Abstract
Methylation at the 5-position of cytosine is a well-studied epigenetic pathway. In addition to 5-methylcytosine (5mC), substantial amounts of 5-hydroxymethylcytosine (5hmC) also referred to as the sixth DNA base have been detected in certain tissues, most notably the brain. However, the genomic distribution of this cytosine modification is unknown. Here, we have used an immunoprecipitation technique (5hmC-IP) to examine the occurrence of 5hmC in DNA from human brain frontal lobe tissue. The distribution of 5hmC was compared to that of 5mC. We show that 5hmC is more selectively targeted to genes than is 5mC. 5hmC is particularly enriched at promoters and in intragenic regions (gene bodies) but is largely absent from non-gene regions. 5hmC peaks at transcription start sites did not correlate with gene expression levels for promoters with intermediate or high CpG content. However, the presence of 5hmC in gene bodies was more positively correlated with gene expression levels than was the presence of 5mC. Promoters of testis-specific genes showed strong 5mC peaks in brain DNA but were almost completely devoid of 5hmC. Our data provide an overview of the genomic distribution of 5hmC in human brain and will set the stage for further functional characterization of this novel DNA modification.
INTRODUCTION
In mammalian cells, methylation of DNA at the 5-position of cytosine bases is an enzymatic process targeted mainly to CpG dinucleotides. DNA cytosine methylation is generally copied during DNA replication in somatic tissues, is reversible during certain stages of early development and is in some cases correlated with modulation of gene expression (1,2). The initial formation and copying of 5-methylcytosine (5mC) patterns is catalyzed by DNA methyltransferases (DNMT1, DNMT3A and DNMT3B) (3,4). Patterns of 5mC, as a stable epigenetic modification of the genome, have been investigated in many tissues using a variety of techniques (5–7). Whereas the presence of 5mC at promoter CpG islands is most often incompatible with gene transcription, the reverse is true for gene bodies where the presence of 5mC is positively correlated with gene expression levels in both plants (8,9) and mammals (10,11).
In 2009, Kriaucionis and Heintz and Tahiliani et al. made the seminal discovery that another specific modified DNA base, 5-hydroxymethylcytosine (5hmC) is present in mouse Purkinje and granule neurons and in embryonic stem (ES) cells (12,13). An enzymatic activity involved in producing 5hmC from 5mC was identified as the TET1 5mC oxidase (13). In the meantime, 5hmC also has been detected at substantial levels in other mammalian tissues and cell types (14–16). Furthermore, two mammalian homologues of TET1, TET2 and TET3 have been characterized and shown to possess similar catalytic activities (17).
5hmC might serve unique biological roles. For example, 5hmC may be recognized by specific proteins that translate a functional role of the modified base in gene control mechanisms. In addition, it was shown that 5hmC inhibits the binding of the methyl-CpG binding domain of MeCP2 (18), and of full-length MBD1, MBD2 and MBD4 proteins to DNA (19). Thus, 5hmC counteracts the role of several 5mC-targeted transcriptional repressors, suggesting a potential gene regulatory function of 5hmC. Others have suggested that 5hmC might be an intermediate in direct DNA demethylation (20,21), although direct evidence for this pathway is currently not available (16).
Since 5hmC is present in mammalian DNA at physiologically relevant levels and in a tissue-specific manner (12–16), there is an important need to determine the genomic location of 5hmC. Here, we have used immunoprecipitation with a 5hmC-specific antibody to map the distribution of this modified base in human brain DNA.
MATERIALS AND METHODS
DNA samples
DNA from two frontal lobe brain tissues of accident victims was obtained from Capital Biosciences (Gaithersburg, MD, USA). Genomic DNA from mouse ES cells was extracted by using the DNeasy Blood and Tissue kit (Qiagen; Valencia, CA, USA). Genomic DNA was fragmented by sonication to an average size of ∼400 bp using a Sonifier cell disruptor 350 (Branson; Danbury, CT, USA).
