Abstract
Actin is the most abundant protein in most eukaryotic cells. It is highly conserved and participates in more protein-protein interactions than any known protein. These properties, along with its ability to transition between monomeric (G-actin) and filamentous (F-actin) states under the control of nucleotide hydrolysis, ions, and a large number of actin-binding proteins, make actin a critical player in many cellular functions, ranging from cell motility and the maintenance of cell shape and polarity to the regulation of transcription. Moreover, the interaction of filamentous actin with myosin forms the basis of muscle contraction. Owing to its central role in the cell, the actin cytoskeleton is also disrupted or taken over by numerous pathogens. Here we review structures of G- and F-actin and discuss some of the interactions that control the polymerization and disassembly of actin.
Keywords: X-ray crystallography, electron microscopy, fiber diffraction, actin-binding-proteins
THE ACTIN MONOMER
Vertebrates express three main actin isoforms, including three α-isoforms of skeletal, cardiac, and smooth muscles and the β- and γ-isoforms expressed in nonmuscle and muscle cells. Actin isoforms differ by only a few amino acids, with most variations occurring toward the N terminus (23). Actin also undergoes various forms of posttranslational modifications. For instance, His73 of skeletal muscle α-actin is methylated, the N-terminal methionine and cysteine residues are acetylated and cleaved, and the resulting N-terminal aspartic acid is then reacetylated.
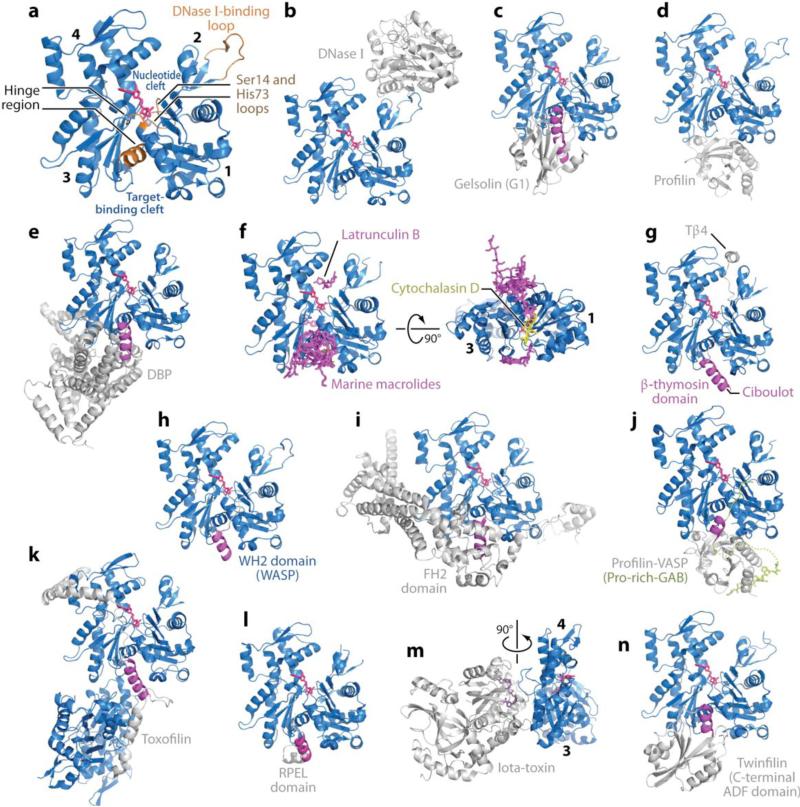
Since the original determination of the crystal structure of G-actin in complex with DNase I (28), over 80 structures of actin have been reported (Figure 1). These are listed in Supplemental Table 1 (follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org). The majority of these structures have been obtained as complexes with actin-binding proteins (ABPs) and small molecules, or by chemically modifying or mutating actin in order to prevent polymerization. Remarkably, irrespective of the bound molecule or nucleotide state, the conformation of the actin monomer is basically the same. Actin belongs to a structural superfamily with sugar kinases, hexokinases, and Hsp70 proteins (3). The Arp proteins (49) and the prokaryotic actin-like proteins MreB (54) and ParM (56) are also now part of this superfamily. Common to these proteins, the 375-amino-acid (aa) polypeptide chain of actin folds into two major α/β-domains (Figure 1a). Because of their location within the actin filament, the two major domains of actin are known as the outer and inner domains, and because of their apparently different sizes in electron microscopy (EM) images, they have also been called the small and large domains, respectively. Traditionally, however, a four-subdomain nomenclature has been adopted (28). Subdomains 1 and 3 are structurally related and probably emerged from gene duplication, whereas subdomains 2 and 4 can be viewed as large insertions into subdomains 1 and 3, respectively. For brevity we refer to these as domains 1--4.The actin monomer is rather flat, fitting into a rectangular prism with dimensions 55 Å × 55 Å × 35 Å.
Figure 1.
Structures of actin and actin complexes. The structures of actin complexes are shown to scale and in chronological order of publication. (a) Classical view of the structure of the actin monomer. The structure shown was derived from the complex with DNase I, with completion of the C terminus from the complex of actin with profilin. Highlighted in orange are the Ser14 and methylated His73 loops, the DNase I-binding loop, and the hinge between domains, consisting of helix Gln137-Ser145 and the loop centered at residue Lys336. Subdomains 1--4 are labeled. (They are also labeled in panels f and m, which show rotated views of the structure.) Together, subdomains 1 and 2 form the outer (or small) domain, whereas subdomains 3 and 4 constitute the inner (or large) domain. Two large clefts are formed between these domains: the nucleotide- and target-binding clefts. Most actin-binding proteins (ABPs) and small molecules bind in the target-binding cleft, and the interaction frequently involves an α-helix (magenta). (b) DNase I (1ATN). (c) Gelsolin segment 1 (G1) (1EQY). See also Supplemental Figure 1 for structures of actin with Gelsolin fragments G1--G3 and G4--G6. (d) β-actin-profilin (2BTF). (e) Vitamin D-binding protein ( DBP ) (1KXP). (f) Two perpendicular views of a superimposition of structures of actin complexes with small molecules, including marine toxins (1QZ5, 1QZ5, 1S22, 1YXQ, 2ASM, 2ASO, 2ASP, 2FXU, 2Q0R, 2Q0U, 2VYP), Latrunculin B [DOUG: Add “A/B’ – missing.] (2Q0U) and cytochalasin D (3EKS). The marine toxins (magenta) bind at the ends of the target-binding cleft, whereas cytochalasin D binds in the middle, and Latrunculin (both A and B) binds in the nucleotide cleft. All these molecules compromise actin polymerization. (g) β-thymosin domain. A complete structure of this complex is not available, but combined, the structures of the N-terminal portion of a β-thymosin domain from Drosophila ciboulot (1SQK) and the C-terminal end of b-thymosin peptide (Tβ4) (1T44) provide a model of this complex. (h) WASP homology domain 2 (WH2) domain of WASP (2A3Z). The WH2 domain, present in many cytoskeletal proteins in the form of tandem repeats, is related to the β-thymosin domain but lacks the C-terminal pointed end capping helix. (i) Formin homology 2 (FH2) domain (1Y64). See also Supplemental Figure 2 for a more detailed representation of this structure. (j) Ternary complex with profilin and the Pro-rich G-actin-binding (Pro-rich-GAB) domains of VASP (2PBD). The GAB domain is related to the WH2 domain but presents a shorter N-terminal helix and adopts a slightly different orientation when bound to actin, possibly because it is designed to co-bind with profilin. (k) Toxofilin from Toxoplasma gondii bound to an antiparallel actin dimer (2Q97). (l) RPEL (RPxxxEL-containing motif)[AU: Does this need to be spelled out for the general reader?] domain from the serum response factor coactivator MAL (2V52). (m) Arginine ADP-ribosylation iota-toxin from Clostridium perfringens (3BUZ). View rotated 90° relative to the other complexes. (n) C-terminal ADF/cofilin domain of twinfilin (3DAW). See Supplemental Table 1 for a complete list of references.
