Abstract
Upon endoplasmic reticulum (ER) stress, mammalian cells induce the synthesis of a transcriptional activator XBP1s to alleviate the stress. Under unstressed conditions, the messenger RNA (mRNA) for XBP1s exists in the cytosol as an unspliced precursor form, XBP1u mRNA. Thus, its intron must be removed for the synthesis of XBP1s. Upon ER stress, a stress sensor IRE1α cleaves XBP1u mRNA to initiate the unconventional splicing of XBP1u mRNA on the ER membrane. The liberated two exons are ligated to form the mature XBP1s mRNA. However, the mechanism of this splicing is still obscure mainly because the enzyme that joins XBP1s mRNA halves is unknown. Here, we reconstituted the whole splicing reaction of XBP1u mRNA in vitro. Using this assay, we showed that, consistent with the in vivo studies, mammalian cytosol indeed had RNA ligase that could join XBP1s mRNA halves. Further, the cleavage of XBP1u mRNA with IRE1α generated 2′, 3′-cyclic phosphate structure at the cleavage site. Interestingly, this phosphate was incorporated into XBP1s mRNA by the enzyme in the cytosol to ligate the two exons. These studies reveal the utility of the assay system and the unique properties of the mammalian cytosolic enzyme that can promote the splicing of XBP1u mRNA.
INTRODUCTION
The endoplasmic reticulum (ER) is a cellular compartment where secretory and membrane proteins are folded. When cells are exposed to stresses such as glucose starvation and viral infection, unfolded proteins accumulate in the ER and interfere with ER function. These conditions are called ER stress. Prolonged ER stress can lead to fatal damage for living cells. To reduce the ER stress, cells activate intracellular signaling pathways termed unfolded protein response (UPR). When a sensor protein of a UPR system senses unfolded proteins in the ER, it leads to the activation of the signaling pathway that causes the upregulation of a subset of proteins including ER molecular chaperones or to the inhibition of protein synthesis. Both of them result in the reduction of the ER stress. In mammalian cells, three UPR branches have been identified. One of them is the IRE1α (inositol requiring enzyme 1)-XBP1 (X-box-binding protein-1) pathway (1,2).
In this pathway, IRE1α is an ER-located transmembrane protein that senses the ER stress (3,4). When IRE1α senses the ER stress, the signal is transduced by several steps of a reaction cascade, leading to the translation of a transcriptional activator XBP1s (5–7). Importantly, under unstressed conditions, the mRNA for XBP1 exists in the cytosol as a precursor containing a 26-base intron (this unspliced form of XBP1 mRNA is called XBP1u mRNA). Thus, the intron must be removed for the production of XBP1s. Under ER stress, IRE1α activates its ribonuclease domain and cleaves the XBP1u mRNA at two specific sites on the conserved double stem–loop to initiate the splicing of this transcript. Then, the two exons are joined by an unknown RNA ligase to form the mature XBP1s mRNA (spliced XBP1 mRNA). This splicing reaction creates a translational frameshift to produce a functional XBP1s transcription factor that induces the UPR target genes. The splicing occurs only when IRE1α senses the ER stress, allowing the regulated activation of this pathway. An analogous system is widely conserved among eukaryotes. In yeast, Ire1, a yeast homolog of IRE1α, cleaves the HAC1 mRNA at two sites during ER stress (8,9). Then, the fragments are joined by tRNA ligase Rlg1 to express a transcriptional activator Hac1 (10).
The mechanism of the HAC1 mRNA splicing, which resembles that of pre-tRNA splicing, is well understood. Like tRNA endonuclease, Ire1 cleaves the HAC1 mRNA, leaving a 2′, 3′-cyclic phosphate terminus at the 3′-end of the 5′-exon and a hydroxyl terminus at the 5′-end of the 3′-exon (11). Then, two exons are joined in multistep reactions by Rlg1 as follows. First, Rlg1 opens the 2′, 3′-cyclic phosphate terminus at the 3′-end of the 5′-exon by hydrolyzing it, resulting in the formation of 2′-phosphate terminus. Rlg1 also phosphorylates the hydroxyl terminus at the 5′-end of the 3′-exon using guanosine triphosphate (GTP) or adenosine triphosphate (ATP). Rlg1 then adenylates the phosphorylated 5′-end of the 3′-exon using ATP. Following these reaction, the resulting two RNA fragments are ligated by Rlg1 (Supplementary Figure S1) (11–13). The ligation by Rlg1 leaves 2′-phosphate at the splice junction. This 2′-phosphate is removed by a 2′-phosphotransferase Tpt1 (14).
In contrast to the mechanism of the HAC1 mRNA splicing, that of XBP1u mRNA splicing is only poorly understood. This is partly due to the lack of our knowledge on the enzyme that ligates the two IRE1α-cleaved fragments of XBP1u mRNA to form the XBP1s mRNA. Here, we show that the whole splicing reaction of the XBP1u mRNA can be reconstituted in vitro. This system allowed us to study the properties of the XBP1u mRNA splicing in vitro.
