Abstract
Translation initiation on most eukaryotic mRNAs occurs via a cap-dependent scanning mechanism and its efficiency is modulated by their 5′-untranslated regions (5′-UTR). The human immunodeficiency virus type 1 (HIV-1) 5′-UTR contains a stable TAR hairpin directly at its 5′-end, which possibly masks the cap structure. In addition, the 5′-UTR is relatively long and contains several stable RNA structures that are essential for viral replication. These characteristics may interfere with ribosomal scanning and suggest that translation is initiated via internal entry of ribosomes. Literature on the HIV-1 5′-UTR-driven translation initiation mechanism is controversial. Both scanning and internal initiation have been shown to occur in various experimental systems. To gain further insight in the translation initiation process, we determined which part of the 5′-UTR is scanned. To do so, we introduced upstream AUGs at various positions across the 5′-UTR and determined the effect on expression of a downstream reporter gene that was placed under control of the gag start codon. This strategy allowed us to determine the window of ribosomal scanning on the HIV-1 5′-UTR.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) particles contain two identical full-length positive-strand RNA molecules as genome. The full-length RNA not only serves as the viral genome, but also functions as an mRNA to encode the Gag and Gag-Pol polyproteins. The highly structured 5′-UTR is the most conserved part of the HIV-1 genome and is involved in several steps of the viral replication cycle (1). Distinct functions have been assigned to individual sequence and/or structure motifs (presented in different colours in Figure 1A). The 5′-UTR starts with the stable TAR and the polyadenylation (polyA) hairpins. The well-characterized TAR hairpin mediates transcription activation by binding the viral Tat protein and the cellular protein cyclin T (2–10). The polyA hairpin inhibits premature polyadenylation of the nascent RNA by masking the AAUAAA polyadenylation signal (11,12). The U5 region is located downstream of the polyA signal and contains important signals for reverse transcription, such as the primer binding site (PBS) and the primer activation signal (PAS) (13). Additional motifs are located further downstream in the 5′-UTR. These include the RNA dimer initiation signal (DIS), the major splice donor site (SD) required for the generation of subgenomic mRNAs and the RNA packaging signal (Ψ) (14–22). Previous work identified an evolutionary conserved long-distance interaction between U5 sequences and the Gag initiation codon known as the U5-AUG duplex (23,24). This duplex does not modulate the level of HIV-1 mRNA translation (25), but affects RNA dimerization and packaging into virus particles (26,27).
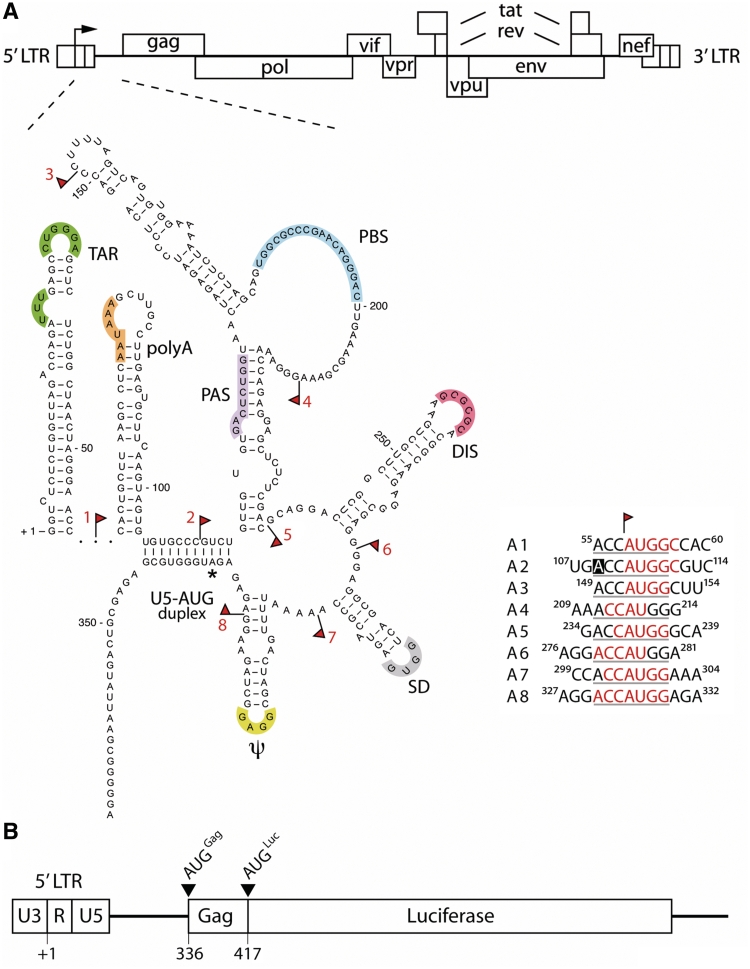
Figure 1.
Insertion of uAUGs in the HIV-1 5′-UTR. (A) The full-length HIV-1 5′-UTR encompasses nt +1 to +335 of the HIV-1 genomic RNA and harbours several structure and sequence motifs that are essential for viral replication. The RNA structure model was published previously (24). The regulatory motifs are marked in colours and further explained in the text. An asterisk indicates the Gag start codon. Red flags indicate where uAUGs or UAGs are inserted. The insertions are indicated A1–A8 for the uAUG insertions. The uAUG insertions are shown in detail next to the RNA structure model; the flanking Kozak sequences are underlined. The nucleotide position of the 5′-UTR is indicated and red nucleotides indicate the insertions whereas the boxed nucleotide indicates a mutation. (B) The luciferase reporter construct is shown with the 5′-LTR promoter elements U3, R and U5; nt positions of transcription start site (+1) and the Gag coding sequences are shown. The reporter constructs encode a fusion protein of the Gag N-terminal 25 amino acids and the firefly luciferase protein. As a result of the cloning strategy, the original AUG start codon that marks the beginning of the luciferase open reading frame (AUGLuc) is in frame with the Gag start codon (AUGGag).
The high structural complexity in the 5′-UTR is expected to interfere with the efficiency of one very important step of the viral life cycle: protein synthesis. Translation initiation on eukaryotic mRNAs is generally initiated by ribosomal scanning [reviewed in ref. (28)]. In this process, translation initiation factors interact with the 40S ribosomal subunit and the mRNA 5′-cap structure. Subsequently, the ribosomal subunit migrates along the 5′-UTR, until it encounters a favourable AUG start codon that marks the start of an open reading frame (ORF). Several mRNA features determine translation efficiency: the presence and accessibility of a 5′-cap structure (29–31), the length and secondary structure of the 5′-UTR (31,32) and the nucleotide context of the initiation codon (33,34). The optimal context of the initiation codon in mammalian cells is the so-called Kozak sequence: RCCAUGG, in which a purine at position −3 and the G at position +4 are most influential on translation initiation efficiency (35).
