Abstract
Some viruses exploit internal initiation for their propagation in the host cell. This type of initiation is facilitated by structured elements (internal ribosome entry site, IRES) upstream of the initiator AUG and requires only a reduced number of canonical initiation factors. An important example are IRES of the virus family Dicistroviridae that bind to the inter-subunit side of the small ribosomal 40S subunit and lead to the formation of elongation-competent 80S ribosomes without the help of any initiation factor. Here, we present a comprehensive functional and structural analysis of eukaryotic-specific ribosomal protein rpS25 in the context of this type of initiation and propose a structural model explaining the essential involvement of rpS25 for hijacking the ribosome.
INTRODUCTION
During translation, the information encoded by the sequence of the messenger RNA (mRNA) codons is converted into a polypeptide chain. The main player in this complex process is the ribosome, a large macromolecular assembly essential for all living organisms from bacteria to human (1,2). The ribosome along with mRNA, transfer RNAs (tRNAs) and many translation factors constitutes the functional module for translation (3). Importantly, translational control occurs mainly during the initiation phase of protein synthesis contributes to the general regulation of gene expression (4,5).
Ribosomes from all kingdoms of life (70S for prokaryotes, 80S for eukaryotes) are divided into two subunits, which are different in their size, structure and function. Both subunits are composed of large ribosomal RNA molecules (rRNA) and ribosomal proteins, whereas the exact number of the latter varies among different species. The small ribosomal subunit, 30S for the prokaryotic and 40S for the eukaryotic ribosome, organizes the decoding step (6) and controls the interactions between the anticodons of the tRNAs and the codons in the mRNA. The large ribosomal subunit, 50S for the prokaryotic and 60S for the eukaryotic ribosome, catalyses the formation of peptide bonds (7). The solution of the structure of prokaryotic ribosomes and ribosomal subunits by X-ray crystallography (7–10) contributed immensely to our understanding of the basic mechanism of translation and, supported by previous biochemical data (11), indicated that the main activities of the ribosome are dominated by its RNA component. Nevertheless, ribosomal proteins appear to play important roles in protein synthesis, especially for both tuning the specific folding of the rRNA and ribosome dynamics.
The 80S ribosome comprises four rRNAs—25S rRNA, 5S rRNA and 5.8S rRNA in the large subunit and 18S rRNA in the small subunit—and at least 78 proteins. Phylogenetic studies suggest that only about one-third of all ribosomal proteins are conserved between the prokaryotic 70S and the eukaryotic 80S ribosome. The additional proteins as well as insertion elements of the rRNA, known as expansion segments, increase the size of the eukaryotic ribosome by ∼50% relative to its prokaryotic counterpart (12,13). Until recently mainly cryo-EM has contributed to the structural analysis of eukaryotic ribosomes (14–21). Structural comparison with the prokaryotic counterparts indicated an evolutionary conserved core of the 80S and 70S ribosome. The eukaryotic-specific ribosomal proteins and the expansion segments build an additional layer of density around that conserved core. While the majority of the expansion segments had been localized so far, the position of the majority of the eukaryotic-specific ribosomal proteins remained largely unknown (16–18), although an attempt has been made to localize all proteins in a cryo-EM map at subnanometer resolution (21). Also the recent crystal structure of the yeast 80S ribosome (22) does not contain a complete set of ribosomal proteins. Only in the very recent crystal structure of the Tetrahymena thermophilia 40S–eIF1 complex all proteins of the 40S subunit have been assigned (23).
The specific functions of most of these additional proteins and nucleotides are not sufficiently characterized yet, but at least one expansion segment and most specific eukaryotic ribosomal proteins are required for viability of the cell (24). Generally, most densities assigned to eukaryotic-specific ribosomal proteins are located on the solvent exposed side of the ribosome, suggesting a possible role as primary targets for interactions with cellular regulatory factors. Moreover, some of these proteins lie at the inter-subunit space between both ribosomal subunits where they are likely involved in the formation of additional eukaryotic intersubunit bridges (16–18,21,22). So far, only a functional role has been suggested for few eukaryotic-specific proteins. Most of them are required during the biogenesis of ribosomal subunits, and are therefore essential, as they work as chaperones during the maturation and folding process of ribosomal RNAs (25,26). In this aspect, several ribosomal proteins have been implicated in human diseases such as Diamond–Blackfan anaemia (27). Furthermore, eukaryotic-specific proteins located at the ribosomal surface can be exploited by viruses for hijacking the ribosome during host-cell infection.
Initiation of translation in eukaryotes is a highly complicated process (4). The canonical pathway of translation initiation is 5′-cap dependent, requires ∼11 translation initiation factors and an AUG start codon. However, there is the alternative pathway of internal initiation, which utilizes structured RNA elements, so-called internal ribosome entry sites (IRES), present in the 5′-untranslated region (28,29). Many viruses use IRES elements to hijack the host translation machinery and IRES elements can be also found in a subset of cellular mRNAs for regulatory purposes. IRES-driven translation initiation is 5′-cap independent and requires only a reduced set of initiation factors. The intergenic IRES of dicistroviruses, such as Plautia stali intestine virus (PSIV) and Cricket paralysis virus (CrPV), use the simplest mechanism of internal initiation. This virus family can start the initiation without any initiation factor and even without the AUG start codon and initiator tRNA (30). Biochemical data have shown, that Dicistroviridae IRES directly bind the 40S and start the translation process from the ribosomal A-site, that is highly unusual since the canonical translation initiation starts from the P-site of the ribosome (31–33).
Cryo-EM studies of CrPV (15,34) and hepatitis C virus (HCV) IRES (35,36) in complex with ribosomal particles have shown that IRES elements adopt a defined RNA-fold and undergo specific interactions with the ribosome. The RNA-fold of the ribosome bound CrPV IRES has been determined by cryo-EM (34) and is good overall with the X-ray structure of the PSIV IGR IRES (37). Importantly, HCV IRES RNA (35) and CrPV IRES RNA (15) induce a conformational change in the ribosomal 40S subunit; that is similar to the conformational change induced by canonical initiation factors eIF1 and eIF1A (38).
Cryo-EM maps of ribosome-bound IRES RNAs have indicated that eukaryotic-specific ribosomal components are involved in IRES RNA binding (15,34–36). The eukaryotic-specific ribosomal protein rpS25 has particularly been implicated in internal initiation of at least two classes of IRES RNAs. rpS25 can be crosslinked to a conserved loop region in dicistroviral IRES elements (39) and has been shown to be essential for internal initiation of CrPV and HCV IRES RNAs (40). In this study, we present a comprehensive structural and functional analysis of rpS25 and its role in internal initiation. We could successfully localize and characterize rpS25 from Saccharomyces cerevisiae by means of cryo-EM in combination with biochemical techniques and tertiary-structure modelling. The estimated position at the head of the 40S subunit and the predicted fold of rpS25 provide the structural basis to discuss the important functional role of rpS25 in internal translation initiation used by viruses to hijack the ribosome.