MATERIALS AND METHODS
Plasmid construction and recombinant protein expression
To generate the plasmid pBS-hXBP1u which was used for the in vitro transcription of the unspliced XBP1, the 0.27-kb KpnI–BglII fragment of pCAX-F-XBP1ΔDBD-venus (15) was cloned into the KpnI, BamHI sites of pBluescript II SK(–). To construct the plasmid IVT-SL85 which was utilized for the synthesis of SL85 RNA, a DNA fragment encoding SL85 RNA under T7 promoter was amplified using both SL85-template-sense (5′-CTCACTATTAGGGAAGAGGTCAGTGGTCGGATCTGTTGAGTCCGCAGTACTCAAACTACGTGTACCTCTGCAGTAGGTACAGGTCCAGTTATCACCCCTC-3′) and SL85-template-antisense (5′-GAGGGGTGATAACTGGACCTGTACCTACTGCAGAGGTACACGTAGTTTGAGTACTGCGGACTCAACAGATCCGACCACTGACCTCTTCCCTAATAGTGAG-3′) as a template and both SL85-sense (5′-CGGGATCCCGCAGTAATACGACTCACTATTAGGGAAGAGGTCAGTG-3′) and SL85-antisense (5′-GGAATTCCGATATCGAGGGGTGATAACTGGACCTGTACC-3′) as primers. The resulting DNA fragment was cut with BamHI and EcoRI in primers (the restriction sites are shown in bold letters) and ligated into the corresponding restriction sites of pEBFP (Clontech). Recombinant IRE1α was expressed in SF9 cells and purified as previously described (16).
In vitro RNA transcription
To prepare the unspliced XBP1 RNA (XBP1u RNA), pBS-hXBP1u was cleaved with SpeI, purified by phenol–chloroform extraction and ethanol precipitation, and then used as a template for the in vitro transcription reaction. The reaction was performed at 30°C for 1 h with the Riboprobe systems (Promega) using T7 RNA polymerase. To synthesize radio-labeled SL85 RNA, the IVT-SL85 plasmid was cleaved with EcoRV and used as a template for the in vitro transcription reaction (see above). To specifically radio-label the RNA, the reaction was performed in the presence of [α-32P]CTP (3000 Ci/mmol). All in vitro transcribed RNAs were purified using RNAiso Plus (TaKaRa) and resuspended in water.
Preparation of rabbit erythrocyte lysate
The rabbits (New Zealand White strain, female) were used as a source to prepare erythrocyte lysate. Blood from the rabbits, collected in a heparinized tube, was centrifuged at 120g for 10 min at 2°C to remove plasma cells in the supernatant. The leukocytes and erythrocytes in the pellet fraction were washed six times by centrifugation at 650g for 5 min with the equal volumes of ice-cold saline buffer (0.14 M NaCl, 1.5 mM MgCl2 and 5 mM KCl). The washed cells were then resuspended in the equal volumes of saline and subjected to centrifugation at 1020g for 15 min at 2°C. After aspirating off leukocytes in the supernatant, erythrocytes in the precipitate were lysed by the addition of the equal volumes of ice-cold distilled water. The suspension was centrifuged at 16 000g for 18 min at 2°C to obtain the supernatant (rabbit erythrocyte lysate) and to remove the pellet (membranes and cell debris). The erythrocyte lysate thus obtained was frozen under liquid nitrogen and stored at −80°C.
In vitro splicing of XBP1u mRNA
The splicing reactions (40 µl) were carried out at 30°C for 1 h in kinase buffer [20 mM HEPES, 10 mM Mg(OAc)2 and 50 mM KOAc (pH 7.3 at 4°C)] containing XBP1u RNA (0.05 pmol), recombinant IRE1α (2.5 pmol), 30 µl cell lysate, 20 U RNasin (Promega), 0.25 mM DTT, 1.25 mM ATP and 0.75 mM GTP. After the splicing reaction, the RNA was purified using RNAiso Plus (TaKaRa) and resuspended in water. To detect the spliced and unspliced XBP1 RNA, we performed RT-PCR as follows. The spliced product was subjected to reverse transcription with the Superscript first-strand synthesis system (Invitrogen) using an oligonucleotide (5′-GGATCTTGAATCTGAAGAGTC-3′) as a primer. The resulting cDNA was then subjected to PCR to amplify the cDNA fragments that represent the spliced and unspliced XBP1 RNA. A three-step reaction (incubation at 96°C for 30 s; 55°C, 30 s; 72°C, 30 s) was repeated for 25 cycles in a reaction mixture (20 µl) containing 0.5 U of KAPATaq Extra (KAPA Biosystems), 0.5 µM each of the sense and antisense primers and 0.5 µl of the product form the reverse transcription. The sense and antisense primers used were 5′-GAACCAGGAGTTAAGACAGC-3′ and 5′-AGTCAATACCGCCAGAATCC-3′. The final RT-PCR products, representing the spliced and unspliced XBP1 RNA, were separated on a 5% acrylamide gel and visualized with ethidium bromide.
The relative ligation activity was defined as the splicing efficiency per 1 µg of the protein. The splicing efficiency was calculated by the following formula: spliced RNA/(unspliced RNA + spliced RNA). For the analysis of the data, we used Image Gauge software (Fuji Film).
Partial purification of RNA ligase for the XBP1u mRNA splicing
Ammonium sulfate precipitation
Ten milliliters of rabbit erythrocyte lysate (∼2500 mg of protein) was diluted 3-fold with buffer A [20 mM Tris–HCl (pH 8.9 at 4°C), 20% v/v glycerol, 0.2 mM ethylenediaminetetraacetic acid (EDTA), 0.5 mM phenylmethanesulfonylfluoride PMSF, 0.5 mM dithiothreitol (DTT)], to which ammonium sulfate was added to 45% saturation. The solution was left for 1 h and then centrifuged at 25 000g for 30 min. The supernatant was treated again with ammonium sulfate to 65% saturation, left for 1 h, and centrifuged at 25 000g for 30 min. The precipitate was dissolved in 2.5 ml of buffer A and then passed to a PD-10 column (GE Healthcare), previously equilibrated with buffer A, to remove any residual ammonium sulfate and change the buffer to buffer A.
Mono Q chromatography
The peak fraction from ammonium sulfate precipitation (precipitate of 45–65% fraction) was loaded onto a Mono Q HR 5/5 column (GE Healthcare) previously equilibrated with buffer A. After washing the column with 10 ml of buffer A, the proteins were eluted by a linear gradient from 0 mM to 1 M KCl. The fractions (1 ml each) were dialyzed for 2 h against buffer A and stored at −80°C. All purification procedures were performed at 4°C.
