Abstract
miR-7 (microRNA-7) has been characterized as a tumour suppressor in several human cancers. It targets a number of proto-oncogenes that contribute to cell proliferation and survival. However, the mechanism(s) by which miR-7 suppresses tumorigenesis in TSCC (tongue squamous cell carcinoma) is unknown. The present bioinformatics analysis revealed that IGF1R (insulin-like growth factor 1 receptor) mRNA is a potential target for miR-7. Ectopic transfection of miR-7 led to a significant reduction in IGF1R at both the mRNA and protein levels in TSCC cells. Knockdown of miR-7 in TSCC cells enhanced IGF1R expression. Direct targeting of miR-7 to three candidate binding sequences located in the 3′-untranslated region of IGF1R mRNA was confirmed using luciferase-reporter-gene assays. The miR-7-mediated down-regulation of IGF1R expression attenuated the IGF1 (insulin-like growth factor 1)-induced activation of Akt (protein kinase B) in TSCC cell lines, which in turn resulted in a reduction in cell proliferation and cell-cycle arrest, and an enhanced apoptotic rate. Taken together, the present results demonstrated that miR-7 regulates the IGF1R/Akt signalling pathway by post-transcriptional regulation of IGF1R. Our results indicate that miR-7 plays an important role in TSCC and may serve as a novel therapeutic target for TSCC patients.
Keywords: Akt (protein kinase B), insulin-like growth factor 1 receptor (IGF1R), microRNA-7 (miR-7), proliferation, tongue squamous cell carcinoma (TSCC), tumour suppressor
INTRODUCTION
OSCC (squamous cell carcinoma of the oral cavity) is one of the undertreated and understudied cancer types. According to the American Cancer Society [1,2], although overall new cancer cases increased by approx. 8 % during the past 5 years, new cases for OSCC increased by approx. 21 %. New cases for TSCC (tongue squamous cell carcinoma), one of the most common subtypes of OSCC, increased by more than 37 % in the same period. More strikingly, although the number of deaths associated with all cancers decreased, the number of deaths associated with OSCC increased by 4 %, and those associated with TSCC increased by over 10 %, during the past 5 years. These statistics indicate a major health problem and emphasize the immediate need for a better understanding of this disease process. Although attempts have been made to identify molecular mechanisms that contribute to the tumorigenesis of TSCC, most studies have focused on protein-coding genes. Our knowledge of genomic aberrations associated with non-coding genes [e.g. miRNA (microRNA)] and their contributions to the onset and propagation of TSCC is relatively limited.
miRNAs are small endogenous non-coding RNAs that control target-gene expression at post-transcriptional levels in a sequence-specific manner. They are believed to be part of a network wherein a modest change in the level of one miRNA will set off a chain reaction involving multiple genes of the same or different pathways [3]. It is currently estimated that the human genome may have 800–1000 miRNAs [4]. Although they account for only a minor fraction of the expressed genome, miRNAs are essential regulators of diverse cellular processes, including proliferation, differentiation, apoptosis, survival, motility, invasion and morphogenesis. Many miRNAs are implicated as proto-oncogenes or as tumour suppressors and are aberrantly expressed in various cancer types, including TSCC [5–9].
miR-7 (miRNA-7) has been characterized as a tumour suppressor in several human cancers. It targets a number of proto-oncogenes, including IRS1 (insulin receptor substrate 1), IRS2, EGFR (epidermal growth factor receptor), RAF1 (v-raf-1 murine leukaemia viral oncogene homologue 1) and PAK1 (p21/CDC42/RAC1-activated kinase 1) [10–12]. Our previous studies have indicated that miR-7 is down-regulated in advanced TSCC [7,8]. However, the potential role(s) of miR-7 in TSCC is unknown. Given the diversity and overlapping nature of the identified targets in terms of function, it is likely that additional miR-7 targets exist that may contribute to the tumorigenesis of TSCC. The present study determined that IGF1R [IGF1 (insulin-like growth factor 1) receptor] is a novel target of miR-7 in TSCC. Furthermore, we were able to demonstrate that miR-7-mediated down-regulation of IGF1R attenuated the IGF1-induced activation of Akt (protein kinase B) and led to reduced cell proliferation and cell-cycle arrest, and to an increase in apoptosis rate.
EXPERIMENTAL
Cell culture and transfection
The TSCC cell lines UM1 and UM2 were used in the present study. These are paired cell lines that were generated from a single patient with TSCC [13]. UM1 has been shown to exhibit enhanced proliferation, invasion and migration, and reduced apoptosis compared with UM2 cells [7,14]. These cells were maintained in Dulbecco’s modified Eagle’s medium/F12 supplemented with 10 % (v/v) fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin (Gibco) at 37 °C in a humidified incubator containing 5 %CO2. For functional analysis, miR-7 mimics and non-targeting miRNA mimics (Dharmacon), anti-miR-7 PNA (peptide nucleic acid) and negative-control PNA (Panagene), and gene-specific siRNA (small interfering RNA) (On-TargetPlus SMARTpool™; Dharmacon) were transfected into cells using DharmaFECT® transfection reagent 1 as described previously [5,7,8]. The resultant changes in miR-7 levels were confirmed by quantitative real-time PCR (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/432/bj4320199add.htm).
