Abstract
Group II introns are found in bacteria and cell organelles (plastids, mitochondria) and are thought to represent the evolutionary ancestors of spliceosomal introns. It is generally believed that group II introns are selfish genetic elements that do not have any function. Here, we have scrutinized this assumption by analyzing two group II introns that interrupt a plastid gene (ycf3) involved in photosystem assembly. Using stable transformation of the plastid genome, we have generated mutant plants that lack either intron 1 or intron 2 or both. Interestingly, the deletion of intron 1 caused a strong mutant phenotype. We show that the mutants are deficient in photosystem I and that this deficiency is directly related to impaired ycf3 function. We further show that, upon deletion of intron 1, the splicing of intron 2 is strongly inhibited. Our data demonstrate that (i) the loss of a group II intron is not necessarily phenotypically neutral and (ii) the splicing of one intron can depend on the presence of another.
INTRODUCTION
Expression of a number of genes in plant mitochondria and plastids (chloroplasts) is dependent on removal of intervening sequences (introns) that interrupt the coding sequences. Intron excision from the primary transcript by splicing represents a prerequisite for translation of the messenger RNA (mRNA) into the correct full-length protein. The vast majority of introns in the organellar genomes of seed plants are so-called group II introns (1,2), but a few group I introns have also been found (3–5). Both group I and group II introns represent catalytically active RNAs (ribozymes) and are often also referred to as self-splicing introns (6–10). At least some group I (6,8) and group II (11,12) introns have the ability to undergo self-splicing in vitro in the absence of any help from protein factors. However, this appears not to be the case for most, if not all, group II introns in the organellar genomes of vascular plants. Their splicing seems to be strictly dependent on the assistance of proteinaceous splicing factors (13–15). Most splicing factors for group II introns in plant organellar genomes are encoded in the nuclear genome and post-translationally imported into plastids or mitochondria (14,16–21). However, a few splicing factors, also called intron maturases, are encoded by the plastid or mitochondrial genomes themselves (22,23). Typically, these maturase ORFs reside within intronic sequences suggesting that the splicing factor is produced by translation of the excised intron. In addition to their splicing activity, some group II introns behave as mobile genetic elements and are capable of inserting themselves into intron-less alleles, a process referred to as intron homing (24).
Group II introns adopt a typical six-domain secondary structure (1,25,26,10) and one assumed function of splicing factors is to help fold the intron RNA into the catalytically active secondary and tertiary structures. Splicing involves two sequential transesterification reactions. First, the 2′OH group of the branch-point nucleotide in domain VI performs a nucleophilic attack on the 5′ splice site forming a circular RNA intermediate called the ‘lariat’. Subsequently, the free 3′OH group of the released 5′ exon performs a nucleophilic attack at the 3′ splice site resulting in exon ligation and release of the intron lariat. This chemical mechanism is conserved in the splicing of spliceosomal introns present in the nuclear genomes of all eukaryotes and, therefore, group II introns are thought to represent the evolutionary ancestors of spliceosomal introns (27,28).
Group II introns are considered to be selfish genetic elements that have no other function besides their own removal from the pre-mRNA. Here we show that this assumption is not generally correct. We demonstrate that elimination of a group II intron from a plastid (chloroplast) gene is associated with a severe decline in plant fitness. Our finding that at least some group II introns have a selective value has important implications for the origin and maintenance of introns in evolution.
MATERIALS AND METHODS
Plant material and growth conditions
Tobacco plants (Nicotiana tabacum cv. Petit Havana) were grown under aseptic conditions on agar-solidified Murashige and Skoog medium containing 30 g/l sucrose (29). Transplastomic lines were rooted and propagated on the same medium. For seed production and analysis of plant phenotypes, transplastomic and wild-type plants were grown in soil under standard greenhouse conditions. Seedling phenotypes were analyzed by germination of surface-sterilized seeds on Murashige and Skoog medium containing 500 mg/l spectinomycin. Growth tests were performed by raising wild-type and mutant plants from seeds in soil at 26°C under light intensities ranging from 30 μE m−2 s−1 to 1000 μE m−2 s−1 (photoperiod: 16 h light, 8 h dark). To assess chilling tolerance, seeds were germinated in soil and grown for 21 days at 26°C, before they were transferred to 10°C and further cultivated under a light intensity of 150 μE m−2 s−1.
Construction of plastid transformation vectors
A 6.6-kb EcoRI/XhoI restriction fragment from a previously constructed vector containing the aadA marker gene and a FLAG-tagged ycf3 (30,31) was cloned into a similarly cut pMCS5-derived vector (MoBiTec GmbH, Göttingen, Germany), in which a 1-kb EcoRI/MluI fragment had been integrated to extend the homologous region upstream of ycf3, generating plasmid pKMP21. To modify the ycf3 coding region, a ycf3-containing 2.5-kb EcoRI/SacI fragment was cloned into a pBluescript II KS(+) vector (Stratagene). Intron-free fragments of ycf3 were obtained via polymerase chain reaction (PCR) with primer pairs Pex1EcoRIfor (5′-GCCGCGAATTCTAGAATGGA-3′)/Pex2XcmIrev (5′-TCCACACAAGTAATGGAGAACA-3′) for the Δ intron 1 construct, Pex2XcmIfor (5′-TGTTCTCCATTACTTGTGTGGA-3′)/Pex3BsmBIrev (5′-TAGAAACGTCTCGTGATCTTCAA-3′) for the Δ intron 2 construct, and Pex1EcoRIfor / Pex3BsmBIrev for the Δ intron 1+2 construct using complementary DNA (cDNA) as template. The purified PCR products were first cloned into a TOPO vector (pCR 2.1-TOPO, Invitrogen). The Δ intron 1 and Δ intron 2 fragments were then used to replace the wild-type ycf3 allele in the pBluescript clone using the restriction sites introduced with the PCR primers and finally integrated into pKMP21 as BsmBI/EcoRI fragments. The Δ intron 1+2 fragment was directly transferred from the TOPO vector into pKMP21 as BsmBI/EcoRI fragment. Correctness of the cloning procedures was verified by DNA sequencing.
