Abstract
Yeast co-expressing rat APOBEC-1 and a fragment of human apolipoprotein B (apoB) mRNA assembled functional editosomes and deaminated C6666 to U in a mooring sequence-dependent fashion. The occurrence of APOBEC-1-complementing proteins suggested a naturally occurring mRNA editing mechanism in yeast. Previously, a hidden Markov model identified seven yeast genes encoding proteins possessing putative zinc-dependent deaminase motifs. Here, only CDD1, a cytidine deaminase, is shown to have the capacity to carry out C→U editing on a reporter mRNA. This is only the second report of a cytidine deaminase that can use mRNA as a substrate. CDD1-dependent editing was growth phase regulated and demonstrated mooring sequence-dependent editing activity. Candidate yeast mRNA substrates were identified based on their homology with the mooring sequence-containing tripartite motif at the editing site of apoB mRNA and their ability to be edited by ectopically expressed APOBEC-1. Naturally occurring yeast mRNAs edited to a significant extent by CDD1 were, however, not detected. We propose that CDD1 be designated an orphan C→U editase until its native RNA substrate, if any, can be identified and that it be added to the CDAR (cytidine deaminase acting on RNA) family of editing enzymes.
INTRODUCTION
The best characterized mRNA editing mechanisms are adenosine to inosine (A→I) and cytidine to uridine (C→U) base modifications, catalyzed by adenosine deaminases (ADARs) (1) and the cytidine deaminase (CDA) APOBEC-1 (2,3), respectively. Both classes of enzyme share a highly conserved zinc-dependent deaminase motif (2,4). It is on this basis that a number of ADARs and putative C→U editases have been identified (4,5). Of the ADARs, ADAR1 and ADAR2 have critical physiological functions and well-defined substrates (6). Other ADARs (RED2, ADAR3, TENR and T20H4.4) (1) have no known substrates and have not been characterized for their nucleotide/nucleoside deaminase activity.
AID (7), APOBEC-2 (8), Phorbolin and Phorbolin-1 (9) contain a deaminase domain and have homology to APOBEC-1 but have no known mRNA substrate nor a demonstrated mRNA editing activity. APOBEC-1 is therefore currently the only CDA that can edit defined RNA substrates (10–12). CDAs do not need cofactors to deaminate free cytidine (13–15). However, unlike ADARs, APOBEC-1 cannot edit apoB mRNA in the absence of other proteins collectively referred to as auxiliary proteins (10,16–20) and alone has only weak binding affinity for AU-rich RNA sequences (21,22). Biochemical purification of editing activity from tissues or mammalian cell cultures identified a multiprotein complex ranging in size from 11 to 60 S, depending on the tissue studied (16,23–26). Recently a cDNA encoding ACF, a 66 kDa RNA-binding protein, was identified (27). This protein, together with APOBEC-1, was sufficient to stimulate editing in vitro and thus constitutes the minimal editosome. However, for appropriate regulation in vivo and/or subcellular distribution of editing factors (28) it is likely that other factors are required.
CDAs hydrolytically deaminate cytidine nucleotides at the C4 position to generate the corresponding uracil nucleotides as part of nucleotide scavenging pathways. CDAs can be classified by their size and sequence as either homodimers, such as Escherichia coli CDA (EcCDA) and APOBEC-1, or homotetramers, such as the CDAs from Bacillus subtilis, Homo sapiens and Saccharomyces cerevisiae (13). The crystal structure of EcCDA has been determined (29). Molecular modeling of the APOBEC-1 primary amino acid sequence onto the tertiary structure of EcCDA suggests a high degree of structural conservation, despite a somewhat low amino acid identity between the two proteins. Modeling of APOBEC-1 upon the crystal structure of EcCDA suggested that APOBEC-1 forms dimers. Dimerization has been reported to be essential for catalytic function of APOBEC-1 (30,31).
We have previously shown that late log phase yeast were capable of editing ectopically expressed apoB mRNA when co-expressed with APOBEC-1, suggesting that sufficient auxiliary factors exist in yeast to facilitate RNA recognition and editing (32). We show here that of the eight yeast open reading frames (ORFs) encoding a ZDD motif, only CDD1 was capable of significant editing of apoB mRNA. Consistent with earlier findings on APOBEC-1 editing in yeast, CDD1-dependent editing was most efficient in late log phase cultures, suggesting growth phase regulation of auxiliary proteins.
MATERIALS AND METHODS
Plasmids
Yeast ORFs were amplified by PCR from genomic DNA or from cosmids (ATCC) using gene-specific primers, which introduced an EcoRI site proximal to the start codon and an XbaI site distal to the stop codon. The genes CDD1, TAD2 and TAD3 were cloned from the original genetic loci L9672, YJD5 and L8543, respectively, from cosmids obtained from ATCC. These PCR products were cloned into a modified version of pYES2 (InVitrogen), permitting galactose-inducible expression of these His6-HA-tagged ORFs. The editing substrate human apoB was expressed from plasmid pGD308 as described previously (32).
Strains, growth and transformation
The yeast strain used was CL51 (33). All yeast strains were grown and maintained as previously described (32).
Hidden Markov model (HMM) sequence analysis
The published ZDD motif HMM (4) was based on the active site residues and substrate-binding region present in the crystal structure of E.coli CDA (28) (an α–β–α structure). An updated HMM was estimating using an alignment of 20 representative ZDD motifs and version 3.2 of the SAM (Sequence Alignment and Modeling Software System) suite (34,35). In order to identify new ZDD motif-containing sequences the revised HMM was used to perform a database search against the October 2000 release of SwissProt.
RNA isolation
Total RNA was prepared from 10 ml aliquots harvested from yeast cultures at early and late log phase, using TriReagent (MRC Inc.) according to the manufacturer’s instructions. RNAs were digested with RQ DNase I (Promega) and an appropriate restriction enzyme for which there is a site between the PCR primer annealing sites to ensure the removal of contaminating DNA.
Immunofluorescence microscopy
Yeast cells were fixed, permeabilized and immunostained as described (36). Confocal microscopy was performed with a Leica confocal microscope using an SP scan head, with a 100× objective and zoom set to 4×. Images were processed with Leica TCS NT and Adobe Photoshop software.
