Abstract
TALENs are important new tools for genome engineering. Fusions of transcription activator-like (TAL) effectors of plant pathogenic Xanthomonas spp. to the FokI nuclease, TALENs bind and cleave DNA in pairs. Binding specificity is determined by customizable arrays of polymorphic amino acid repeats in the TAL effectors. We present a method and reagents for efficiently assembling TALEN constructs with custom repeat arrays. We also describe design guidelines based on naturally occurring TAL effectors and their binding sites. Using software that applies these guidelines, in nine genes from plants, animals and protists, we found candidate cleavage sites on average every 35 bp. Each of 15 sites selected from this set was cleaved in a yeast-based assay with TALEN pairs constructed with our reagents. We used two of the TALEN pairs to mutate HPRT1 in human cells and ADH1 in Arabidopsis thaliana protoplasts. Our reagents include a plasmid construct for making custom TAL effectors and one for TAL effector fusions to additional proteins of interest. Using the former, we constructed de novo a functional analog of AvrHah1 of Xanthomonas gardneri. The complete plasmid set is available through the non-profit repository AddGene and a web-based version of our software is freely accessible online.
INTRODUCTION
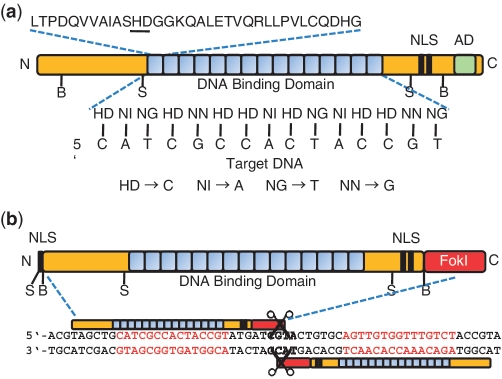
Transcription activator-like (TAL) effectors are a newly described class of specific DNA binding protein, so far unique in the simplicity and manipulability of their targeting mechanism. Produced by plant pathogenic bacteria in the genus Xanthomonas, the native function of these proteins is to directly modulate host gene expression. Upon delivery into host cells via the bacterial type III secretion system, TAL effectors enter the nucleus, bind to effector-specific sequences in host gene promoters and activate transcription (1). Their targeting specificity is determined by a central domain of tandem, 33–35 amino acid repeats, followed by a single truncated repeat of 20 amino acids (Figure 1a). The majority of naturally occurring TAL effectors examined have between 12 and 27 full repeats (2). Members of our group and another lab independently discovered that a polymorphic pair of adjacent residues at positions 12 and 13 in each repeat, the ‘repeat-variable di-residue’ (RVD), specifies the target, one RVD to one nucleotide, with the four most common RVDs each preferentially associating with one of the four bases (Figure 1a) (3,4). Also, naturally occurring recognition sites are uniformly preceded by a T that is required for TAL effector activity (3,4). These straightforward sequence relationships allow the prediction of TAL effector binding sites (3–6) and construction of TAL effector responsive promoter elements (7), as well as customization of TAL effector repeat domains to bind DNA sequences of interest (8–11).
Figure 1.
TAL effector and TALEN structure. (a) Structure of a naturally occurring TAL effector. A consensus repeat sequence is shown with the repeat-variable di-residue (RVD) underlined. The sequence of RVDs determines the target nucleotide sequence. The four most common RVDs, on which our designs and plasmids are based, are shown with their most frequently associated nucleotide. Some evidence suggests that the less common RVD NK (not displayed) has greater specificity for G than NN does and for that reason our plasmid set also includes NK modules. (b) Structure of a TALEN. Two monomeric TALENs are required to bind the target site to enable FokI to dimerize and cleave DNA. NLS, nuclear localization signal(s); AD, transcriptional activation domain; B, BamHI; S, SphI.
As a result, TAL effectors have attracted great interest as DNA targeting tools. In particular, we and other groups have shown that TAL effectors can be fused to the catalytic domain of the FokI nuclease to create targeted DNA double-strand breaks (DSBs) in vivo for genome editing (8,10,12,13). Since FokI cleaves as a dimer, these TAL effector nucleases (TALENs; 8) function in pairs, binding opposing targets across a spacer over which the FokI domains come together to create the break (Figure 1b). DSBs are repaired in nearly all cells by one of two highly conserved processes, non-homologous end joining (NHEJ), which often results in small insertions or deletions and can be harnessed for gene disruption, and homologous recombination (HR), which can be used for gene insertion or replacement (14,15). Genome modifications based on both of these pathways have been obtained with high frequency in a variety of plant and animal species using zinc-finger nucleases (ZFNs) and homing endonucleases. However, for each of these platforms, engineering novel specificities has generally required empirical and selection-based approaches that can be time and resource intensive. Despite a significant recent advance for ZFNs that takes finger context into account to achieve high success rates (16), targeting capacity (the diversity of sequences that can be recognized) still suffers limitations (17–19). TALENs thus far appear not to be subject to these constraints. In at least one study, mutagenesis frequency was estimated to be as high as 25% of transfected cells, on par with or better than ZFNs (10).
The TAL effector repeat domain also has been successfully customized to make targeted transcription factors, both in plants in the native protein context and in human cells with the TAL effector activation domain replaced by VP64 (9,11). Fusions to other protein domains for chromatin modification, gene regulation, or other applications can also be envisioned. Thus, an efficient method for assembling genetic constructs to encode TAL effectors and TAL effector fusions to other proteins, with repeat arrays of user-defined length and RVD sequence, is highly desirable.