bThere is relatively little contact between the two major domains of actin; the polypetide chain passes twice between these domains: at the loop centered at residue Lys336 and at the linker helix Gln137-Ser145, which functions as the axis of a hinge between the domains. As a result, two clefts are formed between the domains. The upper cleft binds the nucleotide and associated divalent cation (Mg2+ in cells), which together provide the other important linkage between domains (Figure 1a). The lower cleft between domains 1 and 3 is lined by residues Tyr143, Ala144, Gly146, Thr148, Gly168, Ile341, Ile345, Leu346, Leu349, Thr351, and Met355, which are predominantly hydrophobic. This cleft constitutes the major binding site for most ABPs and also mediates important longitudinal contacts between actin subunits in the filament (16, 36), as suggested previously (9). The communication between the two clefts provides the structural basis for how nucleotide-dependent conformation changes modulate the binding affinities of ABPs and the strength of intersubunit contacts in the filament.
G-actin is not an effective ATPase, whereas F-actin is and most crystal structures have been solved in the ATP-bound form. The differences between the ATP- and ADP-bound states are relatively minor and involve primarily two loops (Figure 1a): the Ser14 β-hairpin loop and the sensor loop carrying the methylated His73 (21). The Ser14 loop is located in actin subdomain 1 and is structurally equivalent to the loop containing Asp157 in subdomain 3. These two loops engulf the phosphates of the nucleotide. Nucleotide-dependent conformational changes begin with Ser14, which in the ATP state makes a hydrogen-bonding contact with the γ-phosphate. After hydrolysis and γ-phosphate release, Ser14 changes orientation to form a hydrogen-bonding contact with the β-phosphate of the nucleotide. In the ATP state, Ser14 also forms a hydrogen-bonding contact with the main chain of the loop carrying His73. When the side chain of Ser14 rotates in the ADP state, the His73 loop moves toward the nucleotide to occupy some of the space emptied by the γ-phosphate. In this way, this loop appears to sense the state of the nucleotide.
The sensor loop marks the C-terminal end of domain 2, which can be viewed as an insertion into domain 1. At the top of domain 2, residues 39--51 are disordered in most crystal structures. This sequence is referred to as the DNase I-binding loop (or D-loop) because it mediates important interactions in the complex with DNase I (28). When visible in crystal structures this loop takes on a variety of confirmations, including in one case a short a-helix (40). Domain 2, and specifically the D-loop, plays a critical role in longitudinal intersubunit contacts in the filament. Thus, changes in the Ser14 and sensor loops appear to propagate to domain 2, whose conformation changes only slightly between nucleotide states (21), but probably enough to explain the decreased stability of the actin filament in the ADP state. Because the nucleotide sits at the interface between the two major actin domains, another important consequence of nucleotide hydrolysis and γ-phosphate release is a weakening of the linkage between domains, which can then rotate more freely with respect to one another. Two types of rotations have been described: a scissors-like opening and closing of the nucleotide cleft within the plane of the figure (Figure 1a) and a perpendicular propeller-like rotation of one domain with respect to the other (18, 36, 53). Gln137, at the beginning of the hinge helix, is thought to play a critical role in nucleotide hydrolysis (27), which is activated in the filament (7), presumably by interdomain rotation that brings the side chain of Gln137 in contact with the γ-phosphate of the nucleotide (16, 36).
STRUCTURES OF ACTIN BOUND TO ACTIN-BINDING PROTEINS AND SMALL MOLECULES
Nucleotide hydrolysis by F-actin is one of the main factors regulating the transition between G- and F-actin. The ATP state is more stable than the ADP state. In vitro, actin monomers join the fast-growing barbed (or +) end of the filament in the ATP state, hydrolysis takes place in the filament, and ADP-actin monomers dissociate faster from the pointed (or –) end. This steady-state mechanism of actin polymerization/depolymerization is known as actin filament treadmilling (57). However, treadmilling cannot account for the dynamics of filament assembly observed in cells. Numerous ABPs are involved in the regulation of actin cytoskeleton dynamics (45). These proteins have diverse functions, including actin monomer sequestration, filament barbed- and pointed-end capping, filament severing, and filament cross-linking. The structures of various complexes of G-actin with ABPs have been determined by X-ray crystallography (Figure 1 and Supplemental Table 1). Numerous F-actin-binding proteins have been analyzed by EM at limited resolution.
G-actin-binding proteins have diverse functions, belong to different structural families, and present different nucleotide preferences. Despite this diversity, most of these proteins bind in the target-binding cleft of actin and typically insert an amphiphilic helix into this cleft (9). Whereas the overall position of the interacting helices is well conserved (Figure 1), the directionality of their polypeptide chains is not. The directionality of the helices of toxofilin and of the FH2 and WH2 domains is from back to front (according to the classical view of actin), whereas the helices of gelsolin, vitamin D-binding protein ( DBP), ADF/cofilin, and the RPEL domain bind in the opposite direction.