MATERIALS AND METHODS
Ribosome preparation
For the preparation of 80S ribosomes from yeast, the wild-type and mutant strain were grown at 30°C until OD600 reach to 0.8 in YPD media containing 2% glucose. The cells were harvested by centrifugation in a Beckman TA-10.250 rotor at 4000g for 10 min and the cell pellet was washed with a 1% KCl solution. The cell digestion was done in lysis buffer (50 mM HEPES pH 7.6, 6 mM magnesium acetate, 100 mM potassium acetate, 2 mM DTT, 0.5 mM AEBSF, 1 mg/ml heparin) using glass beads (SIGMA, size 425–600 µm) followed by a centrifugation in a Beckman TA-10.250 rotor at 4000g for 5 min. The supernatant was centrifuged in a Beckman TA-10.250 rotor at 9800g for 15 min to remove all cell debris and subsequently in a Beckman MLA-80 rotor at 33 000g for 30 min yielding the 80S ribosomes containing S30 supernatant. S30 was layered onto a 2.4 ml cushion of 30% sucrose in lysis buffer and spun in a Beckman MLA-80 rotor at 70 000g for 18 h. The crude 80S pellet was washed with 1 ml Y80S buffer (20 mM HEPES pH 7.6, 6 mM magnesium acetate, 100 mM potassium acetate, 2 mM DTT) and resuspended in a total volume of 300 μl in Y80S buffer. The sample was then applied on to a 10–30% sucrose gradient prepared in Y80S buffer and spun in a Beckman SW28 rotor at 18 000 rpm for 17 h. Fractions containing 80S ribosomes were pooled, diluted 1:1 with Y80S buffer and centrifuged in a Beckman 50.2 Ti rotor at 45 000 rpm for 21 h. The resulting ribosomal pellet was resuspended in Y80S buffer.
Cryo-EM, image processing and modelling
Ribosomes were diluted to 30 pmol/ml final concentration in Y80S-buffer. Cryo-EM grids were prepared following standard procedures. Micrographs were collected under low-dose conditions on a TECNAI Polara F30 microscope at 39-fold magnification. The final three-dimensional (3D) maps were computed using SPIDER/WEB software package (41) and visualized with CHIMERA (42).
A set of 33 000 and 41 528 particles was used for the final ΔrpS25 and wild-type reconstruction, respectively.
Plasmid
The Plautia stali intestine virus (PSIV) sequence regions 5961–6230 containing the IRES was amplified from pT7CAT-5375 (43). The amplified fragment was digested with HindIII and EcoRI, and ligated into the corresponding sites of pT7Blue (Novagen), generating pT7Blue IRES Domain 1-2-3. PCR fragment was amplified from pT7Blue IRES Domain 1-2-3 using primers according to the T7 promoter and the vector specific U-19-mer sequnece (as indicated in the Novagen vector-sheet) for RNA synthesis to have enough space at the 3′-region for primer extension. Yeast rpS25A gene was amplified from genome by PCR and cloned into the pQE80L vector by BamHI and PstI. Escherichia coli GroES gene was amplified from genome by PCR and cloned into the pET24a vector using NdeI and XhoI.
RNA synthesis
The three vectors pT7Blue IRES Domain 1-2-3, pT7Blue IRES Domain 1-2 (44) and pT7-IRES6240-Fluc (45) were linearized with EcoRI. RNAs were synthesized from linearized pT7Blue IRES Domain 1-2-3, Domain 1-2 and PCR-amplified Domain 1-2-3 containing the U-19-mer region and purified as described (46). IRES6240-Fluc was synthesized using the RiboMAX large-scale RNA production system (Promega). The various IRES RNAs were incubated at 70°C for 3 min before use.
Expression and purification of recombinant proteins
The recombinant rpS25A was produced in BL21 (DE3) cells by growing to an OD600 of 0.6 on 500 ml LB-medium containing ampicillin (50 µg/ml), followed by the addition of IPTG (0.1 mM) for 3 h. Bacterial cells were collected by centrifugation, resuspended in 20 ml buffer containing 10 mM MgCl2, 100 mM NH4Cl, 1 mM dithiothreitol, 20 mM Tris–HCl, pH 7.6 and broken using a sonication. The non-soluble fraction was collected by centrifugation for 30 min at 15 000g and resuspended in 5 ml lysis buffer containing 8 M urea, 50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazol. This extraction of non-soluble protein was repeated for three times. The solubilized protein was incubated in lysis buffer with Ni–NTA–agarose pre-equilibrated same buffer for 1 h 4°C. The material was applied to a column, washed with 20 ml lysis buffer containing 20 mM imidazol. The protein was eluted by 5 ml lysis buffer containing 250 mM imidazol. After confirmed purification at SDS–PAGE, rpS25A was renaturated (47). The recombinant GroES was grown at 30°C for overnight after the addition of IPTG (0.1 mM). The purification step was the same procedure like rpS25A and purified from soluble fractions with buffers without urea.
Filter binding assay
32P-labelled IRES Domain 1-2-3 RNA was mixed with various concentration of S. cerevisiae 80S ribosomes in 50 µl of a solution containing 3 mM MgCl2, 100 mM KCl, 0.2 mM dithiothreitol and 50 mM Tris–HCl, pH 7.6 and incubated at 30°C for 5 min. Filtration procedure was as described (48).
In vitro translation assays
Saccharomyces cerevisiae translation extracts were prepared as described previously (49). After the extract was treated with micrococcal nuclease, in vitro translation was performed with 6.25 µl S. cerevisiae extract in the presence of 12 µg in vitro transcribed IRES6240-Fluc in 12.5 µl reaction and incubated at 25°C for 90 min. Luciferase activity was determined by using a luciferase assay kit (PiccaGene) from Wako Pure Chemical (Osaka).
Pull-down assay
Three hundred and seventy-two picomol of his-S25A or GroES in 100 µl of buffer containing 50 mM Tris–HCl, pH 7.6, 10 mM MgCl2, 50 mM KCl was mixed with a 25 µl of Ni–NTA–agarose beads (Qiagen) pre-equilibrated with buffer and gentle mixing continued at 4°C for 1 h on a rotary mixer. After mixing, the protein bound beads were recovered by centrifugation. The beads were washed five times with 500 µl of buffer. After the addition of Domain 1-2 RNA (500 pmol) in 100 µl buffer, beads were mixed at 4°C for 1 h, and beads were recovered by same method and washed. The beads were then resuspended in 25 µl sample buffer and denatured at 70°C for 2 min. Twenty-five microlitre was subjected to PAGE–urea electrophoresis. The gel was stained with toluidine blue.
