Abstract
The knowledge that potential guanine quadruplex sequences (PQs) are non-randomly distributed in relation to genomic features is now well established. However, this is for a general potential quadruplex motif which is characterized by short runs of guanine separated by loop regions, regardless of the nature of the loop sequence. There have been no studies to date which map the distribution of PQs in terms of primary sequence or which categorize PQs. To this end, we have generated clusters of PQ sequence groups of various sizes and various degrees of similarity for the non-template strand of introns in the human genome. We started with 86 697 sequences, and successively merged them into groups based on sequence similarity, carrying out 66 clustering cycles before convergence. We have demonstrated here that by using complete linkage hierarchical agglomerative clustering such PQ sequence categorization can be achieved. Our results give an insight into sequence diversity and categories of PQ sequences which occur in human intronic regions. We also highlight a number of clusters for which interesting relationships among their members were immediately evident and other clusters whose members seem unrelated, illustrating, we believe, a distinct role for different sequence types.
INTRODUCTION
The occurrence of potential guanine quadruplex sequence motifs (PQs) within non-telomeric nucleic acids has been the subject of a number of studies (1–14) (for reviews, see refs 15 and 16) and several databases and web resources are available (17–21). Most of the emphasis of these surveys has been to examine the number of PQs and the genomic regions in which they occur. Several studies of individual and specific sequences at a small number of loci have been carried out. In particular, PQs associated with the promoter regions of the c-kit (22–25) and c-myc (26,27) genes have been examined in detail, as well as the 5′-untranslated region (UTR) region in several other genes including N-ras (28) and zic-1 (29). Apart from our initial analysis describing loop sequences within PQ sequences (1), there has been no systematic classification of PQs in terms of their primary sequence. Crystallographic, nuclear magnetic resonance (NMR) and modelling studies have demonstrated that the topology of guanine quadruplexes is very dependent on their primary sequence, as found, for example, in various human telomeric sequences (30–33), and the two c-kit sequences (22–25). Biophysical studies of loop size (34–36) and analyses of the effects of sequence in single-base loops (37) also confirm this conclusion.
From the outset of sequence-based studies into potential quadruplex sequences in non-telomeric nucleic acids, it has been clear that there are more sequences than can be experimentally studied, and to date only a very small fraction of the individual sequences have been examined, although there have been attempts to establish some more general rules governing the energetics of quadruplexes (38). Our initial survey of PQs in the human genome showed that there are 226 157 unique sequences that concur with our search criterion (1). In the same study, we carried out a detailed examination of loop sequences and established that in terms of sequence space, the distribution of loop sequences is far from random, with some being very common and many others not appearing at all. However, examining loop sequences in this way is problematic since, in instances with variable numbers of guanines in the G-tracts and/or isolated guanines in loop sequences, it is not currently possible to determine which guanines are part of the loop and which are part of the G-quartet core, in the absence of relevant experimental data. When more than four G-tracts are present in a sequence, we have the additional problem of determining which of them would participate in a more stable quadruplex structure. We thus need a more practical and robust way of studying quadruplex sequences in detail than trying to derive information from loop sequences alone. In this study, we consider the sequences of potential quadruplex-forming regions as a whole rather than their component parts (G-tracts and loop regions) and describe a method for finding groups of similar sequence. This removes any need to make prior assumptions about topology. Finding many examples of a complex sequence is compelling evidence of positive selection. The possibility therefore exists that quadruplex structure is the reason for such selections. Of the clusters which contain sequence that are proven to form G-quadruplex structures, there is also the possibility that similar sequences may also form similar folding topologies. We have used the non-template strand of introns in the human genome to develop our method and at the same time to produce new data on quadruplex sequences within introns. Our goals are therefore to develop a method to find groups of similar short sequence, apply it to potential guanine quadruplex sequences and, subsequently, determine whether one can find correlations within these groups or something to link the genes in which the sequences occur. In addition, the application of this method can be seen as a hypothesis-generating exercise, as the potential guanine quadruplex clusters can be used as a starting point for further analyses. We have therefore chosen a number of clusters to illustrate that different types of correlation can be found in the clusters.
Hierarchical agglomerative clustering is a method with which one starts with the individual data and merges the most similar (39). The resulting clusters are then successively merged until only one cluster remains. One ends up with a grouping of data in a dendrogram where, at successive levels, the cluster members are less similar. This is schematically represented in Figure 1. In order to cluster nucleic acid sequences in such a way, a similarity metric is needed, and for this, pair-wise sequence alignments were carried out and a similarity score was obtained. Once a similarity metric has been established, there are a number of ways to compare clusters. In this instance, the complete linkage method has been used, where the distance between two clusters is the score of the least-similar, longest distance between any member of one cluster to any member of the other.
Figure 1.
The clustering process begins at the top, where the individual data are treated as clusters. The most similar data are merged, the similarity between the new clusters is derived and the most similar of those are merged until all of the data are in the same cluster.
METHODS
All genomic data were taken from the ENSEMBL database (40) homo_sapiens_core_57_37b and the non-template strand sequences were extracted from the intronic regions for all genes with status ‘known’. The same method was used in earlier studies (1,5) to gather the G-rich sequences with the pattern: G3−5L1−7G3−5L1−7G3−5L1−7G3−5, where G represents guanine bases and L represents any base including guanine. The ENSEMBL perl API was used to extract the genomic regions of interest and in-house software written in C++ used to extract the PQ regions. Regions that had more than four G-tracts were treated as a single sequence. A total of 101 926 potential quadruplex-forming regions were extracted; however, a number of identical sequences were identified in this set, giving 86 697 unique sequences. The sequences were then collated into a mySQL database, along with information about their genomic locations.