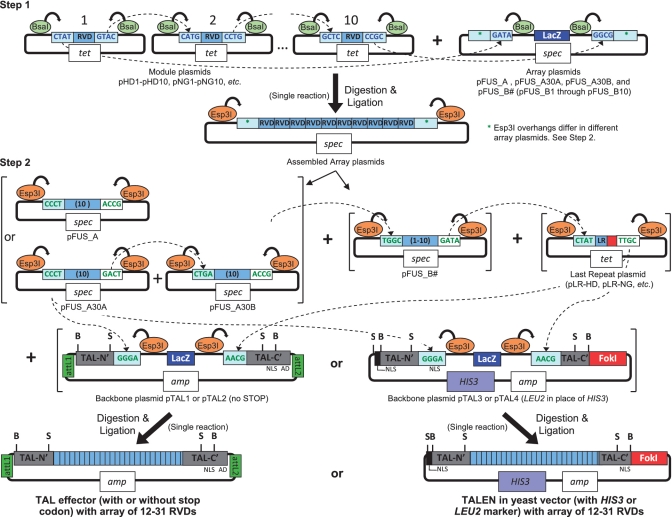
In our previous work, we constructed TALENs with customized repeat arrays through sequential cloning of sequence-verified single, double and triple repeat modules (8). We sought a more rapid approach that would not rely on commercial synthesis, which is expensive, or PCR-based methods, which can result in mutations or recombined repeats. We opted for Golden Gate cloning, a recently developed method of assembling multiple DNA fragments in an ordered fashion in a single reaction (20,21). The Golden Gate method uses Type IIS restriction endonucleases, which cleave outside their recognition sites to create unique 4 bp overhangs (sticky ends) (Figure 2). Cloning is expedited by digesting and ligating in the same reaction mixture because correct assembly eliminates the enzyme recognition site.
Figure 2.
Golden Gate assembly of custom TAL effector and TALEN constructs using module, array, last repeat and backbone plasmids. By using the type IIS restriction endonucleases BsaI and Esp3I, modules containing the desired RVDs can be released with unique cohesive ends for ordered, single-reaction assembly into array plasmids in a first step, and those arrays subsequently released and assembled in order in a second step into a backbone plasmid to create full length constructs with custom repeat arrays (see text for details). NLS, nuclear localization signal(s); AD, transcriptional activation domain; tet, tetracycline resistance; spec, spectinomycin resistance; amp, ampicillin resistance; attL1 and attL2, recombination sites for Gateway cloning; B, BamHI, and S, SphI, useful for subcloning custom repeat arrays. Unique restriction enzyme sites flanking the coding sequences, useful for subcloning the entire constructs into other vectors, are not shown but can be found in the sequence files (Supplementary Data).
We report here a complete set of plasmids for assembling novel repeat arrays for TALENs, TAL effectors or TAL effector fusions to other proteins using the Golden Gate method in two steps. We also describe software for TALEN-targeting based on guidelines we developed to reflect naturally occurring TAL effector binding sites and on our previous TALEN study. We show that TALENs targeted with this software and constructed using the plasmid set are active in a yeast DNA cleavage assay and effective in gene targeting in human cells and Arabidopsis thaliana (hereafter Arabidopsis) protoplasts. Finally, we demonstrate successful construction of a functional analog of the avrHah1 TAL effector gene of Xanthomonas gardneri (22).
MATERIALS AND METHODS
Protocol for assembly of custom TALEN, TAL effector or TAL effector fusion-ready constructs
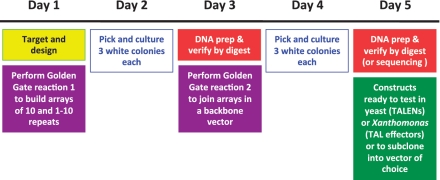
Assembly of a custom TALEN or TAL effector construct is accomplished in 5 days (Figure 3) and involves two steps: (i) assembly of repeat modules into intermediary arrays of 1–10 repeats and (ii) joining of the intermediary arrays into a backbone to make the final construct. A schematic representation is shown in Figure 2 and the complete set of required plasmids is displayed in Supplementary Figure S1. Construction and features of the plasmids themselves are described in the following section. The assembly protocol differs slightly for arrays of 12–21 modules versus arrays of 22–31 modules. We use an example here for construction of a TALEN monomer with a 16 RVD array and note differences in the protocol where they occur for making constructs with arrays of 22–31 RVDs.
Figure 3.
TALEN or TAL effector construct assembly timeline.
Day 1. Consider the RVD array NI HD HD NN HD NI NI NG HD NG HD NI NI NG HD NG, targeting the sequence 5′-AGCCCAATCTCACTCT-3′. Note that the 5′-T preceding the RVD-specified sequence is not shown and need not be considered in the assembly, although based on evidence to date (3,4), it should be considered during site selection. Select from the module plasmids those that encode RVDs 1–10 in the array using plasmids numbered in that order. For example, the plasmid for the first RVD would be pNI1, the second pHD2, the third pHD3, etc. Modules from these plasmids will be cloned into array plasmid pFUS_A. Next, select modules for RVDs 11–15 in the 16 RVD array again starting with plasmids numbered from 1. Thus for RVD 11 pHD1 would be used, for RVD 12 pNI2, etc. Note that the 16th and last RVD is encoded by a different, last repeat plasmid and is added later, in the second step (see Day 3). Modules encoding RVDs 11–15 are cloned into a pFUS_B array plasmid. The pFUS_B plasmids are numbered 1–10 and should be selected according to the number of modules going in. Thus, in our example, pFUS_B5 should be used. If arrays of 22–31 modules are to be assembled, the first 10 modules are cloned into pFUS_A30A, the second 10 modules into pFUS_A30B and the remaining modules into the appropriate pFUS_B plasmid, again according to the number of modules going in.
The module and array plasmids (150 ng each) are subjected to digestion and ligation in a single 20 µl reaction containing 1 µl BsaI (10 U, New England BioLabs) and 1 µl T4 DNA Ligase (2000 U, New England BioLabs) in T4 DNA ligase buffer (New England BioLabs). The reaction is incubated in a thermocycler for 10 cycles of 5 min at 37°C and 10 min at 16°C, then heated to 50°C for 5 min and then 80°C for 5 min. Then, 1 µl 25 mM ATP and 1 µl Plasmid Safe DNase (10 U, Epicentre) are added. The mixture is incubated at 37°C for 1 h, then used to transform Escherichia coli cells. Cells are plated on LB agar containing 50 µg/ml spectinomycin, with X-gal and IPTG for blue/white screening of recombinants, as described (23). Treatment with Plasmid Safe DNase is an important step to prevent linear DNA fragments, including partial arrays, from recombining into and circularizing the linearized array plasmids following transformation, due to the presence of partial repeat sequences at the termini of the array plasmids.