The HIV-1 5′-UTR displays several characteristics that may interfere with efficient scanning and hence translation initiation. Firstly, secondary structures close to the 5′-end of the message can inhibit translation initiation (30,36). The 5′-cap structure may be inaccessible to 40S ribosomal subunits due to the TAR and polyA hairpins at the 5′-end of the RNA (Figure 1) (30,31,37). Disruption of the TAR hairpin stimulated translation efficiency up to 190-fold (30), indicating that the intact TAR hairpin strongly inhibits translation. Second, the 5′-UTR of 335 nt is relatively long and highly structured (18,23,24,38–40). Obviously, viral replication occurs in spite of these potentially inhibitory features, and significant levels of the Gag structural proteins are expressed from the viral RNAs that contain the full-length 5′-UTR.
It has been suggested that the HIV-1 RNA contains an internal ribosomal entry site (IRES) downstream of these inhibitory structure motifs (41,42). IRES elements were initially described in picornaviruses and have since been identified in many viral and cellular mRNAs (43–47). IRES elements usually function under circumstances in which regular cap-dependent scanning is impaired, for example, following expression of a viral factor or in response to cell stress signals, such as cell-cycle arrest or heat shock (48,49). In fact, HIV-1 has been suggested to modulate host cell translation. The viral protease may cleave the translation initiation factor eIF4GI, which is essential for cap-dependent translation initiation (50–53). In addition, the viral Vpr protein causes a G2/M cell cycle arrest (54–57), which is essential for optimal viral replication and which may affect translation efficiency in general (58,59). Thus, the presumed HIV-1 IRES was suggested to allow optimal viral gene expression under these scanning-impaired conditions (41). It has been reported though that the host shut off observed during HIV infection is caused mainly by degradation of cellular mRNAs and less so by impairment of their translation (60).
The mechanism of HIV-1 translation initiation remains under debate (61). Potential IRES elements have been identified in the 5′-UTR and Gag sequences (41,42). In the latter case, ribosomal scanning in the 3′–5′ direction was proposed to allow synthesis of the Gag and Gag-Pol proteins from the upstream Gag start codon. The two studies seem to contradict each other: the Brasey paper described no IRES activity in the Gag ORF and the Buck paper did not find such activity in the 5′-UTR. Moreover, other groups have not been able to confirm either data (62,63).
We used a different approach to gain further insight into the translation initiation mechanism on HIV-1 mRNAs. We addressed ribosomal scanning in the 5′-UTR by introducing upstream AUGs (uAUGs). Scanning ribosomal 40S subunits will recognize the uAUG if scanning initiated upstream. Translation will commence on the introduced ORF followed by ribosomal release. When scanning occurs over the entire 5′-UTR, translation from the downstream authentic Gag start codon will be reduced irrespective of the location of the introduced uAUG codons (Kozak, 1984). In the case that HIV-1 5′-UTR harbours an IRES, the introduced uAUGs will affect gene expression only if they are located downstream of the IRES, similarly as was shown for picornaviruses (64–66). By placing uAUG codons at different positions across the 5′-UTR, the window of ribosomal scanning and the position of a putative IRES element can thus be determined.
MATERIALS AND METHODS
DNA constructs
The plasmid pLTR-gag-flag-luc was used for HIV-1 5′-UTR mutagenesis (25). The protein expressed from this luciferase reporter is a fusion product of the amino-terminal 25 Gag amino acids, the Flag peptide and the firefly luciferase protein. Protein expression is under control of the HIV-1 LAI 5′-LTR promoter and 5′-UTR sequences. To optimize cloning procedures we deleted the Flag sequences and the NcoI site at the luciferase start codon with a polymerase chain reaction (PCR) on pLTR-gag-flag-luc with primers TA033 and TA056 (Table 1). The PCR fragment was digested with HindIII and BspHI and ligated in the HindIII–NcoI-digested pLTR-gag-flag-luc. This PCR mutagenesis created a SalI site upstream of the luciferase initiation codon and generated the plasmid pLTR-25-gag-luc that was used for subsequent mutagenesis. uAUG and uUAG mutations were introduced by PCR mutagenesis. Oligonucleotide primers are listed in Table 1. Forward primers were used in combination with TA056 and reverse primers with TA033. PCR products were used as overlapping templates in a PCR reaction with primers TA033 and TA056. The resulting PCR products were digested with HindIII and SalI, cloned into the pLTR-25-gag-luc vector and sequenced. The constructs with eight individually inserted uAUG codons were designated A1–A8 and those with a stop codon at the same position U1–U8.
Table 1.
Oligonucleotide primers used for the HIV-1 5′-UTR mutagenesis
| DNA | Primer | Primer sequence 5′→3′ |
|---|---|---|
| wt | TA033 | CCCCTCGAGTAATACGACTCACTATAGGGTCTCTCTGGTTAGACC |
| wt | TA056 | GGGTCATGAGTCGACCCCCTGGCCTTAACCG |
| A1 r | TA057 | GGCCATGGTTCCCTAGTTAGCCAG |
| A1 f | TA058 | AGGGAACCATGGCCACTGCTTAAGCCTCA |
| A2 f | TA059 | AATAAAGCTTGCCTTGAGTGCTTCAAGTAGTGTGTGACCATGGCGTCTGTTGTGTGACTCTGG |
| A3 r | TA060 | AAAAGCCATGGTCTGAGGGATCTCTAGTTACC |
| A3 f | TA061 | CAGACCATGGCTTTTAGTCAGTGTGGAAAATC |
| A4 r | TA062 | CCCATGGTTTCGCTTTCAAGTCCC |
| A4 f | TA063 | GAAAGCGAAACCATGGGAAACCAGAGGAGC |
| A5 r | TA064 | GCCCATGGTCGAGAGAGCTCCTCTGGTTTCCC |
| A5 f | TA065 | CTCTCGACCATGGGCAGGACTCGGCTTGCTGAAGC |
| A6 r | TA066 | ACTCACCAGTCGCCTCCATGGTCCTCGCCTCTTGCCGTGCGC |
| A6 f | TA067 | GGAGGCCGACTGGTGAGTACG |
| A7 r | TA068 | CCATGGTGGCGTACTCACCAGTCGCCTCCCC |
| A7 f | TA069 | GAGTACGCCACCATGGAAAATTTTGACTAGCGGAGGC |
| A8 r | TA070 | CTCTCTCCATGGTCCTTCTAGCCTCCGCTAG |
| A8 f | TA071 | AGAAGGACCATGGAGAGAGATGGGTGCGAGAGCG |
| U1 r | TA122 | GGCCTAGGTTCCCTAGTTAGCCAG |
| U1 f | TA123 | AGGGAACCTAGGCCACTGCTTAAGCCTCA |
| U2 f | TA104 | AATAAAGCTTGCCTTGAGTGCTTCAAGTAGTGTGTGACCTAGGCGTCTGTTGTGTGACTCTGG |
| U3 r | TA232 | AAAAGCCTAGGTCTGAGGGATCTCTAGTTACC |
| U3 f | TA233 | CAGACCTAGGCTTTTAGTCAGTGTGGAAAATC |
| U4 r | TA234 | CCCTAGGTTTCGCTTTCAAGTCCC |
| U4 f | TA235 | GAAAGCGAAACCTAGGGAAACCAGAGGAGC |
| U5 r | TA082 | GCCCTAGGTCGAGAGAGCTCCTCTGGTTTCCC |
| U5 f | TA083 | CTCTCGACCTAGGGCAGGACTCGGCTTGCTGAAGC |
| U6 r | TA105 | ACTCACCAGTCGCCTCCTAGGTCCTCGCCTCTTGCCGTGCGC |
| U6 f | TA067 | GGAGGCCGACTGGTGAGTACG |
| U7 r | TA084 | CCTAGGTGGCGTACTCACCAGTCGCCTCCCC |
| U7 f | TA085 | GAGTACGCCACCTAGGAAAATTTTGACTAGCGGAGGC |
| U8 r | TA086 | CTCTCTCCTAGGTCCTTCTAGCCTCCGCTAG |
| U8 f | TA087 | AGAAGGACCTAGGAGAGAGATGGGTGCGAGAGCG |
| A2-LF f | TA110 | AAGTAGTGTGTGACCATGGCGTCTGTTGTGTGGACTCTGGTAACTAGAGATCCC |
| U2-LF f | TA231 | CTCAATAAAGCTTGCCTTGAGTGCTTCAAGTAGTGTGTGACCTAGGCGTCTGTTGTGTGGACT CTGGTAACTAGAGATCCC |
f = forward, r = reverse primer.