Sequence alignments and clustering were carried out using in-house software written in C++. The Smith–Waterman method was used as described by Durbin et al. (41) to carry out the individual alignments. The scoring scheme is quite simple since all mismatches are considered equal. Match = 1, mismatch = 0, gap = −0.5 edge-gap = 0. Edge-gaps are the over-hanging part at the end of the sequences which arise from the fact that the sequences are often of different lengths, so edge-gaps are inevitable and therefore less costly than gaps within the sequence.
The scoring for the clustering was carried out in a different way from that of the pair-wise sequence alignments, since there is a different purpose for each of these. It was done by counting the number of gaps and mismatches dividing by the number of potential matches. The maximum number of matches in any alignment is the length of the shortest sequence. Since gaps in the longer sequence lead to fewer matches, we only penalize gaps on the shorter sequence.
The scoring scheme used is described in the following.
Details of this scheme and our rationale behind it can be found in the Supplementary Data. To avoid confusion these are not being called ‘alignment scores’ because they were not obtained when the alignments were carried out, but rather they are ‘similarity scores’. This now provides a metric of sequence similarity which is independent of the lengths of the sequences involved.
We also obtained more clear-cut results for the clustering by separating the two scoring schemes. The pair-wise sequence alignments were done to find the ‘best’ alignment between the sequences, and the scores for the clustering were calculated so that one pair-wise sequence alignment can be compared with another. It was necessary to score them independently of size since the pair-wise alignments can be of varying size. (Supplementary Figure S1 and Table S4 illustrate how the clustering was biased towards longer sequences being clustered first when using the alignment scores for the clustering from a subset of 1000 sequences chosen at random.)
The scoring scheme that we developed was effectively our definition of sequence similarity and had to compare alignments of various lengths. There are many ways in which we could score our alignments, depending on how we define sequence similarity, which would possibly produce differing results, e.g. scoring mismatches higher than gaps might be sensible if we decided that guanine quadruplex loop length was more important to stability than base composition. However, we wish to assume as little as possible and so have kept the scoring scheme as simple as we can. We believe that the results from the method that we settled on indicates that it fits the purpose well.
To compare whole clusters, full-linkage hierarchical agglomerative clustering was used. When attempting to use single-linkage and mean linkage hierarchical agglomerative clustering, it was found that the clusters were subject to an unacceptable amount of chaining, where unrelated sequences can end up belonging to the same cluster. This method was very computer-intensive since to measure the similarity score between two clusters, every pair of sequences between the two clusters must be aligned. The similarity matrix was too large to be held in computer memory (∼28 Gb of data). It was found that it was faster to pre-compute all of the sequence alignments (3 758 141 556 alignments), calculate the comparison scores and store them on a hard drive, since looking up the scores from the hard drive was faster than carrying out the alignment and obtaining the similarity scores on the fly.
The clustering process went as follows:
Set similarity threshold to 1 and consider each sequence as a cluster.
Compare all clusters and when a pair is found which has a score equal to or better than the similarity threshold, merge them together.
Repeat stage 2 until there are no longer any pairs of clusters at or above the similarity threshold.
Decrease the similarity threshold by 0.05 and go back to stage 2.
The process at stage 2 is traditionally performed by merging the best pair of clusters and re-calculating the similarity scores between the newly formed cluster and the remaining clusters. However, this would have taken an impossibly long time with such a large number of sequences, since cluster comparisons are very costly in terms of computer time. To expedite the process, a coarse-grained approach was adopted which merged many of the clusters in a single cycle and greatly reduced the number of cluster comparisons that were carried out. By decreasing the similarity threshold by 0.05 increments every time, the process was greatly speeded up; we suggest that the outcome was not significantly different from what would have happened if it were practical to cluster by re-calculating the score matrix after every merging. (A comparison of the performance of both methods can be found in Supplementary Figure S2.) The clusters formed at the last cycle before the similarity threshold was dropped, thereby being of most interest. The degree of similarity of the cluster members can be derived from the similarity threshold and hence is related to the cycle number. The earlier in the clustering, the more similar are the cluster members.
Several prominent clusters were chosen and dendrograms drawn with software that was developed in-house, using the Python Imaging Library and the aggdraw module, in the Python programming language. We also carried out multiple sequence alignments between the cluster members for the purposes of illustration using ClustalW (42).
We used FuncAssociate 2.0 (43) which employs the Fisher’s exact test to determine the probability that gene ontology (44) (GO) terms are over-represented [the null hypothesis is that it is unsurprising that the number of any particular GO term appears in the test set (Cluster) by chance]. Since it is not impossible to find false positives when looking for correlations in large sets of data, FuncAssociate calculates an adjusted P-value that includes an estimation of the probability of obtaining at least one false positive. The list of genes belonging to each of the clusters produced by Cycles 5, 9, 13, 17, 20, 23, 26, 29, 32, 36, 39, 42, 45 and 48 whose sequences were associated with more than 10 different genes were sent to the FuncAssociate server. A P-value cut-off of 0.05 was used to determine which clusters were over-represented in any GO term.
RESULTS
Cluster size distribution
Figure 2 shows how the number of clusters decreases at each cycle. The threshold level is also shown and it can be seen at which cycles the similarity threshold was decreased and how this affects the number of clusters. For example, between Cycles 7 and 8 the largest drop in the number of clusters occurs, from 72 106 to 56 049 clusters. The next significant drop, between Cycles 11 and 12 (55 802–39 852 clusters), is almost as large. These are, therefore, the stages with the largest number of clusters merging and coincide with the similarity threshold decreasing from 0.95 to 0.9 and from 0.9 to 0.85.
Figure 2.
The relationship between the similarity threshold and the number of clusters during the progression of the clustering process.