Editing assay
RT–PCR was performed using 500 ng total RNA and oligo(dT) for first strand cDNA synthesis. Oligonucleotides MS2 and MS3 were used for PCR amplification of apoB cDNAs. Poisoned primer extension analysis was performed using 50 ng RT–PCR product and deoxyoligonucleotide primer DD3 or JBΔ85 as previously described (32,37). The regions of the yeast expressed sequences amplified by RT–PCR were: IRA1 (nt 4770–5066, with respect to the start codon); PRP8 (2810–3134); RAD50 (431–909); YLL015W (3212–3624); YIL037C (121–540). The primers used for poisoned primer extension of the yeast genes were: for IRA1, GCGCTATTATTACATTTTCATTAAGTATACTGATC; for PRP8, CGTTGGTCAAGCTTCGTACCATAAATACTGATCAAG; for RAD50, GATTCAATTTCAGACACTTCTTCATTATACTGATC; for YLL015W, GGACATAATCGGAGAGTAAGATATACTGATCAATC; for YIL037C, ACTCCGTAGTTTGCTTTCGAAATACTG.
In vitro CDA assay
His6-HA-CDD1 was subcloned into pET24a (Novagen) and transformed into E.coli strain BL21 codon plus (Novagen). Protein expression was induced at 37°C by addition of IPTG to the growth medium (LB) to a final concentration of 1 mM for 2 h. Recombinant His6-HA-CDD1 was purified by binding to Talon resin (Clontech) according to the manufacturer’s instructions and eluted with editing buffer (24). The deaminase reaction was performed essentially as described (38). Aliquots of 2.5 µg recombinant CDD1 were incubated with varied concentrations of cytidine in 50 mM Tris pH 7.5 in a 100 µl reaction volume at 37°C. Reactions were stopped by addition of 900 µl of 0.5 M perchloric acid. Extinction of the reaction was measured at 280 nm. For the Lineweaver–Burk plot the Δ[S] values were taken at 5 and 10 min incubation times.
Western blot analysis
Aliquots of cells were lysed by disruption with glass beads and treated as described (32). Lysates from 0.75 A600 units of cells were resolved by 12% SDS–PAGE, transferred to nitrocellulose and reacted with the anti-HA monoclonal antibody (BAbCo) under blotting conditions described previously (32). Detection was by enhanced chemiluminescence (Amersham) according to the manufacturer’s instructions. All blots were exposed to film for the same amount of time.
RESULTS
HMM sequence analysis
Figure 1 shows an HMM-generated alignment of yeast genes and selected other genes identified as containing putative ZDD motifs. These include human and rat APOBEC-1, a CDA known to edit mRNA and the APOBEC-1 homologs human and mouse APOBEC-2 (8), human AID (7), human Phorbolin and Phorbolin-1 (9). Also shown are two double-stranded ADARs, whose active sites are structurally and catalytically related to CDAs and which edit RNA by adenosine deamination (1,2). The large number of known ZDD motif-containing sequences had significant scores against the updated HMM (data not shown). Interestingly, human, yeast and viral members of the ERV1/ALR family also had significant scores, and these are also shown in Figure 1. Yeast ERV1 encodes a FAD-linked sulfhydryl oxidase (39) and human ALR (40) may have a role in liver regeneration and spermatogenesis. Since FAD contains an adenine moiety, it is biologically plausible for the ERV1/ALR family to possess a motif structurally and possibly functionally related to the ZDD motif.
Figure 1.
The HMM-generated multiple sequence alignment of the yeast genes containing a ZDD domain. Also included for comparison are APOBEC-1 and APOBEC-1 homologs. The GenBank accession nos are as follows: CDD1, AAD04031; DCD1, AAB68985; FCY1, CAA89179; RIB2, CAA99076; TAD1, CAA07438; TAD2, CAB60629; TAD3, CAB60630; Rn_APOBEC-1, L07114; Hs_APOBEC-1, AB009422; Hs_APOBEC-2, NM_006789; Mm_APOBEC-2, NM_009694; Hs_AID, AF132979; Hs_Phorbolin-1, NM_004900; Hs_Phorbolin, NM_004900; Hs_DRADA, U10439; Hs_RED1, NM_001112; Tc_ADAT1h, AAG08958; Ce_ADAR, Q22618; Hs_ALR, NP_005253; Sc_ERV1, P27882. Columns containing . correspond to insert states, and numbers indicate the lengths of insertions in sequences at that position (if present). Highlighted in gray are the cysteine and histidine zinc-coordinating residues, the glutamate proton shuttle and the substrate-binding proline. Open triangles identify the locations of the internal free insertion modules employed to model regions L1 and L2 referred to in Mian et al. (4).
The alignments show that five of the eight yeast genes contain perfect ZDD motifs. DCD1 is a deoxycytidylate deaminase (41), FCY1 a cytosine deaminase (42), RIB2 is implicated in both riboflavin biosynthesis (MIPS database, http://speedy.mips.biochem.mpg.de/htbin/search_contig/FIFTEEN?string=YOL066c&elemtype=all) and pseudouridine synthesis (43) and TAD1 and TAD2 are ADARs specific for tRNA, known generically as ADATs (44,45). All have the conserved zinc coordinating residues His, Cys, Cys, the essential Glu proton donor and the Pro that coordinates with the amino leaving group. In CDD1 the Zn-coordinating His is replaced by Cys, but this has been shown to have no effect on catalytic activity in the related CDA from B.subtilis (2). The ZDD in TAD3 (an ADAT factor) (45) and ERV1 is potentially non-catalytic, because of the proton donor Glu→Val change.
Recombinant CDD1 has CDA activity in vitro
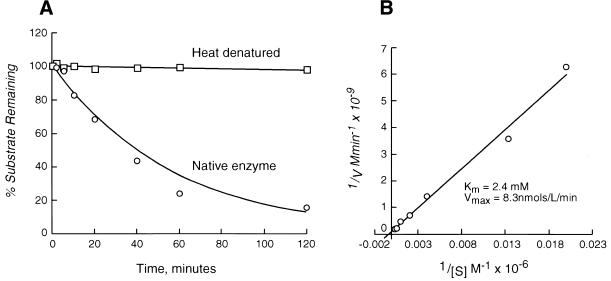
Yeast CDD1 has been previously implicated in purine metabolism in a pathway that converts cytidine to uridine (46). The gene is non-essential and cell extracts from a strain bearing a disruption of the chromosomal gene show a loss of CDA activity in an in vitro assay. The kinetic properties of CDD1 had not been determined. Consequently, CDD1 was overexpressed and purified from E.coli and used for in vitro CDA studies. Recombinant CDD1 was active as a CDA and was sensitive to thermal denaturation (Fig. 2A). The Km of the enzyme for cytidine was ∼2.4 mM (Fig. 2B), a value consistent with those previously reported for other CDAs (Table 1), and Vmax was ∼8.3 nmol/l/min.