Western blot analysis
Western blotting was performed using specific antibodies against EGFR (Cell Signaling Technology), IGF1R (Cell Signaling Technology), IRS1 (Cell Signaling Technology), IRS2 (Cell Signaling Technology), PAK1 (Cell Signaling Technology), PIK3CD [PI3K (phosphoinositide 3-kinase) p110 catalytic subunit δ] (Santa Cruz Biotechnology), RAF1 (Santa Cruz Biotechnology), total Akt (Cell Signaling Technology), p-Akt (phospho-Akt; Thr308) (Cell Signaling Technology), p-Akt (Ser473) (Cell Signaling Technology) and p-ERK1/2 [phospho-ERK1/2 (extracellular-signal-regulated kinase 1/2)] (Cell Signaling Technology). β-Actin (Sigma) was used as an internal control. In brief, after washing with ice-cold PBS three times, cells were lysed in RIPA buffer (150 mM NaCl, 10 mM Tris/HCl, pH 7.4, 0.5 % Triton X-100 and protease inhibitors (Sigma)], homogenized on ice, and centrifuged at 13 500 g at 4 °C for 15 min. The supernatant was collected and stored at −80 °C until use. Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories). Extracted protein (25 μg) was loaded on to 12 % Tris-polyacrylamide gels (SDS/PAGE, Bio-Rad Laboratories). The proteins were then transferred on to nitrocellulose membranes (Whatman). The membranes were blocked using 5 % (w/v) non-fat dried skimmed milk powder, washed in Tris-buffered saline containing 0.05 % Tween 20, and incubated with primary antibodies at room temperature (25 °C) for 1 h or overnight at 4 °C. The membranes were washed three times in PBST (PBS containing 0.05 %Tween 20), incubated for 1 h with appropriate secondary antibodies conjugated to HRP (horseradish peroxidase), washed three times in PBST, and treated with the Immun-Star™ HRP Chemiluminescence Kit peroxide buffer and luminol/enhancer (Bio-Rad Laboratories) for detection of protein bands.
Dual-luciferase reporter assay
An 85-bp fragment from the 3′-UTR (3′-untranslated region) of the IGF1R gene (position 7635–7720, GenBank® accession number NM_000875, containing the miR-7-binding site E1), an 81-bp fragment from the 3′-UTR of IGF1R (position 7835–7916, GenBank® accession number NM_000875, containing the miR-7-binding site E2) and an 78-bp fragment from the 3′-UTR of IGF1R (position 10054–10132, GenBank® accession number NM_000875, containing the miR-7-binding site E3) were cloned into the XbaI site of the pGL3 firefly luciferase reporter vector (Promega). The corresponding mutant constructs were created by mutating the seed regions of the miR-7-binding sites. The constructs were then verified by sequencing. Cells were transfected with the reporter contructs containing the targeting sequence from the IGF1R 3′-UTR (named pGL-E1, pGL-E2 and pGL-E3) or its mutant (named pGL-E1m, pGL-E2m and pGL-E3m) using Lipofectamine™ 2000 (Invitrogen). The pRL-TK vector (Promega) was co-transfected as an internal control for normalization of the transfection efficiency. The luciferase activities were then determined as described previously [5,8] using a Lumat LB 9507 luminometer (Berthold Technologies).
Real-time RT–PCR (reverse transcription–PCR) analysis
The relative level of miR-7 was determined using a mirVana™ qRT-PCR miRNA Detection Kit (Ambion) as described previously [7]. The relative mRNA levels of IGF1R were examined using a quantitative two-step RT–PCR assay with gene-specific primer sets (OriGene) as described previously [5]. The relative expression level was determined using the 2−Δ ΔCt analysis method [15], where actin was used as an internal reference.
Proliferation assay
Cell proliferation was measured using the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide] assay as described previously [16], with minor modifications. In brief, at 48 h post-transfection, the transfection medium in each well was replaced by 100 μl of fresh serum-free medium with 0.5 g/l MTT. After incubation at 37 °C for 4 h, the MTT medium was removed by aspiration and 50 μl of DMSO was added to each well. After incubation at 37 °C for a further 10 min, the A540 of each sample was measured using a plate reader.