Plastid transformation and selection of homoplasmic transplastomic tobacco lines
Young leaves from aseptically grown tobacco plants were bombarded with plasmid-coated 0.6 µm gold particles using a PDS1000He Biolistic gun (Bio-Rad). Primary spectinomycin-resistant lines were selected on regeneration medium containing 500 mg/l spectinomycin (32). Spontaneous spectinomycin-resistant plants were eliminated by double selection on medium containing spectinomycin and streptomycin (500 mg/l each). For each transformation construct, several independent transplastomic lines were subjected to two to three additional rounds of regeneration on spectinomycin-containing medium to enrich the transplastome and select for homoplasmic tissue.
Isolation of nucleic acids and hybridization procedures
Total plant DNA was isolated from fresh leaf tissue by a rapid cetyltrimethylammoniumbromide-based mini-prep procedure (33). RNA was extracted using the peqGOLD TriFast™ reagent (Peqlab, Erlangen, Germany) according to the manufacturer’s protocol. For Southern blot analysis, DNA samples (5 μg total DNA) were digested with the restriction enzymes KpnI and SacI, separated by gel electrophoresis in 1% agarose gels, and transferred onto Hybond XL membranes (GE Healthcare) by capillary blotting using standard protocols. A 479-bp PCR product generated by amplification of the psaA coding region using primers PK52psaA (5′-CCAGTTGAGATGGGATATGATTG-3′) and P18psaA (5′-CAGTAACTGGGGGTCTGTGG-3′) and an 895-bp PCR product generated by amplification of the non-coding region upstream of ycf3 using primers P5′ycf3UTR (5′-TTTGTTGTTGAGAACTCCAAAACC-3′) and P3′rps4 (5′-GGGTCGGTTTGAAAATAAATGA-3′) were used as RFLP probes to verify chloroplast transformation. Total cellular RNA samples (3.5 μg total RNA) were electrophoresed in formaldehyde-containing 1% agarose gels and blotted onto Hybond XL membranes. Probes for detection of intron 1 (Pycf3intron1for: 5′-CCTCGATTAATGCAACCTC-3′; Pycf3intron1rev: 5′-TCCAGGAATTAGTCACTTC-3′) or intron 2 (Pycf3intron2for: 5′-ACCTCATACGGCTCAGCAG-3′; Pycf3intron2rev: 5′-CCGTAAAGATCAATTAGCGAG-3′) containing transcripts and for ndhA (PndhAforEx1: 5′-GAAGTCTATGGGATCATATGGATGC-3′; PndhArevEx1: 5′-TGATTGAGCAGCTGCCCGTAGACC-3′) were generated by amplification of the corresponding DNA sequences. An rps12 probe covering all three exons was obtained by PCR amplification with primer pair P184 (5′-CGGTTAGGATCAATCTAAACCAACCC-3′) and P185 (5′-GAGCGTGAAAGGGGTTCAAGAATC-3′) using cDNA as template. PCR-generated probes were purified by agarose gel electrophoresis following extraction of the DNA fragments of interest from excised gel slices using the Nucleospin Extract II kit (Macherey-Nagel) and radiolabeled with α-32P-dCTP using the MegaPrime kit (GE Healthcare). 5′end-labeling of single-stranded oligonucleotides with polynucleotide kinase (PNK) was done by incubation (30 min at 37°C) of 10 pmol oligonucleotide, 30 µCu γ-32P-ATP and 5 U T4 PNK. Oligonucleotides for end-labeling were derived from ycf3 exon 1 (5′-CTCTGTAATAGGTAAATGCCTCTTTTTCTCCTGAAGTTGTCGGAATTACTCGTAATAAGATATTGGCTACAATTGAAAAGGTCTTATCAATAAAATTTCCATTTATCCGTGATCTAGGC-3′), ycf3 exon 2 (5′-GCCCTATATTATAGAGTATATAACTTCGATCATAGGGATCAATTTCTAGTCGCATAGCTTCATAATAATTCTGCAAAGCTTCCGCGTAATTTCCTTCGGATTGAGCCGAC-3′), and atpF exon 2 (5′-GGTACGTAAATGTAACTCGTTGTTCAAACAACTATTCAGAGTTCCTAGAGCTCCTCGTAAGGC-3′). Hybridizations were performed for 4–12 h at 65°C according to standard protocols.
cDNA synthesis and PCR
Two micrograms total leaf RNA was reverse transcribed using random hexanucleotide primer and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. The resulting cDNA templates were amplified according to standard PCR protocols (30 s at 94°C, 30 s at 54°C, 45 s at 72°C; 27 cycles). Primers for semiquantitative RT-PCR analyses were: Pycf3E1for (5′-CCTAGATCACGGATAAATGGA-3′); Pycf3E2rev (5′-TTCGATCATAGGGATCAATTTC-3′); Pycf3E2for (5′-TCGAGCACTAGAACGAAACC-3′); Pycf3E3rev (5′-ACGTCTCGTGATCTTCAACC-3′); Prpl2for (5′-CGTAACCATAGAATACGACC-3′); Prpl2rev (5′-GCTGCTCTAGCTAATTGTCC-3′).
Thylakoid isolation and immunoblot analyses
To detect the FLAG-tagged Ycf3 protein, thylakoid proteins from wild-type and transplastomic plants were isolated from total leaf material using published procedures (34). Equal amounts of thylakoid proteins were electrophoretically separated in 15% sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to Hybond-P polyvinylidene fluoride (PVDF) membranes (GE Healthcare) using the Trans-Blot Cell (BioRad) and a standard transfer buffer (192 mM glycine, 25 mM Tris pH 8.3). Immunoblot detection was performed with anti-FLAG M2 monoclonal antibody (F3165, Sigma–Aldrich) using the enhanced chemiluminescence system (ECL® PLUS system; GE Healthcare).