Enrichment of 5hmC-containing DNA fragments with anti-5hmC antibody
To characterize the anti-5hmC antibody, 76-mer oligonucleotides (sequence 5′-CCTCACCATCTCAACCAATATTATATTACGCGTATATCGCGTATTTCGCGTTATAATATTGAGGGAGAAGTGGTGA-3′) containing C, 5mC or 5hmC at the six 5′-CG sequences on each strand were prepared as described previously (19). Twenty-five picograms of each double-stranded 76-mer were mixed with 0.4 µg of sonicated genomic DNA from mouse ES cells, denatured in 10 mM Tris–HCl, 1 mM ethylenediaminetetraacetic acid buffer, pH 7.5 (TE buffer), for 10 min at 98°C and immediately chilled on ice for 10 min. The denatured DNA was immunoprecipitated with control rabbit IgG or with a polyclonal anti-5hmC antibody from Active Motif (Carlsbad, CA, USA) in a final volume of 200 μl immunoprecipitation buffer (10 mM sodium phosphate, pH 7.0, 140 mM NaCl and 0.05% Triton X-100) by incubation for 2 h at 4°C on a rocking platform. To allow selective collection of immuno-captured 76-mers, the mixtures were then incubated with 10 μl of magnetic Dynabeads M-280 sheep antibody against rabbit IgG (Dynal Biotech) for 2 h at 4°C on a rocking platform and washed three times with 700 μl of immunoprecipitation buffer for 10 min at room temperature. The enriched DNA was purified using QIAquick PCR purification kit (Qiagen). The levels of immuno-captured 76-mers were measured using quantitative real-time PCR using the following conditions: 95°C for 3 min followed by 50 cycles at 95°C for 10 s and 50°C for 45 s with 0.6 U iTaq polymerase in an iQ5 real-time PCR cycler (Biorad; Hercules, CA, USA), using the forward primer 5′-CCTCACCATCTCAACCAATA-3′, the reverse primer 5′-TCACCACTTCTCCCTCAAT-3′ and the probe 5′-CGCGTATATCGCGTATTTCGCG-3′ with 5′-Cy5 and 3′-Iowa Black RQ-Sp modifications (IDT; Coralville, IA, USA). Data were analyzed with the iQ5 optical system software and displayed as percent by referring to the level of 5hmC immuno-captured DNA.
Immuno dot blot assay
Genomic DNAs from human brain and mouse ES cells were denatured in TE buffer for 10 min at 98°C, immediately chilled on ice for 10 min and spotted onto a charged nylon-based membrane. The membrane was blocked with 5% non-fat milk at 4°C overnight. After washing, the amount of 5hmC in genomic DNA was detected using rabbit polyclonal anti-5hmC antibody (Active Motif; 1:10 000) followed by peroxidase-conjugated anti-rabbit IgG (Jackson Laboratory; Bar Harbor, ME, USA; 1:7000) secondary antibody. The signal was visualized by using ECL-Plus (Amersham Pharmacia Biotech).
Mapping of 5mC
Brain DNA (0.4 µg) was fragmented by sonication to an average size of ∼400 bp. Enrichment of the methylated DNA fraction by the methylated-CpG recovery assay (MIRA) was performed as described previously (22). NimbleGen tiling arrays were used for analysis of 5mC and 5hmC genomic distribution (720 k human CpG island plus promoter arrays). The labeling of amplicons, microarray hybridization and scanning were performed according to the NimbleGen protocol. These arrays cover all UCSC Genome Browser-annotated CpG islands and the promoter regions for all RefSeq genes. The promoter region covered is from −2440 to +610 bp relative to the transcription start sites (TSSs). For all samples, the MIRA-enriched DNA was compared with the input DNA.
Mapping of 5hmC
To map 5hmC in the human brain genome, at first, adaptor-ligated DNA fragments were prepared as described previously with some modifications (22). Two micrograms of sonicated DNA was end-repaired with T4 DNA polymerase (New England Biolabs; Ipswich, MA, USA) to create blunt-ended DNA fragments by incubation for 20 min at 12°C and purified using phenol/chloroform extraction and ethanol precipitation with 1 μl of 20 mg/ml glycogen as carrier (Invitrogen; Carlsbad, CA, USA). The blunt-ended DNA was incubated for 30 min at 37°C with T4 polynucleotide kinase to phosphorylate the 5′-ends. The DNA samples were purified again by phenol/chloroform extraction. The end-repaired DNA fragments were linker-ligated with 6 μl of 50 μM double-stranded adaptors as used in the MIRA protocol (22), 10× T4 DNA ligation buffer, 1 μl of 400 U/μl T4 DNA ligase (New England Biolabs) in 50 μl of total volume and overnight incubation at 16°C, and then purified using QIAquick PCR purification kits (Qiagen). To fill in the overhangs, the adaptor-ligated DNA was incubated at 72°C for 10 min in a reaction containing dNTP mix, 10× PCR buffer, 1.5 μl of 5 U/μl Taq polymerase (Qiagen) and purified using QIAquick PCR purification kit (Qiagen). The DNA was denatured in 200 μl of TE buffer for 10 min at 98°C and immediately chilled on ice for 10 min. An aliquot (20 μl) of denatured DNA was saved as input. The adaptor ligated and denatured DNA was immunoprecipitated with anti-5hmC antibody as described above in the 5hmC-enrichment assay with some modifications. The DNA was incubated with 2 μl of rabbit polyclonal anti-5hmC antibody (Active Motif) and incubated for 14 h at 4°C on a rocking platform. To allow selective collection of immuno-captured DNA, the mixtures were then incubated with 15 μl of magnetic Dynabeads M-280 sheep antibody to rabbit IgG (Dynal Biotech) for 2 h at 4°C on a rocking platform and washed three times with 700 μl of 1× immunoprecipitation buffer for 10 min at room temperature. Washed beads were resuspended in 200 μl of TE buffer with 0.25% sodium dodecyl sulfate and 0.25 mg/ml proteinase K (NEB) for 2 h at 55°C and purified by phenol/chloroform extraction and ethanol precipitation with 1 μl of 20 mg/ml glycogen as carrier. The immuno-captured DNA was PCR amplified in parallel with the input DNA, and then analyzed on NimbleGen CpG island plus promoter arrays as described above for the 5mC mapping assay. For all samples, the 5hmC-enriched DNA was compared with the input DNA. The microarray data were deposited in the GEO database (accession number GSE27051).