Flow-cytometry-based apoptosis and cell-cycle analysis
Cells were grown in six-well plates to approx. 60 % confluence and transiently transfected with the desired miRNA or siRNA reagents. At 48 h post-transfection, cells were harvested for apoptosis and cell-cycle analysis. For the apoptosis assay, cells were resuspended in 1 × binding buffer (Clontech), and 5 μl of annexin–FITC conjugate and 10 μl of propidium iodide solution were added to each cell suspension separately. For cell-cycle analysis, cells were resuspended in PBS and then fixed in ethanol at −20 °C overnight. Cells were washed with PBS and resuspended in staining solution (50 μg/ml propidium iodide, 1 mg/ml RNase A and 0.1 % Triton X-100 in PBS). The stained cells (1 × 105) were then analysed with a flow cytometer (FACScalibur; Becton Dickinson).
Statistical analysis
Statistical analysis was performed using a Student’s t test. Differences with P < 0.05 were considered statistically significant.
RESULTS
The candidate targets for miR-7 were predicted using TargetScan-Human 5.1 [17] (Table 1). Among the predicated miR-7 targets, several are signalling molecules that are involved in the Akt and EGFR signalling pathways. These include the previously identified miR-7 targets such as IRS1, IRS2, EGFR, RAF1 and PAK1 [10–12], as well as the novel targets IGF1R and PIK3CD. The effects of miR-7 on the expression levels of these predicted targets were examined in a pair of TSCC cell lines (UM1 and UM2) that originated from a single TSCC patient [13]. Our previous studies have indicated that UM1 exhibits a reduced miR-7 level compared with UM2 cells [7,8]. As illustrated in Figure 1, when UM1 cells were treated with miR-7 mimic, reductions in the protein levels of IGF1R, IRS2 and PAK1 were consistently observed. No apparent change in the expression of IRS1 and EGFR was observed. PIK3CD was minimally expressed in UM1 cells, and RAF1 was not detectable in UM1 cells. Conversely, knockdown of miR-7 in UM2 cells by anti-miR-7 PNA treatment resulted in a significant increase in the expression of IGF1R and IRS2. The PAK1 level was also slightly elevated upon miR-7 knockdown. However, no apparent change in the IRS1 and EGFR levels was observed, and, similar to the UM1 cells, PIK3CD and RAF1 were not detectable in UM2 cells.
Table 1. Predicted miR-7 targets involved in Akt and EGFR signalling.
Targets for miR-7 were predicted using Target Scan Human 5.1. PCT, probability of conserved targeting.
| Gene symbol | Gene name | Conserved sites
|
Poorly conserved sites
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 8-mer | 7-mer-m8 | 7-mer-1A | Total | 8-mer | 7-mer-m8 | 7-mer-1A | Aggregate PCT | Total context score | References | ||
| IGF1R | Insulin-like growth factor 1 receptor | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 2 | 0.63 | −0.17 | |
| IRS1 | Insulin receptor substrate 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.59 | −0.52 | [10] |
| IRS2 | Insulin receptor substrate 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0.94 | −0.60 | [10] |
| PIK3CD | Phosphoinositide 3-kinase catalytic subunit δ | 1 | 1 | 0 | 0 | 3 | 1 | 1 | 1 | 0.79 | −0.69 | |
| EGFR | Epidermal growth factor receptor | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 0.57 | −0.78 | [10,11] |
| RAF1 | v-raf-1 murine leukaemia viral oncogene homologue 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0.80 | −0.68 | [11] |
| PAK1* | p21/CDC42/RAC1-activated kinase 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0.14 | −0.29 | [12] |
No conserved miR-7-targeting site was predicted by TargetScan in PAK1 mRNA sequence. A poorly conserved miR-7-target site was predicted in the 3′-UTR of the PAK1 gene. The miR-7 interaction with this target site was experimentally validated previously [12].
Figure 1. Effects of miR-7 on expression of key genes in the Akt and EGFR signalling pathways.