Physiological measurements
Chlorophyll contents were determined in 80% (v/v) acetone (35). Chlorophyll fluorescence was recorded with a pulse-amplitude modulated fluorimeter (Dual-PAM-100; Heinz Walz, Effeltrich, Germany) on intact plants (grown under either 30 μE m−2 s−1 or 1000 μE m−2 s−1) at room temperature after dark adaptation. The chlorophyll-a fluorescence parameter qL was calculated based on a lake model for PSII-LHCII (36). The contents of photosystem II (PSII), the cytochrome b6f complex (cyt b6f), plastocyanin (PC) and photosystem I (PSI) were determined in thylakoids prepared as described previously (37). Contents of PSI, PSII, PC and cyt b6f were determined by difference absorption spectroscopy as described in detail previously (38,39).
RESULTS
Elimination of introns from the plastid ycf3 gene
The ycf3 gene is present in the plastid (chloroplast) genome of green algae and all vascular plants and encodes an essential assembly factor for photosystem I (PSI;30). Its reading frame is interrupted by two group II introns, whose removal is essential for synthesis of a functional Ycf3 polypeptide and thus for photosynthetic activity. The introns do not contain a reading frame for an intron maturase (splicing factor) as some other group II introns in plant organellar genomes do (40).
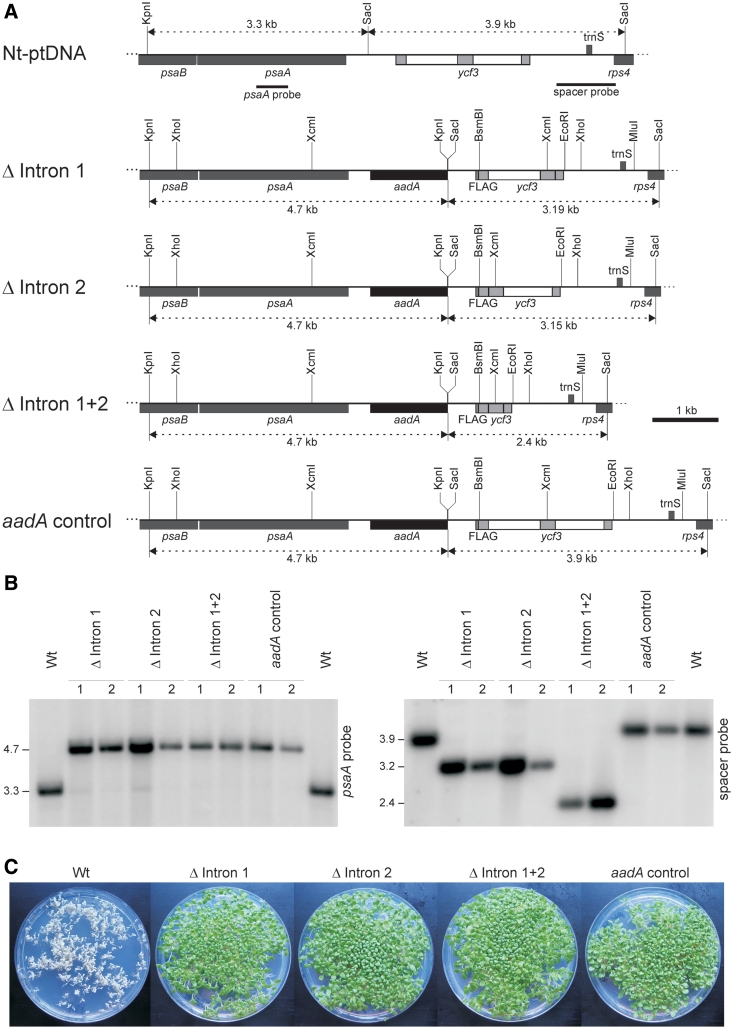
To test if the introns have any function besides their own removal from the pre-mRNA, we constructed three intron deletion alleles of ycf3 (Figure 1A): an allele lacking intron 1 (Δ intron 1), an allele lacking intron 2 (Δ intron 2) and an allele lacking both introns (Δ intron 1+2). The mutant alleles were linked to a selectable marker gene for plastid transformation (aadA) and, as a control, an additional construct carrying the aadA and the wild-type ycf3 allele (aadA control; Figure 1A) was generated. To facilitate immunological detection of the Ycf3 protein, the sequence for a C-terminal FLAG epitope tag was fused to the ycf3 gene. Previous work has shown that tethering the FLAG sequence to the C-terminus of Ycf3 is phenotypically neutral and does not alter Ycf3 function or stability (31).
Figure 1.
Elimination of introns from the chloroplast ycf3 gene. (A) Physical map of the ycf3-containing region in the tobacco plastid genome (Nt-ptDNA) and the four plastid transformation vectors constructed in this study. Genes above the line are transcribed from the left to the right, genes below the line are transcribed in the opposite direction. ycf3 exons are shown as gray boxes, introns as open boxes. Relevant restriction sites, hybridization probes and sizes of DNA fragments appearing in RFLP analyses are indicated. (B) RFLP analysis of transplastomic lines. Fragment sizes are given in kilobasepairs and correspond exactly to the expected sizes [cf. (A)]. Two independently generated transplastomic lines are shown for each construct. (C) Seed assays confirming homoplasmy of the transplastomic lines and maternal inheritance of the plastid-encoded spectinomycin resistance conferred by the aadA transgene. Seeds were germinated on medium containing 500 µg/ml spectinomycin.
All four constructs were introduced into tobacco plants by stable transformation of the chloroplast genome (32). Transgene integration into the plastid genome by homologous recombination was confirmed by DNA gel blot analyses (Figure 1B) and homoplasmy (i.e. the absence of residual wild-type copies of the highly polyploid plastid genome) was additionally confirmed by inheritance assays (Figure 1C; 32,41). For each construct, two independently generated plastid-transformed (transplastomic) lines were selected for in-depth analysis. To exclude the presence of secondary mutations, all transformation vectors and the manipulated region in the plastid genomes of the transplastomic lines were resequenced.