Identification and annotation of 5mC and 5hmC peaks
NimbleGen array data were normalized by the Loess method and log2 ratios were calculated using the Bioconductor ‘Limma’ package. Technical replicates of 5hmC arrays produced Pearson's correlation coefficients between R = 0.65–0.70; P < 2.2 × 10e-16. Probes were selected as positive if their log2 ratio was above the 95th percentile of the ratios on the entire array (P < 0.05). For our analysis, we defined a methylation peak as a region with at least four consecutive positive probes allowing one gap and covering a minimum length of 350 bp. This stringent peak definition will give few false-positive results, with false discovery rate < 0.05 estimated by repeating the peak identification using randomized log2 ratios on the array. Identified peaks were mapped relative to known transcripts defined in the UCSC Genome Browser HG18 RefSeq database. Methylation peaks falling between −2.4 kb upstream and +0.5 kb downstream of TSSs were defined as promoter peaks; those falling within gene bodies (from 500 bp downstream of transcription start to 500 bp upstream of transcript end) were defined as ‘intragenic’ peaks. Methylation peaks that are >2440 bp upstream and >500 bp downstream of any known transcripts were defined as ‘intergenic’.
Identification of 5mC-specific and 5hmC-specific peaks
Methylation peaks in 5mC and 5hmC samples were identified as above. Specific peaks for NB1 and NB2 (two normal brain samples from different individuals) were identified separately. To identify 5mC-specific peaks, the average log2 ratios of probes within each peak in the 5mC sample were compared with the average log2 ratios of the same probes in the 5hmC sample, and the peaks were considered as specific if the difference was more than 1. A similar method was used to identify 5hmC-specific peaks.
Correlation between 5mC or 5hmC and gene expression
Promoters were classified into three categories reflecting their CpG frequency, high-CpG (HCP), intermediate CpG (ICP) and low CpG (LCP) according to Weber et al.'s approach (23). To determine the relationship between methylation or hydroxymethylation of cytosine at promoters and expression levels of genes, we separated each HCP, ICP and LCP promoter category into two subcategories, containing 5mC or 5hmC or not, based on whether there was a peak overlapping with the promoter region (−2400 to +500 relative to the TSS). Gene expression data for frontal cortex tissue were downloaded from Gene Expression Omnibus (GEO, accession number GSE3790). The raw CEL files were processed by the robust multi-array average (RMA) method implemented in the Bioconductor ‘Affy’ package and the average log2 intensity of each gene across all 30 samples was calculated. For each promoter class, Student's t-test was applied to compare whether the expression values of the genes with promoter cytosine modification are different from those of the genes without the modifications.
Gene ontology analysis
Gene ontology analysis was performed using DAVID functional annotation tools with Biological Process FAT and Molecular Function FAT datasets. The enriched Gene Ontology terms were reported as clusters to reduce redundancy. The P-value for each cluster is the geometric mean of the P-values for all the GO categories in the cluster. The gene list in each cluster contains the unique genes pooled from the genes in all the GO categories in the cluster. We report all the clusters with P < 0.05 except for 5mC intragenic peaks, in which P < 0.01 was used to save space.