DNase I
Actin and DNase I form a high-affinity complex (34). Although this complex occurs in vivo (50), its physiological significance is still unclear. Nevertheless, it was the actin--DNase I complex that allowed the first crystal structure of actin to be determined (Figure 1b; references to the actin structures and PDB accession codes are given in Supplemental Table 1).
Gelsolin
Gelsolin belongs to a family of actin-severing and actin-capping proteins, which includes adseverin, villin, capG, advillin, and supervillin (52). Gelsolin consists of six homologous domains (G1--G6) and is regulated by Ca2+, phosphoinositides, and tyrosine phosphorylation. In the inactive, Ca2+-free state, gelsolin adopts a compact conformation stabilized by intramolecular interactions in which actin-binding sites are masked (Supplemental Figure 1). Domains G1 and G4 contain the two major actin-binding sites. These domains bind similarly to actin, and as first revealed by the structure of the G1-actin complex (Figure 1c), they insert a helix into the target-binding cleft of actin. On the basis of subsequent structures of complexes of G1-G3 and G4-G6 with G-actin (Supplemental Figure 1), Robinson and colleagues (4) have proposed a three-step model of filament severing by Ca2+-activated gelsolin. According to this model, Ca2+ binding produces a conformational change that partially exposes the actin-binding sites of gelsolin. Proceeding through a transitional complex in which gelsolin binds to the sides of the actin filament, the actin-binding sites of gelsolin become fully exposed, allowing G1 and G4 to engage actin protomers on two different strands of the actin filament and to sever the filament through a coordinated pincer movement.
Gelsolin is primarily cytoplasmic, but a slightly longer isoform circulates in plasma, where it forms part of a homeostatic mechanism termed the actin-scavenger system (31). Under conditions involving cell death or tissue injury, actin can be released into the bloodstream. The actin-scavenger system is responsible for the depolymerization and removal of actin from the circulation. During the first phase of this mechanism, gelsolin severs the actin filaments, followed by the formation of a high-affinity complex of monomeric actin with DBP. This complex is cleared from the circulation rapidly, primarily through the liver. The structure of the DBP-actin complex has also been determined (Figures 1e), and despite the absence of any relationship with gelsolin, DBP also inserts a helix into the target-binding cleft of actin.
Profilin
Profilin (together with Tβ4) contributes to maintaining a large fraction (~50%) of the cellular actin in the unpolymerized pool, at a concentration (~20--100 μM) that is 200- to 1000-fold higher than the critical concentration for barbed end polymerization (~0.1 μM). Whereas free ATP-actin is present in low amounts (0.1--1 μM) and Tβ4-actin is polymerization incompetent, profilin-actin (present at an intracellular concentration of 5--40 μM) constitutes the main source of actin monomers for polymerization (45). Multiple properties allow profilin to play such a central role in filament assembly (12): (a) Profilin catalyzes the exchange of ADP for ATP on actin, which replenishes the pool of ATP-actin monomers ready for polymerization; (b) it inhibits filament nucleation, which explains in part the need for filament nucleators; (c) profilin-bound actin monomers cannot add to pointed ends but can elongate filament barbed ends at approximately the same rate as free actin monomers; (d) owing to its greater affinity for actin, profilin competes effectively with Tβ4 for binding to actin; and (e) profilin can bind simultaneously to Pro-rich sequences and actin, and it binds to both with higher affinity as a ternary complex than to either one separately (14). Pro-rich sequences are extremely abundant among cytoskeletal proteins and may serve to channel profilin-actin complexes from the cellular pool onto actin filaments in a regulated manner. Thus, recruitment of profilin-actin complexes though the Pro-rich FH1 domain of formin increases its elongation rate (29). Like most ABPs, profilin binds in the target-binding cleft of actin, but it occupies the back of the cleft (Figure 1d), allowing it to bind simultaneously with WH2-related sequences characterized by the presence of a short N-terminal helix, such as the GAB domain of Ena/VASP, which is positioned immediately C-terminal to a Pro-rich profilin-binding sequence (Figure 1j).
ADF/Cofilin
Members of this family are expressed in all eukaryotes, where they are generally involved in the recycling of actin monomers, mainly by filament depolymerization, during processes involving rapid cytoskeleton turnover, such as membrane ruffling and cytokinesis (2). This family includes actin-depolymerizing factor (ADF), cofilin-1 (nonmuscle cells), cofilin-2 (muscle cells), and twinfilin, which contains two ADF domains. Despite the importance of this family, the first structure of a complex with actin was determined only recently (41), corresponding to a complex with the C-terminal ADF domain of twinfilin (Figure 1n). Contrary to profilin, which as a nucleotide exchange factor has a clear preference for ATP-actin, ADF/cofilin binds ADP-actin with higher affinity than ATP-actin (6). However, the twinfilin-actin structure was determined in the ATP state, and twinfilin has a different function than other members of this family by sequestering actin monomers and capping filament barbed ends. Therefore, this structure may not be fully representative of the ADF/cofilin-ADP-actin complex, but it represents the best model currently available of this interaction.
RPEL Domain
While the cytoplasmic functions of actin are generally well established, its role in the nucleus is less well understood. One such role is the regulation of the myocardin transcriptional coactivator MAL [AU: Please spell out.] (35, 38). MAL shuttles between the cytoplasm and the nucleus, where it regulates the activity of the transcription factor serum response factor (SRF). The N-terminal portion of MAL contains a tandem repeat of three G-actin-binding RPEL domains (so named because of the presence of the consensus motif RPxxxEL). The binding of actin to the RPEL domains inhibits the activity of MAL by preventing its accumulation in the nucleus and thereby repressing transcriptional activation by the MAL-SRF complex. The system is ultimately regulated through RhoA-induced alterations of G-actin to F-actin ratios; increased amounts of cytoplasmic G-actin lead to the formation of the inactive actin-MAL complex, whereas reduced G-actin levels resulting from polymerization may release MAL for nuclear accumulation (35). Structures of actin complexes with two of the RPEL domains of MAL have been determined (Figure 1l). The RPEL motif also binds in the actin-binding cleft, inserting a helix into this cleft, but the orientation of this helix is opposite that of the WH2 domain (which is also the case for DBP, gelsolin, and ADF/cofilin).
Structure of Actin Bound to Small Molecules
Recently, a series of macrolides that interact with actin and disrupt polymerization have been isolated from marine organisms (60). These molecules have diverse structures. Moreover, the strong antitumor activities of some of these compounds have sparked interest in their use as molecular probes to help elucidate cellular functions of actin and as scaffolds in the design of anticancer drugs. Numerous structures of actin complexes with marine macrolides have been determined (Supplemental Table 1). Despite their stereochemical diversity, most of these molecules bind in the target-binding cleft of actin (Figure 1f).