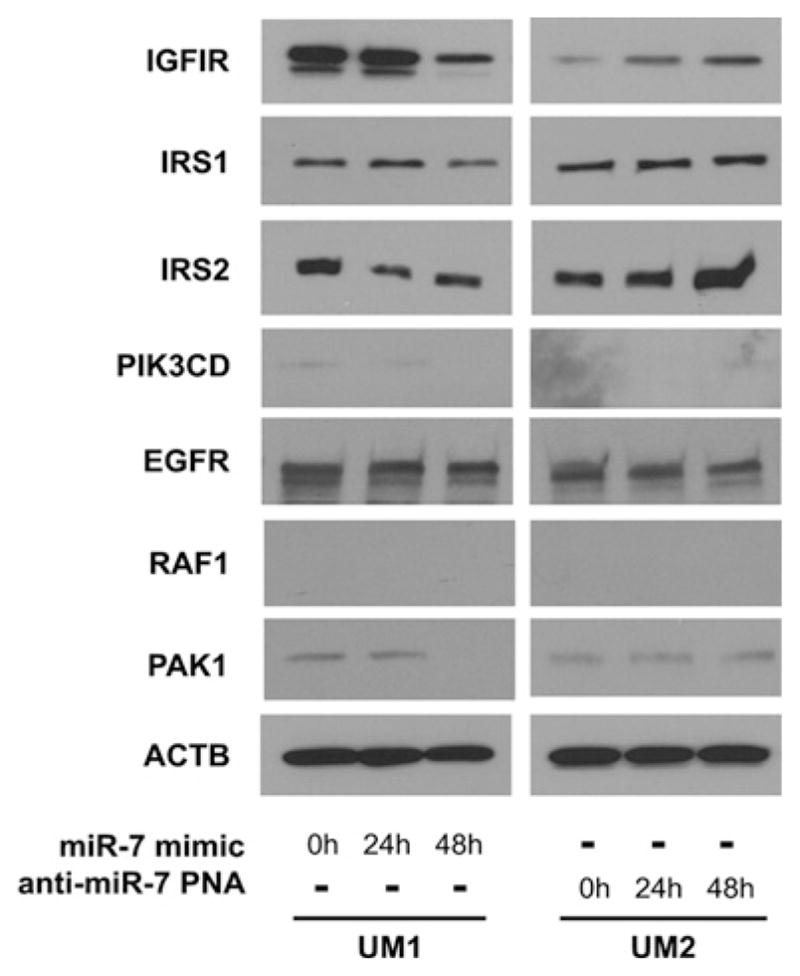
UM1 cells were transfected with miR-7 mimic for 0, 24 or 48 h (left-hand panels). UM2 cells were treated with anti-miR-7 PNA for 0, 24 or 48 h (right-hand panels). The expression levels of IGF1R, IRS1, IRS2, PIK3CD, EGFR, RAF1 and PAK1 were examined by Western blotting as described in the Experimental section. Results shown are representative of at least three independent experiments. ACTB, β-actin.
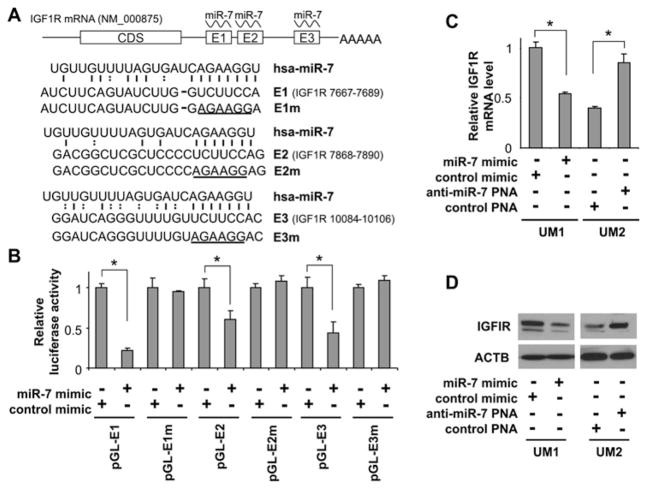
Three miR-7-targeting sequences were identified in the 3′-UTR region of IGF1R mRNA (Figure 2A). The first two targeting sequences (E1 located at position 7667–7689 and E2 located at position 7868–7890) are poorly conserved targeting sites. The third sequence (E3 located at 10084–10106) is a highly conserved targeting site for miR-7 (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/432/bj4320199add.htm). To confirm that miR-7 directly targets these sequences, dual-luciferase reporter assays were performed using the construct in which these targeting sites were cloned into the 3′-UTR of the reporter gene (pGL-E1, pGL-E2 and pGL-E3). As illustrated in Figure 2(B), for cells transfected with all three reporter-gene constructs and treated with miR-7 mimic, the luciferase activities were significantly reduced as compared with the cells treated with control mimic. Similar results were also obtained from the construct containing both E1 and E2 sites (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/432/bj4320199add.htm). When the seed regions of the targeting sites were mutated (pGL-E1m, pGL-E2m and pGL-E3m), the miR-7 effects on luciferase activity were abolished. Furthermore, as shown by quantitative real-time RT–PCR (Figure 2C) and Western blotting (Figure 2D), ectopic transfection of miR-7 reduced the level of IGF1R at both the mRNA and protein levels. Knockdown of miR-7 led to increases in both IGF1R mRNA and protein. Thus these results demonstrated that miR-7 regulates IGF1R expression by directly targeting the 3′-UTR of the IGF1R mRNA.
Figure 2. miR-7 direct targeting of IGF1R mRNA.