RESULTS
Mapping of 5hmC in mammalian DNA
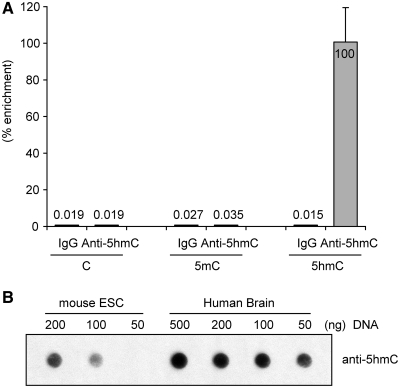
We used an antibody against 5hmC in immuno dot blots and in immunoprecipitation experiments. Initially, we verified that this antibody specifically immunoprecipitates oligonucleotides containing 5hmC but not the same sequences containing cytosine (C) or 5mC (Figure 1A). Immunoprecipitation was most effective when the DNA was denatured. Using immuno dot blotting with the same antibody, we verified that human frontal cortex DNA contains about four-times higher levels of 5hmC than mouse ES cells (Figure 1B) consistent with previous results showing that 5hmC is abundant in brain tissue, most notably in cortex DNA (15,16). We then proceeded to map 5hmC in two different human brain DNA samples derived from the frontal cortex. DNA was sonicated and linker ligated, then denatured and immunoprecipitated with anti-5hmC antibody. Immunoprecipitated fragments and aliquots of input DNA were PCR amplified using linker primers and analyzed on NimbleGen 720 k CpG island plus promoter arrays interrogating ∼28 000 human CpG islands and all Refseq promoters spanning regions of −2.44 kb upstream from the TSS to +0.61 kb downstream of the TSS. In parallel, we mapped the distribution of 5mC using the MIRA, a technique, which was established in our laboratory earlier (10,22,24). This method is based on the MBD2 protein as the methyl-CpG binding entity. MBD2-based DNA methylation mapping techniques and those based on immunoprecipitation with an antibody specific for 5mC have very similar sequence coverage and are highly concordant (25–27) although the exact concordance may depend on experimental parameters such as salt concentration used in the washing steps (28,29). We used a medium salt concentration of 700 mM. Log2 scatter plots of the array data (Supplementary Figure S1) show that the Pearson's correlation coefficient between the two different brain samples (NB1 and NB2) is higher for 5mC (R = 0.81) than for 5hmC (R = 0.61) but both data sets are statistically highly correlated (P < 2.2 × 10e-16). The reason for this difference could be related to technical issues but it is also possible that variability is inherently greater for 5hmC than for 5mC because 5hmC is a secondary modification that depends on the first one (5mC).
Figure 1.
Immunoprecipitation and immuno dot blot analysis of 5hmC with a specific antibody. (A) Twenty-five picograms of 76-mer oligonucleotides containing C, 5mC or 5hmC at identical CpG positions were mixed with 400 ng of sonicated mouse genomic DNA from ES cells, incubated with anti-5hmC antibody and immunoprecipitated. After immunoprecipitation and washing, the spiked target sequences were amplified by quantitative real-time PCR and the specificity of the immunoprecipitation reaction was determined. Normal rabbit IgG was used as a control. (B) Immuno dot blot analysis of 5hmC in DNA from mouse ES cells and in DNA from human brain frontal cortex. A dilution series was used for semi-quantitative analysis of 5hmC levels.