Chemical footprints
The IRES-Domain 1-2-3 (10 pmol) was incubated with different amounts of rpS25A for 5 min at 30°C in 50 µl containing 5 mM MgCl2, 100 mM KCl, 50 mM potassium cacodylate, pH 7.2. One aliquot of the reaction-mixtures was incubated with 0.1 U RNase T1 for 10 min at 37°C, the same samples were reacted with 1-cyclohexyl-3-(2-morpholinoethyl), carbodiimidemetho-p-toluene (CMCT) and RNA extraction, primer extension with the U-19-mer primer in the presence of AMV reverse transcriptase, and gel electrophoresis were as described in ref. (50).
Molecular modelling
The appropriate template structure was detected searching the PDB (October 2010) with PSI-Blast using the scoring matrix BLOSUM62. The three top hits (alignment score 32, E-value 0.063) are different crystal structures (PDB accession codes: 2ve8, 2j50, 2iut) of the FtsK Motor Domain from Pseudomonas aeruginosa. The following hits also consist of helix–turn–helix (HTH) motifs, but are significantly less suitable as modelling templates according to the significantly lower alignment scores (25–27) and E-values (2.0–7.7) obtained. The alignment with the top-ranked template (2ve8) covers the residues 43–104 with 25% identical and 50% positive matches. Based on the results of a FUGUE search, the alignment with 2ve8 could be plausible extended at the N- and C-terminus by several residues, so that our final model covers the residues 42–108 of rpS25 (Supplementary Figure S1) (51). We used the Swiss PDB viewer to generate the hypothetical model based on the alignment shown in Supplementary Figure S1. The model was energetically minimized using the GROMOS96 force field. The crossing angles between helix axes of the final rpS25 model were calculated using MPlot (52). The figures were produced with PyMOL Molecular Graphics System (http://www.pymol.org) and CHIMERA (42).
RESULTS
Structural analysis of 80S ribosomes lacking rpS25
This study is addressed to the localization, functional and structural analysis of the unique eukaryotic ribosomal protein S25 (rpS25) of yeast S. cerevisiae. The protein is encoded by two genes, RPS25A and RPS25B, leading to proteins rpS25A and rpS25B, which differ only by 1 amino acid at their C-terminus. Why does two RPS25 genes exist is not known. Like all ribosomal proteins, except P1/P2 (orthologues of E. coli rpL12), only one copy of rpS25 is present in the ribosome. The exact position of the protein on the ribosome and its functional role remained an intriguing questions for a long time since biochemical and genetic studies have shown that rpS25 is not essential for cell viability but is highly conserved in yeast and mammals (26,53) and the attempts to localize the protein did not succeed (17). Recently, the protein has been shown to play an essential role in the cap-independent initiation of translation suggesting its important functional role as a regulatory element of translation (40).
We took advantage of the fact that the deletion of the two RPS25 genes does not affect the assembly of the 80S ribosomes and their viability, assuming that the ribosome lacking rpS25 would be still active but has an empty space or gap at the site where the protein should have been. On the basis of the cryo-EM reconstruction of such rpS25 deficient ribosomes we would be able to determine the exact position of rpS25 by finding the gap in the ribosome when compared with the control wild-type ribosome.
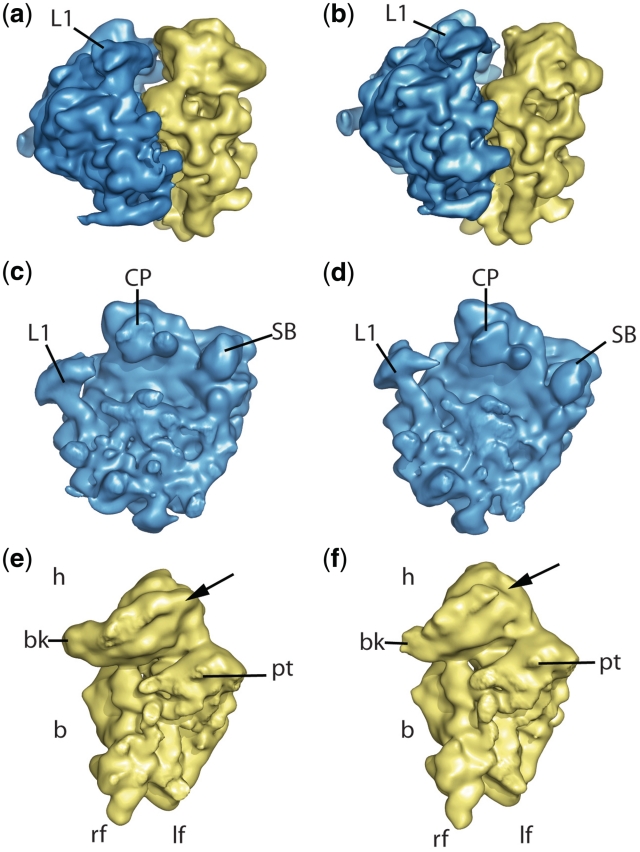
To this end, we purified 80S ribosomes from the S. cerevisiae strain, where both rpS25 genes (RPS25A and RPS25B) were deleted (26) and from the corresponding wild-type strain. To prevent any unspecific structural artefacts, both strains were grown under the same conditions as described in ‘Material and Methods’ section. The deletion mutant strain showed a slightly slower cell growth compared to the wild-type, in agreement with recent results (40). The 80S ribosomes were purified from ribosomal subunits by sucrose gradient centrifugation and used for cryo-EM microscopy. For structural analysis, we collected initial datasets of 47 and 51 micrographs (51 185 and 48 014 projections) of deletion mutant and wild-type ribosomes, respectively. The cryo-EM reconstructions were done using multiparticle approach (54,55) and therefore only a subset of 33 000 particles in the case of the deletion mutant and 41 528 particles in the case of the wild-type ribosome were used for the final reconstruction. Both reconstructions reached a resolution of ∼13 Å, as estimated with the 0.5 cutoff criteria in the Fourier shell correlation curve. Maps showed the same overall shape with the common features like L1 stalk, stalk base and central protuberance for the 60S subunit and the beak, shoulder, left and right foot for the 40S subunit (Figure 1), thus suggesting that no major assembly defects of the ribosome have occurred in the ΔrpS25 strain.
Figure 1.
The cryo-EM maps from the wild-type and rpS25-deletion mutant. (a and b) 80S ribosomes from wild-type and mutant, respectively, seen from the L1 side. (c–f) The corresponding 60S (blue, c and d) and 40S (yellow, e and f) subunits from the interface side. Landmarks of the 40S subunit are head (h), body (b), beak (bk), platform (pt), left and right foot (lf and rf, respectively). Landmarks for the 60S subunit are central protuberance (CP), L1 protuberance and stalk base (SB). The arrow at the 40S subunit points the estimated localization of the protein rpS25.