Synthesis of 32P-labeled SL27
To introduce 32P to the phosphodiester linkage at the IRE1α cleavage site of the model substrate SL27, an RNA oligonucleotide (corresponding to the 3′-terminal 14 nt of SL27: 5′-CAGCACUCAGACUA-3′; its 3′-end was modified with amine) was synthesized and its 5′-hydroxyl group was phosphorylated with [γ-32P]ATP (7000 Ci/mmol) using T4 polynucleotide kinase (TaKaRa) to obtain the 3′-part of SL27 that was radio-labeled with 32P at the 5′-end. After purification using RNAiso Plus (TaKaRa), the 32P-labeled 3′-part of SL27 was ligated with an equal amount of a synthetic RNA oligonucleotide (corresponding to the 5′-terminal 13 nt of SL27: 5′-UAGUCUGAGUCCG-3′) using T4 RNA ligase (TaKaRa). This ligation reaction led to the synthesis of SL27 that was labeled with 32P at the IRE1α cleavage site. For purification, the ligation product (SL27) was excised from a denaturing polyacrylamide gel after electrophoresis and eluted from the gel slice using RNAiso plus (Takara).
Analysis of the terminal structure of the IRE1α cleavage product
To study the terminal structure generated by cleavage of XBP1u mRNA with IRE1α, the SL27 RNA (5000 c.p.m.) was cleaved with 100 pmol of recombinant IRE1α at 30°C for 1 h in the kinase buffer (see above) (40 µl) supplemented with 20 U of RNasin (Promega), 0.25 mM DTT, 1.25 mM ATP and 0.75 mM GTP. The IRE1α-cleaved SL27 was then subjected to the enzymatic reaction with calf intestine alkaline phosphatase or T4 polynucleotide kinase for 1 h at 37°C. The reaction products were separated by electrophoresis on a 10% denaturing polyacrylamide gel. The signals were detected by autoradiography using the BAS 2500 system (Fuji Film) to examine the susceptibility of 32P at the IRE1α-cleavage site to the enzymatic treatments. Removal of 32P from the IRE1α-cleaved SL27, with T4 polynucleotide kinase but not with calf intestine alkaline phosphatase, indicates that the cleavage by IRE1α creates 2′, 3′-cyclic phosphate structure at the 3′-end of the cleavage site.
Discrimination of the two splicing mechanisms
To study the splicing mechanism of XBP1u mRNA, we radio-labeled the model substrate SL85 by in vitro transcription in the presence of [α-32P]CTP (see above). The radio-labeled SL85 RNA was then subjected to in vitro splicing reaction by incubating the RNA with 20 pmol of recombinant IRE1α, and 30 µl of rabbit erythrocyte lysate at 30°C for 1 h in 40 µl of kinase buffer supplemented with 20 U RNasin (Promega), 0.25 mM DTT, 1.25 mM ATP and 0.75 mM GTP. For purification, the re-ligated product of SL85 RNA was excised from a denaturing polyacrylamide gel after electrophoresis and eluted from the gel slice using RNAiso plus (Takara). The purified RNA (15 000 c.p.m.) was then digested with 200 U of RNase I (Ambion) at 37°C for 3 h in a buffer solution containing 10 mM Tris–HCl (pH 7.6) and 0.1 M NaCl. To discriminate the two splicing mechanisms, the digested products (32P-labeled nucleotide 3′-monophosphates) (3000 c.p.m.) were separated by two-dimensional thin-layer chromatography on PEI cellulose (Merck), and visualized and quantified by autoradiography using the BAS 2500 system and Image Gauge software (Fuji Film). For the separation, the first dimension was run in solvent A (isobutyric acid/saturated ammonia/water [66/1/3]), and the second dimension in solvent B (isopropanol/concentrated HCl/water [68/18/14]) (17). 3′-monophosphorylated nucleotide standards were visualized by ultraviolet light.
RESULTS
Reconstitution of the splicing reaction of XBP1u mRNA in vitro
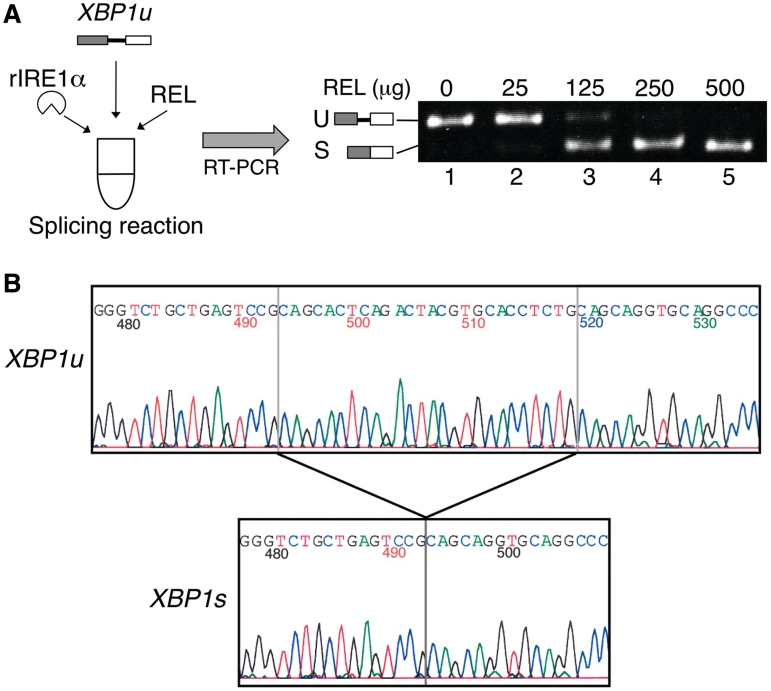
To study the splicing mechanism of XBP1u mRNA, we wanted to develop an in vitro assay for the splicing reaction of XBP1u mRNA. Although the RNA ligase for the splicing reaction of XBP1u mRNA has not been identified, recent results indicate that XBP1u mRNA splicing likely occurs in the cytosol (18, 19, unpublished data by Shinya,S. and Kohno,K.). Thus, we have used the erythrocyte lysate as a source of ligase activity, because erythrocytes of mammals do not contain the nucleus (20). To reconstitute the reaction, we incubated the in vitro transcribed XBP1u RNA, recombinant human IRE1α that cleaves the XBP1u RNA at the correct site (16), and rabbit erythrocyte lysate, a source of the RNA ligase. After incubation, the reaction products were subjected to RT-PCR using primers that can amplify both the XBP1u and XBP1s RNA. The RT-PCR products were then separated by acrylamide gel electrophoresis and visualized with ethidium bromide (Figure 1A). The addition of the rabbit erythrocyte lysate to the reaction caused the appearance of a fast migrating band that corresponds to the XBP1s RNA (Figure 1A, lanes 2–5). The determination of the nucleotide sequence of the fast migrating band revealed that this band represents the RT-PCR product of the XBP1u RNA that has been spliced at the correct position (Figure 1B). Thus, we have successfully reconstituted the splicing reaction of XBP1u mRNA in vitro.