Genomic location of 5hmC peaks
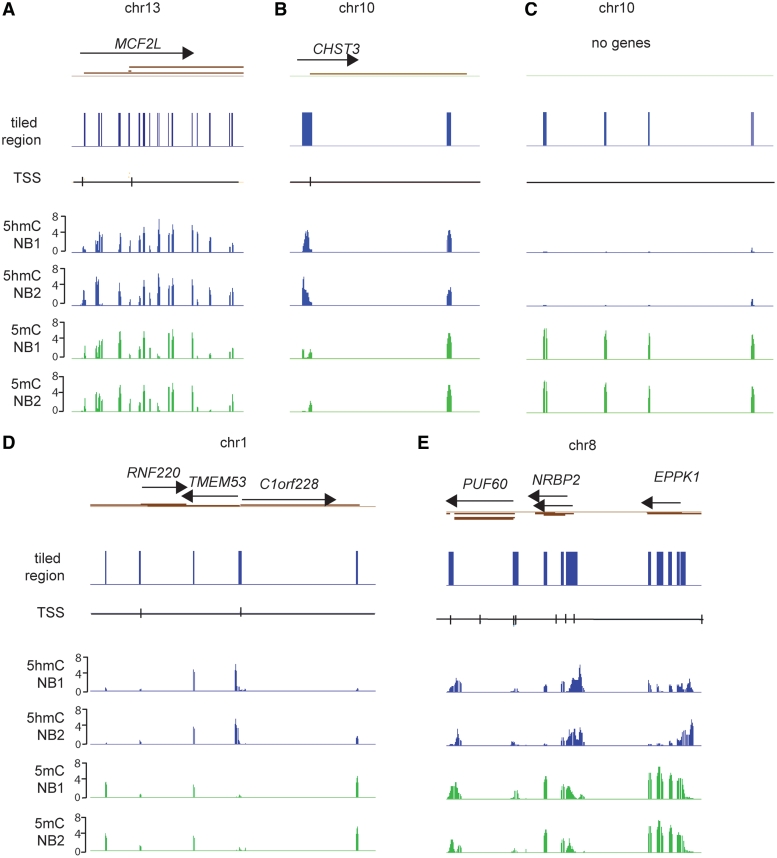
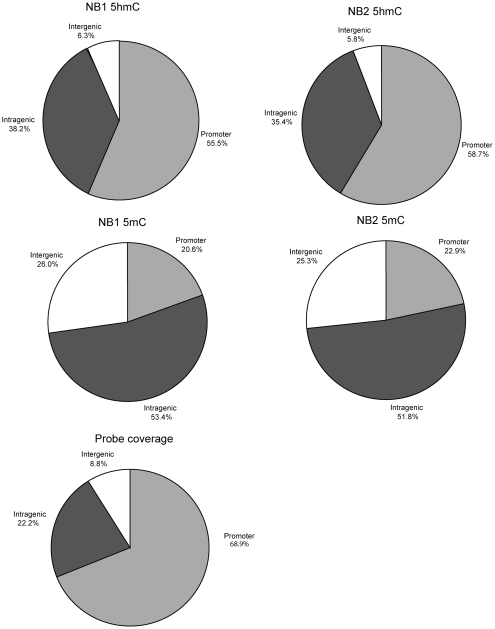
Figure 2 shows several snapshots of 5hmC mapping in human brain DNA. Examples are presented where 5mC and 5hmC either overlap or are more unique for each modified base at specific locations. After Loess normalization, peaks were designated as such when they had at least four consecutive probes above the 95th percentile of the log2 ratios allowing a one-probe gap. In total, we identified about 2600 peaks for 5mC and between 1800 and 2100 peaks for 5hmC in each of the two brain samples. A list of the peaks is shown in Supplementary Tables S1–S4. Peaks were assigned according to their genomic location. For 5mC, slightly >20% of the peaks were at promoter regions, defined as −2.4 kb upstream to 0.5 kb downstream of the TSS, about 52–53% were intragenic and approximately 25–26% were not associated with genes (intergenic). For 5hmC, a higher percentage (55–59%) of the peaks were at promoters, approximately 35–38% were intragenic, i.e. in gene bodies, and only ∼6% were intergenic (Figure 3). Although this distribution is influenced by the array design, the result clearly shows that 5hmC is targeted to genes, in particular promoters, and is more selectively depleted from non-gene regions than is 5mC.
Figure 2.
Mapping of 5hmC in human brain DNA. Several examples of mapping of 5hmC and 5mC in human brain DNA are shown. The blue columns indicate the tiled regions. Green peaks reflect 5mC and blue peaks show the distribution of 5hmC peaks in each of two different normal brain samples (NB1 and NB2). The TSSs and transcripts are indicated. The data are presented as P-value scores (-log10 of the P-value). (A) The MCF2L gene on chromosome 13; (B) the CHST3 gene on chromosome 10; (C) an area of chromosome 10 is shown where several CpG islands are located in a gene-free region. Such regions are often enriched for 5mC peaks but lack 5hmC. (D) The RNF220, TMEM53 and C1orf228 genes on chromosome 1; (E) the PUF60, NRBP2 and EPPK1 genes on chromosome 8.
Figure 3.
Distribution of 5hmC and 5mC peaks in the genome. The location of the respective peaks was assigned as being within the promoter, intragenic, i.e. within gene bodies, or intergenic, i.e. not associated with genes. Two normal brain samples (NB1 and NB2) were analyzed for 5hmC and 5mC. The pie chart at the bottom of the Figure indicates what percentage of sequences covered by the microarray probes fall into promoters, intra- and intergenic categories.