Localization of rpS25
In order to localize rpS25-gap, the two cryo-EM reconstructions were compared. For a detailed analysis of the reconstructions, we computationally separated the 80S ribosomes into their 40S and 60S subunits and aligned them, respectively. As expected, the comparison of the 60S subunits does not show any significant differences and the high degree of similarity is confirmed by a high cross-correlation coefficient (0.96).
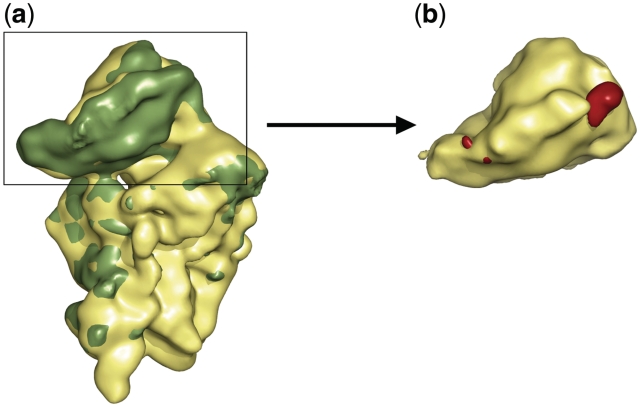
In contrast, a prominent lack of density is present in the head domain of the mutant 40S subunit, which is located adjacent to protein rpS5 (bacterial orthologue S7) at the inter-subunit side of the 40S subunit and above the platform of the 40S subunit (Figures 1e, f and 2a). Thus, the deletion of the genes for rpS25 resulted in a void space in the ribosome density map as expected. In order to analyse the differences between the wild type and the deletion mutant reconstruction in a more quantitative manner, we calculated a difference map of both the structures. As the position of the head appeared somehow changed, we calculated separate difference maps for the head domains and body/platform domains. The presence of significant difference density at the head but not at the body/platform domains confirmed a location of rpS25 in the head domain (Figure 2a).
Figure 2.
Superposition of the 40S subunits from wild-type and the rpS25-deletion mutant. (a) The deletion (yellow) and the wild-type ribosome (green) is shown from the interface side. (b) The difference map (red) between the wild-type and mutant reconstruction shows two peaks at the head of the ΔrpS25 subunit. The most part of the beak side difference overlaps with density in the mutant ribosome, i.e. density is present but weaker than in the control indicative for higher flexibility of this region. In contrast, the difference above the body platform is due to a hole in the density of mutant ribosome indicating the position of the deleted rpS25.
However, the difference map exhibits two difference peaks in the 40S head. The location of one difference peak is in excellent agreement with the density hole observed by a direct comparison of the cryo-EM maps (Figure 2b). The second difference peak is located at the opposite site of the 40S subunit head close to the beak. The presence of two difference peaks is unexpected. One of these difference densities is caused by the lack of rpS25, because only one copy of the protein is present in the ribosome. The pronounced distinctions in the appearance of the cryo-EM map of the mutant ribosome versus that of the wild-type ribosome suggest that the position near the beak is not a site for a missing protein. First of all,-the difference density close to the beak is not caused by the complete lack of density in the mutant ribosome. The density is weaker but can be observed at lower contour level. Such a feature in a cryo-EM map indicates a region with high flexibility. In contrast, the hole in the density located near rpS5 (S7p) is present even at very low contrast levels suggesting the absence of a ribosomal component.
This assignment is corroborated by a comparison of the difference map with previous structural analysis of the yeast 80S ribosome. According to previous models of the yeast 80S ribosome (16,17) the difference peak close to the beak corresponds to helix 33 (h33) of 18S rRNA. The other difference peak, however, corresponds exactly to density that has been previously assigned to a ribosomal protein of unknown identity (16,34). In conclusion, rpS25 can be unequivocally located in the head of the 40S subunit between proteins rpS5 (S7p) and rpS18 (bacterial orthologue S13). This assignment is in excellent agreement with independent structural analysis (21–23). It is likely that the lack of rpS25 in the mutant ribosome leads to a slight destabilization and increased flexibility of the 40S head domain that in turn leads to the weaker density causing the second difference peak close to the beak.
Functional analysis of rpS25
We have previously shown that the protein, which we have identified here as being rpS25, is involved in direct interaction with the CrPV IRES RNA (34). Accordingly, rpS25 interacts with stem loop SL2.3 of the CrPV IRES RNA and stem loop SL2.1 interacts with the adjacent rpS5 (S7p). Such neighbourhood between the two IRES stem loops and rpS25 has been also proposed based on a crosslinking study (39) and both stem loops have been strongly implicated in IRES binding to 40S subunit (48,56). In order to analyse the importance of the observed contact between rpS25 and the IRES RNA, we performed several functional tests with ribosomal particles isolated from a ΔrpS25 strain.
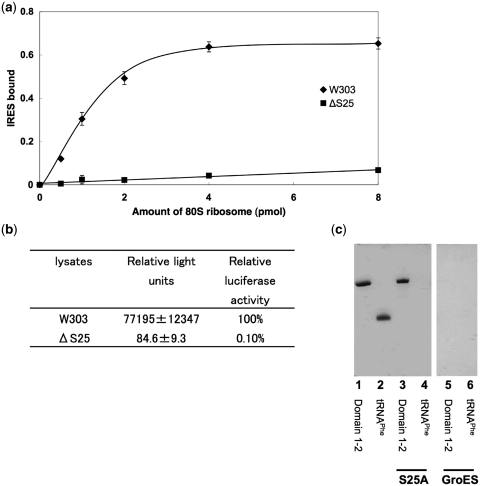
We first tested the contribution of rpS25 to the ability of ribosomes to bind IRES RNA from PSIV, which belongs to the same class of IRES RNAs as the CrPV IRES. Filter binding assays were performed with 32P-labelled PSIV-IRES and increasing amounts of 80S ribosomes isolated either from the yeast strain lacking rpS25 or from the wild-type yeast strain (Figure 3a). The results show that the binding of the IRES RNA is markedly diminished when the reaction was performed in the presence of ribosomes lacking rpS25. At the highest ribosome concentration <10% of the IRES RNA was bound to the rpS25 deficient ribosomes. This result implies that rpS25 deletion should have a direct effect on the PSIV IRES-driven translation initiation, which was tested in the next experiment. We performed in vitro translation assay in yeast lysates prepared either from the wild-type or rpS25 lacking strain (Figure 3b). The measured luciferase activity of the deletion mutant was only 0.1% compared with that of the wild-type (100%) suggesting an essential role of rpS25 on the PSIV IRES mediated translation. Thus, rpS25 is essential for efficient PSIV IRES binding and function. Our results confirms the crosslinking study of Nishiyama et al. (39) where the strongest crosslinking signal to the PSIV IRES was identified for rpS25 and are in excellent agreement with the recently published study by ref. (40), where the essential role of rpS25 for the CrPV IRES and even hepatitis C virus IRES activity has been shown on yeast and mammalian ribosomes, respectively.
Figure 3.