Day 2. Pick up to three white colonies from each transformation and start overnight cultures.
Day 3. Isolate plasmid DNA and identify clones with the correct arrays by restriction enzyme digestion and agarose gel electrophoresis. AflII and XbaI will release the repeat arrays, which will be 1048 bp for pFUS_A, 1052 for pFUS_A30A, 1040 for pFUS_A30B and of varying sizes for pFUS_B plasmids.
The next step is to join the intermediary arrays, along with a last repeat, into the desired context, using one of the four backbone plasmids. A 20 µl digestion and ligation reaction mixture is prepared as in the first step, but with 150 ng each of the pFUS_A and pFUS_B plasmids containing the intermediary repeat arrays (or the pFUS_A30A, pFUS_A30B and pFUS_B plasmids carrying the intermediaries for final arrays of 22–31 RVDs), 150 ng of the backbone plasmid, in this case pTAL3 or pTAL4 for constructing a TALEN monomer, and importantly, 150 ng of the appropriate last repeat plasmid. In our example, pLR-NG, for the 16th and last RVD, would be used. The reaction is treated and used to transform E. coli as above, except that Plasmid Safe DNAse treatment is omitted because the backbone plasmid termini have no homology with the array. Also, in this step, ampicillin (100 µg/ml) is used in place of spectinomycin for selection of transformants.
Day 4. Pick up to three white colonies from each transformation and start overnight cultures.
Day 5. Isolate plasmid DNA and identify clones containing the final, full-length repeat array. Array length can be verified by digestion with BstAPI (or StuI) and AatII, which cut just outside the repeats, or with SphI, which cuts farther out. Array integrity can be checked using BspEI, which cuts only in HD modules 2–10. The array can also be characterized by DNA sequencing.
Construction of module, last repeat, array and backbone plasmids
Repeat modules with the RVDs HD, NG, NI, NK and NN, across 10 staggered positions and with a BsaI site added to each end, were synthesized. The modules were cloned between the unique XbaI and XhoI sites of pTC14, replacing the spectinomycin resistance gene in that plasmid, to create a set of 50 module plasmids (pHD1 through pHD10, pNG1 through pNG10, etc.). pTC14 is a derivative of the Gateway entry and TOPO cloning vector pCR8 (Invitrogen) in which the Gateway cassette was replaced with a gene for tetracycline resistance using the flanking EcoRV and HpaI sites. Aside from the RVD codons, the modules at each position are identical, except for a BspEI site introduced into HD modules 2–10 for testing full-length array integrity by digestion. The modules are based on the first repeat of tal1c of X. oryzae pv. oryzicola strain BLS256 (3), which matches the consensus repeat and is made up of common codons.
Similarly, one module for each of the five RVDs containing the last, truncated repeat of the TAL effector repeat domain was synthesized and cloned in plasmid pCR8 (carrying the spectinomycin resistance gene) using ApaI and XbaI and replacing the Gateway cassette, to create five last repeat plasmids (e.g. pLR-HD).
Next, array plasmids pFUS_A, pFUS_A30A and pFUS_A30B were created by cloning, using AflII and XbaI, synthesized fragments into pCR8 that contain two internal BsaI sites oriented to cut outward into flanking sequences such that linearizing the vector with the enzyme leaves the appropriate overhangs to accept an array of 10 repeat modules (i.e. complementary on one side to the 5′-end of position 1 modules and on the other to the 3′-end of position 10 modules). The series of array plasmids pFUS_B1 through pFUSB10 were made similarly to be complementary on one side to the 5′-end of position 1 modules, but complementary on the other to the 3′-end of modules in position 1–10, respectively, to accept arrays ranging from 1 to 10 modules. A DNA fragment containing the lacZ gene for blue/white screening (23) was cloned between the two BsaI sites. For this, the multiple cloning sites between the HincII and Eco53kI sites in phagemid pBCSK+ (Stratagene) was deleted and the lacZ gene PCR amplified with primers carrying KasI and AgeI overhangs. These sites were included in the synthesized fragments, allowing the lacZ gene to be placed between the BsaI sites, maintaining the overhang sequences for accepting modules. The inserts in the array plasmids all contain terminal Esp3I (another type IIS enzyme) sites positioned to cut inward and release the arrays with appropriate overhangs for ordered ligation into a backbone plasmid for complete arrays of 12–21 (a pFUS_A array with a pFUS_B array, plus a last repeat) or 22–31 repeats (a pFUS_A30A with a pFUS_A30B and a pFUS_B array, plus a last repeat). These sites, or flanking AflII and XbaI sites in the vector (enzymes that are generally less expensive), can also be used to screen assembled clones for the correct size.
Backbone plasmid pTAL3 was derived from pFZ85, a precursor to the TALEN yeast expression vector we created previously (8). Derived from pDW1789 (24), pFZ85 contains the counter-selectable ccdB gene flanked by BamHI sites downstream of the yeast TEF promoter and a sequence encoding a nuclear localization signal and upstream of a sequence encoding a linker and the FokI nuclease catalytic domain. For our previous TALEN constructs, we used tal1c as a context for custom repeat arrays. First, solely for expediency of later adding the lacZ gene, the SphI fragment of tal1c was replaced with the SphI fragment of TAL effector gene pthXo1 (25), which has minor polymorphisms flanking the repeat region that create convenient restriction enzyme sites. The spanning BamHI fragment of the resulting gene was then cloned between the BamHI sites of pFZ85. Finally, the repeat region within the SphI fragment was deleted by digestion with BstAPI and AatII and replaced with a fragment carrying the lacZ gene for blue/white screening (cloned into this fragment as described above), flanked by outward cutting Esp3I sites and the necessary sequences to create a specific overhang on either end to accept final arrays and reconstitute a complete TAL effector domain. Importantly, the SphI sites, which are highly conserved among TAL effectors and are useful for swapping the entire repeat region into other TAL effector constructs, are preserved. The architecture of the constructs is the same as reported in our earlier work (8), encoding 287 and 230 amino acids of the TAL effector upstream and downstream of the repeats, respectively, with an additional six amino acids linking the TAL effector and FokI domains. To create pTAL4, which is identical to pTAL3 except that it carries LEU2 in place of HIS3, first the LEU2 gene was PCR amplified using primers having 20 bp extensions with homology to the region at the 5′-end of the Bpu10I and 3′-end of the AfeI site in pDW1789. Then, pDW1789 was linearized with Bpu10I and AfeI (removing the HIS3 gene) and the PCR-amplified LEU2 gene was inserted by in vivo recombination in E. coli (26). Finally, into this plasmid, the XbaI–SacI fragment of pTAL3 containing the TALEN backbone construct was introduced at the corresponding sites.