Figure 3 shows the cluster size distribution changing with each cluster cycle, for clusters containing between 1 and 400 sequences and on the final column for clusters larger than 400. The clusters were arranged in bins depending on the number of sequence members which they contained, starting with clusters with 0–10 members, then 10–20 members and so on until the clusters along the right-hand side with 400–86 696 sequence members. The heights represent the total number of sequences within the clusters in a particular bin and each coloured row represents a cluster cycle. It can be seen that, as expected, the sequences are initially distributed among the small clusters (0–10) and it is not until Cycle 5 that there are clusters with greater than 10 sequences in them. By Cycle 27 the number of clusters containing 0–10 sequences is dropping significantly and the sequences are distributed among larger clusters and by Cycle 31 there are clusters which contain more than 400 sequences. As the process of merging clusters continues, the distribution moves to the right until by Cycle 56 there are no longer any clusters below 400 sequences and finally at Cycle 65 the clustering has converged into a single cluster.
Figure 3.
The distribution of cluster members by cluster size and the progression of the clustering process.
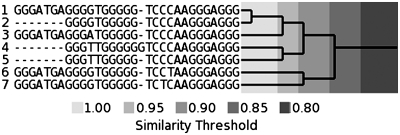
There are a very large number of clusters and to carry out a detailed manual analysis of all of them would be unfeasible. We have therefore taken several clusters and highlighted some interesting features within them. Diagrams of these clusters show the multiple sequence alignments calculated using the program ClustalW next to the dendrograms generated from the clustering data. Many of the groupings in the dendrogram on the right can be correlated to features of the ClustalW-aligned sequences even though they were derived through different means. For example, in Figure 5a sequences 8–10, which share very similar sequences over the first 17 bases, are grouped together much earlier in the clustering process than they are with the rest of the sequences in the cluster which differ in this region.
Figure 5.
(a) Cycle 18 cluster 202 zinc finger type genes 1. (b) Cycle 21 cluster number 4672 zinc finger genes.
Figure 4 and Table 1 show Cycle 27 cluster number 4470, which contains a cluster of human telomere and human telomere-like sequences with the potential to form quadruplex structures. Azzalin et al. (45) and Schoeftner and Blasco (46) showed that telomeres are not transcriptionally silent and that the C-rich strand is transcribed more than the G-rich strand, resulting in r(UUAGGG)n being more abundant than r(CCCUAA)n. These G-rich RNAs can interact with telomeric DNA and also with the telomerase RNA template and thus inhibit the catalytic action of the telomerase enzyme complex. They can also interact with other gene products such as that of SMG which are also involved in the maintenance of telomeres. The clusters that we have here are examples of an area where this new class of RNA could also be transcribed. Although these sequences are within introns, it is not inconceivable that they can exist alone or as part of smaller molecules after splicing and digestion. For example, it was been observed (47) that while in the quadruplex form, G-rich telomeric RNA is immune to digestion by T1 nuclease, which normally cleaves RNA after a single-stranded guanine residue. Further detail on these clusters is given in Supplementary Tables 1S and 2S. Locating telomeric repeats in non-telomeric DNA has been previously observed, albeit not at the sequence level—for example Meyne et al. (48) discussed their distribution in 100 vertebrate species.
Figure 4.
Telomeric like quadruplex sequences.
Table 1.
Telomeric sequence clusters
| Leaf no. | Gene | EnsemblID | From start | To end | Feature |
|---|---|---|---|---|---|
| 1 | BET1L | ENSG00000177951 | 4603 | 6519 | Intron 4–5 |
| 2 | ST8SIA1 | ENSG00000111728 | 7933 | 39 013 | Intron 4–5 |
| MRVI1 | ENSG00000072952 | 28 383 | 31 060 | Intron 1–2 | |
| 3 | BET1L | ENSG00000177951 | 4408 | 6697 | Intron 4–5 |
| 4 | EHD4 | ENSG00000103966 | 2730 | 7842 | Intron 2–3 |
| ARNT2 | ENSG00000172379 | 5495 | 6719 | Intron 3–4 | |
| ARFGAP3 | ENSG00000242247 | 5564 | 18 179 | Intron 1–2 | |
| 5 | BET1L | ENSG00000177951 | 4512 | 6588 | Intron 4–5 |
| BET1L | ENSG00000177951 | 4713 | 6387 | Intron 4–5 | |
| 6 | BET1L | ENSG00000177951 | 4839 | 6279 | Intron 4–5 |
| 7 | NLGN4X | ENSG00000146938 | 39 962 | 81 579 | Intron 2–3 |
| 8 | CBFA2T3 | ENSG00000129993 | 458 | 762 | Intron 8–9 |
| 9 | RP11-40F6.1 | ENSG00000237523 | 8711 | 1055 | Intron 1–2 |
| 10 | RP11-416N4.2 | ENSG00000230506 | 17 796 | 5299 | Intron 1–2 |
| 11 | AC004490.2 | ENSG00000234432 | 18 343 | 8360 | Intron 1–2 |
| 12 | BET1L | ENSG00000177951 | 3916 | 7179 | Intron 4–5 |
| 13 | FAM157C | ENSG00000233013 | 5783 | 6648 | Intron 3–4 |
| 14 | ZNF275 | ENSG00000063587 | 329 | 846 | Intron 3–4 |
| 15 | BET1L | ENSG00000177951 | 3830 | 7298 | Intron 4–5 |
| 16 | BET1L | ENSG00000177951 | 3753 | 7335 | Intron 4–5 |
| 17 | FAM157C | ENSG00000233013 | 5638 | 6804 | Intron 3–4 |
| 18 | CALN1 | ENSG00000183166 | 26 860 | 7885 | Intron 1–2 |
| 19 | RPL23AP82 | ENSG00000184319 | 967 | 2391 | Intron 3–4 |
| RPL23AP7 | ENSG00000226019 | 967 | 2391 | Intron 2–3 | |
| 20 | RP11-218L14.1 | ENSG00000225393 | 8722 | 9458 | Intron 1–2 |
| 21 | ARHGEF3 | ENSG00000163947 | 9469 | 66 708 | Intron 2–3 |
| 22 | BET1L | ENSG00000177951 | 13 701 | 11 400 | Intron 3–4 |
| 23 | KCNJ6 | ENSG00000157542 | 4819 | 120 673 | Intron 2–3 |
| RP1-207H1.1 | ENSG00000231150 | 8043 | 8579 | Intron 1–2 | |
| 24 | CFDP1 | ENSG00000153774 | 60 850 | 29 018 | Intron 5–6 |
| 25 | AL078621.1 | ENSG00000228003 | 969 | 12 303 | Intron 2–3 |
| 26 | AC068541.3 | ENSG00000233897 | 51 142 | 108 760 | Intron 3–4 |