Binding and translation efficiency of IRES RNA. (a) Binding of 32P-labelled IRES RNA (domains 1–3) to yeast wild-type (W303) and ΔS25 80S ribosomes. The fraction of IRES RNA bound is the ratio of [32P]RNA retained on the filter to that of the input [32P]RNA. (b) Luciferase activity of IRES-Fluc with lysates; standard deviations are indicated. The Fluc coding region was preceded by 48 nts of the PSIV capsid coding region in order to increase the Fluc activity [see ref. (45)]. (c) His-tag rpS25 or GroES pull down-assay against RNAs. Control lanes 1 and 2: 25 pmol of IRES RNA domains 1 and 2; and 25 pmol of E. coli tRNAPhe, respectively. RNAs co-precipitated by rpS25 (lanes 3 and 4) and GroES (lanes 5 and 6).
The structural analysis indicated a strong bimolecular interaction between rpS25 and the IRES RNA. This made us wonder, if also isolated rpS25 can directly interact with the IRES RNA or if this interaction is only stable in the context of the whole ribosomal subunit. To test this, the RPS25A gene was cloned into a His-tag containing vector and the binding of the PSIV IRES to the purified His6-rpS25A protein was examined via a pull-down assay. rpS25A protein accounts for ∼66% of the rpS25 in the cell (57), therefore we used only the rpS25A form in this assay. The results clearly show that IRES RNA coprecipitate with rpS25A, whereas the tRNAPhe or combination of RNAs and GroES does not, indicating a direct and specific binding of the PSIV IRES to rpS25A (Figure 3c).
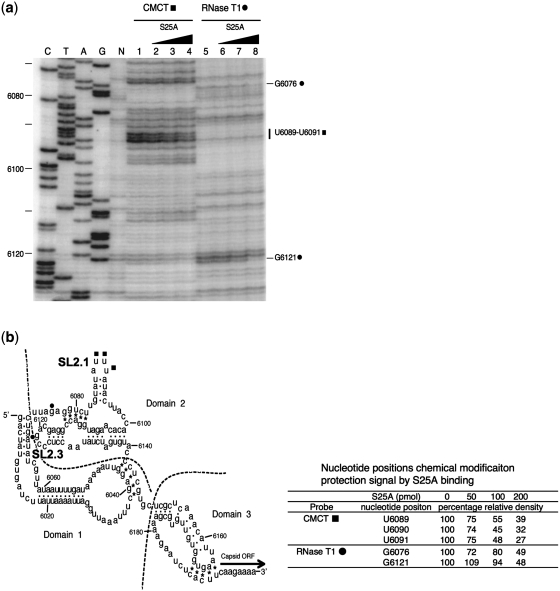
Does the binding mode of IRES to the isolated rpS25 resembles that of IRES to rpS25 on the ribosome? We addressed this question by applying two chemical probing experiments: i.e. chemical foot-printing with CMCT (specific for G and U) and enzymatic protection with RNase T1 (specific for G). Figure 4 reveals that rpS25 protects U6089, U6090 and U6091 of SL2.1 against CMCT and G6076, and G6121 (SL2.3) against T1 RNase. Interestingly, the loop region of SL2.3 of domain 2 of the CrPV IRES was predicted by our previous cryo-EM analysis (34) belong to the attachment site with an unknown protein rpSX, which we identify here as rpS25. The three uracil bases protected against CMCT modification are present in the adjacent loop SL2.1, and both the protected regions were identified previously as contact sites with the small 40S ribosomal subunit (48,56). Our results demonstrate that the isolated rpS25 interacts in a similar way with the IRES as the 40S subunit strongly demonstrating that rpS25 is a main interaction partner with the IRES within the ribosomal subunit. Interestingly, in the cryo-EM map only binding of SL2.3 has been attributed to rpS25 (rpSX). SL2.1 has been seen to interact with rpS5 (S7p), whereas the globular domain of rpS25 is adjacent but not directly in contact with SL2.1 (34). Our present chemical probing experiments, however, suggest that rpS25 can also interact with SL2.1. As we discuss below, the apparent discrepancy could be resolved by a potential interaction of SL2.1 with the extended N-terminal tail of rpS25, which is more flexible and therefore not well resolved in the cryo-EM map.
Figure 4.
Footprint experiments showing protected bases of PSIV-IRES by the binding of isolated rpS25A. (a) To detect protected bases, CMCT (lanes 1–4) and RNase T1 (lanes 5–8) treated RNAs were analysed by primer extension. Lanes 1 and 5, no rpS25; lanes 2 and 6, 5-fold molar excess rpS25A over PSIV-IRES; lanes 3 and 7, 10-fold molar excess rpS25A; lanes 4 and 8, 20-fold molar excess rpS25A. (b) Secondary structure of PSIV IRES; (filled square), bases protected against CMCT by rpS25; (filled circle), bases protected against RNase T1.
Modelling the tertiary structure of rpS25
Knowledge of the exact localization of rpS25—and thus the corresponding density in the 80S•CrPV IRES map—allows us both to propose and evaluate an atomistic model of the protein using a combination of several bioinformatic tools. The structures of nearly all eukaryotic ribosomal proteins modelled so far were generated in homology (sequence identity between template and target structure >30%) to those of their prokaryotic counterparts, an approach known as homology modelling. Since rpS25 does not have any prokaryotic homologue, we could not model the structure of the protein based on sequence similar to another ribosomal protein. In order to find another appropriate template structure to model rpS25, we searched the Protein Data Bank (PDB) using PSI-Blast, an algorithm that detects distant evolutionary relationships between proteins (58).
As a result of the PSI-Blast search, the core and the C-terminal part of rpS25 which account for more than half of the entire sequence are evolutionary related to the γ-domain of the DNA translocase FtsK (PDB accession code 2ve8; Supplementary Figure S1). Within these sections 25% of the amino acids are identical and 51% are similar in both proteins according to the scoring matrix BLOSUM62. Accordingly, it is very likely that the core and the C-terminal part of rpS25 adopt a similar fold as the γ-domain of the DNA translocase FtsK. This is a winged helix composed of three helices in the HTH conformation and a ‘wing’ of two anti-parallel β-sheets (59) that in case of the FtsKγ domain is in β sheet-like conformation, since there is no regular β-sheet main-chain hydrogen-bonding network between the two strands (60).
Winged helix folds are found in different nucleic acid binding proteins such as transcription and elongation factors (59,61–63), but also in proteins which undergo protein–protein interactions (64,65). The FtsKγ domain that binds to the major and minor groove of DNA was chosen as structural template for the amino acids 42–108 of rpS25 (see ‘Materials and Methods’ section). The rpS25 core (amino acid 42–86) was modelled as HTH motif, consisting of one short α-helix (H2) and two longer ones (H1 and H3), preceding the wing (amino acid 87–108) that was modelled in a β-sheet-like conformation (Figure 5a and b). The helices H1 and H3 are connected by a 12 amino acid long-loop comprising the short helix H2, which crosses H1 by exactly 55° (Supplementary Figures S2a). The N-terminal region (amino acid 1–41) could not be modelled based on the selected template structure at this stage. However, this part containing a noticeable amount of basic residues is predicted to consist two α-helices (Supplementary Figure S3a) by Jpred, a consensus secondary-structure prediction server (66).