Figure 1.
Reconstitution of the XBP1u mRNA splicing in vitro. (A) Splicing assay. The reaction mixtures (40 µl) contained recombinant human IRE1α (rIRE1α), transcribed XBP1u RNA (XBP1u) and rabbit erythrocyte lysate (REL). After the reaction, the relative ligase activity was examined as described in ‘Materials and Methods’ section. The amounts of REL added to the reactions are: 0 µg (lane 1), 25 µg (lane 2), 125 µg (lane 3), 250 µg (lane 4), 500 µg (lane 5). The positions of the RT-PCR products representing the XBP1u (U) and XBP1s (S) RNAs are indicated. (B) Determination of the nucleotide sequence of the band that corresponds to the XBP1u (U) and XBP1s (S) RNAs in (A). The sequence around the splice junction revealed that two halves of XBP1s RNA were joined at the correct position after the in vitro splicing reaction. Numbers in (B) denote nucleotide positions. The first A of the initiation methionine codon is set at 1.
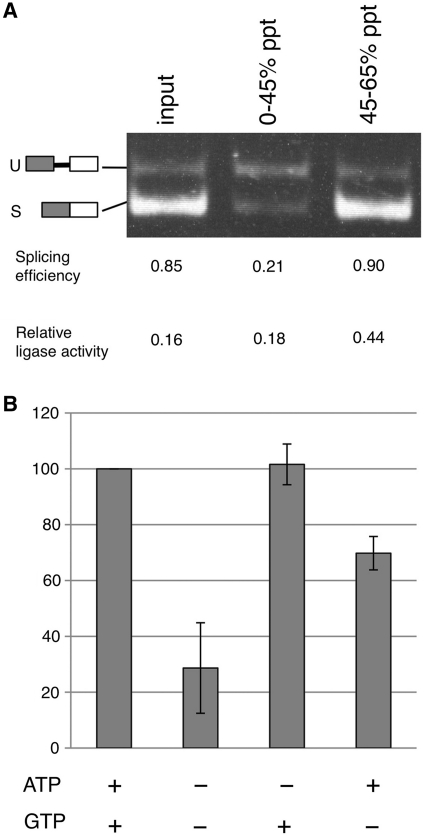
In yeast, ATP and GTP play important roles in the ligation of HAC1 mRNA: Rlg1, the ligase for the splicing of HAC1 mRNA, requires GTP or ATP for the phospharylation of the 5′-terminus of 3′-exon and ATP for the subsequent adenylation of the 5′-phosphorylated terminus of 3′-exon during its catalysis (Supplementary Figure S1) (11). We thus investigated the nucleotide requirements for the splicing of XBP1u mRNA. To remove the endogenous nucleotide, we collected proteins by ammonium sulfate precipitation. The RNA ligase activity was enriched in the precipitate of the 45–65% saturated fraction (Figure 2A, 45–65% ppt). Thus, we used this fraction to analyze the nucleotide requirements. The results (Figure 2B) show that both ATP and GTP can stimulate this ligase activity.
Figure 2.
Nucleotide requirements for the ligase activity. (A) To remove nucleotides from the rabbit erythrocyte lysate (REL) and partially purify the ligase for XBP1u mRNA splicing, we performed ammonium sulfate precipitation. REL was first saturated to 45% with ammonium sulfate and the protein precipitate (0–45% ppt) was collected by centrifugation. The resulting supernatant was then saturated to 65% with ammonium sulfate and the precipitate (45–65% ppt) was obtained by centrifugation. Each of these protein precipitates was dissolved in buffer A and dialyzed against the same buffer. The samples were then subjected to the splicing assay and their relative ligation activity was calculated as described in ‘Material and Methods’ section. U and S indicate the positions of the RT-PCR products representing the XBP1u and XBP1s RNAs. (B) After cleaving the XBP1u RNA with IRE1α, the cleaved product was purified and subjected to in vitro splicing reaction with the 45–65% fraction from the ammonium sulfate precipitation of REL. The activity observed in the presence of both 1.25 mM ATP and 0.75 mM GTP was taken as 100%. Error bars indicate standard deviations from the average of three independent experiments.