(A) Schematic representation of IGF1R mRNA showing the positions and sequences of the three predicted miR-7-binding sites located in its 3′-UTR. CDS, coding sequence. (B) Dual-luciferase reporter assays were performed as described in the Experimental section on cells transfected with constructs containing the predicted targeting sequences (pGL-E1, pGL-E2 and pGL-E3) or targeting sequences with mutated seed regions [pGL-E1m, pGL-E2m and pGL-E3m; mutations are underlined in (A)] cloned into the 3′-UTR of the reporter gene, and treated with miR-7 mimic or control mimic. Quantitative real-time RT–PCR (C) and Western blot (D) analyses were performed to examine the effects of miR-7 on IGF1R expression in UM1 cells that were treated with miR-7 mimic or control mimic, and UM2 cells that were treated with anti-miR-7 PNA or scrambled LNA (locked nucleic acid). Results shown in (B) and (C) are means ± S.D.; all results are representative of at least three independent experiments. *P < 0.05.
Since IGF1R is a major player in the activation of the Akt signalling pathway, we investigated the effect of miR-7-mediated down-regulation of IGF1R on Akt phosphorylation. As illustrated in Figure 3(A), although the total Akt level was constant, the levels of p-Akt at both residues Thr308 and Ser473 were reduced after cells were treated with miR-7 mimic for 48 h. On the contrary, the level of p-ERK1/2 was not significantly altered after miR-7 mimic treatment. This is in agreement with our early observation that EGFR was not significantly affected by miR-7 in TSCC cell lines (Figure 1). As shown in Figure 3(B), Akt phosphorylation can be induced by IGF1 stimulation in a time-dependent manner. This IGF1-induced Akt phosphorylation can be attenuated by pre-treating the cells with miR-7 mimic (Figure 3C). This IGF1-induced Akt phosphorylation can be also blocked by the PI3K inhibitor LY294002.
Figure 3. Effects of miR-7 on IGF1-induced activation of Akt.
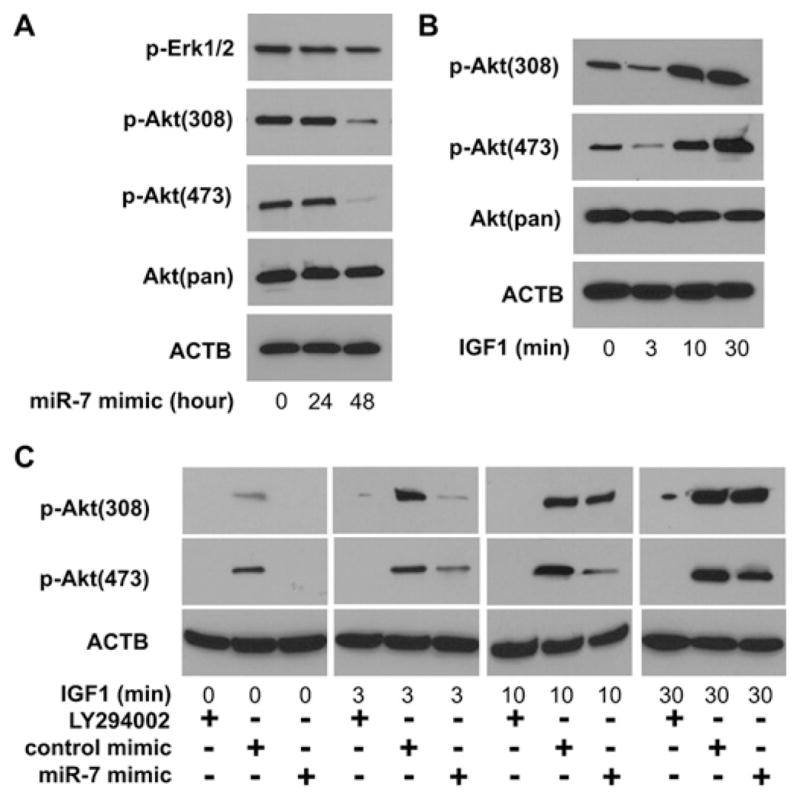
(A) UM1 cells were treated with miR-7 mimic for 0, 24 or 48 h. The levels of total Akt [Akt(pan)], p-Akt (at Thr308 and Ser473) and p-ERK1/2 were examined by Western blotting as described in the Experimental section. (B) UM1 cells were stimulated with IGF1 for 0, 3, 10 and 30 min. The levels of total Akt and p-Akt (at Thr308 and Ser473) were examined by Western blotting using specific antibodies. (C) Cells were pre-treated with the PI3K inhibitor LY294002 (50 μM for 1 h), miR-7 mimic (100 nM for 48 h) or control mimic (100 nM for 48 h) and then stimulated with IGF1 for 0, 3, 10 and 30 min. The levels of p-Akt (at Thr308 and Ser473) were examined by Western blotting. ACTB, β-actin.