| 27 | BET1L | ENSG00000177951 | 3715 | 7412 | Intron 4–5 |
| 28 | FAM157C | ENSG00000233013 | 5594 | 6887 | Intron 3–4 |
| 29 | SLC8A2 | ENSG00000118160 | 2410 | 762 | Intron 6–7 |
Figure 5a and b and Tables 2 and 3 show clusters which are mainly composed of closely related zinc-finger genes. The members of the cluster in Figure 5a all belong to the same interpro (49) families, IPR001909 Krueppel-associated box and IPR007087 Znf_C2H2. They occur at 13 different locations, with 10 unique sequences. The location of the sequences within the genes is similar for most of these genes, usually about 200–300 bases from the beginning of the first intron. The variable parts of the sequence tend to be outer ‘loops’ while the central GGGAGGG core appears to be conserved. This is also conserved in another very similar cluster shown in Figure 5b. The majority of those genes belong to the same interpro families, IPR001909 Krueppel-associated box and IPR007087 Znf_C2H2. Sequence 7 is shared by two genes which overlap, AC010300.1 and ZNF91. ZNF91 being contained entirely within an intron of AC010300.1. Almost all of these genes are found in the same area of chromosome 19; however, two genes are found in entirely different locations, ZNF107 is found on chromosome 7 and MAP1B is found on chromosome 5. Although MAP1B is an unrelated gene, its expression has been shown to be controlled by the zinc finger gene BCL11A which also belongs to interpro family IPR007087 Znf_C2H2 (50). When looking at the variable and conserved regions, we need to be aware that the search criterion may have an effect on what we see, i.e. be cognizant of the fact that we will always have conserved runs of guanines in the sequences. It may be more instructive to look at the conservation of the loop sequences; however, if the guanine runs are longer than three bases, there is scope for variability around the edges, under our search criterion. The cluster in Figure 5a appears to be more variable in the region of the first loop while the central ‘A’ loop is the same throughout and the third loop ‘TCAT’ has only one difference, a substitution of an adenine for a cytosine. Since the final G-runs are longer than three bases, we see two cases where the guanines are substituted for an adenine and for a thymine.
Table 2.
Cycle 18 cluster 202 zinc finger type genes 1
| Leaf no. | Gene | EnsemblID | From start | To end | Feature |
|---|---|---|---|---|---|
| 1 | ZNF844 | ENSG00000223547 | 300 | 8830 | Intron 1–2 |
| 2 | ZNF491 | ENSG00000177599 | 289 | 5499 | Intron 1–2 |
| 3 | ZNF833 | ENSG00000197332 | 286 | 4031 | Intron 1–2 |
| 4 | ZNF709 | ENSG00000242852 | 247 | 46 621 | Intron 1–2 |
| ZNF564 | ENSG00000196826 | 37 833 | 46 621 | Intron 1–2 | |
| 5 | ZNF709 | ENSG00000242852 | 29 380 | 17 489 | Intron 1–2 |
| ZNF564 | ENSG00000196826 | 66 966 | 17 489 | Intron 1–2 | |
| 6 | ZNF69 | ENSG00000198429 | 279 | 15 324 | Intron 1–2 |
| 7 | ZNF627 | ENSG00000198551 | 228 | 16 643 | Intron 1–2 |
| 8 | ZNF791 | ENSG00000173875 | 211 | 12 383 | Intron 1–2 |
| 9 | ZNF20 | ENSG00000132010 | 290 | 3960 | Intron 1–2 |
| ZNF625 | ENSG00000213297 | 290 | 3960 | Intron 5–6 | |
| 10 | ZNF44 | ENSG00000197857 | 6496 | 15 598 | Intron 4–5 |
Table 3.
Cycle 21 cluster number 4672 zinc finger genes
| Leaf no. | Gene | EnsemblID | From start | To end | Feature |
|---|---|---|---|---|---|
| 1 | AC011477.1 | ENSG00000245381 | 31 177 | 25 473 | Intron 2–3 |
| 2 | ZNF100 | ENSG00000197020 | 279 | 1336 | Intron 1–2 |
| ZNF681 | ENSG00000196172 | 264 | 2906 | Intron 1–2 | |
| 3 | ZNF431 | ENSG00000196705 | 288 | 1051 | Intron 1–2 |
| 4 | ZNF493 | ENSG00000196268 | 286 | 7549 | Intron 1–2 |
| 5 | ZNF492 | ENSG00000229676 | 290 | 18 517 | Intron 1–2 |
| 6 | ZNF85 | ENSG00000105750 | 282 | 10 313 | Intron 1–2 |
| 7 | AC010300.1 | ENSG00000235694 | 71 073 | 70 836 | Intron 9–10 |
| ZNF91 | ENSG00000167232 | 287 | 20 248 | Intron 1–2 | |
| 8 | ZNF254 | ENSG00000213096 | 272 | 18 305 | Intron 1–2 |
| 9 | ZNF738 | ENSG00000172687 | 289 | 2308 | Intron 1–2 |
| 10 | ZNF724P | ENSG00000196081 | 287 | 17 634 | Intron 1–2 |
| 11 | MAP1B | ENSG00000131711 | 41 788 | 26 124 | Intron 2–3 |
| 12 | ZNF588 | ENSG00000196247 | 322 | 12 544 | Intron 1–2 |
Within Cycle 18, several clusters were found to be over-represented in the GO term GO:0003823 ‘antigen binding’. Cycle 18 cluster 13 461 (Figure 6 and Table 4) is one such cluster, which consists mainly of LIR genes (leucocyte immunoglobulin-like receptor). These genes are all found in the same genomic location: region 19q13.4. All but one of the genes in the cluster occur at this location; however, since some of the genes are overlapping, the total number of locations is 11. Cycle 18 cluster 448 (Figure 7 and Table 5) contains a number of sequences which occur within two immunoglobulin genes, IGHA2 and IGHM which contain a number of very similar sequences. A third IGH gene IGHV3-6 is a pseudogene; however, since certain pseudogenes may play an important role in regulation (51,52), this may still be a biologically relevant locus. Three other genes which appear in this cluster, TRIM29, ZNF831 and BRSK2, are unrelated to the immunoglobulins. A similar cluster, Cycle 18 cluster 1086, (Figure 8 and Table 6), contains the same immunoglobulin genes and similar sequence motifs. This also contains three non-immunoglobulin genes KCNK2, SMAD and the same kinase gene as found in the aforementioned cluster, BRSK2. Closer examination of the regions in which these sequences occur in both the immunoglobulin genes and the BRSK2 suggests that they are all part of a larger region of similarity. That we have closely related genes with similar sequences within their introns is perhaps no great surprise; however, the existence of similar sequences within the introns of unrelated genes is an unexpected observation.