Figure 2.
Purified recombinant CDD1 shows CDA activity in vitro. His6-HA-tagged CDD1 was overexpressed and purified as described. An aliquot of 2.5 µg protein was used per assay point. (A) The decrease in cytidine concentration with time when incubated with enzyme. Also shown is the activity of the heat-denatured enzyme. (B) The Lineweaver–Burk reciprocal plot from which Km and Vmax were calculated.
Table 1.
| Enzyme |
Km |
Reference |
| Saccharomyces cerevisiae CDD1 | 2.4 mM | This study |
| Aspergillus niger NRRL3 CDA | 2.6 mM | (47) |
| Arabidopsis thaliana CDA1 | 150 µM | (14) |
| Escherichia coli CDA | 110 µM | (48) |
| Homo sapiens CDA | 39 µM | (15) |
Overexpression of ZDD-containing ORFs
Ectopic expression of APOBEC-1 was sufficient to induce editing of the apoB substrate RNA in stationary phase yeast cultures (32). To evaluate the possibility that yeast deaminases may have the capacity to edit mRNA, seven yeast ORFs shown to contain a ZDD motif were cloned into an inducible yeast expression vector and co-transformed into yeast along with a plasmid constitutively expressing the apoB mRNA editing substrate. These strains were cultured such that the cloned deaminases were expressed and total RNA was isolated from these cultures at early and late log phase. In all cases, overexpression of the deaminases had no adverse effect on cell growth, as determined by the doubling time of the cultures or colony morphology on solid medium.
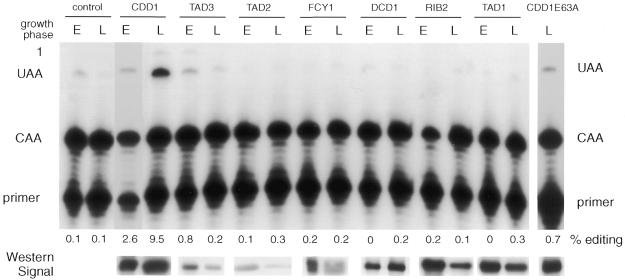
All samples were assayed for editing of the apoB RNA test substrate by RT–PCR and poisoned primer extension. The presence of the second poisoned primer extension product demonstrated that the target cytidine had been edited to uridine. CDD1 was the only ZDD-containing protein capable of significant C→U deamination editing of apoB RNA (Fig. 3). A much lower level of C→U editing was observed when TAD3 was overexpressed during log phase growth. The amount of target apoB mRNA edited (10%) was equivalent to that demonstrated previously in yeast for APOBEC-1 (32). RNA editing by CDD1 was significantly higher in the late log phase culture than in a culture in early log phase (Fig. 3), suggesting a growth phase regulation similar to that observed for APOBEC-1-dependent editing in yeast (32). Editing of cytidines other than C6666 (known as promiscuous editing and reflected by the occurrence of a third primer extension product) was extremely low. This contrasted with APOBEC-1 editing in yeast, where significant promiscuous editing was observed.
Figure 3.
Assay of yeast deaminase gene products for mRNA editing activity. The deaminases identified in the HMM search were overexpressed in a yeast strain that also expressed the apoB editing substrate (as described). Cells were harvested at early (E) and late (L) log phase (as indicated) and total RNA isolated. The control lanes are cells expressing only the apoB substrate RNA. Editing was assayed by RT–PCR and poisoned primer extension. The CDD1 mutant CDD1E63A was overexpressed and assayed for editing at late log phase only. Shown underneath each lane is a segment from an immunoblot showing the signal generated by each overexpressed protein. Promiscuous editing is evident as a third primer extension stop indicated as ‘1’.
We have confirmed by immunoblotting analysis (Fig. 3) that each of the ORFs was expressed as full-length protein. The level of expression of the selected ORFs was not uniform, although the vector and growth conditions used were identical. We cannot rule out the possibility that one or more of the ORFs other than CDD1 may have displayed editing activity had they been expressed at higher levels.
To confirm that CDD1 was the catalytic activity responsible for the increase in editing, the conserved Glu residue within the ZDD motif was replaced with Ala, a mutation that has been shown to dramatically reduce deaminase activity of B.subtilis CDA (49). A Glu63Ala mutant (CDD1E63A) was co-expressed with apoB mRNA in yeast at a similar level to that observed for wild-type overexpressed CDD1 (Fig. 3) and assayed for editing. The E63A mutation suppressed mRNA editing ∼14-fold (Fig. 3), thus confirming that the catalytically active form of CDD1 was responsible for editing of the apoB mRNA.
CDD1-dependent editing is sequence dependent
We have previously shown that apoB RNA editing by APOBEC-1 is dependent on the tripartite motif in both mammalian cells (37,50) and yeast (32). We tested the ability of CDD1 to edit C6666 in the context of either an inversion of the mooring sequence (MI) or a change in the upstream enhancer element (Δ3′TL). Both of these alterations reduced editing to almost background levels (Fig. 4). This is consistent with the effects seen for APOBEC-1 in both mammalian and yeast systems (32,37,50).
Figure 4.
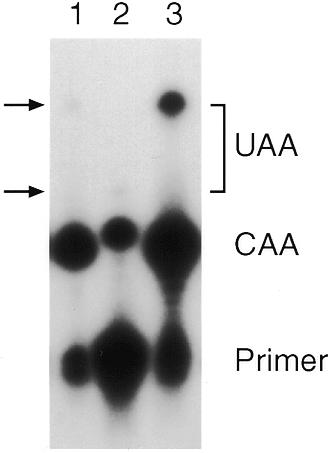
Editing is sequence dependent. Lanes 1–3 show editing assays for strains expressing CDD1 and either MI RNA, Δ3′TL RNA or apoB RNA, respectively. Primer extension products CAA and UAA correspond to unedited and edited RNA, respectively. Editing of apoB and MI give rise to a product extended by a further 11 nt, Δ3′TL gives rise to a product extended by 3 nt.