pTAL1 was created by replacing the SphI fragment of tal1C in pCS691 with the corresponding SphI fragment of pTAL3, containing the lacZ gene and the Esp3I sites and flanking sequences for accepting final arrays. pCS691 is a derivative of Gateway entry vector pENTR-D (Invitrogen) containing between the attL sites, the complete tal1c gene preceded by both Kozak and Shine–Dalgarno consensus sequences for efficient translation in eukaryotic or bacterial cells, respectively. In pCS691, the kanamycin resistance gene of pENTR-D is replaced by the BspHI fragment of pBlueScript SK(-) (Stratagene) for ampicillin resistance. To create pTAL2, the stop codon of tal1c in pTAL1 was deleted using the QuickChange mutagenesis kit (Stratagene) to allow translational fusion to other protein domains following Gateway recombination into a destination vector.
A schematic representation of all modules, last repeat, array and backbone plasmids (Supplementary Figure S1) and a folder containing complete sequences are included in Supplementary Data.
Software to identify candidate TALEN target sites
The software used to design TALENs in this study was written in Python 2.6.4. and runs in Linux (Ubuntu 10.10). It is available for use as an online tool (TAL Effector-Nucleotide Targeter, TALE-NT; http://boglabx.plp.iastate.edu/TALENT/). The tool provides a window to input DNA sequences (Supplementary Figure S2a), which are then scanned for sites based on TALEN design guidelines we established, described in the ‘Results’ section. The software identifies sets of TALEN recognition sites between 15 and 30 bp in length and separated by a spacer. The default spacer lengths are 15 bp and 18–30 bp (8), but other lengths can be specified by the user. In addition, buttons allow users to exclude design guidelines individually. The output is tab-delimited text, which can be imported into standard spreadsheet software (Supplementary Figure S2b). It provides coordinates and sequences of identified targets indicating the recognition sites for the left and right TALEN monomers and the spacer sequence. Since naturally occurring TAL effector recognition sites are uniformly preceded by a T, which is required for TAL effector activity (3,4), only TALEN monomer recognition sites preceded by a T are included. The T itself is not part of the output. Finally, the software provides the RVD sequences needed to construct the corresponding custom TALENs.
Testing TALEN function in yeast
The yeast assay for TALEN function was adapted from one we developed previously for ZFNs (8,24) in which cleavage of the target, positioned between partially duplicated fragments of the lacZ gene, reconstitutes the gene via subsequent recombination to provide a quantitative readout (Supplementary Figure S3a). For typical heterodimeric target sites (i.e. such as would typically occur in a native DNA sequence), paired TALEN constructs, in pTAL3 and pTAL4, are transformed together into yeast strain YPH500 (α mating type) using histidine and leucine prototrophy for selection. Individual TALEN monomers can be tested on homodimeric sites using just one of these plasmids. The target is made using synthesized complementary oligonucleotides that produce BglII- and SpeI-compatible ends and cloned between the lacZ fragments in the high copy DNA cleavage reporter plasmid pCP5 (24) cut with those enzymes (Supplementary Figure S3b). The target plasmid is transformed into yeast strain YPH499 (α mating type), using tryptophan prototrophy for selection, but also excluding uracil from the growth medium: in addition to the target cloning site, pCP5 carries also the URA3 gene between the lacZ fragments so that selection for URA3 ensures that the strain has not undergone spontaneous recombination (and loss of URA3) prior to the assay.
Three transformants each of YPH500 carrying the TALEN construct(s) and of YPH499 carrying the target plasmid are cultured overnight at 30°C, with rotary shaking at 800 rpm, in synthetic complete medium lacking histidine and/or leucine (TALENs) or tryptophan and uracil (target). TALEN and target transformants are next mated (three pairs) by combining 200–500 µl of the overnight cultures, adding 1 ml of YPD medium and incubating for 4–6 h at 30°C, shaking at 250–300 rpm. Cells are harvested by centrifguation, washed in 1 ml synthetic complete medium lacking histidine and/or leucine and tryptophan, but now containing uracil, then resuspended in 5 ml of that medium and incubated overnight again at 30°C, with shaking (800 rpm), to an OD600 between 0.1 and 0.9. Cells are harvested by centrifugation, then resuspended and lysed using YeastBuster Protein Extraction Reagent (Novagen) according to the manufacturer's protocol for small cultures. A total of 100 µl of lysate is transferred to a microtiter well plate and β-galactosidase activity measured and normalized as previously described (24). For high-throughput, yeast may be cultured and mated (using a gas permeable seal) as well as lysed in 24-well blocks. We typically express activity relative to a Zif268 ZFN (24).
Expression of custom TALENs in human cells and Arabidopsis protoplasts and detection of site-specific mutations
One of the pairs of TALENs targeting the human HPRT1 gene was subcloned into the mammalian expression vector pCDNA3.1(-) (Invitrogen) using XhoI and AflII. These enzymes excise the entire TALEN from pTAL3 or pTAL4 and place the coding sequence under control of the CMV (cytomegalovirus) promoter. The resulting plasmids were introduced into HEK293T cells by transfection using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Cells were collected 72 h after transfection and genomic DNA isolated and digested with Hpy188I, which cuts in the spacer sequence of the TALEN target site. After digestion, a chromosomal fragment encompassing the target site was amplified by PCR. Upon completion, the reactions were incubated for 20 min at 72°C with 4 µl of Taq DNA polymerase. PCR products then were digested with Hpy188I and cloned in a TOPO TA vector (Invitrogen). Independent clones containing the full-length PCR product were sequenced to evaluate mutations at the cleavage site.