Figure 6.
Cycle 18 cluster 1346. Cluster containing sequences that occur chiefly within leukocyte immunoglobulin-like receptor (LIR) genes.
Table 4.
Cycle 18 cluster 13 461. Cluster containing sequence which occur chiefly within leucocyte immunoglobulin-like receptor (LIR) genes
| Leaf no. | Gene | EnsemblID | From start | To end | Feature |
|---|---|---|---|---|---|
| 1 | LILRA6 | ENSG00000244482 | 77 | 147 | Intron 5–6 |
| LILRB3 | ENSG00000204577 | 77 | 147 | Intron 5–6 | |
| LILRB3 | ENSG00000204577 | 18 899 | 1733 | Intron 7–8 | |
| 2 | LILRA1 | ENSG00000104974 | 617 | 3193 | Intron 5–6 |
| LILRB1 | ENSG00000104972 | 21 906 | 35 443 | Intron 2–3 | |
| LILRP2 | ENSG00000240258 | 84 | 146 | Intron 3–4 | |
| AC006293.1 | ENSG00000170858 | 84 | 146 | Intron 4–5 | |
| 3 | LILRB1 | ENSG00000104972 | 77 | 148 | Intron 5–6 |
| 4 | KCNH2 | ENSG00000055118 | 446 | 30 | Intron 8–9 |
| 5 | LILRA2 | ENSG00000239998 | 84 | 147 | Intron 6–7 |
| LILRB1 | ENSG00000104972 | 1525 | 55 824 | Intron 2–3 | |
| 6 | LILRA4 | ENSG00000239961 | 79 | 147 | Intron 5–6 |
| LILRA3 | ENSG00000170866 | 77 | 146 | Intron 9–10 | |
| 7 | AC011515.1 | ENSG00000225370 | 77 | 153 | Intron 2–3 |
Figure 7.
Cycle 18 cluster 448. Cluster containing sequences that occur chiefly in immunoglobulin genes IGHA2 and IGHM.
Table 5.
Cycle 18 cluster 448. Cluster containing sequences which occur in immunoglobulin genes IGHA2 and IGHM
| Leaf no. | Gene | EnsemblID | From start | To end | Feature |
|---|---|---|---|---|---|
| 1 | IGHA2 | ENSG00000211890 | 1459 | 1736 | Intron 1–2 |
| 2 | TRIM29 | ENSG00000137699 | 8326 | 383 | Intron 1–2 |
| 3 | IGHA2 | ENSG00000211890 | 979 | 2231 | Intron 1–2 |
| 4 | IGHA2 | ENSG00000211890 | 472 | 2763 | Intron 1–2 |
| 5 | IGHA2 | ENSG00000211890 | 844 | 2321 | Intron 1–2 |
| 6 | IGHA2 | ENSG00000211890 | 549 | 2701 | Intron 1–2 |
| 7 | IGHA2 | ENSG00000211890 | 1084 | 2106 | Intron 1–2 |
| 8 | IGHA2 | ENSG00000211890 | 1749 | 1486 | Intron 1–2 |
| 9 | IGHV3-6 | ENSG00000233855 | 5773 | 86 881 | Intron 10–11 |
| IGHM | ENSG00000211899 | 2872 | 2301 | Intron 1–2 | |
| 10 | IGHV3-6 | ENSG00000233855 | 3982 | 88 648 | Intron 10–11 |
| IGHM | ENSG00000211899 | 1081 | 4068 | Intron 1–2 | |
| 11 | BRSK2 | ENSG00000174672 | 517 | 3328 | Intron 12–13 |
| 12 | IGHV3-6 | ENSG00000233855 | 5943 | 86 716 | Intron 10–11 |
| IGHV3-6 | ENSG00000233855 | 5983 | 86 676 | Intron 10–11 | |
| IGHM | ENSG00000211899 | 3042 | 2136 | Intron 1–2 | |
| IGHM | ENSG00000211899 | 3082 | 2096 | Intron 1–2 | |
| 13 | ZNF831 | ENSG00000124203 | 15164 | 30 732 | Intron 3–4 |
| 14 | IGHV3-6 | ENSG00000233855 | 4833 | 87 826 | Intron 10–11 |
| IGHV3-6 | ENSG00000233855 | 4884 | 87 775 | Intron 10–11 | |
| IGHV3-6 | ENSG00000233855 | 5274 | 87385 | Intron 10–11 | |
| IGHM | ENSG00000211899 | 1932 | 3246 | Intron 1–2 | |
| IGHM | ENSG00000211899 | 1983 | 3195 | Intron 1–2 | |
| IGHM | ENSG00000211899 | 2373 | 2805 | Intron 1–2 |
Figure 8.