Cytochalasin D is a fungal toxin used extensively in cell biology as an agent that binds to the barbed end of actin filaments and inhibits both the association and dissociation of actin monomers. Cytochalasin D binds in the target-binding cleft of actin (Figure 1f).
Latrunculin A and B are also marine macrolides derived from sponges and nudibranchs (59). Contrary to other marine macrolides, both latrunculin A and B bind in the nucleotide-binding cleft of actin, but they have a similar effect in that they inhibit actin polymerization. Both toxins have been used to prevent actin polymerization in the determination of numerous structures of actin (Supplemental Table 1 and Figure 1f).
Pathogens and Actin Assembly
Certain human pathogens, such as Salmonella, Shigella, and Listeria, disrupt or hijack the cytoskeleton of host cells during infection (passive invasion mechanism), whereas others, such as Toxoplasma gondii, have evolved their own actin cytoskeletal systems (active invasion mechanism) (20). One of the most abundantly expressed proteins of T. gondii is toxofilin, a G-actin-binding protein that is secreted into host cells during invasion. Toxofilin forms a high-affinity complex with an antiparallel actin dimer and inserts a helix into the target-binding pocket of one of the bound actins (Figure 1f).
Other pathogenic bacteria produce toxins that ADP-ribosylate actin, resulting in cytoskeleton disorganization and cell death (1, 30). Such is the case of Clostridium botulinum C2 toxin and Clostridium perfringens iota-toxin that modify actin Arg177. The structure of an iota-toxin-actin complex with a nonhydrolyzable NAD analog provides a rare look into an intermediate enzyme-substrate complex implicated in the modification of actin (Figure 1m).
ACTIN-BINDING DOMAINS INVOLVED IN THE DE NOVO POLYMERIZATION OF ACTIN
An important group of ABPs are those that regulate the de novo formation of actin filaments, which include actin filament nucleation and elongation factors (5, 11, 44). Known filament nucleators include the Arp2/3 complex and its large family of nucleation-promoting factors, formins, Spire, Cobl, VopL/VopF, TARP, and Lmod. These molecules control the time and location for polymerization and influence the structures of the actin networks that they generate. Filament nucleators are generally unrelated but, with the exception of formins, they all use the WASP-homology 2 (WH2) domain for interaction with actin. Formins are unique among actin filament nucleators in that they use the forminhomology 2 (FH2) domain for interaction with actin and promote not only nucleation, but also processive barbed-end elongation (19).
The β-Thymosin/WH2 Domain
The WH2 domain is perhaps the most abundant and functionally diverse actin-binding fold (10, 42). It is found in proteins involved in actin monomer sequestration and cytoskeleton scaffolding, but it is particularly abundant among proteins that mediate actin filament nucleation (11). A common architecture, found in Spire, Cobl, and VopL/VopF, consists of tandem WH2 domains that bind three to four actin subunits to form a nucleus. The WH2 domain is short, 17--27 aa long. Its N-terminal portion forms a helix that binds in the target-binding cleft of actin (Figure 1h). After this helix, the WH2 domain presents an extended region that contains the conserved four-residue motif LKKT(V), which is also found in the linker between gelsolin domains G1 and G2.
Tβ4, a short 43-aa polypeptide, is the best-known member of the β-thymosin family and is described in this section because of its structural relationship with the WH2 domain (10, 42). The best model of the Tβ4-actin interaction currently available results from the combination of two structures, corresponding to the complexes of actin with an N-terminal fragment of a β-thymosin domain from ciboulot and with a C-terminal 19-aa fragment of Tβ4 (Figure 1g). Like the WH2 domain, the N-terminal portion of the β-thymosin fold consists of a helix that binds in the target-binding cleft of actin, followed by an LKKT-containing linker. The C-terminal portion, which is missing in the WH2 domain, consists of a helix that binds atop actin subdomains 2 and 4. The combination of N- and C-terminal helices that block monomer addition to both the barbed and pointed ends of the actin monomer makes Tβ4 a remarkably simple but effective actin-monomer-sequestering protein. Tβ4-actin complexes cannot participate in actin filament nucleation or elongation. Instead, Tβ4 is thought to function as an actin buffer, losing actin in competitive equilibrium to profilin, which has higher affinity for actin monomers and functions as the main reservoir of actin monomers for filament assembly (45).
The FH2 Domain
In cellular processes ranging from cell locomotion and morphogenesis to cytokinesis, formins mediate the assembly of unbranched actin networks, such as filopodia, stress fibers, and actin cables (19). Diaphanous-related formins , which are the most extensively studied, contain a profilin-binding Pro-rich region known as the FH1 domain toward the middle of their polypeptide chain. Recruitment of profilin-actin through the FH1 domain accelerates formin-mediated filament elongation (29). As often observed among cytoskeletal proteins, the actin-binding FH2 domain is positioned immediately C-terminal to the Pro-rich region. The FH2 domain is responsible for the two filament assembly functions, nucleation and elongation. The structure of the FH2 dimer consists of two rod-shaped domains connected in head-to-tail fashion by highly flexible linkers (58) (see also Supplemental Figure 2). The structure of an actin complex with the FH2 domain of Bni1 was also determined (39) (Figure 1i). Formin also contains a helix that inserts into the target-binding cleft of actin (Figure 1i).
WH2 Domain and Filament Elongation
While formins promote both nucleation and elongation, none of the WH2-based nucleators seems to promote elongation. However, one family of WH2-containing proteins, Ena/VASP (13), appears to be involved in elongation 13a, although it plays no significant role in nucleation. Like formins, Ena/VASP proteins contain a central Pro-rich region and WH2-related sequences, known as the G-actin-binding and F-actin-binding (GAB and FAB) domains. Analogous to the stimulation of formin-mediated elongation produced by profilin (29), various studies indicate that profilin--actin may stimulate Ena/VASP-dependent elongation (reviewed in Reference 11). The ternary complex of profilin--actin with a fragment of VASP comprising a Pro-rich segment and GAB domain suggests that profilin--actin complexes recruited to the Pro-rich region can be delivered directly to the barbed end of elongating filaments (Figure 1j). This structure also demonstrated that the GAB domain binds actin in a similar manner to the WH2 domain but its N-terminal helix is shorter and its orientation slighlty different from that of the WH2 domain, such that it can bind actin simultaneously with profilin.