The TALENs targeting the Arabidopsis ADH1 gene were subcloned into the plant expression vector pFZ14 (27) using XbaI and SacI. These enzymes excise the entire TALEN from pTAL3 or pTAL4 and place the coding sequence under control of the CaMV (cauliflower mosaic virus) 35S promoter. Recombinant plasmids were transformed into Arabidopsis protoplasts as previously described (27). Forty-eight hours after transformation, DNA was prepared and digested with PflFI, which cuts in the spacer sequence of the TALEN target site. After digestion, a chromosomal fragment encompassing the target site was amplified by PCR and the reaction products were once again digested with PflFI and run on an agarose gel. The band corresponding in size to undigested product was excised and cloned and individual clones were sequenced to evaluate mutations at the cleavage site.
Expression of a custom TAL effector in Xanthomonas and in planta activity assay
An analog of avrHah1 was assembled into pTAL1 using the Golden Gate method with HD, NI, NG and NN modules, ordered to match the AvrHah1 binding site in the promoter of the Bs3 gene (22). A native avrHah1 construct was made by replacing the BamHI fragment of tal1c in pCS495 with that of avrHah1. pCS495 is tal1c preceded by Shine–Dalgarno and Kozak consensus sequences in pENTR-D (Invitrogen). The analog and native avrHah1 constructs and tal1c were moved into pKEB31 by Gateway cloning (LR reaction). pKEB31 is a derivative of pDD62 (28) that contains a Gateway destination vector cassette (Invitrogen) between the XbaI and BamHI sites and a tetracycline resistance gene in place of the gene for gentamycin resistance. The resulting plasmids were introduced into X. campestris pv. vesicatoria strain 85–10 by electroporation and transformants were inoculated to 6-week-old pepper plants by syringe infiltration, as described (22). After 48 h, infiltrated leaves were cleared in 70% ethanol and 10% glycerol and photographed.
RESULTS
Efficient assembly of custom repeat arrays into TALEN and other TAL effector-based constructs
Our implementation of the Golden Gate method accomplishes custom TAL effector construct assembly in two steps (Figure 2 and Supplementary Figure S1). In the first step, it uses five sets of 10 staggered repeat clones, one for each of the four most common RVDs HD, NI, NG and NN, which associate most frequently with C, A, T and G, respectively and one for the less common NK, which at least in some contexts appears to have higher specificity for G than NN does (9,10). Inserts in these ‘module’ plasmids carrying the desired RVDs are released and assembled in order in one or two sets of 10 and one set of 1–10 into ‘array’ plasmids, using a type IIS enzyme. In the second step, the resulting array fragments are joined, along with a final, truncated repeat from a collection of five ‘last repeat’ plasmids (one for each RVD), into any of four different ‘backbone’ plasmids, using a different type IIS enzyme, for a final array of 12 (10+1+the last) to 31 (10+10+10+the last) RVDs. Counting the 5′-T that precedes the RVD specified sequences in TAL effector binding sites, the corresponding target ranges from 13 to 32 nt.
The backbone plasmids include (i) pTAL1 for assembling a custom TAL effector gene preceded by Shine–Dalgarno and Kozak sequences for efficient translation in bacteria and eukaryotes, respectively, (ii) pTAL2, identical to pTAL1, but without a stop codon so that the effector can be fused to other protein domains, (iii) pTAL3 for assembling a custom TALEN and expressing it in yeast using the selectable marker HIS3 and (iv) pTAL4, identical to pTAL3 but containing the marker LEU2, so that two TALEN monomers can be paired in the yeast assay (see subsequently). The TAL effector constructs are flanked by attL sites for transfer by Gateway recombination (Invitrogen) into destination vectors of choice. The TALEN constructs, though not Gateway compatible, are flanked by restriction enzyme sites convenient for subcloning into different expression vectors. All constructs retain the internal SphI sites flanking the repeat domain as well as the BamHI sites farther out that are conserved in most TAL effectors and can be used to readily swap a custom array into other TAL effector-based constructs.
All of the array and backbone plasmids contain within the cloning site the lacZ gene for blue/white screening to identify recombinants (23). For the work presented here, we successfully assembled >30 custom TALENs (Supplementary Table S1) and one custom TAL effector, ranging in array length from 15 to 30 RVDs. We never failed to obtain the correctly assembled array plasmid clone or the correctly assembled, final backbone plasmid clone for any of these by screening only three white colonies per cloning reaction transformed into E. coli. We routinely pick just two colonies and usually both are correct (not shown). Assembly of one or more constructs takes just 5 days (Figure 3 and refer ‘Materials and Methods’ section).