Cycle 18 cluster 1086. Cluster containing sequences that occur chiefly in immunoglobulin genes IGHA2 and IGHM.
Table 6.
Cycle 18 cluster 1086. Cluster containing sequences which occur chiefly in immunoglobulin genes IGHA2 and IGHM
| Leaf no. | Gene | EnsemblID | From start | To end | Feature |
|---|---|---|---|---|---|
| 1 | IGHV3-6 | ENSG00000233855 | 6237 | 86 417 | Intron 10–11 |
| IGHM | ENSG00000211899 | 3336 | 1837 | Intron 1–2 | |
| 2 | IGHA2 | ENSG00000211890 | 2392 | 848 | Intron 1–2 |
| 3 | KCNK2 | ENSG00000082482 | 61 113 | 19 276 | Intron 1–2 |
| 4 | IGHV3-6 | ENSG00000233855 | 4247 | 88 402 | Intron 10–11 |
| IGHM | ENSG00000211899 | 1346 | 3822 | Intron 1–2 | |
| 5 | BRSK2 | ENSG00000174672 | 363 | 3468 | Intron 12–13 |
| 6 | IGHA2 | ENSG00000211890 | 1654 | 1591 | Intron 1–2 |
| 7 | IGHV3-6 | ENSG00000233855 | 4052 | 88 592 | Intron 10–11 |
| IGHM | ENSG00000211899 | 1151 | 4012 | Intron 1–2 | |
| 8 | IGHV3-6 | ENSG00000233855 | 4197 | 88 447 | Intron 10–11 |
| IGHM | ENSG00000211899 | 1296 | 3867 | Intron 1–2 | |
| 9 | SMAD1 | ENSG00000170365 | 10 591 | 14 155 | Intron 2–3 |
Cycle 18 cluster 2 (Figure 9 and Table 7) contains 27 sequences; however, many occur in more than one locus and the sequences in the cluster appear 140 times. Sequences 1 and 5 are the most common, occurring 45 and 51 times, respectively. We used biomart (53) in the ENSEMBL website to retrieve the interpro (49) IDs for the genes involved (full details are given in the Supplementary Data). Of the 88 genes which had interpro mappings, several were related; however, none occurred more than seven times and the genes are distributed over a range of gene families. Among them are kinases, zinc-finger genes, RAB/RAS genes, WD-40 domains and catenins. Many are known to be associated with signal-transduction pathways (RIN3, TBC1D19, RASGRF3, CDK14, ARHGAP6, CTNNA3 and CTNND2, to name a few) and many are involved in mitosis (CENPQ, PARD3B, ALMS1, SPTLC1, etc.). This cluster demonstrates a significant number of genes both related and unrelated, which contain similar and often identical PQ sequences.
Figure 9.
Cycle 18 cluster 2. This shows sequences that occur in unrelated genes.
Table 7.
Cycle 18 cluster 2. Sequences which occur in unrelated genes. List of genes in which each sequence in this cluster occurs
| 1 | CHST9 CTNND2 PDZD2 TBC1D19 PDE4D TRIM5 PLCB1 LRRC9 TMEM170B AP000705.4 LRRK2 SUMF1 NELL1 MEGF10 FBN2 AP003355.2 VPS13B AC090922.1 AC096733.1 RNF150 WDFY4 ALK SLC16A7 GLIPR1L2 SEMA3D EIF4G3 PFTK1 C2orf34 DLG2 F8 RP11-451L9.1 FRMD4B ZNF28 ZNF665 PDE4B AC010132.1 CTB-111H14.1 AC009264.1 COL24A1 RP11-457K10.1 DYTN KCNE4 AC007254.1 RAB3GAP2 RP11-479J7.2 |
| 2 | RIN3 |
| 3 | PTPRD |
| 4 | NBEA FBXL5 KLF12 ADAMTS3 SLC24A3 RASGRF2 BICD1 BEND7 AP000235.2 LACTB2 FAM190A C11orf74 TBCK AMBRA1 PSMC1 TEX11 PPP2R2B KIAA2022 SPTLC1 MAGT1 CTNNA3 ODZ3 UQCRFS1 AC008413.1 MON2 C11orf80 CENPQ NRCAM TRIM77 AC003050.1 ATRNL1 FXYD6 RP11-702L6.4 SLC9A10 STXBP5L RP11-310E22.1 CASC2 AC005582.1 ST6GALNAC3 RP4-630C24.1 LRP1B GALNT13 SLC25A24 RP11-439L18.3 AC018359.2 PTH2R AC079613.1 AC093865.2 RP11-542C10.1 RP11-202K23.1 RP11-479J7.2 |
| 5 | RP11-735B13.1 |
| 6 | PK4P |
| 7 | DLG2 |
| 8 | PTPRD |
| 9 | AF127577.3 AGBL1 AFF2 CYP4B1 AC003090.1 FAM19A3 PARD3B ALMS1P |
| 10 | HERC2 |
| 11 | SLC26A7 |
| 12 | NRSN1 |
| 13 | EFCAB5 TFAP2D |
| 14 | NAV3 |
| 15 | XKR4 |
| 16 | BBOX1 ALOX5 AL592494.3 |
| 17 | C2orf34 |
| 18 | TRPC4 |
| 19 | THSD4 |
| 20 | ARHGAP6 ALMS1 RP11-615J4.4 AC009499.1 MRPL33 |
| 21 | ARL15 ACCN1 PDSS2 JAK1 PDE4B RP3-433F14.1 |
| 22 | RP4-781K5.2 |
| 23 | KIAA0146 |
| 24 | COL5A3 |
| 25 | PDE3B |
| 26 | MBD5 |
| 27 | RP11-202P11.1 |
Using the FuncAssociate tool to characterize gene sets, we discovered that a number of clusters were over-represented in certain GO terms. The results are summarized in Table 8, which shows, for each cycle, the number of clusters whose sequences fell in more than 10 ENSEMBL genes, the number of these which were over-represented in GO terms, the sum of the number of GO terms which were found to be over-represented in each cluster and the percentage of chosen clusters in which were found to be over-represented in GO terms. The percentage of clusters examined which contained over-represented GO terms did not vary dramatically for clusters 5–39 where it began at 12% for Cycle 5 and remained for the most part between 9% and 10%. At cluster 49 it began to rise, 18% for Cycle 49, 24% for Cycle 45 and 48% for Cycle 48. This approximately steady rate at Cycles 5–39 is probably due to the fact that while new clusters are being formed and genes which are associated to common GO terms come together, other clusters which are over-represented in GO terms are being ‘diluted’ and the significance of the over-represented GO terms is being reduced.