Mutant phenotype of transplastomic plants lacking ycf3 intron 1
For phenotypic analysis, transplastomic plants and control plants were raised from seeds and grown side by side in a controlled-environment chamber. Surprisingly, plants lacking ycf3 intron 1 (Δ intron 1) displayed a pronounced mutant phenotype (Figure 2A). Compared to the wild type and all other transplastomic lines, they were strongly retarded in growth and had light-green leaves, suggesting that the Δ intron 1 plants are impaired in photosynthesis. In contrast, none of the Δ intron 2 and Δ intron 1+2 lines showed any discernable phenotype. Interestingly, the phenotype of the Δ intron 1 mutants was much less pronounced under high-light conditions (Figure 2B), indicating that the presence of intron 1 is especially beneficial, when light availability limits plant growth.
Figure 2.
Phenotypes of transplastomic plants. (A) Phenotype under low-light conditions (30 μE m−2 s−1). (B) Phenotype under high-light conditions (1000 μE m−2 s−1). Note the less pronounced phenotype of the Δ intron 1 mutant, which is solely visible by a slightly delayed development and senescence (i.e. a darker color of the old leaves).
To characterize the physiological basis of the mutant phenotype of the Δ intron 1 plants, chlorophyll content, photosynthetic electron transport and the contents of the components of the photosynthetic electron transport chain (photosystem II: PSII, cytochrome b6f complex: cyt b6f, plastocyanin: PC, photosystem I: PSI) were determined (Table 1; Figure 3). Δ Intron 1 mutants grown under low-light conditions reached only ∼70% of the chlorophyll content of the control plants, explaining their light-green phenotype (Table 1; Figure 2A). Moreover, strongly increased non-photochemical quenching (qN) already at low light intensities (Figure 3A) suggested impaired photochemistry in Δ intron 1 mutants. Determination of the chlorophyll fluorescence parameter qL (correlating with the fraction of PSII reaction centers that are ‘open’, 36) in dependence on the light intensity revealed that, already at low light intensities, the qL values in the Δ intron 1 mutants were drastically reduced compared to the wild type and the two other intron deletions (Figure 3B), indicating strong overreduction of the PSII acceptor side and pointing to a deficiency in one of the downstream components of the electron transport chain. As expected, the differences in qN and qL between the Δ intron 1 mutant and the wild type were less pronounced in plants grown under low-light conditions (Figure 3A and B), due to the earlier saturation of the electron transport system in plants adapted to low light intensities.
Table 1.
Photosynthetic parameters of ycf3 intron deletion mutants grown at low-light conditions (30 µE m−2 s−1; upper part) or high-light conditions (1000 µE m−2 s−1; lower part)
| Parameter | Wild type | aadA control | Δ Intron 1 | Δ Intron 2 | Δ Intron 1+2 | |
|---|---|---|---|---|---|---|
| Chlorophyll per leaf area (mg m−2) | 209.2 ± 9.7 | 221.5 ± 18.7 | 142.6 ± 9.1 | 208.2 ± 13.3 | 210.5 ± 24.8 | 30 µE m−2 s−1 |
| Leaf absorbance (%) | 77.1 ± 1.1 | 77.8 ± 2.4 | 73.0 ± 2.8 | 79.5 ± 2.0 | 78.6 ± 2.9 | |
| FV/FM | 0.80 ± 0.01 | 0.79 ± 0.01 | 0.74 ± 0.04 | 0.78 ± 0.01 | 0.78 ± 0.01 | |
| Chlorophyll/leaf area (mg m−2) | 421.9 ± 59.5 | 447.2 ± 34.9 | 346.1 ± 32.7 | 438.7 ± 18.7 | 416.1 ± 32.1 | 1000 µE m−2 s−1 |
| Leaf absorbance (%) | 89.0 ± 1.6 | 88.5 ± 1.3 | 86.5 ± 1.4 | 88.0 ± 1.4 | 88.5 ± 1.3 | |
| FV/FM | 0.77 ± 0.02 | 0.77 ± 0.02 | 0.75 ± 0.01 | 0.77 ± 0.01 | 0.76 ± 0.03 |
The values represent averages from at least five independent measurements ± standard deviation. FV/FM: maximum quantum efficiency of PSII.
Figure 3.
Analysis of photosynthesis in ycf3 intron deletion mutants and control plants. Data sets are shown for plants grown under high-light conditions (1000 μE m−2 s−1; left panels) and plants grown under low-light conditions (30 μE m−2 s−1; right panels). The values represent averages from five independent measurements/thylakoid isolations per plant line (± standard deviation). (A) Measurement of non-photochemical quenching (qN) in dependence on the light intensity. (B) Measurement of the photochemical quenching parameter qL. (C) Quantitation of the components of the photosynthetic electron transport chain by difference absorption spectroscopy. Data were subjected to one-way analysis of variance (ANOVA) using a pair-wise multiple comparison procedure (Holm–Sidak method) in SigmaPlot. Highly significant differences in PSI contents between the Δ intron 1 mutant and all other plants were observed in both high light and low light (P < 0.001; indicated by asterisks).
Next, we determined the contents of the components of the photosynthetic electron transport chain by spectroscopic methods (39,42). Interestingly, the Δ intron 1 mutants displayed a specific reduction in PSI contents (Figure 3C), raising the possibility that the mutant phenotype is directly related to ycf3 gene function. Consistent with their more severe phenotype, the reduced PSI accumulation in the Δ intron 1 mutants was even more pronounced in low light-grown plants than in high-light grown plants (Figure 3C).