Partial purification of RNA ligase for XBP1u mRNA splicing
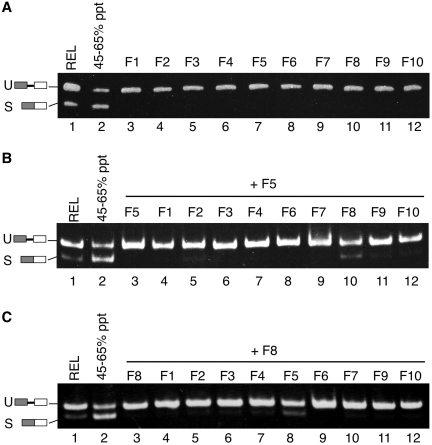
This splicing assay allowed us to partially purify the RNA ligase that can promote XBP1u mRNA splicing. In the first step of purification, we fractionated proteins by ammonium sulfate precipitation as described above (Figure 2A). The active fraction (Figure 2A, 45–65% ppt) was separated by anion exchange chromatography on a Mono Q column. Surprisingly, when we analyzed the ligase activity in the resulting Mono Q fractions, we could not detect the peak of activity (Figure 3A). This result raised a possibility that two or more fractions might be necessary to reconstruct the ligation activity. To test this possibility, we mixed every one fraction with another and measured the RNA ligase activity of the combined two fractions using the splicing assay (Figure 3B and C). Remarkably, when two of the Mono Q fractions (fractions 5 and 8) were mixed, a peak of ligase activity was recovered (Figure 3B, lane 10 and Figure 3C, lane 8). The activity was much higher than those of the original individual fractions. These results suggest that the RNA ligase for XBP1u mRNA splicing may include at least two components.
Figure 3.
Efficient splicing of XBP1u mRNA requires two Mono Q fractions. (A) RNA ligase after ammonium sulfate precipitation was separated by anion-exchange chromatography on a Mono Q column. The total of 10 fractions, obtained by elution with buffer A containing KCl, were subjected to the splicing assay (lanes 3–12). Lanes 1 and 2: assay with REL and the fraction before anion-exchange chromatography. (B) We mixed every one fraction with another and measured the RNA ligase activity of the combined two fractions using the splicing assay. The combinations of fractions in the assays shown in this panel are: fractions 5 and 5 (lane 3), fractions 5 and 1 (lane 4), fractions 5 and 2 (lane 5), fractions 5 and 3 (lane 6), fractions 5 and 4 (lane 7), fractions 5 and 6 (lane 8), fractions 5 and 7 (lane 9), fractions 5 and 8 (lane 10), fractions 5 and 9 (lane 11) and fractions 5 and 10 (lane 12). (C) The combinations of fractions in the assays shown in this panel are: fractions 8 and 8 (lane 3), fractions 8 and 1 (lane 4), fractions 8 and 2 (lane 5), fractions 8 and 3 (lane 6), fractions 8 and 4 (lane 7), fractions 8 and 5 (lane 8), fractions 8 and 6 (lane 9), fractions 8 and 7 (lane 10), fractions 8 and 9 (lane 11) and fractions 8 and 10 (lane 12).
To further characterize the two fractions (fractions 5 and 8), we studied the nucleotide requirements for the ligation reaction promoted by these fractions. The splicing reaction using the mixed-Mono Q fractions was stimulated by ATP or GTP (Supplementary Figure S2). This finding is consistent with the result obtained with the reaction using the ammonium sulfate-precipitated rabbit erythrocyte lysate. We also studied the sensitivity of these fractions to heat treatment. Upon heat treatment, both the rabbit erythrocyte lysate and two Mono Q fractions (fractions 5 and 8) lost their ability to promote the splicing of XBP1u mRNA (Supplementary Figure S3). Thus, some factors involved in the reaction are heat-labile.
IRE1α generates 2′, 3′-cyclic phosphate terminus
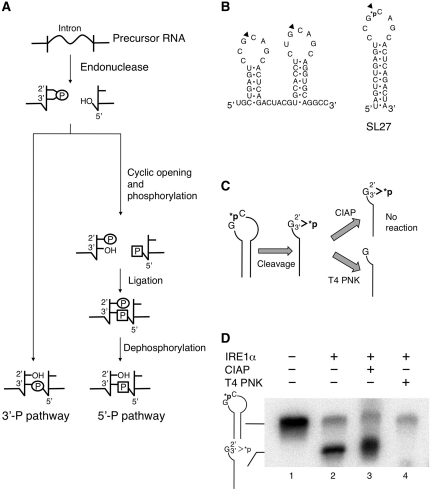
We could not further purify the RNA ligase, possibly because of the low abundance of the enzyme in vivo. Thus, we next proceeded to examine the mechanism of XBP1u mRNA splicing in vitro. Biochemical studies have revealed that mammalian cells have two distinct enzymatic pathways for the ligation of the 5′- and 3′-exons of tRNA (Figure 4A). One is the yeast-like pathway that joins these tRNA halves by using a new phosphate derived from the phosphorylated 5′-end of the 3′-exon at the splice junction (5′-P pathway) (21). Another is the pathway that directly ligates the 2′, 3′-cyclic phosphate at the 3′-end of the 5′-exon and 5′-hydroxyl terminus of the 3′-exon to form a 3′, 5′-phosphodiester bond (3′-P pathway) (22,23). To know which mechanism is employed for the XBP1u mRNA splicing, we have performed the following experiments.
Figure 4.