Table 8.
Number of clusters whose associated GO terms were found to be over-represented using FuncAssociate
| Cycle number | Clusters with over- represented GO terms | Sum of GO terms over- represented in each cluster | Number of clusters occurring in >10 different genes | % Clusters with over represented GO terms |
|---|---|---|---|---|
| 5 | 16 | 94 | 133 | 12.030075188 |
| 9 | 22 | 120 | 219 | 10.0456621005 |
| 13 | 33 | 201 | 389 | 8.48329048843 |
| 17 | 65 | 291 | 642 | 10.1246105919 |
| 20 | 94 | 367 | 1048 | 8.96946564885 |
| 23 | 169 | 532 | 1768 | 9.55882352941 |
| 26 | 268 | 772 | 2921 | 9.17494008901 |
| 29 | 348 | 862 | 3620 | 9.61325966851 |
| 32 | 311 | 670 | 3167 | 9.82001894537 |
| 36 | 196 | 392 | 1824 | 10.7456140351 |
| 39 | 111 | 240 | 1060 | 10.4716981132 |
| 42 | 86 | 230 | 506 | 16.9960474308 |
| 45 | 54 | 207 | 238 | 22.6890756303 |
| 48 | 50 | 331 | 100 | 50.0 |
The raw cluster data will be available upon request from alan.todd@pharmacy.ac.uk and in the future from a webpage.
DISCUSSION
Introns have often been assumed to be mutationally neutral. However, there is growing interest in blocks of intronic regions which are conserved across species and which have been suggested as candidate areas of trans-acting regulatory regions (54–56). Although we have only examined a single species in the present analysis, the same reasoning can be applied to paraloguous regions as well as orthologues. Indeed, genes which are co-expressed and have a common regulatory mechanism do not necessarily have to be paralogues; the same cis-acting promoter binding motifs, for example, often exist upstream of unrelated genes. From a sequence conservation point of view, it is perhaps more remarkable to find large numbers of similar sequences in unrelated genes as in closely related ones. One could argue that it is possible for similar sequences in closely related genes to be simply passenger sequences which have not yet had time to diverge. In less closely related genes, it could even be argued that mutational cold spots (57) are responsible for some of the conserved sequence. Since we have identified clusters in genes whose members have a range of genetic distances from the closely related zinc finger genes in Figure 5 to the unrelated genes in Figure 9, we feel confident in stating that selective pressure is likely to be responsible for many of the sequence clusters observed here. The range of types of cluster and sequence types suggests that they have many different biological roles.
Eddy and Maizels (4) showed that there was a relationship between gene function and the number of PQ sequences found within those genes. By finding clusters which are over-represented in particular GO terms, we have shown that this type of relationship also applies at the sequence level and we can use the clusters to examine it further.
By comparing the sequences in a multiple sequence alignment, we may see which elements are conserved and which are variable. If the sequence group forms a quadruplex structure then some of these conserved and variable regions may not be critical in quadruplex formation but may be critical bases for molecular recognition. In certain cases, this would be more useful than simply finding quadruplex-dependent positions.
Whether one takes the abundance of similar PQs as evidence of selective pressure or not, the clustering data may still be exploited. For example, one of the key areas of G-quadruplex research currently focuses on developing ligands which block transcription by stabilizing a particular quadruplex sequence. It may be important to know how unique that sequence is in order to provide specificity.
Meaningfulness of clusters
Since the clusters were merged using the full-linkage method, then the similarity threshold will be the lowest score between any pair of sequences in a cluster. At Cycle 16 (where most of the examples are from), the similarity threshold was 0.8. For a comparison of sequences where the shortest sequence is around 24 bases long, similar to the majority of cases in the cluster in Figure 4, the worst alignments would have to contain, for example three mismatches and two gaps which would give a similarity score of 0.808. In practice, the majority of alignments in that cluster are much more similar and this generally appears to be the case.
We have derived clusters of varying similarity and size, which raises the question of what represents a biologically relevant cluster. As the clustering progresses, less similar sequences are added to each cluster and at some stage the members will be merged, which do not have a similar biological role. The point at which this occurs is impossible to determine without knowledge of the role of these sequences or without experimental evidence. In the cases where we have discretely grouped clusters, rather than continuous merging through the clustering process, this should be less of a problem. We suggest that sequence types whose significance is determined in the future may have differing roles and so will require different degrees of similarity. Indeed, the cluster examples which we have presented were chosen because they represent a variety of different types of correlation: clusters which had a correlation with gene ontologies, those which correlated with protein families, clusters which belonged to disparate protein families and an example of a cluster which was found because of a particular interest (TERRA). The TERRA cluster is also an example which contains sequences that are known to form stable DNA and RNA quadruplexes.