Defective splicing of ycf3 intron 2 in the absence of intron 1
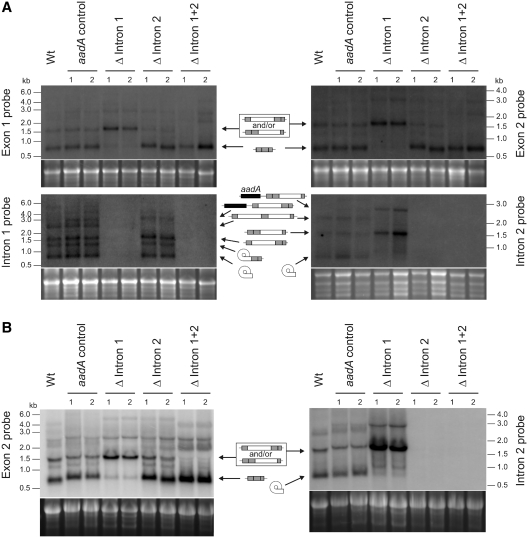
To test if lack of intron 1 affects ycf3 transcript processing or mRNA accumulation, we performed RNA gel blot experiments. Interestingly, hybridizations to ycf3 exon-specific probes revealed that, in the Δ intron 1 mutants, the mature 0.7-kb ycf3 transcript was hardly detectable and, instead, the mutants accumulated a larger transcript of ∼1.5 kb (Figure 4). This larger RNA species corresponds in size to an unspliced precursor that contains intron 2. To test the idea that lack of intron 1 prevents splicing of intron 2, hybridizations with intron-specific probes were conducted. The results confirmed that the Δ intron 1 mutants accumulate the intron 2-containing precursor but not the excised intron 2 and, thus, are defective in intron 2 splicing (Figure 4). The splicing defect was not dependent on the light intensity (Figure 4A and B). This, however, does not contradict the light-dependent phenotype of the mutant plants, because a more severe growth phenotype under low-light conditions is a general property of PSI-deficient mutants (Dominika Bednarczyk,R.B. and M.A.S., manuscript in preparation). Consequently, the phenotypic difference in dependence on the light intensity (Figure 4) is not related to any difference in splicing efficiency, but is solely due to similar reductions in photosystem I levels causing stronger phenotypes in low light than in high light
Figure 4.
Analysis of RNA accumulation and processing patterns of ycf3 in intron deletion mutants and control plants. Northern blots (from denaturing 1% agarose gels) were hybridized to the specific exon or intron probes indicated at the left or right of each autoradiograph. Two independently generated transplastomic lines are shown for each construct. (A) Analysis of plants grown under low-light conditions (30 μE m−2 s−1). The various RNA species accumulating are indicated by the symbols in the middle. Gray boxes denote ycf3 exons, open boxes introns, the black box represents the aadA (present in dicistronic transcripts originating from read-through transcription; 30) and the curved box indicates the lariat intermediate in group II intron splicing; 1). Note that the circularized intermediate resulting from the first transesterification reaction is detectable for intron 1 but not for intron 2, suggesting that in intron 2 splicing, the second transesterification reaction follows more rapidly than in intron 1 splicing. (B) Analysis of intron 2 splicing in plants grown under high-light conditions (1000 μE m−2 s−1).
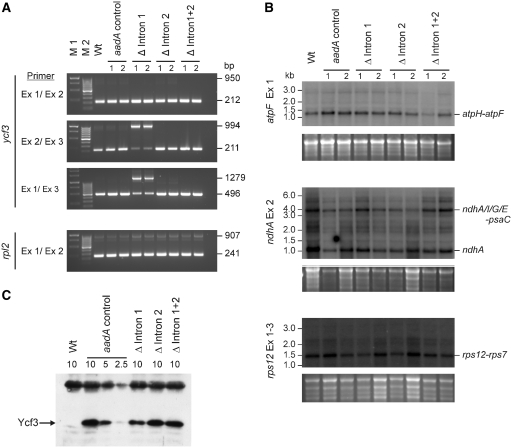
As complete lack of intron 2 excision should be equivalent to a ycf3 knock-out, which would be incapable of photoautotrophic growth (30), we suspected that low levels of spliced ycf3 mRNA accumulate in Δ intron 1 mutants. To confirm this assumption, we used a sensitive semiquantitative RT-PCR assay, in which amplification of the small spliced cDNA product is strongly favored over the amplification of large unspliced molecules. The results demonstrated that indeed some spliced ycf3 mRNA accumulates in the Δ intron 1 mutants (Figure 5A). Control amplifications with rpl2, another plastid gene containing a group II intron, showed that the splicing defect was specific for ycf3 intron 2 (Figure 5A), a conclusion that was further corroborated by the analysis of three additional intron-containing genes by northern blotting: atpF, ndhA and rps12 (Figure 5B).
Figure 5.
Analysis of splicing of intron-containing plastid transcripts and Ycf3 protein accumulation in ycf3 intron deletion mutants. (A) Semiquantitative RT-PCR assays to detect low-level ycf3 intron 2 splicing in Δ intron 1 mutants. Two independently generated transplastomic lines are shown for each construct. cDNA from the intron-containing rpl2 gene was amplified as a control. The specific primers used for RT-PCR are indicated at the left (Ex: exon), sizes of amplification product are given at the right. (B) Analysis of group II intron splicing in the atpF, ndhA and rps12 transcripts by northern blotting. Major spliced RNA species are labeled (with the cistrons they contain) at the right. Note that the mature rps12 mRNA is assembled from three exons (two of which are joined by trans-splicing). (C) Reduced accumulation of the Ycf3 protein in Δ intron 1 mutants. The FLAG-tagged Ycf3 proteins in the transplastomic lines are detected with a monoclonal anti-FLAG antibody. For quantitative comparison, a dilution series of the protein from an aadA control plant is shown (loaded amounts of thylakoid proteins given in µg chlorophyll per lane). The upper cross-reacting band is likely to represent a light-harvesting complex protein and can serve as an additional loading control.