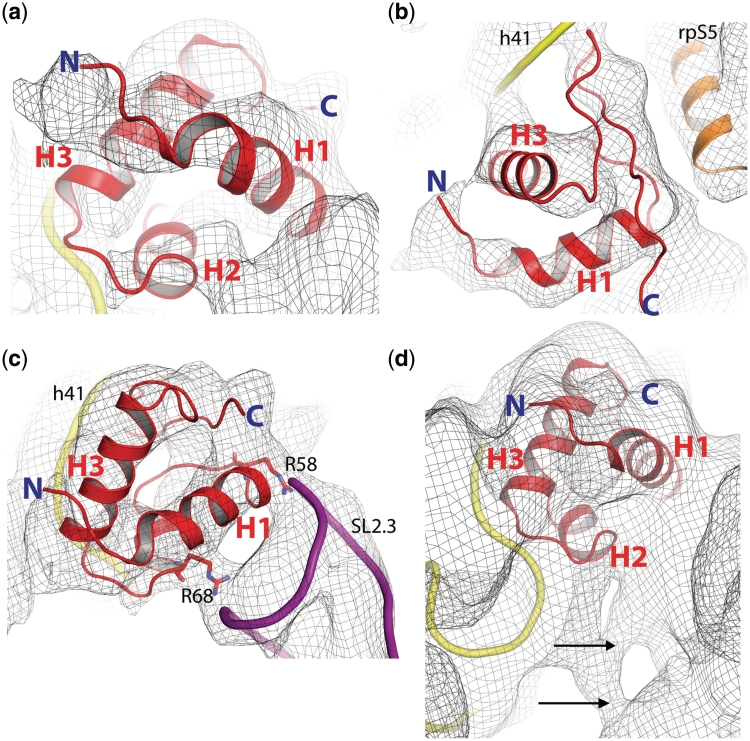
Figure 5.
Tertiary-structure model of rpS25 (comprising residues 42–108) together with yeast 80S•CrPV map at 6.7Å resolution (M. Muhs and C. M. T. Spahn, unpublished). (a) Perspective from the intersubunit space. (b) The C-terminus shows possible interactions of rpS25 with helix 41 (h41) of the 18S rRNA. It is flanked by rpS5. H2 is cut by the clipping plane and is not visible here. (c) The two helices H1 and H3 form the core of a HTH motif. The highly conserved residues Arg58 and Arg68 fit into two well-defined density regions bridging rpS25 to the stem loop SL2.3 of the CrPV IRES. (d) At lower thresholds, the cryo-EM density map indicates further interactions between the CrPV IRES and the ribosome (black arrows). The N-terminus (N) is truncated, because it could not be modelled based on our template structure.
Evaluation of the rpS25 model
To check the quality of the model, the atomic packing density was measured to quantify van der Waals interactions and to detect structural irregularities (67). The overall packing density of the rpS25 model (0.74) is comparable with the atomic packing density of the template structure (0.75) and indicates reasonable packing of all atoms buried inside the protein (Supplementary Figure S2b). This well-packed hydrophobic structural core is mainly composed of the conserved hydrophobic residues Leu51, Val60, Val64, Leu65, Ile71, Leu75, Ala76, Leu80, Leu83 and Ile89 (Supplementary Figures S2c and S3b). Moreover, several conserved polar residues seem to stabilize the tertiary structure of the protein by potential intra- (Supplementary Figure S2d) and inter-molecular hydrogen bonds (Figure 5c). In our model, the H1–H2 turn is stabilized by a potential hydrogen bonding network involving the highly conserved residues Ser61 and Ser63, and the N-terminal cap of H3 by a polar interaction involving Ser74 and Arg77.
Since we have determined the position of rpS25, the homology model can be further evaluated by comparing our model with cryo-EM maps of the yeast 80S ribosome in complex with the CrPV IRES at subnanometre resolution [(34), M. Muhs and C. M. T. Spahn, unpublished data]. Inspection of the cryo-EM density for rpS25 clearly indicates the presence of a rod-like density that is in good agreement with at least two α-helices, which are in a HTH motif like orientation. Indeed, the predicted HTH fold of the core part of our rpS25 model fits well into the cryo-EM density (Figure 5a). Due to the internal symmetry of the HTH motif, six possible orientations of the rpS25 core are possible (Supplementary Figure S4). However, if we include the C-terminal wing, only two of these orientations fit reasonably well into the cryo-EM map. In the first, the wing structure is oriented along the inter-subunit space down to the platform of the 40S subunit (Supplementary Figure S5a). In the second, the wing is directed towards the ribosomal 18S RNA (Supplementary Figure S5b). We favour the second variant, since the model is supported by the presence of unassigned density in the cryo-EM that fits the wing structure. In this orientation, when viewed from the inter-subunit space, one of the two longer helices (H1) occupies the front density, while the other one (H3) is located behind it and rotated by nearly 69° (Figure 5, Supplementary Figure S5b).
Accordingly, the C-terminal wing structure is suggested to occupy the empty density in close vicinity to helix 41 (h41) of the ribosomal 18S RNA indicating a potential interaction (Figure 5b). Moreover, the proposed rpS25 model is supported by two well-defined contact densities between rpS25 and the CrPV IRES present in the cryo-EM map (Figure 5c). These are occupied by two highly conserved arginines (Arg58 and Arg68) of H1, which could perfectly bridge the ribosomal protein to the negatively charged backbone of the CrPV IRES. Thus, we propose that rpS25 interacts with the ribosomal RNA using the C-terminal wing structure and with the IRES RNA per HTH (Figure 5c). This binding mode would strikingly resemble that of the used template structure (FtsKγ), which interacts with the major groove of the DNA via the HTH motif and with the next minor groove via the wing (60).
DISCUSSION
rpS25 is one of the proteins, which is specific for the eukaryotic 80S ribosome, i.e. it does not have a bacterial homologue. Localization, structure and function of eukaryotic-specific ribosomal proteins are largely unknown. Although the fundamental mechanism of protein synthesis is evolutionary conserved, there are pronounced differences between eukaryotes and prokaryotes. In particular, the initiation phase of translation is much more complicated in eukaryotes and eukaryotic ribosomes are subjected to a more intensive regulation. Thus, it is conceivable that the role of at least some of these extra proteins is to play a role in control mechanisms of eukaryotic translation, for example, as possible targets for regulation factors. Interestingly, rpS25 has been suggested to play a crucial role in the cap-independent translation initiation mediated by IRES RNA present in the intergenic region (IGR) of several viruses of the family Dicistroviridae (39,40). In excellent agreement with these studies, we have shown here that rpS25 is essential for IGR–IRES translation activity and in particular for IRES binding. We further demonstrate that the IGR–IRES interact even with the isolated rpS25 outside of the context of the ribosome.