Relationship between 5hmC and 5mC peaks
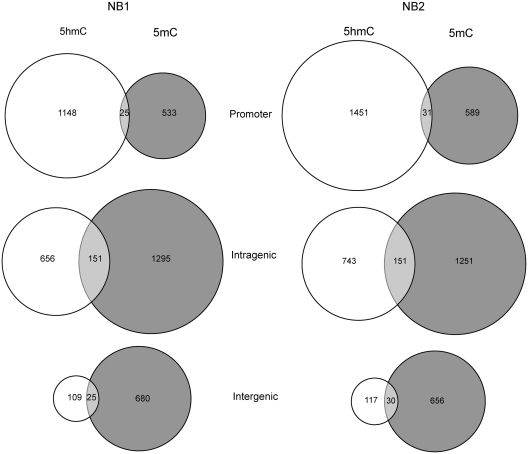
5hmC and 5mC peaks that overlapped by at least one base were considered as common peaks. Figure 4 shows that 5hmC peaks and 5mC peaks overlap infrequently. Intragenic peaks have more overlap between 5mC and 5hmC than promoter peaks and intergenic peaks (chi-square test; P < 2.2e-16 for 5hmC and P < 1e-07 for 5mC). However, peaks at promoter regions rarely overlap (Figure 4).
Figure 4.
Overlap between 5hmC and 5mC peaks. The peaks found in two brain samples NB1 and NB2 were annotated to promoter, intragenic and intergenic regions as described in the ‘Materials and Methods’ section. For each of the regions, peaks were considered as common between 5mC and 5hmC when they overlapped at least by 1 bp. Intragenic peaks of 5hmC overlap more commonly with 5mC than 5hmC peaks in promoters or intergenic regions do (chi-square test, P < 2.2e-16).
Correlation between 5hmC or 5mC at promoters and gene expression
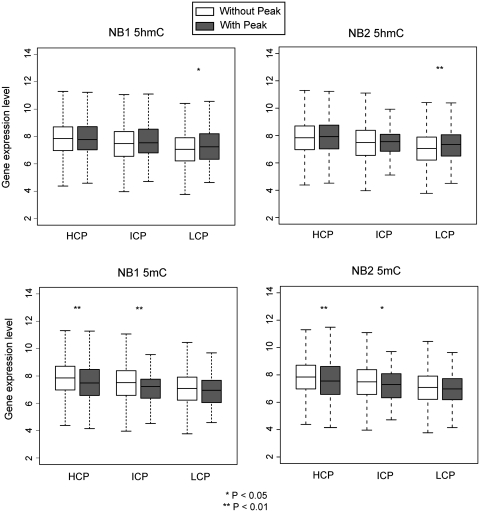
Using gene expression data of brain frontal lobe tissue from public databases, we initially verified that genes with 5mC peaks at promoters have lower gene expression levels than genes with no 5mC peak at the promoter (Figure 5, bottom panels). This is true for promoters with high CpG content (HCP promoters) and intermediate CpG content (ICP promoters) confirming previous results (23,30) but not for promoters with low CpG content (LCP promoters). We then looked at possible correlations between the presence of 5hmC peaks at promoters and expression levels. We did not find any positive or negative correlation for HCP and ICP promoters (Figure 5, top panels) although 5hmC peaks and gene expression levels were positively correlated at LCP promoters.
Figure 5.
Correlation between 5hmC and 5mC peaks at promoters and gene expression levels. Gene expression data were obtained from GEO dataset GSE3790. The raw CEL files were processed by RMA and average log2 intensities among biological replicates were calculated to represent their expression levels. Genes were separated into groups having a peak for 5mC or 5hmC or not having a peak at their promoters. Gene expression levels (log2 scale) were assigned for these two categories and represented as box plots and their difference was determined by Student's t-test. **P-values were significant for 5mC at promoters with high and intermediate CpG contents (HCP and ICP, P < 0.05 for both NB1 and NB2) but not for low CpG (LCP) promoters (P > 0.05 for both NB1 and NB2). For HCP and ICP promoters, there was no significant difference between the expression levels of 5hmC-marked promoters and other promoters (P > 0.05 for both NB1 and NB2) but expression levels were positively correlated for LCP promoters (P < 0.05).
Gene categories marked by 5hmC or 5mC at promoters
We conducted gene ontology analysis using the DAVID interface to identify potentially interesting gene categories characterized by 5hmC or 5mC peaks (but not both) in the promoter. For promoter-associated 5hmC peaks, we identified genes involved in muscle function, ion transport, neuronal development and patterning processes as significantly enriched categories (Supplementary Table S5).