Nucleotide insertions are indicated in bold italics.
The constructs A1–A5 and A2–A5 were created by digesting the A1, A2 and A5 luciferase constructs with NcoI and SalI. The A5 fragment was cloned into the A1 and A2 vectors to construct A1–A5 and A2–A5, respectively. The U1–U5 and U2–U5 were created similarly with the U1, U2 and U5 constructs; however, digestions were performed with AvrII and SalI. The construct A2LF was created by PCR mutagenesis using primers TA110 and TA056, and pLTR-25-gag-luc as the template, resulting in the insertion of a G residue at position 124 of the HIV-1 leader sequence. The PCR fragment was digested with NcoI and SalI, and ligated into pLTR-A2-25-gag-luc as described above. The U2-LF control construct was created similarly using primers TA231 and TA056 in the PCR and HindIII and SalI as restriction sites for further construction. The insertional mutagenesis was confirmed by sequence analysis for all constructs.
Plasmid pRL-CMV expresses the Renilla luciferase reporter gene under the control of the CMV promoter (Promega). pcDNA3-Tat (67) expresses the HIV-1 LAI Tat protein under the control of the CMV promoter and is a derivate of the pcDNA3 vector (Invitrogen). The HIV-1 molecular clone pLAI has been described previously (68).
Cell culture
C33A cells, a human cervix carcinoma cell line, were grown in Dulbecco's MOD Eagle medium (DMEM) supplemented with 10% FBS, non-essential amino acids (Invitrogen), 20 mM glucose, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C and 5% CO2.
Transfection assays
Reporter gene expression driven by the wild-type (wt) and mutant 5′-UTR was analysed in C33A cells. Cells were seeded into 24-wells plates and transfected the next day with calcium phosphate-precipitated luciferase constructs in the presence of pcDNA3-Tat or pcDNA3 (67,69). pRL-CMV expressing Renilla luciferase was co-transfected as internal control. pBlueScript was used to make a total amount of 1 µg DNA for each transfection. Cells were washed with PBS 2 days after transfection and lysed in 125 µl of passive lysis buffer provided by the Dual-luciferase® Reporter Assay System (Promega). The firefly and Renilla luciferase activity was determined according to the manufacturer's protocol. The firefly luciferase activity was normalized to the Renilla luciferase activity.
RNA quantification
C33A cells were transfected at 50% confluence in a 6-well plate with 3 µg CaPO4-precipitated luciferase reporter construct as described. Two days post-transfection RNA was isolated with Trizol as described by the manufacturer. The RNA pellet was dissolved in 50 µl H2O and DNase treated with 1 µl Turbo DNase (Ambion) for 30 min at 37°C. The reaction was stopped by phenol/chloroform extraction with subsequent ethanol precipitation. The RNA was dissolved in 25 µl H2O and analysed and quantified on an ethidium bromide-stained agarose gel (1.5%).
Primer extension reactions were performed on equal amounts of total RNA. The oligonucleotide primer R368 (5′-TCCCCCGCTTAATACTGACGCT) was end-labelled with [γ-32P]-dATP using T4 polynucleotide kinase for 2 h at 37°C and purified using NucAway spin columns (Ambion). Subsequently, the primer was annealed to the RNA and reversed transcribed with Thermoscript reverse transcriptase (Thermoscript kit, Invitrogen). The reaction was stopped with 7 µl of 1 M NaOH for 15 min at 70°C and subsequent neutralization with 7 µl of 1 M HCl. The cDNA products were ethanol-precipitated, dissolved in formamide-containing loading buffer and applied to a denaturing polyacrylamide gel. Products were analysed using a PhosphorImager (Molecular Dynamics).
Sodium dodecyl sulphate–polyacrylamide gel electophoresis and western blot analysis
After determining the firefly and Renilla activities in lysates from transfected cells, equal amounts of Renilla luciferase activity were applied to a 12% sodium dodecyl sulphate (SDS)–polyacrylamide gel electophoresis (PAGE) to analyse the firefly luciferase proteins produced. Proteins were subsequently blotted on a low-autofluorescence membrane, incubated with a polyclonal antibody against luciferase (1:1000) (Promega). Bound antibodies were visualized with horse radish peroxidase-linked (HRP) donkey anti-goat IgG and the ECL+ kit (Amsersham Biosciences). The resulting luminescent signal was analysed after exposure of the blot to X-ray film for between 10 s to 5 min.