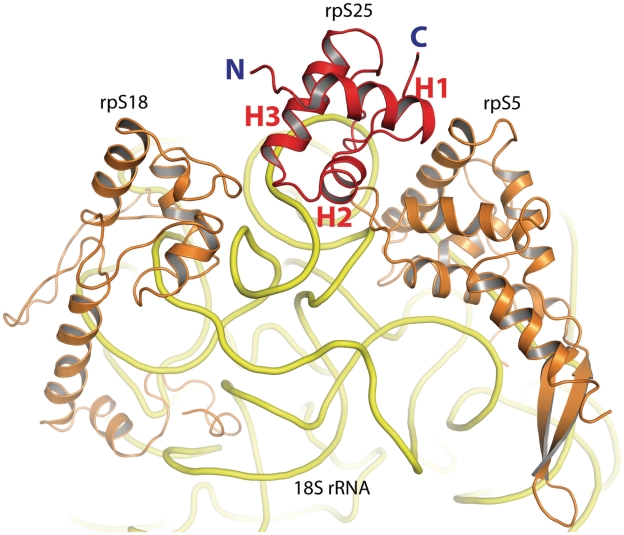
The decisive role of rpS25 for IRES binding can be explained by the present structural analysis. Our results clearly identify the location of the protein in the head domain of the ribosomal 40S subunit. rpS25 is wedged between rpS18, rpS5 and the 18S rRNA (Figure 6). The present resolution of our yeast 80S•CrPV cryo-EM map allowed us to propose a tentative model for the structure of the protein as a winged HTH protein. Remarkably, despite the low-sequence identity to the used template structure, the rpS25 model significantly fits into the cryo density without any adjustments (Figure 5a). Our cryo-EM map clearly indicates helical densities suitable for the HTH fold of the proposed rpS25 core. Surprisingly, if oriented in the selected manner, the C-terminal wing occupies an empty density in close vicinity to helix 41 of the 18S rRNA, which visually coincides with the wing structure (Figure 5b).
Figure 6.
rpS25 with neighbouring proteins and 18S rRNA. rps25 (red) is shown from the side of the N-terminal helix (H1). It is flanked by ribosomal proteins S18 (left) and S5 (right). The 18S rRNA is depicted in yellow.
According to our tertiary-structure model, rpS25 contacts the stem loop 2.3 of the CrPV IRES via two conserved arginines of the HTH motif thereby intercalating between two RNAs, the ribosomal and the IRES RNA (Figure 5c), thus strongly resembling the binding mode of the template structure FtsKγ that binds to the minor and major groove of DNA. Structural studies on different proteins have shown that the winged helix motif, as we propose for the rpS25 structure, is extremely versatile (Supplementary Figure S6). Therefore, at the present resolution we cannot exclude the possibility that the C-terminal wing structure of the protein adopts another conformation or even occupies another location. Also, the internal symmetry of the HTH motif increases the uncertainty, since the helices, especially those of the same length, can fill in other densities as suggested in this study. However, after our analysis had been concluded independent models of rpS25 have been presented based on a cryo-EM map of the 80S ribosome at 5.5 Å resolution (21) and in particular by X-ray structures of the yeast 80S ribosome (22) and the Tetrahymena thermophila 40S ribosomal subunit in complex with initiation factor eIF1 (23). All four models are in good overall agreement, suggesting the same overall fold of rpS25 (Supplementary Figure S7) as well as the same orientation of the protein in the context of the ribosome. Therefore, structural modelling in combination with cryo-EM at subnanometre resolution has the potential to correctly predict the fold of unknown proteins, given that sufficient additional information, i.e. the exact localization of the protein, is available.
Our cryo-EM map indicates two density bridges connecting rpS25 and the SL2.3 of the IRES (Figure 5c). A direct binding of the protein to the viral RNA is also proven by the His-tag pull-down assay performed in this study. The results provide an explanation for the complete abolishment of the IRES-driven translation initiation in the absence of the protein as shown by our filter binding and in vitro translation assays using the related PSIV IRES. The data coincide with previous biochemical results performed in the presence of CrPV and HCV IRES (40). Interestingly, the transcription of the rpS25 mRNA is increased several fold in mammalian and human cells under stress conditions like amino acid starvation or apoptosis (68,69). It is conceivable that a free pool of rpS25 might block the attachment of IRES structures to 40S subunits and thus protects the cell of translation of unfavourable mRNAs.
The structural role of the N-terminal region of rpS25 does not become clear from our modelling studies, at this stage. However, our cryo-EM map of the mammalian 80S ribosome (T. Budkevich and C. M. T. Spahn, unpublished data) or of the wheat germ ribosome (EMD accession code: EMD-1664), which do not comprise the IRES, suggest a potential position of the 41 amino acid long N-terminus straight down towards the platform of the 40S subunit. In the yeast 80S•CrPV map analysed in this study, we see at lower contour level a connection bridging the IRES and the hypothetical N-terminus of rpS25, which may have moved towards the IRES (Figure 5d, Supplementary Figure S7). These observations let us to propose a model, where the N-terminus of rpS25 acts like a hinge and contributes to IRES binding. As the N-terminal tail approaches SL2.1 of the IRES this proposed interaction may result in the clear chemical protection of SL2.1 by rpS25 (Figure 4). Interestingly, there is a lysine-rich region (Supplementary Figure S3a, 8 of 13 residues are lysines), which could potentially accomplish the IRES binding in the middle of the N-teminus. In this model the conserved Lys70 present in the H2–H3 loop (Supplementary Figure S8) would bind the N-terminus and thus act like an anchor keeping the core in the right position and allowing the motion of the N-terminus, at the same time.
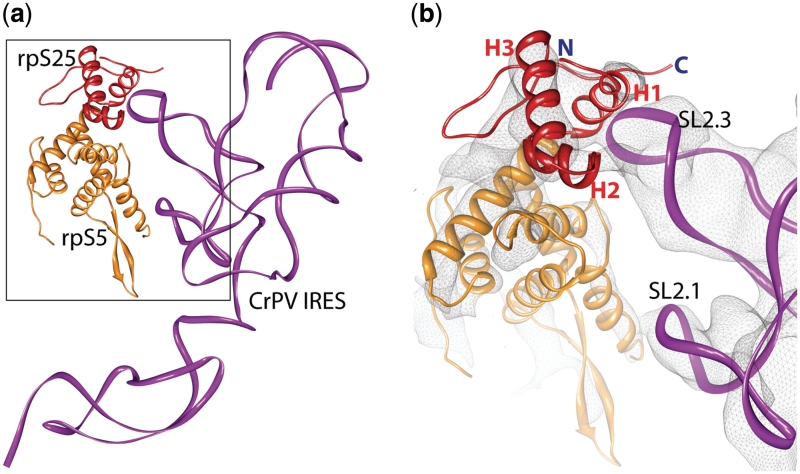
The binding of CrPV IRES and related IRES to the 40S subunit occurs via SL2.1 and SL2.3 of domain 2 of the IRES RNA (34). The contact between domain 3 of the IRES and the decoding centre of the 40S subunit is functionally important but does not contribute in a major way to the affinity of the IRES for the ribosome Thus, rpS25 together with the neighbouring rpS5 (S7p) constitutes the major binding site of the ribosome for the CrPV IRES family (Figure 7).