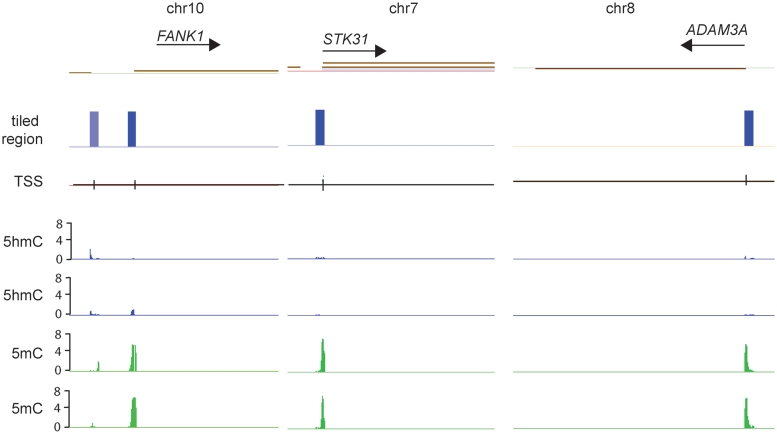
When we characterized genes with 5mC peaks at promoters, we found that genes involved in male gamete function and DNA packaging were most significantly overrepresented (Supplementary Table S6). Testis-specific genes and other germ line-specifically expressed genes are often silenced by promoter DNA methylation in somatic tissues (31). These same genes are remarkably devoid of 5hmC peaks at their promoters. Examples for three testis-specific genes, FANK1, STK31 and ADAM3A, are shown in Figure 6. The data suggest that testis-specific genes are characterized by the absence of 5hmC at their promoters.
Figure 6.
Testis-specific genes are marked by 5mC at their promoters but lack promoter-associated 5hmC peaks. Three examples are shown for the testis-specific genes FANK1, STK31 and ADAM3A, located on three different chromosomes. Green peaks reflect 5mC and blue peaks show the distribution of 5hmC peaks in each of two different normal brain samples (NB1 and NB2). The TSS and direction of transcription are shown. The blue columns indicate the tiled regions.
5hmC and 5mC in gene bodies
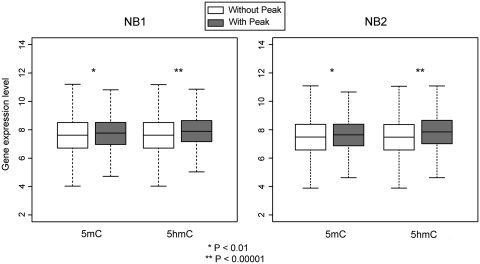
Genes with 5hmC peaks in gene bodies have higher expression levels than those without 5hmC peaks (Figure 7, P < 0.00001). This correlation is even more significant than the positive correlation between 5mC in gene bodies and expression levels (P < 0.00063 and P < 0.00028 for NB1 and NB2, respectively). Gene ontology analysis indicated that 5hmC-marked gene bodies include genes involved in cytoskeletal function, ion transport, regulation of transcription and cell death, as well as other functions (Supplementary Table S7). Data for genes with 5mC peaks in gene bodies showed that genes encoding cell adhesion molecules including the protocadherin family, metal-binding proteins and ion channels were most significantly enriched (Supplementary Table S8). Other significantly enriched categories included proteins involved in neuron development and neuronal differentiation. Several of these groups of genes are expected to be expressed and functionally important in the brain.
Figure 7.
Positive correlation between 5hmC and 5mC in gene bodies and gene expression levels. Gene expression data were obtained from GEO dataset GSE3790. The raw CEL files were processed by RMA and average log2 intensities among biological replicates were calculated to represent their expression levels. Genes were separated into groups having peaks for 5mC or 5hmC or not having peaks in intragenic regions. Gene expression levels (log2 scale) were assigned for these two categories and represented as box plots and their difference was determined by Student's t-test. P-values were highly significant for 5hmC and expression levels (**P < 0.00001) and significant for 5mC and expression levels (*P < 0.01).
DISCUSSION
In this article, we report one of the first attempts to map the genomic distribution of 5hmC in human genomic DNA. We chose brain frontal cortex tissue, because the total level of 5hmC relative to 5mC is relatively high in various regions of the brain, most notably the cortex (12,14–16). The reason for the high levels of 5hmC in this tissue, which has a low cell division rate, is currently unknown. Perhaps it is the lack of cell division per se that prevents dilution of oxidized 5mC by DNA replication, and 5hmC can therefore accumulate over time. We attempted to acquire clues as to the function of 5hmC in brain DNA by mapping its distribution in different parts of the genome using 5hmC-immunoprecipitation.