Native substrates for CDD1-dependent editing
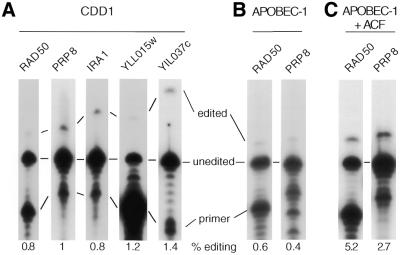
Given that yeast express a bifunctional CDA and APOBEC-1-complementing auxiliary protein(s), we postulated that there may be a yeast substrate for editing. On the basis of homology to the apoB tripartite editing cassette, 42 candidate yeast genes were identified as encoding mRNAs that may be editing substrates (Table 2). The observed deviations in nucleotide sequence in the mooring sequence, spacer and enhancer elements from that present in the tripartite sequence in apoB mRNA were all predicted to support editing based on previous mutagenesis studies (37,50–53). Five mRNAs were selected to test for editing (IRA3, PRP8, RAD50, YIL037C and YLL015W) as these represented the range of sequence diversity observed among the 42 candidates with respect to the tripartite motif of apoB RNA. The candidate mRNAs were isolated from a strain overexpressing CDD1, amplified in the region of the tripartite motif by RT–PCR and the products analyzed for editing. None of the candidate substrates were edited as well as the ectopic apoB RNA (Fig. 5A).
Table 2.
| Gene |
Target coordinate |
Tripartite motif-like sequence |
Codon change due to editing |
| Human apoB | 6666 | tatgataCaattTGATCAGTATA | Q→STOP |
| 1. YLL015W | 3436 | agCtaaaaagatTGATCAGTATA | L→L |
| 2. JEM1 | 406 | gcatCagaaggaTGATCAGTATg | Q→STOP |
| 3. YER186C | 522/524 | tgCtCtgaaattaGATCAGTATA | C→C/S→F |
| 4. YHR115C | 608/609 | ttCCaaaattccaGATCAGTATc | S→F/S→S |
| 5. TRA1 | 3687/3688 | tttCCtaaaaaagtATCAGTATA | F→F/L→L |
| 6. YDR200C | 174 | aaatgtCgatataGATCAGTATA | V→V |
| 7. RAD50 | 692 | gcaaaCtaagataGATCAGTATA | T→I |
| 8. YLR106C | 14577 | gaactaCattccaGATCAGTATg | Y→Y |
| 9. CCT5 | 1384 | gcaaCgtggtatTGATCAGTATg | R→C |
| 10. YJL029C | 78 | aCatagatgaacTGATCAGTATt | N→N |
| 11. PRP8 | 2978 | cgatgCatatctTGATCAGTATt | A→A/L→F |
| 12. YJR119C | 1279 | ttggCatatggaaGATCAGTATA | H→Y |
| 13. FKS1 | 318 | tgatagCtatggTGgTCAGTATA | S→S |
| 14. YIL037C | 326 | tgataataCaatTGATCAGTATt | T→I |
| 15. YLR278C | 484/489 | gaggCaagaCaaaGATCAGTATA | Q→STOP/D→D |
| 16. RDI1 | 48 | ggagagaaaCaacGATCAGTATA | N→N |
| 17. RAV2 | 952/953/956 | gtgatCCaaCttTGATCAGTATA | P→S/P→L/T→I |
| 18. POM152 | 3251/3252 | taccagtaggaCaGATCAGTATA | T→I/T→T |
| 19. IRA1 | 4843 | cattaaggCataaGATCAGTATA | H→Y |
| 20. YBR269C | 191 | gcaagaggCaataGATCAGTATA | A→V |
| 21. FKH2 | 2564 | cggatggtgCaaaGATCAGTATt | A→V |
| 22. IAH1 | 78 | tgaagatggCaaaGATCAGTATg | G→G |
| 23. MTD1 | 314/316 | cggtaatgCtCaaGATCAGTATt | A→V/Q→STOP |
| 24. BPH1 | 3618 | tggaaaaatgCaaGATCAGaATA | Y→Y |
| 25. OST3 | 444 | tggaccacagCgTGATCAGTATt | H→H |
| 26. PRP5 | 1076/1077 | gatacatCCattaGATCAGTATg | S→F/S→S |
| 27. PKH1 | 1375 | aagagcattCtgTGATCAGTATg | H→Y |
| 28. YGR271W | 2489/2490 | atgtattgtCCaTGATCAGTATg | S→F/S→S |
| 29. TOS3 | 1352/1355 | cttttgCatCtcaGATCAGTATt | A→V/S→F |
| 30. YAR009C | 3386 | atagtagatCaacGATCAGTATA | S→L |
| 31. YBL101W-B | 5109 | atagtagatCaacGATCAGTATA | S→L |
| 32. YCL019W | 5109 | atagtagatCaacGATCAGTATA | S→L |
| 33. YER138C | 5064 | atagtagatCaacGATCAGTATA | S→L |
| 34. YFL002W-A | 5109 | atagtagatCaacGATCAGTATA | S→L |
| 35. YJR027W | 5064 | atagtagatCaacGATCAGTATA | S→L |
| 36. YML039W | 5063 | atagtagatCaacGATCAGTATA | S→L |
| 37. SNF1 | 1368 | atcacccttCatgGATCAGTATA | F→F |
| 38. YER010C | 676 | caaaatgatCgaaGATCAGTATt | R→STOP |
| 39. RIF1 | 43 | gatagatCgaatTGATCAGcATA | R→STOP |
| 40. MLP1 | 935 | CtgtaaaagaagaatTGAaCAGTATA | A→V |
| 41. TOM1 | 5438 | CaaataaagttgaggcaGATCAGTAcc | S→L |
| 42. YDR119W | 416 | CagattttagtgataGATCAGTATc | T→L |
The yeast genes identified as containing a mooring sequence-like element through a FASTA sequence identity search are listed. Those assayed are shown in bold. The cytidine(s) predicted to be edited is(are) identified by nucleotide coordinate with respect to the first nucleotide of the ORF and shown underlined in the sequence column. The mooring sequence-like element is highlighted in bold. The human apoB mooring sequence and tripartite motif are shown for comparison. The effect on the codon of editing at the indicated cytidine(s) is also shown. ORFs 30–36 are a Ty element ORF at different loci.
Figure 5.