STRUCTURE OF THE ACTIN FILAMENT
Although closely related, the structures of G-actin and that of the actin subunit in F-actin are in fact different. It has not been possible to crystallize actin filaments, nor do any of the G-actin structures show the conformation of the actin monomer as in the filament. Details of the structure in the filament have been deduced from X-ray fiber diagrams of orientated gels of F-actin and from EM. These studies have now yielded atomic models of the actin filament.
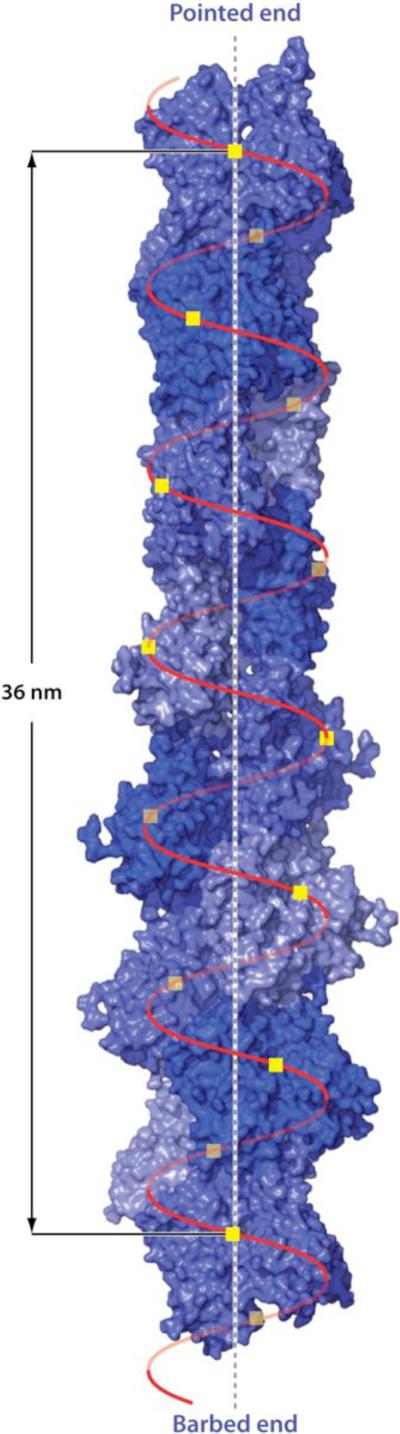
EM of negatively stained actin fibers showed F-actin to be made of two chains that turn gradually around each other to form a right-handed, two-chained long helix (22). The actual symmetry is a single left-handed genetic helix with approximately 13 molecules repeating every six turns in an axial distance of 35.9 nm. The distance between molecules along the helix axis is 27.6 Å. However, because the twist per molecule (–166.6°) is close to 180°, the structure actually appears like two slowly turning right-handed chains (Figure 2).
Figure 2.
The helical structure of F-actin derived from cryo-electron microscopy (16). The molecules are arranged on a single helix with 13 molecules repeating in almost exactly six left-handed turns. The rise per molecule is 2.76 nm and the twist per molecule is –166.6±0.6°(for simplicity of drawing, in the figure the value -166.15 has been used to make the structure repeat exactly after 13 residues). Because -166° is close to 180° the structure takes on the appearance of a two-start right-handed long-pitch helix.
Fiber Diffraction Patterns Give the First Structures of F-Actin
Concentrated solutions of F-actin can be induced to form gels consisting of aligned actin filaments (46). Until recently, the only source of higher-resolution structural data from F-actin was X-ray diffraction from such oriented gels (strictly ‘orientate’ means to point in one direction whereas ‘orient’ means to face east KCH) . In oriented gels, the fibers, while parallel, have a random orientation about the fiber axis. Thus, X-ray diffraction yields essentially a section through the diffraction pattern of a single fibrous molecule that has been averaged by spinning it around the fiber axis (cylindrical averaging). Because the structures are periodic in the axial direction, the fiber diagram is limited to layer lines. To interpret a fiber diagram pattern generally one compares the measured data with the computed fiber diagram of a starting model and then uses a refinement procedure to modify the model and achieve a better fit.
Thus, the first near-atomic resolution structure of F-actin (25) was deduced by combining X-ray fiber diffraction data and the crystal structure of G-actin (25) However, this model was based on the G-actin monomer. Because there is in fact a state-change between the monomer in G- and F-actin, the fit to the fiber diffraction pattern was not optimal. One consequence of this approximate fit was that, although the orientation of the actin monomers in the helix was correct, the two long-pitch helices in this early model were about 3 Å too far apart.
Since this initial calculation, a number of attempts have been made to deduce the nature of the G- to F-actin transformation by refining models against the data from X-ray fiber diagrams (24, 33, 53). The fiber diagram used had limited resolution (~7 Å). Moreover, disorientation in the sample, which smears out the layer lines, loses information. Unique answers were not obtained.
Actin fibers are diamagnetic. Well-oriented X-ray fiber diffraction patterns can be obtained by placing F-actin gels in a very strong magnetic field (36) (Supplemental Figure 3). The resulting higher resolution X-ray fiber diagram indeed made it possible to elucidate the nature of the G- to F- actin transition (36). The main component of the transition from G- to F-actin is a 12°--13° propeller-like rotation of the outer domain with respect to the inner domain about an axis roughly at right angle to the helix axis (Figure 3). Moreover, there are bending movements of domains 2 and 4 in the same direction. The final result is that F-actin is flatter than G-actin by about 17°--18°. Furthermore, the D-loop takes on an open-loop configuration and inserts itself into the target-binding cleft of the subunit immediately above it.
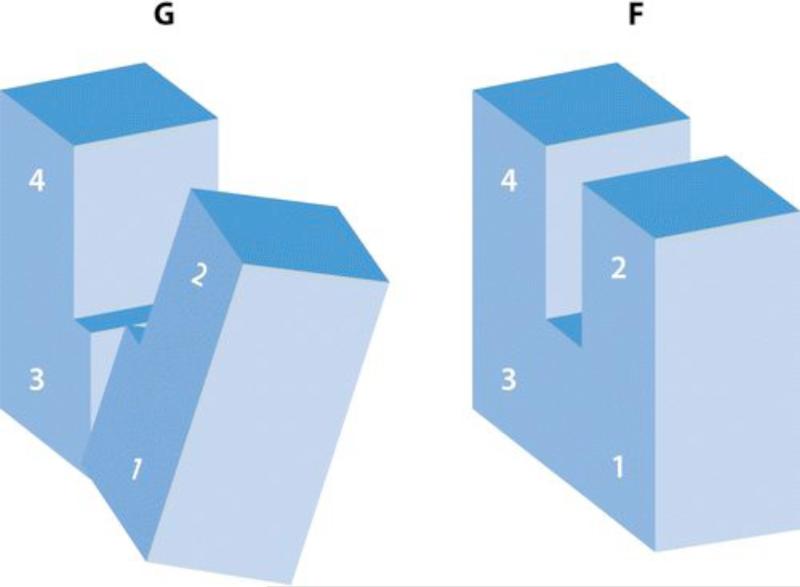
Figure 3.