Guidelines and software for TALEN site selection and repeat array design
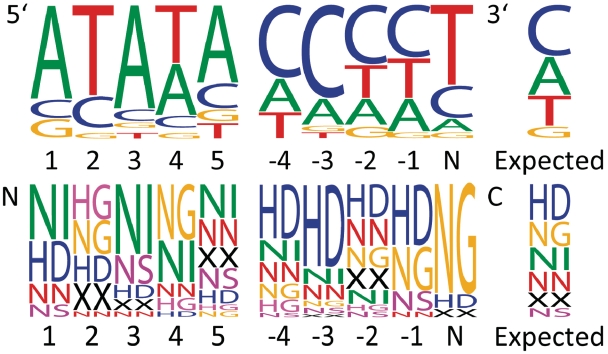
To facilitate TALEN design for genome editing, we wrote a computer program that analyzes DNA sequences, identifies suitable, paired and opposing TAL effector target sites across a spacer and generates corresponding RVD sequences using the four most common RVDs (see ‘Materials and Methods’ section). The software uses guidelines for TAL effector targeting that reflect naturally occurring TAL effectors and their binding sites and spacer lengths that we observed to function well in our previous study using TALENs derived from naturally-occurring TAL effectors (8). We established the targeting guidelines by examining the 20 TAL effector-target pairs identified by Moscou and Bogdanove (3). We looked for positional biases, neighbor effects and overall trends in nucleotide and RVD composition. To examine position effects for sequences of different lengths, we confined the analysis to the five positions at either end. We compared observed nucleotide and RVD frequencies to expected frequencies, taken as the frequencies in the entire set of sequences (Figure 4). The binding sites showed a strong bias against T at position 1 (5′-end), a bias against A at position 2, biases against G at the last (3′) and next-to-last positions and a moderate bias for T at the last position. RVD sequences showed corresponding positional biases: NG was disfavored at position 1; NI was disfavored at position 2 and NG was favored and NN disfavored at the last position. The bias for NG at the last position was particularly striking: NG occurs at this position in 85% of the sequences compared to its overall observed frequency of 18%. No neighbor effects were detected in the binding sites or RVD sequences. Average nucleotide composition of the binding sites was 31±16% A, 37±13% C, 9±8% G, and 22±10% T. To expand on this dataset, we used the weight matrix developed by Moscou and Bogdanove (3) to identify the best-scoring binding sites (preceded by a T) for each of 41 X. oryzae TAL effectors in each of approximately 57 000 rice promoters. We retained those in genes shown by microarray analysis (www.plexdb.org, experiment OS3) to be up-regulated during infection. This analysis yielded close to 100 putative additional TAL effector–target pairs. These reflected the same positional biases (data not shown). The guidelines are therefore as follows: (i) As noted previously for TAL effector binding sites (3,4), TALEN monomer binding sites should be preceded by a 5′-T, (ii) they should not have a T at position 1, (iii) they should not have an A at position 2, (iv) they should end with a T, so that the corresponding TALENs will reflect the strong bias for NG at this position and (v) they should have a base composition within two standard deviations of the averages we observed.
Figure 4.
Nucleotide and RVD frequencies at the termini of 20 target and TAL effector pairs. RVDs that have a frequency of ≥20% at one or more of the positions are shown. ‘XX’ represents all other RVDs.
We did not systematically test the guidelines, but data from intermediate constructs we obtained while building full-length TALENs with our earlier sequential ligation method provide some support (Supplementary Table S2). Of the four intermediate length TALEN–target pairs showing no detectable activity in the yeast assay for DNA cleavage (8), one did not match overall target nucleotide composition, one did not have an RVD sequence ending in NG and another did not meet either of these guidelines. Two out of seven with activity <25% of the Zif268 ZFN used as a control did not match overall target nucleotide composition. One of four with activity 25–50% of Zif268 did not have an RVD sequence ending in NG. TALENs with 50% or greater activity of Zif268 met all of the guidelines. The impact of the number of repeats in a TALEN was also considered. In general, longer TALENs that met all of the guidelines or medium-length TALENs that met all guidelines and had a high percentage of HDs showed the highest activity. Longer TALENs that failed to meet one or more guidelines showed reduced activity when compared to those of the same length that met all guidelines. Thus, in addition to providing preliminary support for the guidelines, the results also suggest that array length positively correlates with activity.
Toward validating our method for making custom TAL effector arrays, we used the software to first identify candidate TALEN sites in seven plant (Arabidopsis, tobacco), animal (human, zebrafish, Drosophila) and protist (Plasmodium) genes as well as in GFP and eGFP. In these genes, the software found unique TALEN sites on average every 35 bp (range = 15–120 bp).
Activity of custom TALENs in a yeast-based DNA cleavage assay
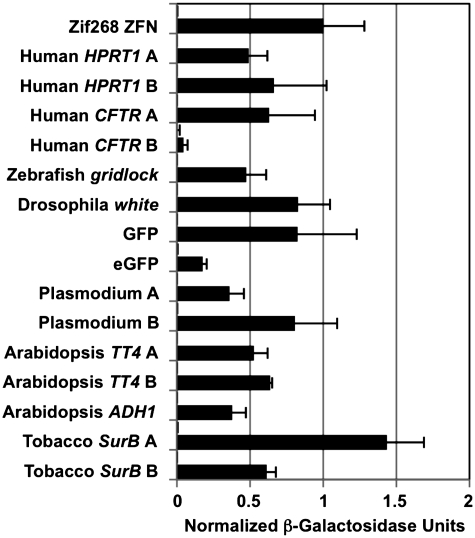
Custom TALEN pairs for 15 target sites (30 TALENs total; Supplementary Table S1) were made using the Golden Gate method and plasmids described above and tested in the yeast-based DNA cleavage assay we described previously (8). All TALEN pairs showed significant activity above the target-only negative controls and 14 of 15 showed activity ≥25% of our positive control, a Zif268 ZFN (Figure 5). We have generally found for ZFNs that this level of activity is sufficient for targeted mutagenesis of endogenous plant loci (24,27).
Figure 5.
Activity of 15 custom TALEN pairs targeting diverse sequences in a reporter-based yeast assay. TALENS were targeted to gene sequences from the indicated organisms and to GFP and eGFP using the software and constructed using the Golden Gate method and plasmids described in the text. Activity was measured in a yeast-based assay in which cleavage and recombination reconstitutes a functional lacZ gene (see text for details). Activity was normalized to a Zif268 ZFN positive control. Activity of target-only controls for each is plotted above the target-plus-TALEN values; in each case the activity was undetectable. Error bars denote s.d.; n = 3.