While 5mC and 5hmC peaks are often mutually exclusive, this is not always the case and peaks of both modified cytosine bases coexist at many genomic locations, for example in gene bodies (Figure 4). However, we noticed that 5hmC is more frequently targeted to promoters. Despite of this, we did not find a positive (or negative) correlation between 5hmC peaks at promoters and gene expression levels for HCP and ICP promoters, although a positive correlation existed for LCP promoters. One potential model for the existence of 5mC oxidation at promoters could be that this pathway may represent a ‘repair’ process that is aimed at keeping promoter CpG islands largely free of DNA methylation introduced by errors due to aberrant de novo methylation of CpG islands by DNA methyltransferases. Interestingly, the TET1 protein contains a CXXC domain (13), a domain known to bind to unmethylated CpG sites, which are present abundantly in CpG islands. 5mC oxidation can neutralize the repressive function of DNA CpG methylation, for example by disallowing the binding of methyl-CpG binding proteins, as we previously reported (19), thus maintaining an open chromatin configuration. Since we did not find a strict correlation between 5hmC peaks at promoters and gene expression levels, this situation may apply to active, poised and inactive genes having unmethylated CpG islands at their promoters.
Gene ontology analysis for 5hmC in promoters revealed a group of genes involved in ion transport, nerve function and several other categories. When analyzing gene categories marked or not marked by 5hmC, we noticed a striking absence of 5hmC from the promoters of testis-specific genes that are highly methylated and have strong 5mC peaks at their promoters. This finding suggests that these sequences, perhaps in association with specific chromatin marks, are refractory to modification by the TET oxidases.
5hmC is largely absent from intergenic regions (Figure 2). Thus, in comparison to 5mC, 5hmC is more selectively targeted to genes. This suggests that there is a mechanism that targets TET proteins to presumably active genes.
We also found substantial amounts of 5hmC in gene bodies, in which 5mC tends to be enriched as well, as previously reported (10,32,33). While this article was in preparation, another group using chemical labeling and biotin enrichment of 5hmC-containing DNA fragments reported similar enrichment of 5hmC in gene bodies of mouse cerebellum (34). The function of 5mC in gene bodies, although found in many tissue types and organisms (32), is currently unknown. It has been proposed that 5mC in gene bodies may prevent inappropriate transcription initiation, for example by suppressing noise transcription and/or antisense transcription (2,10). Interestingly, we found that 5mC is particularly enriched in those genes, which encode proteins with function in neurons and other cells of the nervous system, such as ion channel proteins and cell adhesion molecules (Supplementary Table S8). Why some of the 5mC in gene bodies is further modified to form 5hmC and what the specific role of this process could be is unclear at this time. We observed that there is a stronger positive correlation between 5hmC in gene bodies and transcript levels than there is for 5mC and transcript levels (Figure 7), suggesting that 5hmC may be more potent than 5mC in preventing inappropriate intragenic transcription initiation. Further work will be required to determine how the 5mC oxidases are targeted to genes, including promoters and gene bodies, and what the precise molecular function of 5hmC in these specific genomic locations might be.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health (grants CA084469 and AG036041).
Conflict of interest statement. Under a licensing agreement between City of Hope and Active Motif (Carlsbad, CA, USA), the MIRA technique was licensed to Active Motif, and the author G.P.P. is entitled to a share of the royalties received by City of Hope from sales of the licensed technology.
Supplementary Material
REFERENCES
- 1.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–2113. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 6.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Methods Mol. Biol. 2009;507:3–20. doi: 10.1007/978-1-59745-522-0_1. [DOI] [PubMed] [Google Scholar]
- 7.Hahn MA, Pfeifer GP. Methods for genome-wide analysis of DNA methylation in intestinal tumors. Mutat. Res. 2010;693:77–83. doi: 10.1016/j.mrfmmm.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 10.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc. Natl Acad. Sci. USA. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. Int. Ed. Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 15.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nat. Chem. Biol. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 21.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch TA, Pfeifer GP. DNA methylation profiling using the methylated-CpG island recovery assay (MIRA) Methods. 2010;52:213–217. doi: 10.1016/j.ymeth.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 24.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 25.Beck S. Taking the measure of the methylome. Nat. Biotechnol. 2010;28:1026–1028. doi: 10.1038/nbt1010-1026. [DOI] [PubMed] [Google Scholar]
- 26.Bock C, Tomazou EM, Brinkman AB, Müller F, Simmer F, Gu H, Jäger N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J, Han X, Chen Q, Zhang X, Wang J. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52:203–212. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 29.Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, Robinson MW, Clark SJ. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011;6:34–44. doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, Rauch TA, Zhong X, Bennett WP, Latif F, Krex D, Pfeifer GP. CpG island hypermethylation in human astrocytomas. Cancer Res. 2010;70:2718–2727. doi: 10.1158/0008-5472.CAN-09-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol. Cell. Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 33.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.