The essence of the G- to F-actin transition is a flattening of the actin molecule by a propeller-like twist of the outer and inner domains about an axis roughly at right angles to the actin helix axis. The numbers refer to the subdomains (28) (diagram courtesy of Y. Maeda).
Oda & Maeda (37) have defined the propeller-like twist angle (q) between the two major domains as the angle between two planes, one containing the Cα atom of residue Gly55 and the axis of rotation, and the other containing Glu207 and the axis of rotation. Gly55 is in domain 2 and Glu207 is in domain 4, so that q measures the total twist---a rotation of domains 1 and 3 plus a further bending of domains 2 and 4. The average twist angle is about 20° for all G-actin structures (it is the largest for the two monomeric actin structures 1J6Z and 3HBT at 25°), whereas for F-actin structures the twist angle q is about 7°.
Cryo-Electron Microscopy
Cryo-EM has now yielded an atomic model of F-actin (16). Fujii et al. (16) have produced a 6.6 Å resolution density map of F-actin that allows the disposition of the domains and some of the polypeptide chain to be visualized directly. By controlling humidity and ice thickness during freezing, these authors obtained fields of long, straight actin filaments with nearly perfect helical symmetry. This result made possible the high resolution obtained by these authors. An analysis of their data shows that the angular twist per molecule in the actin helix is %166.6 ± 0.6° and the rise per residue is 27.6 Å.
Structure of F-Actin
The models produced by Fujii et al. (16) and Oda et al. are similar. The most significant difference is in the D-loop, which Fujii et al. observed directly. Moreover, in the method of refinement used by Fujii et al. coordinates were altered as little as possible from the crystallographic starting model. As a result, the Fujii model is likely to be more precise.
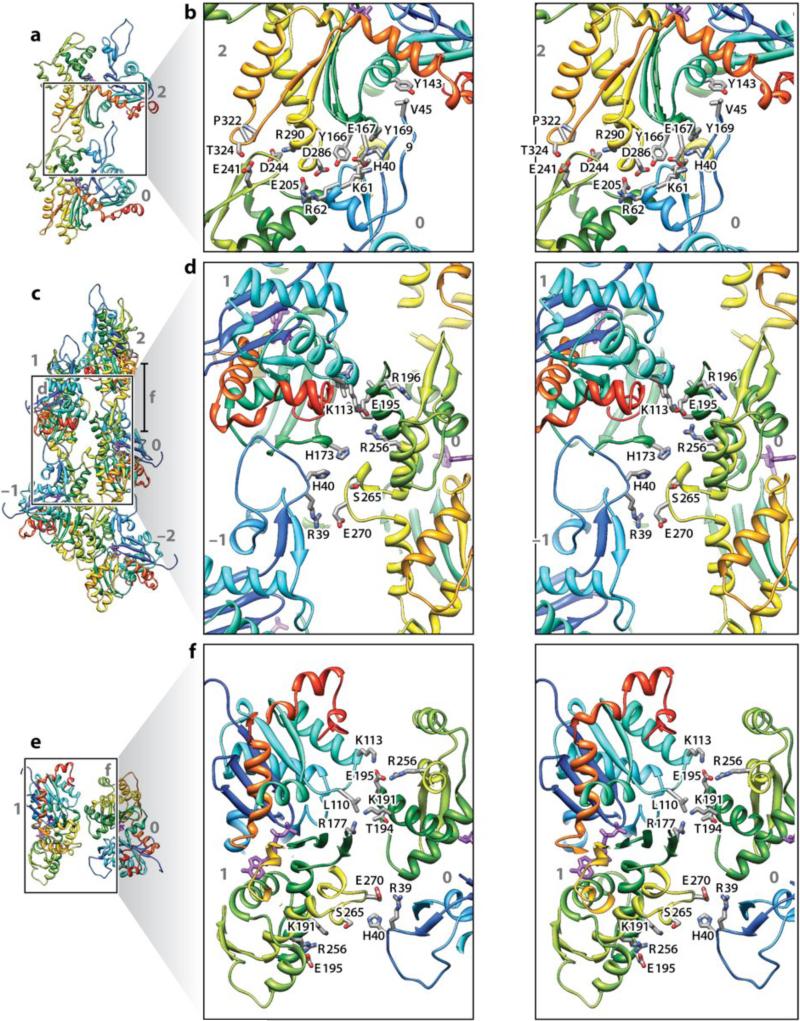
Stereo views of the interface are shown in Figure 4. The stronger bonding between molecules is in the direction of the filament axis and takes place between neighboring molecules along the two-start long-pitch helix. Molecules in Figure 4 are numbered –2, –1, 0, 1, and 2 along the genetic helix (subunit 0 is taken to be the reference). Axial interactions are extensive. One important longitudinal interaction is between domains 4 and 3 in neighboring molecules: The loop Pro243-Asp244-Gly245 in domain 4 interacts with two loops in domain 3 of the molecule above, particularly residues Pro322 and Met325 in one loop and Met283, Ile287, and Asp288 in the adjoining loop. Two previous report have suggested similar interactions (25,51). The second important longitudinal interaction involves the D-loop. The D-loop of subunit 0 extends into the target-binding cleft of subunit 2 where it interacts with domains 1 and 3. In addition, residue Ile64 forms a bond with domain 3 of molecule 2. Fujii et al. (16) remark that the axial interactions are mostly electrostatic in character, but there are also hydrophobic contacts.
Figure 4.
Intermolecule bonding in F-actin. Shown are five actin molecules labeled –2 to +2. The run of the protein chain is shown as a secondary structure cartoon color-coded from blue (N terminus) to red (C terminus). Interacting side chains are shown as sticks. Panels b, d, and f are stereo pairs. Panels a and b show the main longitudinal interface between molecules 0 and 2. Panels c and d show the transverse interaction across the helix axis in the neighborhood of the plug. This interaction also has contributions from the D-loop. Panels e and f show the transverse interaction higher up (see right hand side of panel c for a definition of panel f) involving loops 195--198 and 108--113. (Reprinted by permission from Macmillan Publishers Ltd: Nature 467:724-728, copywrite 2010)
The lateral bonding across the axis between the long-pitch helices is much less extensive than the longitudinal bonding. One interaction is between the plug-like insertion (Gln 263--Gly 273) in domain 3 and the beginning of the D-loop of subunit %1Interactions of the plug across the helix axis with molecules %1 and 1 stabilize the two-stranded F-actin. The other interchain interaction is just above the plug, where domain 4 contacts domains 1 and 3 of molecule 1. Details are given in the original paper (16).