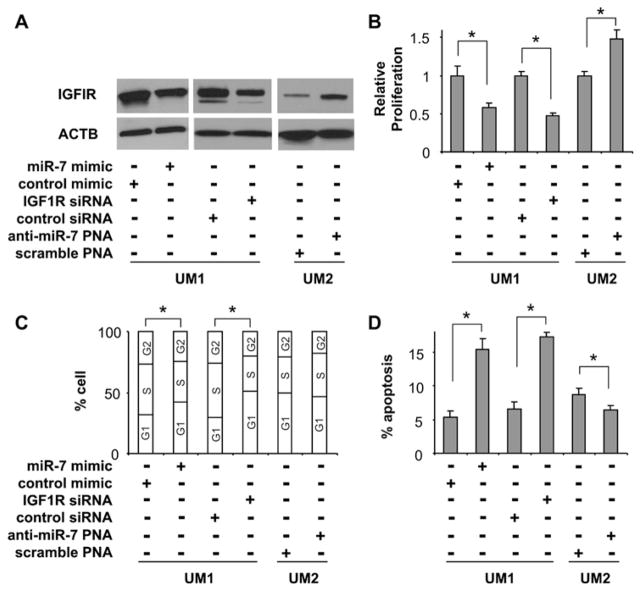
As illustrated in Figure 4, miR-7-mediated down-regulation of IGF1R expression in UM1 cells (Figure 4A) was accompanied by significant inhibition of cell proliferation (Figure 4B) and G0/G1 cell-cycle arrest (Figure 4C), and enhanced cell apoptosis (Figure 4D). Similar effects on proliferation, cell cycle and apoptosis were also observed when cells were treated with siRNA specific for IGF1R. On the other hand, enhanced cell proliferation and reduced apoptosis were observed when UM2 cells were treated with anti-miR-7 PNA. No statistically significant difference in cell cycle was observed in UM2 cells that were treated with anti-miR-7 PNA. Together with the observed miR-7 effects on IGF1-induced Akt activation, these results suggest that miR-7 regulates TSCC cell growth, at least in part, by targeting IGF1R.
Figure 4. The effects of miR-7 on proliferation, the cell cycle and cell apoptosis.
The changes in IGF1R expression (A), cell proliferation (B), cell-cycle distribution (C) and cell-apoptosis rate (D) were examined as described in the Experimental section in UM1 cells that were treated with miR-7 mimic, control mimic, IGF1R-specific siRNA or control siRNA, and UM2 cells that were treated with anti-miR-7 PNA or scrambled LNA. Results shown in (B) and (D) are means ± S.D.; all results are representative of at least three independent experiments. *P < 0.05. ACTB, β-actin.
DISCUSSION
miRNAs are not directly involved in protein coding, but are believed to control the expression of more than one-third of the protein-coding genes in the human genome [18–20]. Each miRNA can potentially regulate hundreds of genes at post-transcriptional levels by binding to the specific sequences in the target mRNA molecules based on ‘imperfect complementarity’. However, the precise interactions between miRNA and its target genes may be further dictated by cell types and the micro-environment. Therefore miRNAs contribute to a newly recognized level of fine-tuning regulation of gene expression. miRNA-7 has previously been characterized as a tumour-suppressor miRNA in several human cancers by targeting a number of key signalling molecules, including IRS1, IRS2, EGFR, RAF1 and PAK1 [10–12]. On the basis of bioinformatics analysis, we further predicted a number of additional miR-7 targets, including IGF1R and PIK3CD. Our present experimental results confirmed that IGF1R, as well as IRS2 and PAK1, are functional targets of miR-7 in TSCC cells. Our results, together with recent reports on miR-7 and Akt and EGFR signalling pathways [10–12], indicate that miR-7 contributes to tumorigenesis through multiple pathways and at multiple levels (Scheme 1).
Scheme 1. Potential roles of miR-7 in tumorigenesis of TSCC.