Determination of the terminal structure generated by specific cleavage of XBP1u mRNA by IRE1α. (A) Proposed mechanisms for the ligation of RNA fragments during RNA splicing in mammalian cells. (B) Predicted secondary structure of the region surrounding the IRE1α cleavage sites of the human XBP1u mRNA (left) and schematic representation of RNA substrate (SL27) used in this assay (right). SL27 contains the stem–loop structure of the 5′-splice site flanked by four nucleotide sequences on both ends. The latter sequences were added to stabilize its stem structure. Black triangles mark the sites of specific cleavage by IRE1α. The phosphate group radio-labeled with 32P is indicted by *p. (C) Scheme for determining the terminal structure generated by specific cleavage of XBP1u mRNA by IRE1α. T4 polynucleotide kinase (T4 PNK), but not calf intestine alkaline phosphatase (CIAP), can remove phosphate from the 2′, 3′-cyclic phosphate structure (here abbreviated G>p). Thus, this assay allows for the identification of 2′, 3′-cyclic phosphate at the terminus generated by specific cleavage of the model substrate by IRE1α. (D) SL27 RNA was cleaved by rIRE1α for 1 h at 30°C, and then incubated with CIAP or T4 PNK for additional 1 h at 37°C. The reaction products were separated by electrophoresis on a 10% denaturing polyacrylamide gel and detected by autoradiography. The positions of the model substrate SL27 and the IRE1α cleaved SL27 fragment containing the 2′, 3′-cyclic phosphate structure are indicated on the left.
As a first step toward the understanding of the ligation mechanism, we determined the terminal structures of 5′- and 3′-exons generated after cleavage of XBP1u mRNA by IRE1α. It has been known that cleavage of precursor RNAs by tRNA endonuclease and yeast Ire1 generates 2′, 3′-cyclic phosphate terminus at the 3′-end of the 5′-exon and 5′-hydroxyl terminus at the 5′-end of the 3′-exon (11). However, the terminus structures generated by cleavage of XBP1u mRNA by mammalian IRE1α have not been studied. To study their structures, we synthesized a radioactive stem–loop substrate (SL27) that had an [α-32P] labeled phosphodiester linkage only at the cleavage site (Figure 4B). After cleaving SL27 RNA with recombinant IRE1α, we studied the terminal structures of the cleaved products using calf intestine alkaline phosphatase (CIAP) and T4 polynucleotide kinase (T4 PNK). Importantly, T4 PNK, but not CIAP, can hydrolyze 2′, 3′-cyclic phosphate and release the phosphate group from the remaining RNA moiety (24). This difference in the activities of these two enzymes has been used to study the terminal structures of the products of RNA cleaving enzymes (Figure 4C).
To analyze the terminal structure, we first cleaved the labeled SL27 RNA with IRE1α. As expected, the cleavage reaction yielded a smaller RNA fragment that gives a radioactive signal (Figure 4D, lane 2), indicating that the latter fragment represents the IRE1α-cleaved fragment with 32P. We next studied the susceptibility of the 32P-labeled phosphate moiety in the fragment to each of the two enzymes. Remarkably, T4 PNK successfully removed 32P from the fragment (Figure 4D, lane 4). However, CIAP failed to do so (Figure 4D, lane 3). This result indicates that IRE1α generates 2′, 3′-cyclic phosphodiester terminus, the same structure generated by cleavage of RNA with Ire1 or tRNA endonuclease.
The 3′-phosphate of 5′-exon is incorporated into the XBP1s mRNA
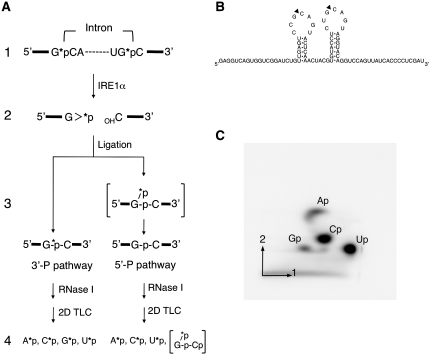
To further study the mechanism of ligation step of the XBP1u mRNA splicing, we next asked whether the enzyme uses the 5′-P pathway or the 3′-P pathway. The difference between these two models is that phosphate at the 2′, 3′-cyclic phosphate structure at the 3′-end of the 5′-exon (Figure 5A, stage 2) remains in the final spliced product in the 3′-P pathway but not in the 5′-P pathway (Figure 5A, stage 3). Thus, to distinguish between two reaction mechanisms, we radio-labeled this phosphate with 32P and then examined whether 32P stays at the junction after the splicing reaction (Figure 5A, stage 3). To radiolabel the phosphate at the cleavage sites in the model substrate SL85 (Figure 5B), we performed in vitro transcription in the presence of [α-32P]CTP (Figure 5A, stage 1). The labeled transcript was then subjected to the in vitro splicing reaction, and the resulting spliced product was purified. To examine the presence or absence of 32P at the splice junction, we treated the spliced product with ribonuclease I (RNase I) that cleaves after all four bases, leading to the formation of nucleotide 3′-monophosphate (Figure 5A, stage 4). Importantly, after digestion of the spliced product with RNase I, 32P at the splice junction will be present in guanosine 3′-monophosphate only when the reaction proceeds via the 3′-P pathway (stage 4). Thus, we examined the presence or absence of 32P-labeled guanosine 3′-monophosphate (Gp) in the resulting nucleotides by separating them by two-dimensional thin-layer chromatography (stage 4). The presence of 32P-labeled Gp spot (G*p) in the final product will support the 3′-P mechanism for the XBP1u mRNA splicing (stage 4).
Figure 5.