Editing assay of yeast mRNAs containing mooring sequence-like elements. The chromosomally expressed mRNAs from five yeast genes identified as containing mooring sequence-like elements were amplified by RT–PCR and assayed for editing by poisoned primer extension. (A) A typical editing assay for the genes identified in Table 2. The distance from the primer to the first and second stops is different for each primer. The lanes have been aligned with respect to the first stop. (B and C) Editing assays for RAD50 and PRP8 mRNAs from strains expressing APOBEC-1 and APOBEC-1 plus ACF, respectively.
The data suggested that the five test mRNAs may not have been appropriate substrates for CDARs. Alternatively, the editing sites may have been occluded in vivo by RNA secondary structure or ribonucleoprotein complexes. To address this, APOBEC-1 (Fig. 5B) or APOBEC-1 together with ACF (Fig. 5C) were expressed in yeast and the candidate mRNAs assayed for editing. Two of the mRNAs (RAD50 and PRP8) revealed significant editing when APOBEC-1 and ACF were co-expressed (Fig. 5C), suggesting that their editing sites were accessible to editases. The data also suggested that there was predictive value in using the mammalian tripartite motif as a model for candidate yeast mRNA editing substrates but that combined overexpression of CDAR and cognate RNA-binding proteins was necessary to reveal those that could potentially serve as substrates. Consequently, each candidate substrate will need to be evaluated for RNA editing in future experiments.
Subcellular localization of CDD1
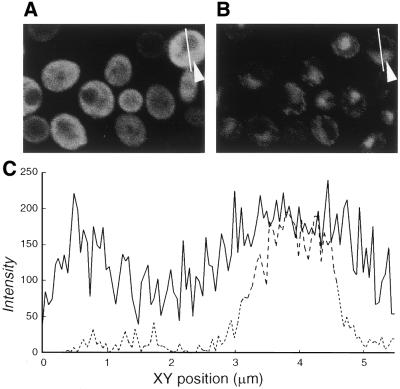
APOBEC-1-dependent apoB editing has been proposed and demonstrated to be a nuclear event in mammals (28,54,55). APOBEC-1 and ACF were both distributed in the nucleus and cytoplasm of editing-competent cells (28,56,57). Evidence has also been presented that APOBEC-1 distributes to the nucleus and cytoplasm of yeast (32). The cellular localization of CDD1 was evaluated in cells expressing His6-HA-CDD1 at late log phase by immunofluorescence confocal microscopy. Typical fields showing FITC detection of the anti-HA antibody and DAPI staining are shown (Fig. 6A and B). The intensities of the fluorescence from both the DAPI staining and the HA-CDD1-specific staining were quantified along a line drawn through the long axis of the cell through the nucleus and the plots overlaid. A typical plot is shown (Fig. 6C). The data demonstrated CDD1 in both the cytoplasm and nucleus.
Figure 6.
Subcellular localization of CDD1. Yeast expressing His6-HA-CDD1 were visualized by immunofluorescence confocal microscopy. A representative field is shown for anti-HA antibody staining (A) and DAPI staining (B). The cell on the right is in the optical plane of the cell nucleus. Arrowheads indicate the positions of nuclei. The line through the cell on the right (A and B) indicates, from top to bottom, the path of the fluorescence quantification scan shown left to right in (C). The relative subcellular distribution of fluorescence was characterized by quantification of pixel intensities for DAPI (broken line) and FITC (solid line).
DISCUSSION
Recently a number of CDAs with homology to APOBEC-1 have been identified (7–9). No natural substrates for these presumptive editing enzymes have been identified nor have these enzymes been shown to have activity on test RNA substrates. CDD1 is therefore the first CDA other than APOBEC-1 to have demonstrable mRNA editing activity. Similar to APOBEC-1, recombinant CDD1 is unable to edit mRNA in the absence of appropriate auxiliary proteins as cell extracts or recombinant factors (unpublished findings). A mutation in CDD1 predicted to inhibit deaminase catalytic activity impaired editing activity.
We have presented evidence that CDD1 is a CDA and is also capable of deaminating mRNA. Dual substrate capacity has been observed for APOBEC-1 as well (58). CDD1 has previously been identified as a CDA involved in nucleotide scavenging (46). We have shown that purified recombinant CDD1 can deaminate free cytidine and have calculated the Km and Vmax for this reaction to be 2.4 mM and 8.3 nmol/l/min, respectively. Although CDD1 can directly catalyze the formation of uridine from cytidine in vitro, the in vivo utilization of cytidine as a UMP source requires cleavage of cytidine into cytosine (46). The primary function of CDD1 may therefore not be restricted to nucleotide scavenging. CDD1 has been shown to be a non-essential gene and, given the apparent lack of redundant editing enzyme activity (this report), mRNA C→U editing may not be essential in yeast under normal growth conditions. We cannot rule out the possibility that RNA editing by CDD1 becomes important to yeast under stressful growth conditions. In this regard it is interesting that CDD1 and APOBEC-1 RNA editing activities were both maximally stimulated as cells became growth arrested, suggesting induction of an essential auxiliary protein and/or removal of an inhibitory factor.
The updated ZDD motif HMM identified all the previously known yeast deaminases, including three genes (TAD1, TAD2 and TAD3) that were subsequently shown to be involved in tRNA adenosine deamination. All three ADAT (adenosine deaminase active on tRNA) genes have significant identity to the mammalian ADARs in the deaminase domain, yet lack the double-stranded RNA-binding domains. TAD1 most likely acts as a monomer (44), whereas TAD2 acts as a heterodimer with the non-catalytic specificity factor TAD3 (45). Interestingly, we observed a slight increase in apoB reporter RNA editing in log phase cells overexpressing TAD3. Although the level of editing activity was close to background, we cannot rule out the possibility that TAD3 had C→U editing activity or that it stimulated endogenous CDD1 editing activity. Editing in strains overexpressing the E63A mutant were slightly above background. It is possible that the E63A mutant does not have a dominant negative effect on editing. Multimers of CDD1 containing wild-type and mutant deaminases may be competent for editing. This would lead to an increased number of editosomes and explain the slight increase above background observed.
The ERV1/ALR family of metazoan and viral proteins appear to be new members of the ZDD family since they score well against the HMM and possess three of the four conserved residues (the Glu residue is absent).