RESULTS
Design of the uAUG blocks
How translation initiation proceeds on HIV-1 genomic RNA has thus far not been answered conclusively. Controversy remains over whether the HIV-1 genomic RNA contains an IRES element (41,42) or not (62,63). This prompted us to address this issue from a different angle. We created a firefly luciferase reporter construct with the HIV-1 genomic 5′-UTR including the Gag start codon. Luciferase gene expression is driven from the HIV-1 LTR promoter, which can be enhanced by the addition of the viral Tat protein. The constructs contain the 5′ 75 nt of the Gag ORF fused to the luciferase ORF, which allows the formation of long distance interactions between sequences in the 5′-UTR and Gag ORF (24). As a consequence, the reporter constructs contain two in frame start codons: AUGGag and AUGLuc (Figure 1B). This allowed the detection of ribosomes that scan past the Gag start codon. We subsequently introduced uAUG blocks at eight positions in the 5′-UTR (A1–A8; indicated with red flags in Figure 1A). In the case of scanning, these uAUGs will reduce the expression of the downstream reporter gene, since the authentic start codon will not be reached by the scanning ribosome. However, if the uAUGs are inserted upstream of an active IRES element they will not affect gene expression. The inserted uAUGs were flanked with the consensus Kozak sequence (ACCAUGG) to allow optimal recognition by scanning ribosomal subunits. Wild-type (wt) HIV-1 nucleotides were used where possible to create the Kozak consensus, thus minimizing the number of inserted nucleotides (marked in red in Figure 1A). A single nucleotide was mutated in the A2 construct to comply with the −3 purine requirement (indicated with a black box in Figure 1A). We introduced the uAUG blocks in regions of the 5′-UTR predicted to be single stranded in order to minimize disruption of the RNA structure, except for A2, in which the U5-AUG duplex is likely affected. However, ablation of this structure does not affect translation (25).
Introduction of uAUGs affects Gag translation in a site-specific manner
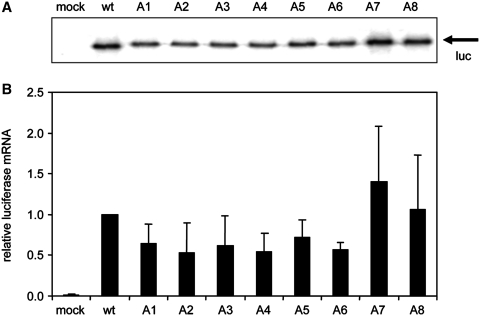
To rule out that the uAUG insertions affected LTR-driven transcription and/or RNA stability in our test system, we first determined the luciferase mRNA levels in C33A cells transfected with the wt and mutant reporter constructs (Figure 2). Total RNA was isolated 2 days after transfection and the luciferase mRNA was reverse transcribed with a 32P end-labelled oligonucleotide primer that anneals to the gag coding sequence. The radio-labelled cDNA products were run on a denaturing sequence gel (Figure 2A). A modest decrease in mRNA levels was observed for the mutants A1–A6, but the mRNA levels did not deviate significantly from the wt mRNA level after quantification of several independent assays (Figure 2B). Therefore, the luciferase mRNA levels were not significantly affected by the introduction of the uAUGs, in agreement with studies on other mRNAs (70,71).
Figure 2.
Luciferase mRNA levels are not significantly affected by the uAUG insertions. Wild-type (wt) and mutant constructs were transfected into C33A cells, total RNA was isolated and subjected to primer extension reactions. cDNA products were analysed on a denaturing polyacrylamide gel (panel A) and quantified (panel B). The wt mRNA level was set at 1. A representative of two independent experiments is shown. The error bars indicate standard deviation.
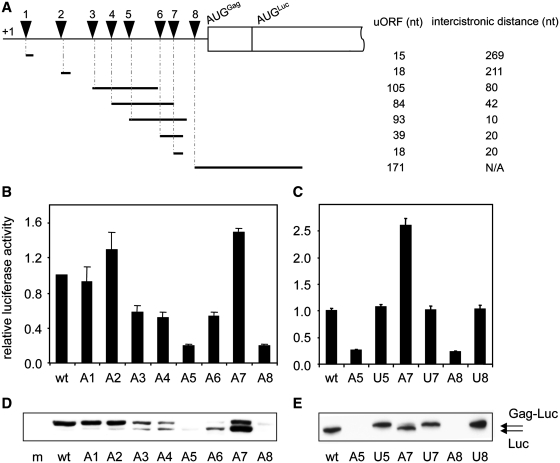
We proceeded to analyse luciferase expression from the A1–A8 mutant luciferase constructs. The constructs were transfected into C33A cells and luciferase activity was measured after 2 days. A construct expressing Renilla luciferase was co-transfected as an internal control. The results show that luciferase expressed from the A5 and A8 constructs decreased ∼5-fold compared to wt (Figure 3B). A reproducible ∼2-fold inhibition of luciferase activity was observed for the A3, A4 and A6 constructs. The A1 and A2 constructs expressed similar luciferase activity as wt, whereas the A7 expression level was reproducibly increased. Luciferase was expressed in the presence of the viral Tat protein to activate LTR-driven transcription. Experiments carried out in the absence of Tat co-expression showed a general 4-fold reduction in luciferase activity, but we observed the same relative expression pattern for the wt and mutants (Supplementary Figure S1).
Figure 3.
Insertion of uAUGs in the HIV-1 5′-UTR differentially affects protein expression levels. (A) Overview of uAUGs and upstream open reading frames in A1–A8. The A1–A8 reporter constructs encode uORFs that differ in size (indicated in nt). Triangles indicate the positions of the uAUGs. The 3′ border of the uORFs is determined by endogenous stop codons in the HIV-1 5′-UTR. The intercistronic distances vary among the constructs and are also indicated (in nt). The A8 uORF and Gag-Luc open reading frame are overlapping. (B and C) C33A cells were transfected with wt or mutant luciferase construct and pcDNA-tat. pRL-CMV was co-transfected as an internal control. The firefly and Renilla luciferase activities in cell lysates were determined two day post transfection. Firefly luciferase activity was corrected for Renilla luciferase activity. Wt expression levels are set as 1. One of the four independent experiments is shown, error bars indicate standard deviation. (D and E) C33A cells were transfected with wt or mutant luciferase reporter, pcDNA-tat and pRL-CMV. Firefly and Renilla luciferase activities were determined and equal amounts of Renilla luciferase activity were applied to SDS–PAGE. Luciferase protein was detected using western blot analysis with anti-luciferase antibodies. The constructs express two luciferase products: the Gag-luciferase fusion protein (Gag-Luc) and the non-fused luciferase (Luc) protein. M is mock-transfected cells.
As the mutational effects could be a consequence of the sequence insertion and not per se of the introduction of the start codon, we also inserted control sequences (ACCUAGG) at all positions. These control inserts are of the same nucleotide composition as the uAUG inserts, but the AUG start codon is inactivated into UAG. Expression of these U constructs was similar to wt (Figure 3C and Supplementary Figure S2), which indicates that the change in luciferase expression from A3 to A8 as compared to U3–U8 is caused by the inserted uAUG start codon. These results are supportive of ribosomal scanning as a means of translation initiation on this region of the HIV-1 5′-UTR. The reproducibly increased A7 expression over the U7 control remains to be explained.
The reporter constructs contain two in frame AUG codons that mark the beginning of the overlapping Gag-Luc and Luc ORF (Figure 1B). We assumed that the Gag start codon was used predominantly. However, the downstream AUGLuc could be used upon leaky scanning over the Gag start codon. Both Gag-Luc and Luc will display luciferase activity, although the amino terminal extension in Gag-Luc may affect the enzymatic activity in an unknown manner. We therefore wanted to confirm the luciferase activity results by western blot analysis that separates the Gag-Luc and Luc proteins. The results are shown in Figure 3D and E.