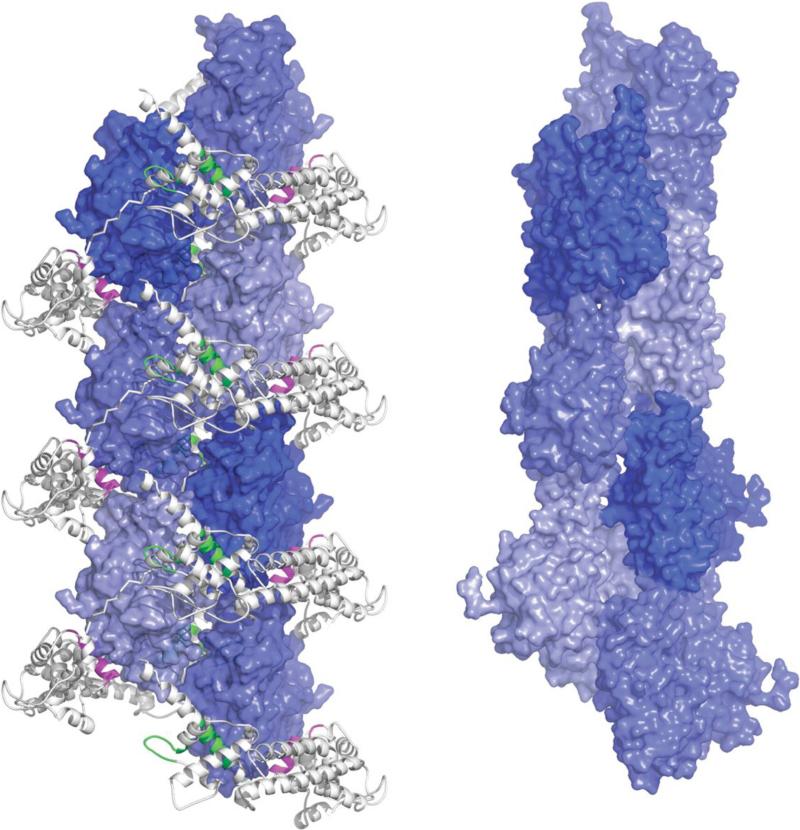
Precursor Actin Helix Stabilized by the Formin FH2 Domain
The precursor helical assembly of G-actin engendered by the formin FH2 domain (39) is related to the F-actin structure. The asymmetric unit of the crystal structure (1Y64) contains a single FH2-actin complex. However, the C2 symmetry positions identical FH2-actin complexes at ±28.1 Å and rotated by 180° with respect to one another, with neighboring FH2 subunits tethered by lasso/post interactions (Figure 5 and Supplemental Figure 2). The actin subunits of two successive FH2-actin complexes are thus held in a relative orientation similar to that of the short-pitch F-actin helix (actin subunits of the filament helix are rotated by 166.6° and translated by 27.6 Å), such that the structure found in the crystal looks like a precursor of the F-actin helix (Figure 5). However, the longitudinal interactions between actin subunits are not the same as in F-actin and the D-loop is disordered. Nevertheless, the G-actin monomers are positioned by the FH2 scaffold in a way that would facilitate a transition to the F-actin helix. For this transition to occur, the FH2 domain would have to move out to let the D-loop bind in the target-binding cleft of the subunit above it, which is occupied in the complex by a helix of the FH2 knob domain.
Figure 5.
Precursor helix assembled by the formin FH2 domain. The left view shows the precursor helix assembled by FH2 subunits along the crystallographic C2 axis. The forming FH2 domain is shown grey as in Figure 1i. Neighboring molecules are positioned by binding respectively to the knob (magenta)[ and the post (green) to form a helix with a twist of 180° per molecule and a rise per residue of 28.1 Å (see Supplemental Figure 2 for details). The right view shows the F-actin helix for comparison. Actin subunits of the filament helix are rotated by -166.6° and translated by 27.6 Å (as in Figure 2)
Bacterial Actin-Like Homologues
A number of actin-like homologues have been discovered in bacteria. These form filaments (see, e.g., 47). The bacterial actin homology MreB forms filaments (54) that are single stranded and straight. The monomer is flat similar to the F-actin monomer. However, it does not appear to have a D-loop. ParM (48) is a prokaryotic actin homolog that segregates plasmids before bacterial cell division. ParM polymerizes with GTP. The ParM filament has a left-handed long-pitch helix, opposed to the right-handed long-pitch helix of actin (48). It has a sequence that appears to bind to domain 3, similar to the D-loop in actin. Furthermore, cryo-EM has visualized a form in which the nucleotide-binding cleft is open (17). AlfA (43) defines a new class of actins that shows an extremely rapid polymerization rate and assembles with ATP or GTP. The helical symmetry of AlfA filaments differs markedly from the helical symmetry of F-actin. Nevertheless, all these polymers show similar values for the rise per residue, approximately 3.0 nm.
THE G- TO F- TRANSITION
There is a mass of data on the inhibition of actin polymerization by binding of proteins or small molecules. Sometimes the bound molecules simply impede polymerization by steric hindrance. Thus, the fluorescence probe tetramethylrhodamine-5-maleimide [AU: Acronym not used in the text.] sits exactly in the binding pocket of the D-loop and inhibits D-loop binding. In most cases it appears that the inhibiting molecule competes with the binding of the D-loop to target-binding cleft of its neighbor along the long-pitch helix. A graded response is sometimes observed. For example, binding of gelsolin leads to severance, whereas binding of cofilin changes the helical pitch of F-actin and increases the filament flexibility (8).
The D-loop breaks into two regions: The sequence 39--43 is involved in stereospecific interaction, whereas the sequence 44--51 lies toward the outside of the F-actin structure [this sequence may be a target primarily for F-actin-binding proteins such as myosin (32)]. The WH2 domain sits in the cleft between domains 1 and 3, where it would collide with one end of the D-loop sequence 44--51 but does not interfere with the sequence 39--43. Perhaps differences in response lie in the behavior of the sequence 44--49 and the specific binding site of each protein in the target-binding cleft. For instance, the FAB domain of Ena/VASP is related in sequence to the WH2 domain but binds F-actin and can presumably co-bind in the cleft together with the D-loop of a neighboring actin subunit, whereas the classical WH2 domain cannot (14).