Targeted mutagenesis in human cells and Arabidopsis protoplasts using custom TALENs
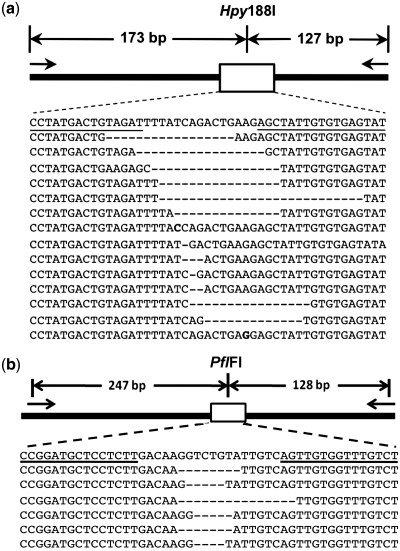
To validate the activity of our custom TALENs outside of yeast, we used one of the TALEN pairs for the human HPRT1 gene (HPRT1 B in Figure 5) and the TALEN pair for the Arabidopsis ADH1 gene to carry out targeted mutagenesis in human embryonic kidney cells and Arabidopsis protoplasts, respectively. In both cases the custom TALENs generated mutations at the recognition site through imprecise repair of the cleaved chromosomes by NHEJ (Figure 6). Our method of detection used an enrichment step, so it was not possible to quantify mutagenesis frequency. However, we obtained for HPRT1, 17 independent mutations including two single base pair substitutions and deletions ranging from 1–27 bp roughly centered on the spacer and for ADH1, 6 independent mutations consisting of deletions ranging from 4 to 15 bp, also centered on the spacer.
Figure 6.
Site-directed mutagenesis in human embryonic kidney cells and Arabidopsis protoplasts using custom TALENs. TALENs targeted to the human HPRT1 gene (pair HPRT1 B in Figure 5) and the Arabidopsis ADH1 gene (Figure 5) were transiently expressed in human embryonic kidney cells and in Arabidopsis protoplasts, respectively and the targets subsequently amplified and sequenced (see text for details). Prior to amplification, genomic DNA was digested with a restriction endonuclease having a site present in the TALEN target site to reduce amplification of wild-type sequences and enrich the amplicon pool for mutated ones. Results for HPRT1 are shown in (a) and ADH1 in (b). For each, the schematic at the top shows the chromosomal locus, short arrows designate primers used for PCR amplification following TALEN transient expression, sequence of the wild-type gene (top line) and unique mutated alleles obtained are shown below, binding sites for the TALEN monomers are underlined and the coincident restriction endonuclease site is indicated.
Replication of AvrHah1 TAL effector activity with a Golden Gate assembled clone
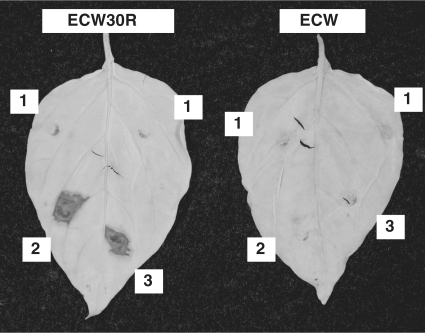
To assess our plasmids for construction of custom TAL effectors, we assembled an analog of the avrHah1 TAL effector gene of X. gardneri, which elicits a hypersensitive reaction in pepper by transcriptionally activating the Bs3 resistance gene (22). We chose AvrHah1 because it is highly divergent relative to other characterized TAL effectors, carrying predominantly 35 amino acid repeats (in contrast to the more common 34 amino acid repeat on which our modules are based) as well as other deviations from the consensus sequences both within and outside the repeat region. Introduced into X. campestris pv. vesicatoria strain 85–10, which lacks AvrHah1, that was then inoculated into pepper leaves, the Golden Gate assembled clone triggered a Bs3 specific hypersensitive reaction indistinguishable from that elicited by the native effector (Figure 7). This recreation of AvrHah1 specificity using our modular reagents demonstrates their utility for making custom transcription factors and underscores the sufficiency of the RVD sequences for targeting.
Figure 7.
Activity of an AvrHah1 analog created using the Golden Gate method and our plasmid set. Shown are leaves of pepper varieties ECW30R, carrying the Bs3 resistance gene and ECW, lacking it, 48 h following spot-infiltration with suspensions of X. campestris pv. vesicatoria strain 85–10 transformed to deliver (1) Tal1c (the effector used to make the backbone plasmids in this study), (2) native AvrHah1 or (3) an AvrHah1 analog encoded by a construct made using the Golden Gate method and our plasmid set. Leaves were cleared with ethanol to reveal the accumulation of phenolic compounds, visible as dark stained areas, indicative of the hypersensitive reaction induced by TAL effector driven transcriptional activation of Bs3.
DISCUSSION
The hallmark feature of TAL effectors that makes them such remarkably powerful tools for DNA targeting, their long arrays of 33–35 amino acid repeats that specify nucleotides in the recognition site in a straightforward and modular fashion, also makes them challenging to engineer. Commercial synthesis is effective (10) but expensive. PCR-based methods (11) carry the risk of artifact and recombination. Assembly by sequential ligation of sequence-verified modules (8) is inexpensive and assures array integrity, but is time consuming. The Golden Gate method using the reagents we describe here, provides a cost-effective, robust and rapid solution. TAL effector constructs with arrays of up to 31 RVDs are assembled in just two cloning steps using a set of sequence-verified modules. Furthermore, the reagents provide great flexibility for cloning arrays in different contexts and expressing them in different organisms, either in our set of backbone plasmids for TALENs, TAL effectors, or TAL effector fusions to additional proteins, or by simple subcloning or Gateway recombination into other vectors.
Zhang et al. (11) recently presented a protocol and set of templates for Golden Gate-like assembly that involves PCR amplification of modules, intermediary arrays and full-length arrays to yield TAL effector DNA binding domains with 13 RVDs fused in a backbone vector to VP64 (see also www.taleffectors.com). This marked a significant advance that enabled the authors to rapidly assemble custom arrays and demonstrate the utility of TAL effector-based proteins as custom transcription factors to activate endogenous genes in human cells. However, the method and plasmids we describe here offer more versatility for broader utility, not only with regard to the available contexts and portability of the arrays, as noted above, but also in array length. The ability with our reagents to construct arrays ranging from 12 to 31 RVDs allows fine-tuning for targeting and will be important for testing the important outstanding question of the relationship of length to affinity and specificity. The broad range in array length also offers greater flexibility to systematically address other important questions including the contributions of individual RVD–nucleotide associations to affinity and specificity, as well as the effect of position on mismatch tolerance (1). This could be accomplished, e.g. by starting with an array of minimal functional length and comparing the effects of adding or interspersing additional RVDs aligned to different nucleotides in the target.