Expression of the wt construct resulted in a protein corresponding to the estimated Gag-Luc size, migrating slower in the gel than the unfused Luc product from a control plasmid in which the Gag start codon was inactivated (data not shown). The A1 and A2 constructs expressed wt levels of Gag-Luc protein, in agreement with the unaltered luciferase activity. However, introduction of the uAUGs at positions 3–8 resulted in a gradual shift from Gag-Luc to Luc protein expression (Figure 3D). The shift was caused by the inserted uAUG and not by the insertional mutagenesis itself as the U control constructs predominantly expressed Gag-Luc (Figure 3E and Supplementary Figure S2). The inhibition of Gag-Luc expression by the inserted uAUG codons in the A3–A8 constructs was thus higher than we concluded from the results of the luciferase activity assays.
It appears that the position of the introduced uAUG codon has an effect on which start codon is selected further downstream. A map of the uORFs in the uAUG constructs (Figure 3A) indicates that the Gag-Luc to Luc shift was most pronounced when the uORF terminated in close proximity of AUGGag, for instance compare A1 and A2 with A5, A6 and A7. This Gag-Luc to Luc shift could possibly be explained by translation reinitiation. This process has been studied in various systems and its efficiency has been reported to largely depend on two variables: the length of the uORF and the distance between the uORF and the downstream start codon. It has been postulated that upon translation of a short uORF, the 40S ribosomal subunit can resume scanning and reinitiate on a downstream start codon (72,73). For this second initiation event, the intercistronic distance needs to be a minimal size to allow the 40S ribosomal subunit to acquire the initiation factors required for a new round of translation (72). The intercistronic distance differs among the mutant reporter constructs, varying from 269 to 10 nt for the A1 and A5 constructs, respectively (Figure 3A). For the A1 and A2 constructs the intercistronic distance between the uORF and the AUGGag is relatively long, which can explain the predominant expression of the Gag-Luc product. In contrast, the AUGGag is bypassed in the A3–A7 constructs with shorter intercistronic distances because the ribosomal subunit may not yet be equipped for a second initiation event when it encounters AUGGag. It is not until after scanning to AUGLuc that the ribosomal subunit has acquired the translation initiation factors and can embark upon a new round of translation. This move to AUGLuc would increase the length of the intercistronic region by 81 nt. It is not surprising that the A8 construct does not express detectable levels of Gag-Luc, since its uORF overlaps with the gag coding sequences. To express the Gag-Luc protein from this construct, reinitiating ribosomal subunits must scan in the 3′–5′ direction. The data, therefore, strongly suggest that scanning occurs on the HIV-1 5′-UTR from nt 152 onwards, as construct A3–A8 all reduce translation initiation on the AUGGag.
Extension of uORF1 and 2 also results in effective inhibition of Gag translation
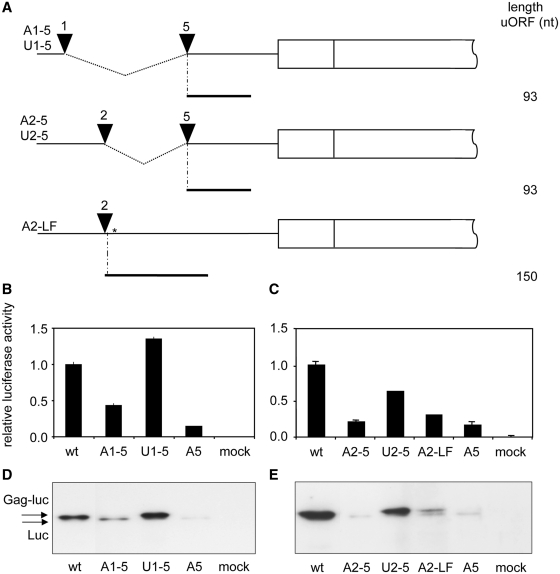
A second variable of reinitiation efficiency is the length of the uORF (72,73). This could explain why the level of reinitiation is not similar for all the constructs. The lengths of the uORFs in the A1–A8 constructs are shown in Figure 4. The A1, A2 and A7 constructs have very small uORFs (15–18 nt) that potentially allow efficient reinitiation, consistent with the high luciferase expression measured. The remaining constructs harbour uORFs of 39–171 nt in size that would decrease the reinitiation efficiency substantially. Overall, these data suggest that the insertion of short uORFs allows high expression levels of the downstream reporter gene, whereas lengthening of the uORFs leads to a significant reduction. Thus, efficient Gag-Luc expression by the A1 and A2 mutants could be due to the combination of short uORFs and long intercistronic distances.
Figure 4.
The length of the uORF determines translation reinitiation efficiency. (A) Extension of the uORFs in A1 and A2 constructs. The A1 and A2 uAUGs were fused to the A5 uAUG, thereby creating constructs A1–A5 and A2–A5 with an uORF size of 93 nt. The U1–U5 and U2–U5 were created similarly with the U1, U2 and U5 constructs, respectively. In addition, the uORF in A2 was extended in A2-LF by insertion of a G at nt +123 (shown by an asterisk), which destroys the original stop codon. As a consequence, the uORF length is extended from 18 to 150 nt, and the intercistronic distance is decreased by 132 nt. (B and C) Transfections were performed as in Figure 3A and B. One of three independent experiments is shown. Error bars indicate standard deviation. (D and E) Western blot analysis of the luciferase proteins. See Figure 3C and D for details.
To investigate these parameters, we extended the uORFs in A1 and A2, which consequently reduced the length of the intercistronic distance, by fusing these uAUGs with that of A5, thereby deleting the 5′-UTR sequences in between (A1–A5 and A2–A5 in Figure 4A). A similar fusion was made between U1/U2 and U5 as control constructs. In a separate A2 extension, we inserted a G nucleotide into the stop codon of the uORF, which causes a frameshift and extension of the uORF from 18 to 150 nt and reduced the intercistronic distance by 132 nt (A2-LF in Figure 4A). For this construct, a U control (U2-LF) was created. All constructs were analysed for luciferase expression in a luciferase activity assay and on western blot (Figure 4B–E and Supplementary Figure S2). Both analyses are in full agreement. Extension of the uORF in A1–A5, A2–A5 and A2-LF significantly reduced luciferase activity and Gag-Luc expression, yet induced the expression of the further downstream Luc ORF. These effects are not observed with the U control constructs. These data strongly suggest that translation reinitiation is very efficient when the uORFs are short as in A1 and A2, thus frustrating our strategy to determine the window of ribosomal scanning with the initial set of A1 and A2 mutants. However, extension of the uORF as in A1–A5, A2–A5 and A2-LF caused a severe Gag-Luc expression defect. These results are in agreement with a model in which the ribosome scans throughout the 5′-UTR across the position of A1–A8.