Ycf3 protein deficiency and stress sensitivity of plants lacking ycf3 intron 1
We next wanted to confirm the significance of inhibited intron 2 splicing in Δ intron 1 mutants at the protein level. To this end, we used a monoclonal anti-FLAG antibody and determined the amount of Ycf3 protein accumulating in the mutants. The data revealed that Δ intron 1 mutants accumulate only approximately half the amount of Ycf3 as all other transplastomic lines (Figure 5C). The effect on Ycf3 protein accumulation was much less severe than what could be expected from the strong reduction in mature ycf3 mRNA (Figures 4 and 5A), confirming previous reports that, in plastids, translational regulation can largely override changes in mRNA levels (43). Recent work has revealed that the Ycf3 protein acts in a protein complex that plays a crucial role in PSI assembly. Knockdown of the expression of an essential component of this assembly complex to 20–30% of wild-type levels resulted in a similarly strong decline in PSI accumulation (31), suggesting that the observed Ycf3 deficiency (Figure 5C) is sufficiently severe to explain the phenotype of the Δ intron 1 mutants.
To ultimately confirm the specific PSI deficiency in Δ intron 1 plants, we performed PSI photoinhibition experiments in the cold. Chilling stress is known to cause photoinhibitory damage to PSI (44–46), presumably caused by inhibited superoxide dismutase (SOD) function. Insufficient SOD activity leads to overaccumulation of the reactive oxygen species that are generated as by-products of PSI activity, which in turn causes irreversible photooxidative damage in PSI. Therefore, reduced amounts of PSI should lead to increased sensitivity to chilling stress. Indeed, when grown under chilling stress at 10°C, Δ intron 1 plants bleached out and the developing leaves nearly fully lost their pigmentation (Figure 6), whereas the old leaves (that had already a fully assembled photosynthetic apparatus prior to cold exposure) were much less affected.
Figure 6.
Chilling sensitivity of Δ intron 1 mutants due to increased susceptibility to PSI photoinhibition. Note loss of chlorophyll in developing leaves of the mutant.
A model for dependence of intron 2 excision on intron 1
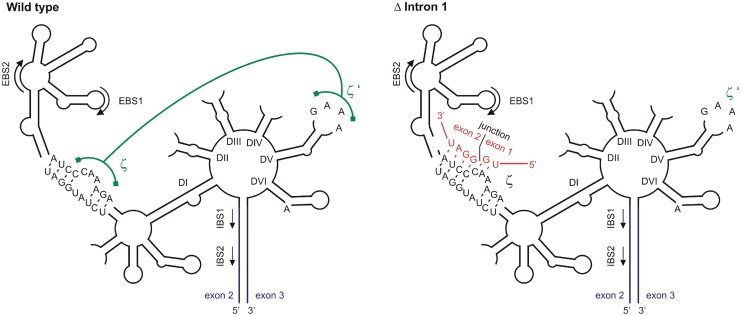
How can lack of a group II intron inhibit the splicing of another, distantly located, intron? The two ycf3 introns are separated by an exon of 230 nt. cis-splicing of group II introns is dependent on the typical six-domain secondary structure of the intron and a few adjacent exonic nucleotides, but does not require distant sequence elements (1,2). We, therefore, considered the conceivable possibility that the lack of intron 1 interferes with the proper folding of intron 2, for example, by inducing the formation of an aberrant secondary structure. If this were the case, the disruptive activity should come from the sequence at the junction between exon 1 and exon 2, because this is the only sequence motif not present in the intron 1-containing (wild-type) ycf3 allele. When we searched for complementarities between the junction sequence and sequences in intron 2, we discovered a 6-nt perfect match with a sequence motif in intron domain I (Figure 7). Such relatively short complementarities are known to be sufficient for long-range base pairing between sequences located on the same RNA molecule (47). Base pairing between the exon junction sequence and the bulge in domain I of intron 2 would disrupt the essential ζ–ζ′ tertiary interaction, a long-range non-Watson–Crick interaction between the GAAA tetraloop in domain V (ζ′) and the ζ bulge in domain I (Figure 7; 48,49). We, therefore, propose that intron 1 is required to prevent masking of the ζ motif in intron 2.
Figure 7.
Model of inhibited intron 2 splicing. The typical six-domain structure of group II introns is shown schematically (1,2,48) for the wild type (left) and the Δ intron 1 mutant (right). The sequence of the junction between exon 1 and exon 2 is shown in red and its possible base pairing with the ζ bulge in domain I (DI) is indicated in the structure for the Δ intron 1 mutant. This base pairing would disrupt the essential tertiary interaction between ζ and ζ′, a GAAA tetraloop in domain V (DV). Exons 2 and 3 are shown in blue, the tertiary interaction between ζ and ζ′ is depicted in green. EBS: exon-binding site; IBS: intron-binding site.
DISCUSSION
In this work, we have tested the idea that group II introns can be required for fitness and are maintained by selective pressure. We have shown that, while deletion of one intron (intron 2) from the plastid gene ycf3 has no apparent effect on plant growth and photosynthetic performance, elimination of another intron (intron 1) had severe consequences for fitness, which were especially pronounced under low light (Figure 2) and under cold stress conditions (Figure 6). As the stability of RNA secondary structures can change with temperature and, moreover, the efficiency of group II intron splicing can be dependent on the growth temperature (50,51), it seems conceivable that the aberrant secondary structure of intron 2 in the Δ intron 1 plants (Figure 7) is even more stable under chilling stress conditions, thus additionally contributing to the severity of the phenotype in the cold. However, although our model provides a plausible mechanistic explanation for the dependence of efficient intron 2 removal on the presence of intron 1, we currently cannot definitively rule out the alternative explanations that (i) a sequence in intron 1 exerts a positive effect on intron 2 splicing or (ii) the negative effect on intron 2 splicing in the absence of intron 1 comes from a different interaction (with other RNA sequences or with proteins, such as the LAGLIDADG-type PPR protein OTP51 shown to be involved in ycf3 splicing; 21).