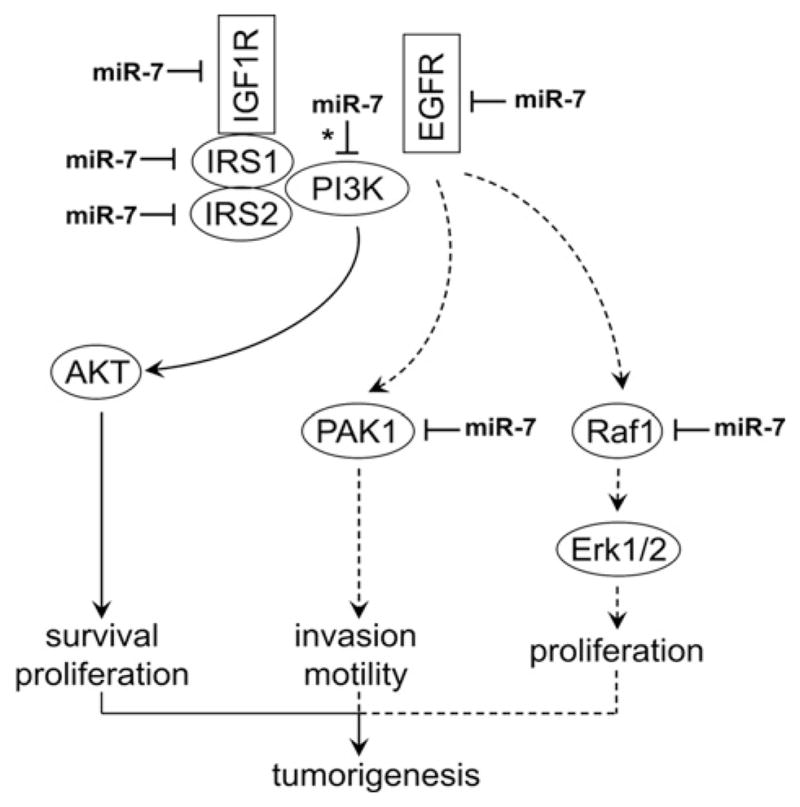
Solid arrows indicate the miR-7-regulated signalling pathway that is confirmed experimentally in the present study. Broken arrows indicate the miR-7-regulated signalling pathway that was suggested by previous studies. * indicates an miR-7 target predicted by a bioinformatics tool, but not validated experimentally in the present system.
IGF1R is a receptor tyrosine kinase that consists of heterotetramers (α2β2) held together by disulfide bridges, and it mediates IGF1-induced signalling events. Our present results clearly demonstrate that miR-7 down-regulates IGF1R by directly targeting the 3′-UTR of IGF1R mRNA. This miR-7-mediated IGF1R down-regulation is accompanied by attenuation of Akt phosphorylation, inhibition of cell proliferation, enhanced apoptosis and cell-cycle arrest. This is consistent with the well-established IGF1R/IRS/PI3K/Akt signalling pathway that plays major roles in cell survival and proliferation. It is worth noting that IRS2, one of the previously identified miR-7 targets in glioblastoma [10], was also down-regulated by ectopic transfection of miR-7 in our TSCC cell lines. Therefore miR-7 concurrently targets at least two major signalling molecules in the IGF1R/IRS/PI3K/Akt signalling pathway in TSCC. Furthermore, one of the PI3K members (PIK3CD) is also predicted by bioinformatics analysis to be a potential target of miR-7. One highly conserved and three poorly conserved binding sites for miR-7 were predicted in the 3′-UTR of PIK3CD mRNA. Although our luciferase-reporter-gene assays suggested that these predicted sequences in the 3′-UTR of PIK3CD are functional binding sites for miR-7 (results not shown), the expression level of PIK3CD was minimal in our cell lines. Furthermore, ectopic transfection or knock down of miR-7 did not change the expression level of PIK3CD to any significant extent. Therefore it appears that PIK3CD may not contribute significantly to the miR-7-mediated regulation of the Akt signalling pathway in our system. Nevertheless, it remains possible that miR-7 may functionally target PIK3CD in other cell types or different biological systems. It is worth noting that the PI3K family includes at least four members: PIK3CA (p110α), PIK3CB (p110β), PIK3CG (p110γ) and PIK3CD (p110δ). PIK3CD is selectively expressed in leucocytes [21] and, together with PIK3CG, these two isoforms of PI3K contribute to the immune processes that underpin inflammatory responses. On a separate note, results from a study by Webster et al. [11] suggested that PI3K was down-regulated by miR-7 overexpression in a lung adenocarcinoma cell line (A549). However, the authors did not specify which PI3K member was affected.
Although our present study has provided evidence indicating the role of miR-7 in the regulation of the IGF1R/IRS/PI3K/Akt signalling pathway, several other functional roles for miR-7 have been previously suggested. It has been reported previously that PAK1, a serine/threonine kinase that plays a pivotal role in cell migration and invasion, is targeted by miR-7 [12]. We confirmed the miR-7 effect on PAK1 in our TSCC cell lines. miR-7 has also been suggested to be a regulator of the EGFR signalling pathway by concurrently targeting EGFR and RAF1 [10,11]. However, we did not observe any apparent change in EGFR expression after the transfection of miR-7 mimic, and RAF1 was not expressed to any significant extent in our cell lines. Furthermore, miR-7 treatment did not alter the phosphorylation status of ERK1/2, one of the major downstream molecules of EGFR signalling. It is worth knowing that miR-7 precursors were used in the previous studies [10,11], whereas we used mature miR-7. Nevertheless, it is possible that this apparent difference in the miR-7 effect on the EGFR signalling pathway may be due to differences in cancer types. Alternatively, this difference may be cell-line specific. The cell lines we tested in the present study (or the cell lines used in the previous study [10,11]) may have specific mutation(s) that dictate the miR-7 effects on EGFR. More in-depth functional analysis will be needed to fully evaluate the effect of miR-7 on the EGFR signalling pathway.