CDD1-catalyzed RNA editing in yeast displays high fidelity. There is only barely detectable promiscuous editing of the apoB substrate, compared to the editing of apoB mRNA seen with APOBEC-1 in yeast (32). CDD1-dependent editing is also highly sequence specific. Yeast mRNAs that might be predicted to be edited by overexpression of CDD1, based on their sequence similarity to the apoB mooring sequence, were not edited. Furthermore, CDD1-dependent editing is sensitive to changes in the mooring sequence and the upstream enhancer element. CDD1 was unable to edit the MI and Δ3′TL substrates, as was also the case with APOBEC-1 when it was assayed for editing of these substrates in yeast (32). These findings suggest specificity and/or regulation of the editing event. The level of fidelity seen with CDD1 was much stronger than that seen when the heterologous deaminase APOBEC-1 was expressed (32), suggesting that CDD1 is better suited to the cognate auxiliary factors expressed in yeast. The factors that confer sequence selectivity and fidelity are functionally homologous to higher eukaryotic auxiliary factors, as APOBEC-1 alone does not display high affinity nor selective RNA-binding capacity (21,22,59,60).
Given that the apoB editing substrate was a polyadenylated RNA polymerase II transcript, we sought yeast mRNA editing substrates and identified 42 candidates based on the occurrence of sequence motifs with a 9+ sequence identity to the 11 nt mammalian mooring sequence. However, none of those tested supported a high level of editing by CDD1. The editing of PRP8 and RAD50 mRNAs following co-expression of APOBEC-1/ACF suggested that these editing sites were accessible to an editosome. The fact that CDD1 was unable to edit them suggests either that they are not CDD1 targets or that expression of the presumptive RNA-binding protein with which CDD1 interacts may be rate limiting. We cannot rule out the possibility that the mooring sequence in apoB mRNA may not be the best editing site for CDD1 and thus that our identity search was sub-optimal. It is possible that RNAs other than mRNAs may be CDD1 substrates, but we have been unable to find a mooring sequence-like element in non-coding transcripts. Finally, we cannot rule out the possibility that editing of the candidate mRNAs may have been prevented by inhibitory factors.
CDD1 has been classified as a homotetrameric enzyme (13). The ability of CDD1 to edit RNA was unexpected, given that APOBEC-1 is a homodimer. It may be that the quaternary structures of CDD1 and APOBEC-1 are quite similar and that their catalytic sites and RNA-binding surfaces may be superimposable. It has been proposed that sequence differences between APOBEC-1 and EcCDA should be diagnostic of CDARs (30,61). Although CDD1 does not conform to this model, molecular modeling of CDD1 may generate a more widely applicable CDAR motif.
Global yeast two-hybrid analysis of the interactions of yeast proteins (62) suggested that CDD1 interacts only with itself. Whilst this supports the proposed tetrameric structure of the holoenzyme, it conflicts with our model that there are essential auxiliary factors which lend specificity and fidelity to the editing reaction in yeast and with which the deaminase must interact. The identification by genetic or physical methods of a yeast ACF ortholog and other interacting factors will be a goal for future studies.
Finally, the cellular localization of overexpressed CDD1 has been determined by immunocytochemistry to be both nuclear and cytoplasmic, as was APOBEC-1 (32). This is consistent with the apparent lack of subcellular localization signals and its small size, allowing it to freely diffuse through the nuclear pore complex. Addition of the SV40 nuclear localization signal to the N-terminus of APOBEC-1 caused an increase in apoB editing (31), suggesting that mRNA editing in yeast may be a nuclear event. The data showing a nuclear distribution of CDD1 are therefore in agreement with the prevailing hypothesis that editing occurs in the nucleus (28).
In summary, we have identified a yeast CDA that can edit RNA. This is only the second example of a CDA that can use RNA as a substrate. CDD1 is thus the second member of what is likely to become a growing family of enzymes that are CDAs that act on RNA. It was recently proposed (5) that, in keeping with the ADAR and ADAT nomenclature, CDAs that act on RNA be termed CDARs. APOBEC-1 and CDD1 are therefore members of the CDAR family, but we suggest that this nomenclature should be a functional definition and thus restricted to editases with a proven capacity to edit RNA. We therefore propose that CDD1 is an orphan CDAR.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Jenny M.L.Smith for preparation of the figures. This work was supported by Public Health Service Grant DK43738 and the Alcoholic Beverage Medical Research Foundation, awarded to H.C.S., and the Life Sciences Division under US Department of Energy Contract no. DE-AC03-76SF00098, awarded to I.S.M.
References
- 1.Bass B.L., Nishikura,K., Keller,W., Seeburg,P.H., Emeson,R.B., O’Connell,M.A., Samuel,C.E. and Herbert,A. (1997) A standardized nomenclature for adenosine deaminases that act on RNA. RNA, 3, 947–949. [PMC free article] [PubMed] [Google Scholar]
- 2.Smith H.C. and Sowden,M.P. (1996) Base-modification mRNA editing through deamination—the good, the bad and the unregulated. Trends Genet., 12, 418–424. [DOI] [PubMed] [Google Scholar]
- 3.Davidson N.O., Innerarity,T.L., Scott,J., Smith,H., Driscoll,D.M., Teng,B. and Chan,L. (1995) Proposed nomenclature for the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme: APOBEC-1. RNA, 1, 3. [PMC free article] [PubMed] [Google Scholar]
- 4.Mian I.S., Moser,M.J., Holley,W.R. and Chatterjee,A. (1998) Statistical modelling and phylogenetic analysis of a deaminase domain. J. Comput. Biol., 5, 57–72. [DOI] [PubMed] [Google Scholar]
- 5.Maas S. and Rich,A. (2000) Changing genetic information through RNA editing. Bioessays, 22, 790–802. [DOI] [PubMed] [Google Scholar]