Interestingly, deletion of both introns abrogated the negative effect of the absence of intron 1, suggesting that the combined loss of both introns could be phenotypically neutral. The loss of the mutant phenotype upon additional elimination of intron 2 supports our model of intron 1 function in intron 2 splicing (Figure 7). It also suggests that the correct folding of group II introns may be more sensitive to changes in remote RNA sequences than previously recognized. This could pose serious restrictions on the spreading of group II introns in organellar and bacterial genomes and may in part explain, why spliceosomal introns, which are considerably less dependent upon RNA folding, have been much more successful in evolution than group II (and group I) introns.
The presence of more than one group II intron in transcripts from a single gene or an operon is quite common in both plastid and mitochondrial genomes. It will be interesting to determine how widespread interdependent intron splicing, sequential splicing and, perhaps, other moonlighting functions of group II introns are. Unfortunately, in the absence of methods for the generation of plants with transformed mitochondrial genomes, stable transformation of the plastid genome is currently the only possible approach to study intron functions in vivo. This involves laborious and time-consuming procedures and thus poses severe restrictions on the systematic probing of group intron function. Preliminary bioinformatics analyses of other pairs of chloroplast introns suggest at least one additional case that could be analogous to the sequential splicing of the two ycf3 introns (Figure 7). The splicing of intron 2 in the clpP gene (encoding the proteolytic subunit of the Clp protease) could be inhibited by prior removal of intron 1 by a very similar aberrant secondary structural interaction with the sequence arising from ligation of exons 1 and 2 (Figure 7). Circumstantial evidence for splicing events occurring in a specific order has also been obtained for the nad5 mRNA in plant mitochondria (52). If trans-splicing of exons c and d does not precede the trans-splicing of exons b and c, extensive mis-splicing takes place that results in the production of aberrant RNAs.
Taken together, our data provide an intriguing example of a group II intron that has a function besides its own removal by facilitating the splicing of another intron. This demonstrates that group II introns can have a selective value in that their loss can cause a decline in fitness. The acquisition of functions that go beyond their own excision may have contributed to the evolutionary maintenance of group II introns, because once an intron has adopted such an additional function, it cannot be lost again without entailing an immediate selective disadvantage. Such selective pressures may have been important, and currently underappreciated, forces not only in the preservation of group II introns, but also in the evolution of spliceosomal introns from group II intron progenitors (28).
FUNDING
Deutsche Forschungsgemeinschaft (SFB 429) and the Max Planck Society. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Stephanie Ruf (MPI-MP) for providing plasmid clones for vector construction, Claudia Hasse and Dr Stephanie Ruf for help with plant transformation and Drs Juliane Neupert, Oliver Drechsel (MPI-MP) and Dr Reimo Zoschke (Humboldt University Berlin) for discussion.
REFERENCES
- 1.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns – a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 2.Bonen L, Vogel J. The ins and outs of group II introns. Trends. Genet. 2001;17:322–323. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- 3.Vaughn JC, Mason MT, Sper-Whitis GL, Kuhlman P, Palmer JD. Fungal origin by horizontal gene transfer of a plant mitochondrial group I intron in the chimeric coxI gene of Peperomia. J. Mol. Evol. 1995;41:563–572. doi: 10.1007/BF00175814. [DOI] [PubMed] [Google Scholar]
- 4.Cho Y, Qiu Y-L, Kuhlman P, Palmer JD. Explosive invasion of plant mitochondria by a group I intron. Proc. Natl Acad. Sci. USA. 1998;95:142244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhsel MG, Stickland R, Palmer JD. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 6.Pyle AM, Cech TR. Ribozyme recognition of RNA by tertiary interactions with specific ribose 2′-OH groups. Nature. 1991;350:628. doi: 10.1038/350628a0. [DOI] [PubMed] [Google Scholar]
- 7.Belfort M. Self-splicing introns in prokaryotes: migrant fossils? Cell. 1991;64:9–11. doi: 10.1016/0092-8674(91)90201-9. [DOI] [PubMed] [Google Scholar]
- 8.Dürrenberger F, Rochaix J-D. Chloroplast ribosomal intron of Chlamydomonas reinhardtii: in vitro self-splicing, DNA endonuclease activity and in vivo mobility. EMBO J. 1991;10:3495–3501. doi: 10.1002/j.1460-2075.1991.tb04913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowira BM, Berzal-Herranz A, Burke JM. Novel RNA polymerization reaction catalyzed by a group I ribozyme. EMBO J. 1994;12:3599–3605. doi: 10.1002/j.1460-2075.1993.tb06033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel F, Ferat J-L. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 11.Winkler M, Kück U. The group IIB intron from the green alga Scenedesmus obliquus mitochondrion: molecular characterization of the in vitro splicing products. Curr. Genet. 1991;20:495–502. doi: 10.1007/BF00334778. [DOI] [PubMed] [Google Scholar]
- 12.Padgett RA, Podar M, Boulanger SC, Perlman PS. The stereochemical course of group II intron self-splicing. Science. 1994;266:1685–1688. doi: 10.1126/science.7527587. [DOI] [PubMed] [Google Scholar]
- 13.Hess WR, Hoch B, Zeltz P, Hübschmann T, Kössel H, Börner T. Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell. 1994;6:1455–1465. doi: 10.1105/tpc.6.10.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins BD, Kulhanek DJ, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel J, Börner T, Hess WR. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003;22:3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watkins KP, Kroeger TS, Cooke AM, Williams-Carrier RE, Friso G, Belcher SE, van Wijk KJ, Barkan A. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell. 2007;19:2606–2623. doi: 10.1105/tpc.107.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small ID. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondria nad1 intron 1 in Arabidopsis thaliana. Plant Cell. 2007;19:3256–3265. doi: 10.1105/tpc.107.054841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asakura Y, Barkan A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell. 2007;19:3864–3875. doi: 10.1105/tpc.107.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 2008;56:157–168. doi: 10.1111/j.1365-313X.2008.03581.x. [DOI] [PubMed] [Google Scholar]