Studies on the mechanism of ligation of the two halves of XBP1s mRNA. (A) Scheme for analyzing the ligation mechanism (see text for detail). The phosphate labeled with 32P is indicated by *p. The 2′, 3′-cyclic phosphate structure was abbreviated as >p. Note that RNase I would not cleave a dinucleotide bond that has a 2′-phosphate. Thus, the cleavage of the spliced product with RNase I would yield a specific set of labeled nucleotides depending on the mechanism of ligation (stage 4). (B) Schematic representation of RNA substrate (SL85) used in this assay. Note that the last three nucleotides of SL85 (GAU) originate from the EcoRV restriction site used to linearize the template plasmid for in vitro transcription. (C) Following in vitro splicing reaction, the ligated product from SL85 RNA was gel-purified, and digested with RNase I. The resulting nucleotide monophosphate was separated by two-dimensional thin-layer chromatography to examine the fate of [α-32P] phosphate at the splice junction. The positions of nucleotide 3′-monophosphate (Ap, Gp, Cp and Up) were indicated. The ratio of radioactivity in each of the nucleotide 3′-monophosphate (Ap, Gp, Cp and Up) to the total radioactivity (the sum of their radio-activities) was determined. The actual values are: 0.119 ± 0.006 (for Ap), 0.078 ± 0.006 (Gp), 0.359 ± 0.004 (Cp) and 0.447 ± 0.007 (Up). The numbers represent mean ± SD of three independent experiments. We also calculated the values expected if the splicing of the model substrate SL85 occurs via the 3′-P pathway: 0.133 (for Ap), 0.0667 (Gp), 0.333 (Cp) and 0.467 (Up). The actual values were in good agreement with the expected values.
It should be noted that, to facilitate the analysis, we have made several changes in the model substrate. The model substrate contains 85-nt sequence corresponding to the region surrounding the IRE1α cleavage sites (SL85) (Figure 5B). In the original sequence (Supplementary Figure S4), there are eight GC sequences in addition to the two GC sequence at the cleavage sites. Thus, guano sine at these eight GC positions will also generate 32P-labeled guanosine 3′-monophosphate when the spliced RNA is digested with RNase I, which makes it difficult to evaluate whether the phosphate at the splice junction still retains 32P or not. To overcome this problem, we removed all GC sequences from the original XBP1u mRNA sequence except the two GC sequences at the IRE1α cleavage sites, by introducing substitution mutations as illustrated in the Supplementary Figure S4. To do this, the substitutions were introduced in a way that allows the formation of the putative secondary structure of the molecule (Supplementary Figure S4).
The separation of the final products by two-dimensional thin-layer chromatography (Figure 5A, stage 4) revealed the presence of 32P-derived radioactivity in the Gp spot (Figure 5C). We determined the ratio of radioactivity in each spot (A*p, G*p, C*p and U*p) relative to the total radioactivity. These values were in good agreement with the values expected if the splicing occurs via the 3′-P pathway (see the legend to Figure 5C). Importantly, we detected a reasonable amount of radioactivity in Gp spot. The latter value should be zero if the splicing occurs via the 5′-P pathway. These findings led us to conclude that the cytosol of mammalian cells contains an RNA ligase that promotes the splicing of XBP1u mRNA via the 3′-P pathway.
DISCUSSION
The splicing of XBP1u mRNA proceeds via two steps of reactions. In the first step, XBP1u mRNA is cleaved in the cytosol by the endonuclease IRE1α that has been activated by ER stress. This reaction liberates the 5′- and 3′-exons from the precursor mRNA. In the second step, the two exons are ligated to form the XBP1s mRNA. However, the mechanism of this unconventional splicing is still unclear, especially in the ligation step, because the ligase required for the XBP1u mRNA splicing has not been identified. Here, we have successfully reconstituted the whole splicing reaction of XBP1u mRNA in vitro (Figure 1). This assay allowed us to characterize the splicing reaction of XBP1u mRNA in vitro.
Our results reveal the similarities and differences in the splicing mechanism between the yeast HAC1 mRNA and the mammalian XBP1u mRNA. It has already been demonstrated that the cleavage of HAC1 mRNA by yeast Ire1 generates 2′, 3′-cyclic phosphate terminus at the 3′-end of the 5′-exon. Human IRE1α takes the same domain structure as yeast Ire1. In addition, its RNase domain shares 33% sequence identity with that of yeast Ire1 (25). Yet, how human IRE1α cleaves XBP1u mRNA remained obscure. Here, we used two RNA processing enzymes to study the terminal structure of the cleavage product of XBP1u mRNA with human IRE1α. Our data clearly show that human IRE1α also generates 2′, 3′-cyclic phosphate terminus at the 3′-end of the 5′-exon, illuminating the remarkable similarities between the yeast Ire1–Hac1 and human IRE1α–XBP1 pathways.
Despite these similarities, our studies also suggest that the mechanism used by the enzyme in the cytsol of mammalian cells to join the two halves of XBP1s mRNA may be different from the mechanism employed by yeast Rlg1 to ligate the two exons of HAC1 mRNA. In yeast, it has been established that tRNA ligase Rlg1 is responsible for the ligation of HAC1 mRNA splicing. For the ligation, Rlg1 uses a new phosphate derived from the phosphorylated 5′-end of the 3′-exon at the splice junction. However, in mammalian cells, the previous biochemical analysis revealed the existence of two distinct pathways for the ligation of RNA. They are the yeast-like 5′-P pathway (the same reaction as Rlg1) and the 3′-P pathway (see above) (Figure 4A). Using our system, we have found that the RNA ligase in the cytosol of mammalian cells can join the two halves of XBP1s mRNA using the 3′-P pathway (Figure 5C).
Importantly, the final product of ligation differs between the two pathways. With the 5′-P pathway, RNA ligase leaves a 2′-phosphate at the splice junction; thus, this 2′-phosphate is removed by a 2′-phosphotransferase. With the 3′-P pathway, however, RNA ligase does not leave a 2′-phosphate at the splice junction. The yeast RNA ligase Rlg1 leaves 2′-phosphate at the splice junction after ligation of the HAC1 mRNA (11), necessitating an enzyme catalyst that removes this phosphate to complete the HAC1 mRNA splicing. In yeast, this reaction is performed by 2′-phosphotransferase Tpt1. Surprisingly, the deletion of Trpt1, the mammalian homolog of Tpt1, from the mouse chromosome, has no effect on the XBP1u mRNA splicing (26). Here, we showed that the mammalian cytosol has an enzyme that uses the 3′-P pathway to ligate the two halves of XBP1s mRNA. Since this reaction does not leave 2′-phosphate at the splicing junction, our finding can explain the non-essentiality of the 2′-phosphotransferase TRPT1 for the splicing of XBP1u mRNA in mouse.