Co-expression of viral proteins does not affect the HIV-1 translation initiation mechanism
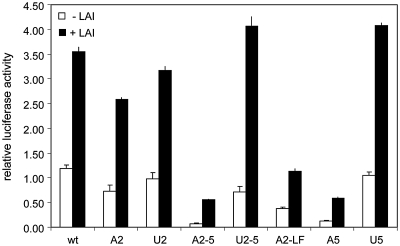
We thus far used a reporter construct driven by an HIV-1 promoter and the viral Tat protein was provided in trans to enhance transcription. In fact, the Tat expression vector also expressed the Rev protein, but neither protein differentially affected the luciferase activity expressed from the set of 5′-UTR mutants. It could be argued that other viral factors modulate the translation process. For instance, it has been suggested that viral factors contribute to a cellular environment that negatively affects cap-dependent scanning, either by cleavage of eIF-4G or by induction of G2-M cycle arrest (50–53,58,59). In order to investigate whether viral trans-acting factors affect ribosomal scanning and thus the expression from the mutant reporter constructs, we repeated the transfection experiments in the presence of the full-length HIV-1 LAI molecular clone that expresses all viral proteins. The results are shown in Figure 5 and Supplementary Figure S3. Co-expression of the molecular clone increased the luciferase expression by ∼3-fold, which we attribute to Tat trans-activation of the LTR promoter (compare Figure 5 with Supplementary Figure S1). The relative luciferase expression from the set of constructs did not change upon expression of HIV-1 proteins. In addition, virus production was unaltered by co-expression of wt versus mutant luciferase expression vectors.
Figure 5.
Luciferase expression is similarly affected by the uAUG insertions in the presence or absence of HIV-1 particle production. C33A cells were transfected with wt or mutant luciferase construct in the absence or presence of 100 ng of the pLAI molecular clone. pRL-CMV was co-transfected as an internal control. The firefly and Renilla luciferase activities in the lysates were determined 2 days post-transfection. Firefly luciferase activity was corrected for Renilla luciferase activity. CA-p24 was determined in the supernatant and was consistent in all transfections (20.3 ng/ml average, 2.37 ng/ml standard deviation). The overall increase observed upon HIV-1 co- expression is similar as upon Tat co-expression (compare with Supplementary Figure S1). The relative representation of the data set is shown in Supplementary Figure S3. One of three independent experiments is shown, error bars indicate standard deviation.
DISCUSSION
In this study, we analysed the window of ribosomal scanning on the HIV-1 genomic RNA by introducing uAUGs throughout its 5′-UTR. Insertion of uAUGs significantly blocked translation initiation on the Gag start codon if the uORF was of sufficient length, independent of its location. This inhibition was not observed when UAG sequences were inserted at the same positions as a control for the insertional mutagenesis. These results are in agreement with the observation that natural HIV-1 isolates do not harbour uAUGs in the 5′-UTR, suggesting that uAUGs are not tolerated (1). Furthermore, the results support previous studies that investigated the effect of an uAUG insertion at a single location in the HIV 5′-UTR, not only in reporter constructs but also in infectious molecular clones (63,74–76). In all of these cases, expression of either viral or reporter genes was severely affected. Unfortunately, the uAUGs were inserted in the proposed IRES domain (41) and control insertions were not analysed in these studies. It was therefore uncertain if the AUG insertions destroyed IRES activity or ribosomal scanning. The findings from our study strongly support the latter explanation.
Western blot analysis showed that insertion of uAUGs in various constructs (A3–A7) resulted in the translation of a Luc product initiated at the in frame Luc start codon downstream of the Gag start codon. This shift in start codon selection from AUGGag to AUGLuc was not observed with any of the control U constructs. This observation strongly suggests that the uORF-translating ribosomes can resume scanning in these constructs, bypass the Gag start codon and reinitiate further downstream. It has been well established that the efficiency of translation reinitiation depends on the size of the upstream ORF and the intercistronic distance (72,73,77). Thus, the constructs A1 and A2 with short ORFs and long intercistronic distances had to be re-evaluated, as the efficient Gag-Luc expression could be due to lack of ribosomal scanning in the region with the inserted uAUGs or to efficient reinitiation and sufficient intercistronic distance to allow a second-translation event on the AUGGag. Indeed, when we increased the length of the short ORFs in constructs A1–A5, A2–A5 and A2-LF, translation of Gag-Luc was severely reduced in a uAUG-dependent manner. This observation not only confirms the correlation between ORF size and reinitiation efficiency (72,73,77), but strongly suggests that reinitiation accounts for the wt levels of Gag-Luc production from the constructs A1 and A2, and thus support ribosomal scanning in this region of the 5′-UTR. The A7 construct also has a very short uORF, but it predominantly expresses Luc, which is likely caused by the short (20 nt) intercistronic distance between uORF7 and AUGGag. The scanning ribosomal subunit is unlikely to regain its full initiation capacity, e.g. by acquisition of essential initiation factors such as the eIF2–GTP–Met–tRNA ternary complex, before it reaches the Gag start codon. The intercistronic distance between uORF7 and AUGLuc is 101 nt and apparently compatible with reinitiation on the Luc start codon.
HIV-1 infection may affect the translation initiation mechanism by disfavouring ribosomal scanning. HIV-1 Vpr has been suggested to induce cell cycle arrest which modestly affects scanning-dependent translation (58,59,78) and IRES activity in the HIV-1 5′-UTR was shown to be increased in cell cycle-arrested lysates (41). Alternatively, the viral protease activity can cleave and inactivate the translation initiation factor eIF4G in vitro using the recombinant HIV-1 protease (50–52). eIF4G is essential for cap-dependent ribosomal scanning; it is cleaved during poliovirus infection upon which translation of cellular mRNAs ceases (79). However, translation of IRES-containing poliovirus RNA continues independent of intact eIF4G (47). A similar scenario has been postulated for HIV-1 infection (50–52). We thus co-expressed the HIV-1 proteins from an infectious molecular clone with the LTR-luc reporter constructs to address ribosomal scanning under conditions that resemble HIV infection. Luciferase expression from the uAUG and UAG constructs was similar with or without HIV-1 proteins (Figure 5 and Supplementary Figure S1). Specifically, the inserted uAUGs inhibited downstream Gag-Luc expression in a similar fashion, indicating that the window of ribosomal scanning on the HIV-1 5′-UTR was not affected by the viral proteins. In addition, co-expression of the poliovirus 2 A protease, which efficiently cleaves and inactivates eIF4G (80), reduced luciferase expression from wt and deletion mutant constructs (Supplementary Figure S4). Gene expression driven by an HCV IRES (81) was increased by ∼1.4-fold under these conditions, whereas a control construct in which the HCV IRES was inactivated was similarly affected by 2A protease as the HIV-1 LTR constructs. In our view, these data strongly suggest that intact eIF4G is required for optimal translation of HIV-1 RNAs and do not support the presence of an efficient IRES element in the 5′-UTR, in agreement with earlier data (63). IRES activities observed in various mRNAs have recently been attributed to artefacts of the experimental set up: cryptic promoter activity in the reporter constructs or aberrant splicing of the bicistronic RNAs resulted in the expression of unwanted subgenomic, monocistronic RNAs that were efficiently translated via the ribosomal scanning mechanism (78,82–88). Careful re-examining of the proposed HIV-1 IRES activity is therefore much desired (78).