- 22.Thomson MC, Macfarlane JL, Beagley CT, Wolstenholme DR. RNA editing of mat-r transcripts in maize and soybean increases similarity of the encoded protein to fungal and bryophyte group II intron maturases: evidence that mat-r encodes a functional protein. Nucleic Acids Res. 1994;22:5745–5752. doi: 10.1093/nar/22.25.5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Schmitz-Linneweber C. An organellar maturase associates with multiple group II introns. Proc. Natl Acad. Sci. USA. 2010;107:3245–3250. doi: 10.1073/pnas.0909400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belfort M, Perlman PS. Mechanisms of intron mobility. J. Biol. Chem. 1995;270:30237–30240. doi: 10.1074/jbc.270.51.30237. [DOI] [PubMed] [Google Scholar]
- 25.Copertino DW, Hallick RB. Group II and group III introns of twintrons: potential relationship with nuclear pre-mRNA introns. Trends Biochem. Sci. 1993;18:467–471. doi: 10.1016/0968-0004(93)90008-b. [DOI] [PubMed] [Google Scholar]
- 26.Scott WG, Klug A. Ribozymes: structure and mechanism in RNA catalysis. Trends Biochem. Sci. 1996;21:220–223. [PubMed] [Google Scholar]
- 27.Hetzer M, Wurzer G, Schweyen RJ, Mueller MW. Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature. 1997;386:417–420. doi: 10.1038/386417a0. [DOI] [PubMed] [Google Scholar]
- 28.Chalamcharla VR, Curcio MJ, Belfort M. Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry. Genes Dev. 2010;24:827–836. doi: 10.1101/gad.1905010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 30.Ruf S, Kössel H, Bock R. Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 1997;139:95–102. doi: 10.1083/jcb.139.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albus C, Ruf S, Schöttler MA, Lein W, Kehr J, Bock R. Y3IP1, a nucleus-encoded thylakoid protein, co-operates with the plastid-encoded Ycf3 protein in photosystem I assembly. Plant Cell. 2010;22:2838–2855. doi: 10.1105/tpc.110.073908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 34.Machold O, Simpson DJ, Moller BL. Chlorophyll-proteins of thylakoids from wild-type and mutants of barley (Hordeum vulgare L.) Carlsberg Res. Com. 1979;44:235–254. [Google Scholar]
- 35.Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta. 1989;975:384–394. [Google Scholar]
- 36.Kramer DM, Johnson G, Kiirats O, Edwards GE. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth. Res. 2004;79:209–218. doi: 10.1023/B:PRES.0000015391.99477.0d. [DOI] [PubMed] [Google Scholar]
- 37.Schöttler MA, Kirchhoff H, Weis E. The role of plastocyanin in the adjustment of the photosynthetic electron transport to the carbon metabolism in tobacco. Plant Physiol. 2004;136:4265–4274. doi: 10.1104/pp.104.052324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirchhoff H, Mukherjee U, Galla HJ. Molecular architecture of the thylakoid membrane: lipid diffusion space for plastoquinone. Biochemistry. 2002;41:4872–4882. doi: 10.1021/bi011650y. [DOI] [PubMed] [Google Scholar]
- 39.Schöttler MA, Flügel C, Thiele W, Stegemann S, Bock R. The plastome-encoded PsaJ subunit is required for efficient photosystem I excitation, but not for plastocyanin oxidation in tobacco. Biochem. J. 2007;403:251–260. doi: 10.1042/BJ20061573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bock R. Transgenic chloroplasts in basic research and plant biotechnology. J. Mol. Biol. 2001;312:425–438. doi: 10.1006/jmbi.2001.4960. [DOI] [PubMed] [Google Scholar]
- 42.Schöttler MA, Flügel C, Thiele W, Bock R. Knock-out of the plastid-encoded PetL subunit results in reduced stability and accelerated leaf age-dependent loss of the cytochrome b6f complex. J. Biol. Chem. 2007;282:976–984. doi: 10.1074/jbc.M606436200. [DOI] [PubMed] [Google Scholar]
- 43.Eberhard S, Drapier D, Wollman F-A. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002;31:149–160. doi: 10.1046/j.1365-313x.2002.01340.x. [DOI] [PubMed] [Google Scholar]
- 44.Kudoh H, Sonoike K. Irreversible damage to photosystem I by chilling in the light: cause of the degradation of chlorophyll after returning to normal growth temperature. Planta. 2002;215:541–548. doi: 10.1007/s00425-002-0790-9. [DOI] [PubMed] [Google Scholar]
- 45.Zhang S, Scheller HV. Photoinhibition of photosystem I at chilling temperature and subsequent recovery in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1595–1602. doi: 10.1093/pcp/pch180. [DOI] [PubMed] [Google Scholar]
- 46.Rogalski M, Schöttler MA, Thiele W, Schulze WX, Bock R. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell. 2008;20:2221–2237. doi: 10.1105/tpc.108.060392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirose T, Sugiura M. Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J. 1997;16:6804–6811. doi: 10.1093/emboj/16.22.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai L, Chai D, Gu S-Q, Gabel J, Noskov SY, Blocker FJH, Lambowitz AM, Zimmerly S. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol. Cell. 2008;30:472–485. doi: 10.1016/j.molcel.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karcher D, Bock R. Temperature sensitivity of RNA editing and intron splicing reactions in the plastid ndhB transcript. Curr. Genet. 2002;41:48–52. doi: 10.1007/s00294-002-0278-y. [DOI] [PubMed] [Google Scholar]
- 51.Landau AM, Lokstein H, Scheller HV, Lainez V, Maldonado S, Prina AR. A cytoplasmically inherited barley mutant is defective in photosystem I assembly due to a temperature-sensitive defect in ycf3 splicing. Plant Physiol. 2009;151:1802–1811. doi: 10.1104/pp.109.147843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elina H, Brown GG. Extensive mis-splicing of a bi-partite plant mitochondrial group II intron. Nucleic Acids Res. 2009;38:996–1008. doi: 10.1093/nar/gkp994. [DOI] [PMC free article] [PubMed] [Google Scholar]