Figure 7.
Interactions of the IRES RNA with the ribosome. (a) Molecular interactions of the CrPV IRES (magenta ribbon) with ribosomal proteins S5 (orange) and S25 (red). (b) A close-up of the interaction partners docked into the cryo-EM density (grey mesh) of the yeast 80S•CrPV complex [(34); M. Muhs and C. M. T. Spahn, unpublished data].
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft DFG (SFB 740 TP A3 and TP Z1 to C.M.T.S.); the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum) and Alexander von Humboldt-Foundation GAN 1127366 STP-2 (to H.Y.). Funding for open access charge: DFG (SFB 740).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sébastien Ferreira-Cerca and Philipp Milkereit for providing the strain Y338.
REFERENCES
- 1.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 2.Frank J, Spahn CM. The ribosome and the mechanism of protein synthesis. Rep. Prog. Phys. 2006;69:1383–1417. [Google Scholar]
- 3.Hofmann KP, Spahn CM, Heinrich R, Heinemann U. Building functional modules from molecular interactions. Trends Biochem. Sci. 2006;31:497–508. doi: 10.1016/j.tibs.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 7.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 8.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 9.Yonath A. Antibiotics targeting ribosomes: resistance, selectivity, synergism and cellular regulation. Annu. Rev. Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 10.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr. Opin. Chem. Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green R, Noller HF. Ribosomes and translation. Ann. Rev. Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 12.Dresios J, Panopoulos P, Synetos D. Eukaryotic ribosomal proteins lacking a eubacterial counterpart: important players in ribosomal function. Mol. Microbiol. 2006;59:1651–1663. doi: 10.1111/j.1365-2958.2006.05054.x. [DOI] [PubMed] [Google Scholar]
- 13.Gerbi SA. Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA. Structure, Evolution, Processing, and Function in Protein Synthesis. New York: CRC Press; 1996. pp. 71–87. [Google Scholar]
- 14.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Spahn CMT, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S Ribosome from Saccharomyces cerevisiae - tRNA-Ribosome and Subunit-Subunit Interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 17.Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halic M, Becker T, Frank J, Spahn CM, Beckmann R. Localization and dynamic behavior of ribosomal protein L30e. Nat. Struct. Mol. Biol. 2005;12:467–468. doi: 10.1038/nsmb933. [DOI] [PubMed] [Google Scholar]
- 20.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl Acad. Sci. USA. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl Acad. Sci. USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 23.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 2010;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 24.Sweeney R, Chen L, Yao MC. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol. Cell. Biol. 1994;14:4203–4215. doi: 10.1128/mcb.14.6.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 26.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 29.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl Acad. Sci. USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 33.Jan E, Goss Kinzy T, Sarnow P. Divergent tRNA-like element supports initiation, elongation and termination of protein biosynthesis. Proc. Natl Acad. Sci. USA. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 35.Spahn CMT, Kieft JS, Grassucci RA, Penczek P, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 36.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 42.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J. Biol. Chem. 2007;282:7770–7776. doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]
- 45.Shibuya N, Nishiyama T, Kanamori Y, Saito H, Nakashima N. Conditional rather than absolute requirements of the capsid coding sequence for initiation of methionine-independent translation in Plautia stali intestine virus. J. Virol. 2003;77:12002–12010. doi: 10.1128/JVI.77.22.12002-12010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uchiumi T, Nomura T, Shimizu T, Katakai Y, Mita K, Koike Y, Nakagaki M, Taira H, Hachimori A. A covariant change of the two highly conserved bases in the GTPase-associated center of 28 S rRNA in silkworms and other moths. J. Biol. Chem. 2000;275:35116–35121. doi: 10.1074/jbc.M004596200. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu T, Nakagaki M, Nishi Y, Kobayashi Y, Hachimori A, Uchiumi T. Interaction among silkworm ribosomal proteins P1, P2 and P0 required for functional protein binding to the GTPase-associated domain of 28S rRNA. Nucleic Acids Res. 2002;30:2620–2627. doi: 10.1093/nar/gkf379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl Acad. Sci. USA. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 51.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 52.Rose A, Goede A, Hildebrand PW. MPlot–a server to analyze and visualize tertiary structure contacts and geometrical features of helical membrane proteins. Nucleic Acids Res. 2010;38:W602–W608. doi: 10.1093/nar/gkq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wool IG, Chan YL, Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- 54.Spahn CM, Penczek PA. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr. Opin. Struct. Biol. 2009;19:623–631. doi: 10.1016/j.sbi.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loerke J, Giesebrecht J, Spahn CM. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- 56.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 57.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 58.Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proc. Natl Acad. Sci. USA. 2009;106:9637–9642. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gajiwala KS, Burley SK. Winged helix proteins. Curr. Opin. Struct. Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 60.Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol. Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong G, Chakshusmathi G, Wolin SL, Reinisch KM. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 2004;23:1000–1007. doi: 10.1038/sj.emboj.7600115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selmer M, Su XD. Crystal structure of an mRNA-binding fragment of Moorella thermoacetica elongation factor SelB. EMBO J. 2002;21:4145–4153. doi: 10.1093/emboj/cdf408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fourmy D, Guittet E, Yoshizawa S. Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J. Mol. Biol. 2002;324:137–150. doi: 10.1016/s0022-2836(02)01030-6. [DOI] [PubMed] [Google Scholar]
- 64.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 65.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 66.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 67.Rother K, Hildebrand PW, Goede A, Gruening B, Preissner R. Voronoia: analyzing packing in protein structures. Nucleic Acids Res. 2009;37:D393–D395. doi: 10.1093/nar/gkn769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lavery WL, Goyns MH. Increased expression of the S25 ribosomal protein gene occurs during ageing of the rat liver. Gerontology. 2002;48:369–373. doi: 10.1159/000065505. [DOI] [PubMed] [Google Scholar]
- 69.Adilakshmi T, Laine RO. Ribosomal protein S25 mRNA partners with MTF-1 and La to provide a p53-mediated mechanism for survival or death. J. Biol. Chem. 2002;277:4147–4151. doi: 10.1074/jbc.M109785200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.