An alternative mechanism for translation initiation could be ribosomal shunting. Shunting is a discontinuous form of scanning in which 40S ribosomal subunits ‘jump’ from an upstream shunt donor region to a downstream shunt acceptor in the 5′-UTR thus bypassing stable RNA structures and uAUGs (89). We showed that all uAUG insertions affected downstream Gag-Luc production if the uORF size was of sufficient length. None of the inserted uAUGs seemed to be skipped by ribosomal subunits via a shunting mechanism. The results therefore do not provide evidence for ribosomal shunting on HIV-1 mRNAs. In conclusion, we propose ribosomal scanning as the main mechanism for the translation initiation process on HIV-1 mRNA, not only in the absence of HIV-1 proteins, but also in the presence of Tat and all other HIV-1 proteins.
This leaves the question of how the relatively long and structured HIV-1 5′-UTR allows ribosomal scanning and translation of the Gag and Gag-Pol structural proteins. The 5′ TAR hairpin has been shown to inhibit translation in rabbit reticulocyte lysates in cis, likely by blocking cap recognition (31). It has been shown in various studies that length and structures in 5′-UTRs are more inhibitory in rabbit reticulocyte lysates than in cells (90–93). In agreement with this, several host factors have been identified that alleviate the TAR-mediated block of translation in cells (94–96). In addition, the cellular RNA helicase DDX3 that is involved in HIV-1 RNA nuclear export (97) was recently implicated in translation regulation: its RNA helicase activity improved translation of mRNAs with long and structured 5′-UTRs, possibly by unwinding secondary structure or facilitating ribosomal scanning (98). This role of DDX3 in HIV-1-driven translation has not been carefully investigated. A role in facilitating translation efficiency on the HIV-1 5′-UTR was recently suggested for RNA helicase A (RHA) also (99). RHA has already been described as a post-transcriptional enhancer for other retroviral mRNAs (100). Furthermore, DHX29 has been reported to promote translation initiation on mRNAs that contain highly structured 5′-UTRs (101). The involvement of the various cellular factors in HIV-driven translation is an intriguing topic that warrants further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: TOP grant from the Netherlands Organization for Scientific Research (NWO) Chemical Sciences and a VICI grant from the Netherlands Organization for Health Research and Development (ZonMW).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We owe many thanks to Wim van Est for artwork. Prosper Rek and Suuske Popma are acknowledged for technical assistance. We owe many thanks to Dr Peter Bredenbeek (Leiden University, the Netherlands) for the gift of the bicistronic HCV constructs and the 2A-expression constructs, to Dr Emily Manktelow for critically reading this manuscript and to Dr Adri A.M. Thomas for fruitful discussions.
REFERENCES
- 1.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 2.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 3.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 5.Ippolito JA, Steitz TA. A 1.3-A resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl Acad. Sci. USA. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. Embo J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA, Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl Acad. Sci. USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 10.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 11.Klasens BI, Das AT, Berkhout B. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 1998;26:1870–1876. doi: 10.1093/nar/26.8.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klasens BI, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbink TE, Berkhout B. HIV-1 reverse transcription: close encounters between the viral genome and a cellular tRNA. Adv. Pharmacol. 2007;55:99–135. doi: 10.1016/S1054-3589(07)55003-9. [DOI] [PubMed] [Google Scholar]
- 14.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 15.Harrison GP, Lever AM. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 19.Clavel F, Orenstein JM. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J. Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Reilly MM, McNally MT, Beemon KL. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–385. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 22.Abbink TE, Berkhout B. RNA structure modulates splicing efficiency at the HIV-1 major splice donor. J. Virol. 2007;82:3090–3098. doi: 10.1128/JVI.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Abbink TE, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 25.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 26.Nikolaitchik O, Rhodes TD, Ott D, Hu W-S. Effects of Mutations in the Human Immunodeficiency Virus Type 1 gag Gene on RNA Packaging and Recombination. J. Virol. 2006;80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song R, Kafaie J, Laughrea M. Role of the 5′ TAR stem–loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry. 2008;47:3283–3293. doi: 10.1021/bi7023173. [DOI] [PubMed] [Google Scholar]
- 28.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 30.Parkin NT, Cohen EA, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 34.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 35.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 36.Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SenGupta DN, Berkhout B, Gatignol A, Zhou AM, Silverman RH. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson RJ. RNA translation. Picornaviruses break the rules. Nature. 1988;334:292–293. doi: 10.1038/334292a0. [DOI] [PubMed] [Google Scholar]
- 44.Pelletier J, Kaplan G, Racaniello VR, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 47.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 49.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 50.Ohlmann T, Prevot D, Decimo D, Roux F, Garin J, Morley SJ, Darlix JL. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 2002;318:9–20. doi: 10.1016/S0022-2836(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 51.Perales C, Carrasco L, Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003;533:89–94. doi: 10.1016/s0014-5793(02)03764-x. [DOI] [PubMed] [Google Scholar]
- 52.Ventoso I, Blanco R, Perales C, Carrasco L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl Acad. Sci. USA. 2001;98:12966–12971. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castelló A, Franco D, Moral-López P, Berlanga JJ, Álvarez E, Wimmer E, Carrasco L. HIV- 1 Protease Inhibits Cap- and Poly(A)-Dependent Translation upon eIF4GI and PABP Cleavage. PLoS ONE. 2009;4:e7997. doi: 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartz SR, Rogel ME, Emerman M. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J. Virol. 1996;70:2324–2331. doi: 10.1128/jvi.70.4.2324-2331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elder RT, Yu M, Chen M, Edelson S, Zhao Y. Cell cycle G2 arrest induced by HIV-1 Vpr in fission yeast (Schizosaccharomyces pombe) is independent of cell death and early genes in the DNA damage checkpoint. Virus Res. 2000;68:161–173. doi: 10.1016/s0168-1702(00)00167-2. [DOI] [PubMed] [Google Scholar]