The clustering in this study was performed on introns of human genes. It is now possible to examine other regions of genomic DNA with this methodology and search for clusters in, for example UTR regions, promoter regions or exons. The sequences which were clustered here are those which we selected using our criteria of four runs of at least three guanines, separated by loop regions. However, guanine quadruplex structures may not necessarily be formed exclusively from this sequence type. Indeed, in light of a recent structural study by Kuryavyi and Patel (58), we feel that a clustering approach using a yet more general rule for which sequences can potentially form quadruplex structures, will in due course bear fruit. This structure is not the only one to report a G-quadruplex with a topology which involves more than a simple sequence containing G-tracts separated by loop sequences; see in particular the molecular structures of the sequence in the promoter region of the c-kit gene (22–25). Clustering methods can be applied to any group of sequences including, for example those which follow a specific template and those which are generally G-rich.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work has been supported by a programme grant (No. C129/A4489) from Cancer Research UK (to S.N.). Funding for open access charges: CRUK grant.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Antonio L, Bagga P. IEEE Computer Society. 2004. Computational methods for predicting intramolecular G-quadruplexes in nucleotide sequences 3rd International IEEE Computer Society Computational Systems Bioinformatics Conference (CSB 2004) pp. 590–591. [Google Scholar]
- 3.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todd AK, Neidle S. The relationship of potential G-quadruplex sequences in cis-upstream regions of the human genome to SP1-binding elements. Nucleic Acids Res. 2008;36:2700–2704. doi: 10.1093/nar/gkn078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eddy J, Maizels N. Selection for the G4 DNA motif at the 5' end of human genes. Mol. Carcinogenesis. 2009;48:319–325. doi: 10.1002/mc.20496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang L-S, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawal P, Kummarasetti VBR, Ravindran J, Kurmar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1330. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd AK, Haider SM, Parkinson GN, Neidle S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007;35:5799–5808. doi: 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Z, Kong P, Gao Y, Li N. Enrichment of G4 DNA motif in transcriptional regulatory region of chicken genome. Biochem. Biophys. Res. Commun. 2007;354:1067–1070. doi: 10.1016/j.bbrc.2007.01.093. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Du Z, Li N. Extensive selection for the enrichment of G4 DNA motifs in transcriptional regulatory regions of warm blooded animals. FEBS Lett. 2007;581:1951–1956. doi: 10.1016/j.febslet.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 15.Todd AK. Bioinformatics approaches to quadruplex sequence location. Methods. 2007;43:246–251. doi: 10.1016/j.ymeth.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Huppert JL. Quadruplexes in the genome. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge: Royal Society of Chemistry; 2006. pp. 208–227. [Google Scholar]
- 17.Zhang R, Lin Y, Zhang C-T. Greglist: a database listing potential G-quadruplex regulated genes. Nucleic Acids Res. 2007;36:D372–D376. doi: 10.1093/nar/gkm787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostadinov R, Malhotra N, Viotti M, Shine R, D'Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. QuadBase: genome-wide database of G4 DNA occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kikin O, Zappala Z, D'Antonio L, Bagga PS. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;39:D141–D148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikin O, D'Antonio L, Bagga P. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernando H, Reszka AP, Huppert JL, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu ST, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 28.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nature Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Patel DJ. Solution structure of the human telomeric repeat D[Ag3(T2Ag3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 31.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 32.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guédin A, Alberti P, Mergny J-L. Stability of intramolecular quadruplexes: sequence effects in the central loop. Nucleic Acids Res. 2009;37:5559–5567. doi: 10.1093/nar/gkp563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rachwal PA, Brown T, Fox KR. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007;581:1657–1660. doi: 10.1016/j.febslet.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 38.Stegle O, Payet L, Mergny JL, MacKay DJ, Huppert JL. Predicting and understanding the stability of G-quadruplexes. Bioinformatics. 2009;25:i374–i382. doi: 10.1093/bioinformatics/btp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson SC. Hierarchical clustering schemes. Psychometrica. 1967;32:241–254. doi: 10.1007/BF02289588. [DOI] [PubMed] [Google Scholar]
- 40.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durbin R, Eddy SR, Krogh A, Mitchison G. Biological Sequence Analysis. Probabilistic Models of Proteins and Nucleic Acids. UK: Cambridge University Press; 2002. [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 46.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 47.Randall A, Griffith JD. Structure of long telomeric RNA transcripts. J. Biol. Chem. 2009;284:13980–13986. doi: 10.1074/jbc.M900631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyne J, Baker RJ, Hobart HH, Hsu TC, Ryder OA, Ward OG, Wiley JE, Wurster-Hill DH, Yates TL, Moyzis RK. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990;99:3–10. doi: 10.1007/BF01737283. [DOI] [PubMed] [Google Scholar]
- 49.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuoa T-Y, Hongb C-J, Yi-Ping Hsueh Y-P. Bcl11A/CTIP1 regulates expression of DCC and MAP1b in control of axon branching and dendrite outgrowth. Mol. Cell. Neurosci. 2009;42:195–207. doi: 10.1016/j.mcn.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Balakirev ES, Ayala FJ. Pseudogenes: are they “junk” or functional DNA? Annu. Rev. Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- 52.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal - unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hare MP, Palumbi SR. High intron sequence conservation across three mammalian orders suggests functional constraints. Mol. Biol. Evol. 2003;20:969–978. doi: 10.1093/molbev/msg111. [DOI] [PubMed] [Google Scholar]
- 55.Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human–mouse genome comparisons to locate regulatory sites. Nat. Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 56.Dermitzakis ET, Reymond A, Lyle R, Scamuffa N, Ucla C, Deutsch S, Stevenson BJ, Flegel V, Bucher P, Jongeneel CV, et al. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature. 2002;420:578–582. doi: 10.1038/nature01251. [DOI] [PubMed] [Google Scholar]
- 57.Clark AG. The search for meaning in noncoding DNA. Genome Res. 2001;11:1319–1320. doi: 10.1101/gr.201601. [DOI] [PubMed] [Google Scholar]
- 58.Kuryavyi V, Patel DJ. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure. 2010;18:73–82. doi: 10.1016/j.str.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.