Our method has the technical advantage of involving no PCR. Although the Zhang et al. (11) repeat templates for different RVDs are codon engineered to guard against slippage and inter-repeat recombination during PCR amplification, this strategy does not prevent recombination between repeats carrying the same RVD, particularly if they are present in tandem. Also, in part because our method involves no PCR, though it is 2 days longer, it is less labor-intensive and time consuming day to day.
Though all of the custom arrays made for this study use just the four most common RVDs, our plasmid set includes modules with NK, which users might opt to substitute for NN to specify G, because NN sometimes associates with A. We note however, based on data presented by Miller et al. (Figure 2e in ref. 10), that NK also associates substantially with A in some contexts. Modules with yet additional RVDs can be generated readily by mutagenesis of an existing set.
Among the genes we selected for targeting with TALENs, we deliberately chose some for which targeting with ZFNs has proven difficult. For example, one of the most common mutations in patients with cystic fibrosis is a deletion of 3 nt (DF508) in CFTR; however, best efforts to engineer a ZFN for this position only succeeded in targeting a site >120 bp away, a distance that would likely compromise gene targeting efficiency (18). For our CFTR TALENs, the DF508 mutation resides within the spacer sequence at the site of TALEN cleavage. Similarly, we previously created herbicide resistant tobacco plants by gene targeting with ZFNs that recognize and cleave the acetolactate synthase gene (24). The nearest ZFN that could be engineered to the desired site of modification was 188 bp away, whereas our TALENs cleave within 10 bp of the desired sequence modification. Finally, AT-rich sequences have been difficult to target with ZFNs; we successfully targeted two sites in the AT-rich (75.5%) Plasmepsin V gene of Plasmodium falciparum, which has an overall genome content of 80.6% AT (29). Generally, the high success rate of TALENs designed using our software, which found sites in diverse sequences on average every 35 bp, suggests that targetability of TALENs will prove superior to the public ZFN platforms, which are estimated to be capable of targeting on average every 500 bp (16,18). Indeed, we anticipate our estimate of targeting range is conservative, as some TALENs that do not follow our design principles still recognize and cleave DNA efficiently (10; Supplementary Table S2).
Activity varied among the TALENs we tested in the yeast assay. The reason for this is not clear. It could relate to expression levels or variability in the assay itself, but more likely, the data reflect inherent differences in the DNA binding affinity of the arrays, possibly related to their length and composition. The relationship of array length and composition to overall affinity is still an open question that must be addressed. The important conclusion for this study is that all of the TALENs were active, demonstrating that the targeting approach as well as the Golden Gate methods and plasmids for assembly are robust. Our results in Arabidopsis protoplasts and human cells, along with recent results from other groups (10,13), indicate that TALENs are likely to be broadly effective for genome engineering.
We have deposited all of our plasmids for constructing and expressing TALENs as well as TAL effectors with or without a stop codon in the non-profit clone repository AddGene (www.addgene.org). To complement our method and reagents, we have also made our software for TALEN site selection and design freely accessible as an online tool, the TAL Effector Nucleotide Targeter at http://boglabx.plp.iastate.edu/TALENT/. Although our success rate was high with TALENs designed using the software, we have not shown that it is ‘necessary’ to follow the guidelines on which the software is based. So, even though the guidelines place only relatively minor constraints on targeting, the online tool allows users to exclude them individually to increase candidate target site frequency. Also, because optimal spacing may differ for different TALEN architectures, the software provides the option to specify desired spacer lengths. In making these resources available, we hope to facilitate further characterization of TAL effector DNA targeting properties, broad adoption of TALENs and other TAL effector-based tools and further development of the utility of these unique DNA binding proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Foundation (DBI 0923827 and MCB 0209818 to D.V., DBI 0820831 to A.B.); the University of Minnesota; the China Scholarship Council (2009104157 to Y.Z.); the National Natural Science Foundation of China (30900779 to Y.Z.). Funding for open access charge: The National Science Foundation (DBI 0923827 to D.V.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Marit Nilsen-Hamilton and Lee Bendickson for assistance with mammalian cell culture, Divya Mistry for assistance with development of the TALE-NT website, and Jeff Jones for providing a native avrHah1 clone.
REFERENCES
- 1.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 3.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 4.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 5.Antony G, Zhou J, Huang S, Li T, Liu B, White F, Yang B. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os11N3. Plant Cell. 2010;22:3864–3876. doi: 10.1105/tpc.110.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romer P, Recht S, Strauss T, Elsaesser J, Schornack S, Boch J, Wang S, Lahaye T. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 2010;187:1048–1057. doi: 10.1111/j.1469-8137.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- 7.Romer P, Recht S, Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl Acad. Sci. USA. 2009;106:20526–20531. doi: 10.1073/pnas.0908812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl Acad. Sci. USA. 2010;1078:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 11.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl Acad. Sci. USA. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 16.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc. Natl Acad. Sci. USA. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid "open-source" engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schornack S, Minsavage GV, Stall RE, Jones JB, Lahaye T. Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol. 2008;179:546–556. doi: 10.1111/j.1469-8137.2008.02487.x. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor (NY): Cold Spring Laboratory Press; 1989. [Google Scholar]
- 24.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl Acad. Sci. USA. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann U, Hecht R, Boldyreff B, Issinger OG. Characterization of the cDNA and three processed pseudogenes from the murine protein kinase CK2 alpha subunit. Biochim. Biophys. Acta. 1995;1260:337–340. doi: 10.1016/0167-4781(94)00243-v. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mudgett MB, Chesnokova O, Dahlbeck D, Clark ET, Rossier O, Bonas U, Staskawicz BJ. Molecular signals required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl Acad. Sci. USA. 2000;97:13324–13329. doi: 10.1073/pnas.230450797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.