- 6.Smith H.C., Gott,J.M. and Hanson,M.R. (1997) A guide to RNA editing. RNA, 3, 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 7.Muramatsu M., Sankaranand,V.S., Anant,S., Sugai,M., Kinoshita,K., Davidson,N.O. and Honjo,T. (1999) Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem., 274, 18470–18476. [DOI] [PubMed] [Google Scholar]
- 8.Liao W., Hong,S.H., Chan,B.H., Rudolph,F.B., Clark,S.C. and Chan,L. (1999) APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem. Biophys. Res. Commun., 260, 398–404. [DOI] [PubMed] [Google Scholar]
- 9.Madsen P., Anant,S., Rasmussen,H.H., Gromov,P., Vorum,H., Dumanski,J.P., Tommerup,N., Collins,J.E., Wright,C.L., Dunham,I., MacGinnitie,A.J., Davidson,N.O. and Celis,J.E. (1999) Psoriasis upregulated phorbolin-1 shares structural but not functional similarity to the mRNA-editing protein apobec-1. J. Invest. Dermatol., 113, 162–169. [DOI] [PubMed] [Google Scholar]
- 10.Teng B., Burant,C.F. and Davidson,N.O. (1993) Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science, 260, 1816–1819. [DOI] [PubMed] [Google Scholar]
- 11.Skuse G.R., Cappione,A.J., Sowden,M., Metheny,L.J. and Smith,H.C. (1996) The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res., 24, 478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamanaka S., Poksay,K.S., Arnold,K.S. and Innerarity,T.L. (1997) A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev., 11, 321–333. [DOI] [PubMed] [Google Scholar]
- 13.Carlow D.C., Carter,C.W.Jr, Mejlhede,N., Neuhard,J. and Wolfenden,R. (1999) Cytidine deaminases from B. subtilis and E. coli: compensating effects of changing zinc coordination and quaternary structure. Biochemistry, 38, 12258–12265. [DOI] [PubMed] [Google Scholar]
- 14.Vincenzetti S., Cambi,A., Neuhard,J., Schnorr,K., Grelloni,M. and Vita,A. (1999) Cloning, expression and purification of cytidine deaminase from Arabidopsis thaliana. Protein Expr. Purif., 15, 8–15. [DOI] [PubMed] [Google Scholar]
- 15.Vincenzetti S., Cambi,A., Neuhard,J., Garattini,E. and Vita,A. (1996) Recombinant human cytidine deaminase: expression, purification and characterization. Protein Expr. Purif., 8, 247–253. [DOI] [PubMed] [Google Scholar]
- 16.Harris S.G., Sabio,I., Mayer,E., Steinberg,M.F., Backus,J.W., Sparks,J.D., Sparks,C.E. and Smith,H.C. (1993) Extract-specific heterogeneity in high-order complexes containing apolipoprotein B mRNA editing activity and RNA-binding proteins. J. Biol. Chem., 268, 7382–7392. [PubMed] [Google Scholar]
- 17.Lau P.P., Zhu,H.J., Nakamuta,M. and Chan,L. (1997) Cloning of an Apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing,. J. Biol. Chem., 272, 1452–1455. [DOI] [PubMed] [Google Scholar]
- 18.Navaratnam N., Shah,R., Patel,D., Fay,V. and Scott,J. (1993) Apolipoprotein B mRNA editing is associated with UV crosslinking of proteins to the editing site. Proc. Natl Acad. Sci. USA, 90, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau P.P., Chen,S.H., Wang,J.C. and Chan,L. (1990) A 40 kilodalton rat liver nuclear protein binds specifically to apolipoprotein B mRNA around the RNA editing site. Nucleic Acids Res., 18, 5817–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schock D., Kuo,S.R., Steinburg,M.F., Bolognino,M., Sparks,J.D., Sparks,C.E. and Smith,H.C. (1996) An auxiliary factor containing a 240-kDa protein complex is involved in apolipoprotein B RNA editing. Proc. Natl Acad. Sci. USA, 93, 1097–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anant S., MacGinnitie,A.J. and Davidson,N.O. (1995) apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J. Biol. Chem., 270, 14762–14767. [PubMed] [Google Scholar]
- 22.Navaratnam N., Bhattacharya,S., Fujino,T., Patel,D., Jarmuz,A.L. and Scott,J. (1995) Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell, 81, 187–195. [DOI] [PubMed] [Google Scholar]
- 23.Smith H.C., Kuo,S.R., Backus,J.W., Harris,S.G., Sparks,C.E. and Sparks,J.D. (1991) In vitro apolipoprotein B mRNA editing: identification of a 27S editing complex. Proc. Natl Acad. Sci. USA, 88, 1489–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith H.C. (1998) Analysis of protein complexes assembled on apolipoprotein B mRNA for mooring sequence-dependent RNA editing. Methods, 15, 27–39. [DOI] [PubMed] [Google Scholar]
- 25.Garcia Z.C., Poksay,K.S., Bostrom,K., Johnson,D.F., Balestra,M.E., Shechter,I. and Innerarity,T.L. (1992) Characterization of apolipoprotein B mRNA editing from rabbit intestine. Arterioscler. Thromb., 12, 172–179. [DOI] [PubMed] [Google Scholar]
- 26.Driscoll D.M. and Casanova,E. (1990) Characterization of the apolipoprotein B mRNA editing activity in enterocyte extracts. J. Biol. Chem., 265, 21401–21403. [PubMed] [Google Scholar]
- 27.Mehta A., Kinter,M.T., Sherman,N.E. and Driscoll,D.M. (2000) Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol., 20, 1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y., Sowden,M.P. and Smith,H.C. (2000) Induction of cytidine to uridine editing on cytoplasmic apolipoprotein B mRNA by overexpressing APOBEC-1. J. Biol. Chem., 275, 22663–22669. [DOI] [PubMed] [Google Scholar]
- 29.Betts L., Xiang,S., Short,S.A., Wolfenden,R. and Carter,C.W.Jr (1994) Cytidine deaminase. The 2.3 Å crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol., 235, 635–656. [DOI] [PubMed] [Google Scholar]
- 30.Navaratnam N., Fujino,T., Bayliss,J., Jarmuz,A., How,A., Richardson,N., Somasekaram,A., Bhattacharya,S., Carter,C. and Scott,J. (1998) Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J. Mol. Biol., 275, 695–714. [DOI] [PubMed] [Google Scholar]
- 31.Oka K., Kobayashi,K., Sullivan,M., Martinez,J., Teng,B.B., Ishimura-Oka,K. and Chan,L. (1997) Tissue-specific inhibition of apolipoprotein B mRNA editing in the liver by adenovirus-mediated transfer of a dominant negative mutant APOBEC-1 leads to increased low density lipoprotein in mice. J. Biol. Chem., 272, 1456–1460. [DOI] [PubMed] [Google Scholar]
- 32.Dance G.S., Sowden,M.P., Yang,Y. and Smith,H.C. (2000) APOBEC-1 dependent cytidine to uridine editing of apolipoprotein B RNA in yeast. Nucleic Acids Res., 28, 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesser C.F. and Guthrie,C. (1993) Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics, 133, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krogh A., Brown,M., Mian,I.S., Sjolander,K. and Haussler,D. (1994) Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol., 235, 1501–1531. [DOI] [PubMed] [Google Scholar]