In summary, our results indicate that miR-7 co-ordinately regulates IGF1R signalling at multiple levels. The findings shown in the present paper, in conjunction with recent reports on miR-7 and other proto-oncogenes (e.g. IRS, PAK1, RAF1 and EGFR) [10,11], suggest that miR-7 plays major roles in tumorigenesis. Given the centrality of the IGF1R/IRS/PI3K/Akt signalling pathway in TSCC, delivery of miR-7 as a therapeutic agent may represent an effective approach for the treatment of this disease.
Supplementary Material
Acknowledgments
We thank Katherine Long for her editorial assistance prior to submission.
FUNDING
This work was supported in part by the National Institutes of Health Public Health Service [grant numbers K22DE014847, RO1CA139596 and RO3CA135992] and the Prevent Cancer Foundation (to X.Z.).
Abbreviations used
- EGFR
epidermal growth factor receptor
- ERK
extracellular-signal-regulated kinase
- HRP
horseradish peroxidase
- IGF1
insulin-like growth factor 1
- IGF1R
IGF1 receptor
- IRS
insulin receptor substrate
- LNA
locked nucleic acid
- miRNA
microRNA
- miR-7
miRNA-7
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide
- OSCC
squamous cell carcinoma of the oral cavity
- PAK1
p21/CDC42/RAC1-activated kinase 1
- p-Akt
phospho-Akt
- PBST
PBS containing 0.05 % Tween 20
- p-ERK1/2
phospho-ERK1/2
- PIK3CD
PI3K (phosphoinositide 3-kinase) p110 catalytic subunit δ
- PNA
peptide nucleic acid
- RAF1
v-raf-1 murine leukaemia viral oncogene homologue 1
- RT–PCR
reverse transcription–PCR
- siRNA
small interfering RNA
- TSCC
tongue squamous cell carcinoma
- 3′-UTR
3′-untranslated region
Footnotes
AUTHOR CONTRIBUTION
Xiaofeng Zhou, Yang Dai, Anxun Wang, Xiqiang Liu and Antonia Kolokythas designed the study. Lu Jiang, Xiqiang Liu, Zujian Chen, Yi Jin and Caroline Heidbreder performed the experiments. Xiaofeng Zhou and Yang Dai performed the bioinformatics analysis. Xiaofeng Zhou, Anxun Wang, Antonia Kolokythas and Xiqiang Liu wrote the paper. All authors discussed the manuscript prior to submission.
References
- 1.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 5.Jiang L, Liu X, Kolokythas A, Yu J, Wang A, Heidbreder CE, Shi F, Zhou X. Down-regulation of the Rho GTPase signaling pathway is involved in the microRNA-138 mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int J Cancer. 2010;127:505–512. doi: 10.1002/ijc.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Chen Z, Yu J, Xia J, Zhou X. MicroRNA profiling and head and neck cancer. Comp Funct Genomics. 2009;2009:1–7. doi: 10.1155/2009/837514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Jiang L, Wang A, Yu J, Shi F, Zhou X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009;286:217–222. doi: 10.1016/j.canlet.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H, Zhou X. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics. 2009;6:131–139. [PMC free article] [PubMed] [Google Scholar]
- 9.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 10.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 11.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 12.Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama S, Sasaki A, Mese H, Alcalde RE, Matsumura T. Establishment of high and low metastasis cell lines derived from a human tongue squamous cell carcinoma. Invasion Metastasis. 1998;18:219–228. doi: 10.1159/000024515. [DOI] [PubMed] [Google Scholar]
- 14.Ye H, Wang A, Lee BS, Yu T, Sheng S, Peng T, Hu S, Crowe DL, Zhou X. Proteomic based identification of manganese superoxide dismutase 2 (SOD2) as a metastasis marker for oral squamous cell carcinoma. Cancer Genomics Proteomics. 2008;5:85–94. [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTmethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Xu JH, Wang AX, Huang HZ, Wang JG, Pan CB, Zhang B. Survivin shRNA induces caspase-3-dependent apoptosis and enhances cisplatin sensitivity in squamous cell carcinoma of the tongue. Oncol Res. 2010;18:377–385. doi: 10.3727/096504010x12644422320663. [DOI] [PubMed] [Google Scholar]
- 17.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3 ′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanhaesebroeck B, Welham MJ, Kotani K, Stein R, Warne PH, Zvelebil MJ, Higashi K, Volinia S, Downward J, Waterfield MD. p110δ, a novel phosphoinositide 3-kinase in leukocytes. Proc Natl Acad Sci USA. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.