.1261/rna.2310406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schluckebier G, Zhong P, Stewart KD, Kavanaugh TJ, Abad-Zapatero C. The 2.2 A structure of the rRNA methyltransferase ErmC’ and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J. Mol. Biol. 1999;289:277–291. doi: 10.1006/jmbi.1999.2788. [DOI] [PubMed] [Google Scholar]
- 25.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 26.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogenhagen DF. Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J. Biol. Chem. 1996;271:12036–12041. [PubMed] [Google Scholar]
- 28.Matsushima Y, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in schneider cells. J. Biol. Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- 29.Matsushima Y, Adán C, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B1 modulates mitochondrial translation but not transcription or DNA copy number in Schneider cells. J. Biol. Chem. 2005;280:16815–16820. doi: 10.1074/jbc.M500569200. [DOI] [PubMed] [Google Scholar]
- 30.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 31.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 32.McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 2003;23:5816–5814. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson N, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 35.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl Acad. Sci. USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metodiev MD, Lesko N, Park CB, Cámara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson N. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 39.Metodiev MD, Lesko N, Park CB, Cámara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson N. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Chiron S, Gaisne M, Guillou E, Belenguer P, Clark-Walker GD, Bonnefoy N. Studying mitochondria in an attractive model: Schizosaccharomyces pombe. Methods Mol. Biol. 2007;372:91–105. doi: 10.1007/978-1-59745-365-3_7. [DOI] [PubMed] [Google Scholar]
- 41.Sun W, Wang Z, Jiang H, Zhang J, Bähler J, Chen D, Murchie AIH. A novel function of the mitochondrial transcription factor Mtf1 in fission yeast; Mtf1 regulates the nuclear transcription of srk1. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1179. Advance access published online 7 December 2010, doi:10.1093/nar/gkq1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bullerwell CE, Leigh J, Forget L, Lang BF. A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 2003;31:759–768. doi: 10.1093/nar/gkg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schäfer B, Hansen M, Lang BF. Transcription and RNA-processing in fission yeast mitochondria. RNA. 2005;11:785–795. doi: 10.1261/rna.7252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 45.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 46.Weser S, Gerlach M, Kwak DM, Czerwinska M, Gödecke A. Detection of TAP-tagged proteins in Western blot, confocal laser scanning microscopy and FACS using the ZZ-domain. J. Biochem. Biophys. Methods. 2006;68:189–194. doi: 10.1016/j.jbbm.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 49.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Matsunaga M, Jang S, Jaehning JA. Expression and purification of wild type and mutant forms of the yeast mitochondrial core RNA polymerase, Rpo41. Protein Expr. Purif. 2004;35:126–130. doi: 10.1016/j.pep.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 51.Pevala V, Kolarov J, Polcic P. Alterations in mitochondrial morphology of Schizosaccharomyces pombe induced by cell-death promoting agents. Folia Microbiol. 2007;52:381–390. doi: 10.1007/BF02932093. [DOI] [PubMed] [Google Scholar]
- 52.MacLachlan TK, El-Deiry WS. Apoptotic threshold is lowered by p53 transactivation of caspase-6. Proc. Natl Acad. Sci. USA. 2002;99:9492–9497. doi: 10.1073/pnas.132241599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller DK, Martin CT, Coleman JE. Processivity of proteolytically modified forms of T7 RNA polymerase. Biochemistry. 1988;27:5763–5771. doi: 10.1021/bi00415a055. [DOI] [PubMed] [Google Scholar]
- 54.Chu Z, Li J, Eshaghi M, Karuturi RKM, Lin K, Liu J. Adaptive expression responses in the Pol-gamma null strain of S. pombe depleted of mitochondrial genome. BMC Genomics. 2007;8:323. doi: 10.1186/1471-2164-8-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mangus DA, Jang SH, Jaehning JA. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J. Biol. Chem. 1994;269:26568–26574. [PubMed] [Google Scholar]
- 56.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paratkar S, Patel SS. Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation. J. Biol. Chem. 2010;285:3949–3956. doi: 10.1074/jbc.M109.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Savkina M, Temiakov D, McAllister WT, Anikin M. Multiple functions of yeast mitochondrial transcription factor Mtf1p during initiation. J. Biol. Chem. 2010;285:3957–3964. doi: 10.1074/jbc.M109.051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.