Notably, we observed that both ATP and GTP stimulated the ligation of the XBP1s mRNA halves in the reaction catalyzed by the enzyme in the mammalian cytosol. This is in contrast to the situation in yeast where ATP works as an essential cofactor for the ligation of two HAC1 mRNA halves in the reaction catalyzed by Rlg1 (Supplementary Figure S1). Importantly, ATP is essential for the adenylation step of HAC1 mRNA ligation. However, this step will not be required if the reaction catalyzed by the mammalian cytosolic enzyme proceeds via the 3′-pathway as has been demonstrated in this study. Thus, the difference in the requirements for nucleotides observed between Rlg1 and the enzyme in the mammalian cytosol may reflect their respective mechanistic properties.
Interestingly, our results suggested that the RNA ligase includes at least two components (Figure 3B and C). Both of them were heat-labile (Supplementary Figure S3). However, the reason for the necessity of the two Mono Q fractions for the efficient ligation of the XBP1s mRNA halves remains unclear. One possibility is that one of the components is used to specifically recognize the two exons and deliver them to a general RNA ligase. Since IRE1α not only cleaves XBP1u mRNA for splicing but also cleaves other mRNA for their degradation (27–29). The promiscuous ligation of the latter RNA fragments will yield unwanted forms of mRNA and thus would be unfavorable for the cells. To avoid the latter reaction, it is possible that the cells may have evolved an adapter that connects the two exons and the RNA ligase.
The RNA ligase for the splicing of XBP1u mRNA has not been identified. Our current findings suggest that there could be two reasons for it. First, the ligation has been thought to occur via the 5′-P pathway as yeast Rlg1 uses this mechanism. Here, we showed evidence that the XBP1 ligase, instead, may use the 3′-P pathway. This discrepancy may have caused difficulty in the search for the enzyme. Second, our results suggest that two or more components may be required for the ligation of the two halves of XBP1s mRNA. If this is the case, the simple overexpression of a cDNA product may not lead to an increase in the activity of the ligase required for the XBP1u mRNA splicing. Our development of the assay system will open up a doorway for the identification of the RNA ligase involved in the XBP1u mRNA splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Grants-in-Aid for Scientific Research on Priority Areas (19058010 to K.K.) and Scientific Research B (20380062 to K.K.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT); the Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (09J55502 to S.S.); and the International research fellowship from the Global Center of Excellence program from the Japan Society for the Promotion of Science (to H.K.). Funding for open access charge: The Uehara Memorial Foundation (to K.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We greatly thank Junko Iida-Hashimoto and Hisayo Masuda for technical assistance; Ikumi Kawahara and Chojiro Kojima for materials and discussion; Rie Kurata, Yoichiro Fukao, and Junko Tsukamoto for mass spectrum analysis.
REFERENCES
- 1.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 2.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 3.Kohno K. Stress-sensing mechanisms in the unfolded protein response: Similarities and differences between yeast and mammals. J. Biochem. 2010;147:27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- 4.Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr. Opin. Cell Biol. 2010 doi: 10.1016/j.ceb.2010.10.008. [Epub ahead of print, 17 November 2010] [DOI] [PubMed] [Google Scholar]
- 5.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 6.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 7.Yanagitani K, Imagawa Y, Iwawaki T, Hosoda A, Saito M, Kimata Y, Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 8.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 9.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 10.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez TN, Sidrauski C, Dorfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greer CL, Peebles CL, Gegenheimer P, Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- 13.Phizicky EM, Schwartz RC, Abelson J. Saccharomyces cerevisiae tRNA ligase. purification of the protein and isolation of the structural gene. J. Biol. Chem. 1986;261:2978–2986. [PubMed] [Google Scholar]
- 14.Culver GM, McCraith SM, Consaul SA, Stanford DR, Phizicky EM. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:13203–13210. doi: 10.1074/jbc.272.20.13203. [DOI] [PubMed] [Google Scholar]
- 15.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 16.Imagawa Y, Hosoda A, Sasaka S, Tsuru A, Kohno K. RNase domains determine the functional difference between IRE1alpha and IRE1beta. FEBS Lett. 2008;582:656–660. doi: 10.1016/j.febslet.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 17.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 18.Back SH, Lee K, Vink E, Kaufman RJ. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J. Biol. Chem. 2006;281:18691–18706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- 19.Uemura A, Oku M, Mori K, Yoshida H. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J. Cell. Sci. 2009;122:2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]
- 20.Testa U. Apoptotic mechanisms in the control of erythropoiesis. Leukemia. 2004;18:1176–1199. doi: 10.1038/sj.leu.2403383. [DOI] [PubMed] [Google Scholar]
- 21.Zillmann M, Gorovsky MA, Phizicky EM. Conserved mechanism of tRNA splicing in eukaryotes. Mol. Cell. Biol. 1991;11:5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- 23.Perkins KK, Furneaux H, Hurwitz J. Isolation and characterization of an RNA ligase from HeLa cells. Proc. Natl Acad. Sci. USA. 1985;82:684–688. doi: 10.1073/pnas.82.3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morse DP, Bass BL. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- 25.Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35:395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Harding HP, Lackey JG, Hsu HC, Zhang Y, Deng J, Xu RM, Damha MJ, Ron D. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA. 2008;14:225–232. doi: 10.1261/rna.859908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oikawa D, Tokuda M, Hosoda A, Iwawaki T. Identification of a consensus element recognized and cleaved by IRE1 alpha. Nucleic Acids Res. 2010;38:6265–6273. doi: 10.1093/nar/gkq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.