- 56.Emerman M. HIV-1, Vpr and the cell cycle. Curr. Biol. 1996;6:1096–1103. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 57.Masuda M, Nagai Y, Oshima N, Tanaka K, Murakami H, Igarashi H, Okayama H. Genetic studies with the fission yeast Schizosaccharomyces pombe suggest involvement of wee1, ppa2, and rad24 in induction of cell cycle arrest by human immunodeficiency virus type 1 Vpr. J. Virol. 2000;74:2636–2646. doi: 10.1128/jvi.74.6.2636-2646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subbramanian RA, Kessous-Elbaz A, Lodge R, Forget J, Yao XJ, Bergeron D, Cohen EA. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J. Exp. Med. 1998;187:1103–1111. doi: 10.1084/jem.187.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gummuluru S, Emerman M. Cell cycle- and Vpr-mediated regulation of human immunodeficiency virus type 1 expression in primary and transformed T-cell lines. J. Virol. 1999;73:5422–5430. doi: 10.1128/jvi.73.7.5422-5430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agy MB, Wambach M, Foy K, Katze MG. Expression of cellular genes in CD4 positive lymphoid cells infected by the human immunodeficiency virus, HIV-1: evidence for a host protein synthesis shut-off induced by cellular mRNA degradation. Virology. 1990;177:251–258. doi: 10.1016/0042-6822(90)90478-a. [DOI] [PubMed] [Google Scholar]
- 61.Yilmaz A, Bolinger C, Boris-Lawrie K. Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr. HIV Res. 2006;4:131–139. doi: 10.2174/157016206776055039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwartz S, Felber BK, Pavlakis GN. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol. Cell. Biol. 1992;12:207–219. doi: 10.1128/mcb.12.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hellen CU, Pestova TV, Wimmer E. Effect of mutations downstream of the internal ribosome entry site on initiation of poliovirus protein synthesis. J. Virol. 1994;68:6312–6322. doi: 10.1128/jvi.68.10.6312-6322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pilipenko EV, Gmyl AP, Maslova SV, Belov GA, Sinyakov AN, Huang M, David T, Brown K, Agol VI. Starting Window, a Distinct Element in the Cap-independent Internal Initiation of Translation on Picornaviral RNA. J. Mol. Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- 66.López de Quinto S, Martínez-Salas E. Parameters influencing translational efficiency in aphthovirus IRES-based bicistronic expression vectors. Gene. 1998;217:51–56. doi: 10.1016/s0378-1119(98)00379-5. [DOI] [PubMed] [Google Scholar]
- 67.Verhoef K, Tijms M, Berkhout B. Optimal Tat-mediated activation of the HIV-1 LTR promoter requires a full-length TAR RNA hairpin. Nucleic Acids Res. 1997;25:496–502. doi: 10.1093/nar/25.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berkhout B, van Wamel J, Klaver B. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J. Mol. Biol. 1995;252:59–69. doi: 10.1006/jmbi.1994.0475. [DOI] [PubMed] [Google Scholar]
- 69.Das AT, Klaver B, Klasens BI, van Wamel JL, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva AL, Pereira FJ, Morgado A, Kong J, Martins R, Faustino P, Liebhaber SA, Romao L. The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context. RNA. 2006;12:2160–2170. doi: 10.1261/rna.201406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maquat LE. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 72.Poyry TAA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luukkonen BG, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Das AT, van Dam AP, Klaver B, Berkhout B. Improved envelope function selected by long-term cultivation of a translation-impaired HIV-1 mutant. Virology. 1998;244:552–562. doi: 10.1006/viro.1998.9124. [DOI] [PubMed] [Google Scholar]
- 75.Hache G, Shindo K, Albin JS, Harris RS. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 2008;18:819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hache G, Abbink TE, Berkhout B, Harris RS. Optimal translation initiation enables Vif-deficient human immunodeficiency virus type 1 to escape restriction by APOBEC3G. J. Virol. 2009;83:5956–5960. doi: 10.1128/JVI.00045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kochetov AV, Ahmad S, Ivanisenko V, Volkova OA, Kolchanov NA, Sarai A. uORFs, reinitiation and alternative translation start sites in human mRNAs. FEBS Lett. 2008;582:1293–1297. doi: 10.1016/j.febslet.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 78.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee KA, Sonenberg N. Inactivation of cap-binding proteins accompanies the shut-off of host protein synthesis by poliovirus. Proc. Natl Acad. Sci. USA. 1982;79:3447–3451. doi: 10.1073/pnas.79.11.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ehrenfeld E. In: Translational Control. Hershey JWB, M. M. B. a. S. N, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 81.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspé G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 82.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baranick BT, Lemp NA, Nagashima J, Hiraoka K, Kasahara N, Logg CR. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc. Natl Acad. Sci USA. 2008;105:4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bert AG, Grépin R, Vadas MA, Goodall GJ. Assessing IRES activity in the HIF-1α and other cellular 5′ UTRs. RNA. 2006;12:1074–1083. doi: 10.1261/rna.2320506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han B, Zhang J-T. Regulation of Gene Expression by Internal Ribosome Entry Sites or Cryptic Promoters: the eIF4G Story. Mol. Cell. Biol. 2002;22:7372–7384. doi: 10.1128/MCB.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S. Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the β-gal/CAT bicistronic reporter system. RNA. 2005;11:1605–1609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vopálenský V, Mašek T, Horváth O, Vicenová B, Mokrejš M, Pospíšek M. Firefly luciferase gene contains a cryptic promoter. RNA. 2008;14:1720–1729. doi: 10.1261/rna.831808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saffran HA, Smiley JR. The XIAP IRES activates 3′ expression by inducing production of monocistronic mRNA in the βgal/CAT bicistronic reporter system. RNA. 2009;15:1980–1985. doi: 10.1261/rna.1557809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ryabova LA, Pooggin MM, Hohn T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006;119:52–62. doi: 10.1016/j.virusres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 90.Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, Shatsky IN. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van der Velden AW, van Nierop K, Voorma HO, Thomas AA. Ribosomal scanning on the highly structured insulin-like growth factor II-leader 1. Int. J. Biochem. Cell Biol. 2002;34:286–297. doi: 10.1016/s1357-2725(01)00116-9. [DOI] [PubMed] [Google Scholar]
- 92.Hensold JO, Stratton CA, Barth D. The conserved 5′-untranslated leader of Spi-1 (PU.1) mRNA is highly structured and potently inhibits translation in vitro but not in vivo. Nucleic Acids Res. 1997;25:2869–2876. doi: 10.1093/nar/25.14.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Andreev DE, Dmitriev SE, Terenin IM, Prassolov VS, Merrick WC, Shatsky IN. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dorin D, Bonnet MC, Bannwarth S, Gatignol A, Meurs EF, Vaquero C. The TAR RNA-binding Protein, TRBP, Stimulates the Expression of TAR-containing RNAs in Vitro and in VivoIndependently of Its Ability to Inhibit the dsRNA-dependent Kinase PKR. J. Biol. Chem. 2003;278:4440–4448. doi: 10.1074/jbc.M208954200. [DOI] [PubMed] [Google Scholar]
- 97.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 98.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010;38:1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complex proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parsyan A, Shahbazian D, Martineau Y, Petroulakis E, Alain T, Larsson O, Mathonnet G, Tettweiler G, Hellen CU, Pestova TV, et al. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc. Natl Acad. Sci. 2009;106:22217–22222. doi: 10.1073/pnas.0909773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.