- 35.Hughey R. and Krogh,A. (1996) Hidden Markov models for sequence analysis: extension and analysis of the basic method. Comput. Appl. Biosci., 12, 95–107. [DOI] [PubMed] [Google Scholar]
- 36.Roberts C.J., Raymond,C.K., Yamashiro,C.T. and Stevens,T.H. (1991) Methods for studying the yeast vacuole. Methods Enzymol., 194, 644–661. [DOI] [PubMed] [Google Scholar]
- 37.Backus J.W., Schock,D. and Smith,H.C. (1994) Only cytidines 5′ of the apolipoprotein B mRNA mooring sequence are edited. Biochim. Biophys. Acta, 1219, 1–14. [DOI] [PubMed] [Google Scholar]
- 38.Neuhard J. (1968) Pyrimidine nucleotide metabolism and pathways of thymidine triphosphate biosynthesis in Salmonella typhimurium. J. Bacteriol ., 96, 1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J., Hofhaus,G. and Lisowsky,T. (2000) Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett., 477, 62–66. [DOI] [PubMed] [Google Scholar]
- 40.Hagiya M., Francavilla,A., Polimeno,L., Ihara,I., Sakai,H., Seki,T., Shimonishi,M., Porter,K.A. and Starzl,T.E. (1994) Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc. Natl Acad. Sci. USA, 91, 8142–8146. [Erratum, Proc. Natl Acad. Sci. USA, 1995, 92, 3076] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIntosh E.M. and Haynes,R.H. (1986) Sequence and expression of the dCMP deaminase gene (DCD1) of Saccharomyces cerevisiae. Mol. Cell. Biol., 6, 1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senter P.D., Su,P.C., Katsuragi,T., Sakai,T., Cosand,W.L., Hellstrom,I. and Hellstrom,K.E. (1991) Generation of 5-fluorouracil from 5-fluorocytosine by monoclonal antibody-cytosine deaminase conjugates. Bioconjug. Chem., 2, 447–451. [DOI] [PubMed] [Google Scholar]
- 43.Koonin E.V. (1996) Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res., 24, 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerber A., Grosjean,H., Melcher,T. and Keller,W. (1998) Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J., 17, 4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerber A.P. and Keller,W. (1999) An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science, 286, 1146–1149. [DOI] [PubMed] [Google Scholar]
- 46.Kurtz J.E., Exinger,F., Erbs,P. and Jund,R. (1999) New insights into the pyrimidine salvage pathway of Saccharomyces cerevisiae: requirement of six genes for cytidine metabolism. Curr. Genet., 36, 130–136. [DOI] [PubMed] [Google Scholar]
- 47.Ali T.H. (1998) Some kinetic studies on cytidine aminohydrolase activity from Aspergillus niger NRRL3. Acta Microbiol. Pol., 47, 365–372. [PubMed] [Google Scholar]
- 48.Vita A., Amici,A., Cacciamani,T., Lanciotti,M. and Magni,G. (1985) Cytidine deaminase from Escherichia coli B. Purification and enzymatic and molecular properties. Biochemistry, 24, 6020–6024. [DOI] [PubMed] [Google Scholar]
- 49.Carlow D.C., Smith,A.A., Yang,C.C., Short,S.A. and Wolfenden,R. (1995) Major contribution of a carboxymethyl group to transition-state stabilization by cytidine deaminase: mutation and rescue. Biochemistry, 34, 4220–4224. [DOI] [PubMed] [Google Scholar]
- 50.Backus J.W. and Smith,H.C. (1992) Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res., 20, 6007–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith H.C. (1993) Apolipoprotein B mRNA editing: the sequence to the event. Semin. Cell Biol., 4, 267–278. [DOI] [PubMed] [Google Scholar]
- 52.Shah R.R., Knott,T.J., Legros,J.E., Navaratnam,N., Greeve,J.C. and Scott,J. (1991) Sequence requirements for the editing of apolipoprotein B mRNA. J. Biol. Chem., 266, 16301–16304. [PubMed] [Google Scholar]
- 53.Backus J.W. and Smith,H.C. (1994) Specific 3′ sequences flanking a minimal apolipoprotein B (apoB) mRNA editing ‘cassette’ are critical for efficient editing in vitro. Biochim. Biophys. Acta, 1217, 65–73. [PubMed] [Google Scholar]
- 54.Lau P.P., Xiong,W.J., Zhu,H.J., Chen,S.H. and Chan,L. (1991) Apolipoprotein B mRNA editing is an intranuclear event that occurs posttranscriptionally coincident with splicing and polyadenylation. J. Biol. Chem., 266, 20550–20554. [PubMed] [Google Scholar]
- 55.Sowden M., Hamm,J.K., Spinelli,S. and Smith,H.C. (1996) Determinants involved in regulating the proportion of edited apolipoprotein B RNAs. RNA, 2, 274–288. [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y., Yang,Y. and Smith,H.C. (1997) Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc. Natl Acad. Sci. USA, 94, 13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siddiqui J.F., Van Mater,D., Sowden,M.P. and Smith,H.C. (1999) Disproportionate relationship between APOBEC-1 expression and apolipoprotein B mRNA editing activity. Exp. Cell Res., 252, 154–164. [DOI] [PubMed] [Google Scholar]
- 58.Navaratnam N., Morrison,J.R., Bhattacharya,S., Patel,D., Funahashi,T., Giannoni,F., Teng,B.B., Davidson,N.O. and Scott,J. (1993) The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem., 268, 20709–20712. [PubMed] [Google Scholar]
- 59.Yang Y., Yang,Y., Kovalski,K. and Smith,H.C. (1997) Partial characterization of the auxiliary factors involved in apolipoprotein B mRNA editing through APOBEC-1 affinity chromatography. J. Biol. Chem., 272, 27700–27706. [DOI] [PubMed] [Google Scholar]
- 60.Anant S., MacGinnitie,A.J. and Davidson,N.O. (1995) The binding of apobec-1 to mammalian apo B RNA is stabilized by the presence of complementation factors which are required for post-transcriptional editing. Nucleic Acids Symp. Ser., 33, 99–102. [PubMed] [Google Scholar]
- 61.Carter C.W. (1998) Nucleoside deaminases for cytidine and adenosine: comparison with deaminases acting on RNA. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 363–375.
- 62.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P., Qureshi-Emili,A., Li,Y., Godwin,B., Conover,D., Kalbfleisch,T., Vijayadamodar,G., Yang,M., Johnston,M., Fields,S. and Rothberg,J.M. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]