ATP-bound G-actin is the normal substrate for filament formation. It may be induced to polymerize to ATP-containing F-actin, which is energetically favored under physiological conditions. However, F-actin is a slow ATPase, such that the filament soon contains just ADP. ADP F-actin is less stable than ADP G-actin. This enables actin filaments to be readily disassembled, an essential part of the actin recycling process in cells, but how is this differential stability controlled? It could come about by movements of the D-loop. Studies cited earlier show that the methylated histidine sensor loop can detect the status of nucleotide and pass this information to the D-loop. Moving the end of the D-loop by 1--2 Å would probably be enough to alter its binding affinity to the domain 1–domain 3 cleft. Unfortunately the data necessary to explore such mechanisms in detail is not available: We still lack the structure of ATP F-actin.
Supplementary Material
FUTURE ISSUES.
There are no accurate atomic models of proteins bound to F-actin.
We do not know why ATP F-actin is more stable than ADP F-actin. We need details of the actin ATPase, detailed structure of ADP F-actin, and detailed structure of ATP F-actin.
More accurate structures of the ends of the actin filament (pointed and barbed), and the binding of capping proteins to these ends are needed.
How much do F-actin-binding proteins change the structure of the filament?
More information is needed regarding the mechano-sensing properties of the actin filament, exerted through interactions with membrane associated proteins and cytoskeletal proteins.
Acknowledgments
R.D. was supported by NIH grants MH087950, GM073791, and HL086655
KEY TERMS AND DEFINITIONS
- G-actin
Monomeric (or globular) actin
- Target-binding cleft
The cleft between domains 1 and 3 of the actin molecule, where most actin-binding proteins interact (also known as the hydrophobic cleft)
- D-loop
DNase I-binding loop of actin (residues 39--51)
- F-actin
Filamentous (or polymeric) actin:
- Filament growth
Refers to the process of addition of actin monomers to the actin filament, which is faster through the barbed or (+) end of the filament
- Filament disassembly
Refers to the process of dissociation of actin subunits from the filament. Disassembly can occur in blocks, as a result of filament severing, or through the dissociation of individual actin subunits, which dissociate preferentially from the pointed end and in the ADP-bound state.
- G-actin to F-actin transition
Refers to the process of actin polymerization
- Propeller-twist
Rotation of the outer domain with respect to the inner domain of actin subunits in the filament, which results in a more flat structure of the actin molecule
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
References
- 1.Barth H, Stiles BG. Binary actin-ADP-ribosylating toxins and their use as molecular Trojan horses for drug delivery into eukaryotic cells. Curr. Med. Chem. 2008;15:459–-69. doi: 10.2174/092986708783503195. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–-95. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA. 1992;89:7290–-94. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burtnick LD, Koepf EK, Grimes J, Jones EY, Stuart DI, et al. The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation. Cell. 1997;90:661–-70. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 5.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010;11:237–-51. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell. Biol. 1997;136:1307–-22. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlier MF, Valentin-Ranc C, Combeau C, Fievez S, Pantoloni D. Actin polymerization: regulation by divalent metal ion and nucleotide binding, ATP hydrolysis and binding of myosin. Adv. Exp. Med. Biol. 1994;358:71–-81. doi: 10.1007/978-1-4615-2578-3_7. [DOI] [PubMed] [Google Scholar]
- 8.De la Cruz EM. How cofilin severs an actin filament. Biophys. Rev. 2009;1:51–-59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominguez R. Actin-binding proteins---a unifying hypothesis. Trends Biochem. Sci. 2004;29:572–-78. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez R. The beta-thymosin/WH2 fold: multifunctionality and structure. Ann. N. Y. Acad. Sci. 2007;1112:86–-94. doi: 10.1196/annals.1415.011. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez R. Structural insights into de novo actin polymerization. Curr. Opin. Struct. Biol. 2010;20:217–-25. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, et al. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83:433–-73. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 13.Drees F, Gertler FB. Ena/VASP: proteins at the tip of the nervous system. Curr. Opin. Neurobiol. 2008;18:53–59. doi: 10.1016/j.conb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008;27:2943–-2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–-606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.—Deleted in proof [AU: Renumbered thruout as Ref. 14]
- 16.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–-28. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 17.Galkin VE, Orlova A, Rivera C, Mullins RD, Egelman EH. Structural polymorphism of the ParM filament and dynamic instability. Structure. 2009;17:1253–-64. doi: 10.1016/j.str.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galkin VE, VanLoock MS, Orlova A, Egelman EH. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin's sequence and structure. Curr. Biol. 2002;12:570–-75. doi: 10.1016/s0960-9822(02)00742-x. [DOI] [PubMed] [Google Scholar]
- 19.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007;76:593–-627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 20.Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 2005;8:35–-45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state: structural basis of nucleotide-dependent actin dynamics. J. Biol. Chem. 2003;278:34172–-80. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 22.Hanson J, Lowy J. The structure of F-actin and the actin filaments isolated from muscle. J. Mol. Biol. 1963;6:46–-60. [Google Scholar]
- 23.Herman IM. Actin isoforms. Curr. Opin. Cell Biol. 1993;5:48–-55. doi: 10.1016/s0955-0674(05)80007-9. [DOI] [PubMed] [Google Scholar]
- 24.Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–-27. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 25.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–-49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 27.Iwasa M, Maeda K, Narita A, Maeda Y, Oda T. Dual roles of Gln137 of actin revealed by recombinant human cardiac muscle alpha-actin mutants. J. Biol. Chem. 2008;283:21045–-53. doi: 10.1074/jbc.M800570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–-44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 29.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–-35. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 30.Lang AE, Schmidt G, Schlosser A, Hey TD, Larrinua IM, et al. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science. 2010;327:1139–-42. doi: 10.1126/science.1184557. [DOI] [PubMed] [Google Scholar]
- 31.Lee WM, Galbraith RM. The extracellular actin-scavenger system and actin toxicity. N. Engl. J. Med. 1992;326:1335–-41. doi: 10.1056/NEJM199205143262006. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz M, Holmes KC. The actin-myosin interface. Proc. Natl. Acad. Sci. USA. 107:12529–-34. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorenz M, Popp D, Holmes KC. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J. Mol. Biol. 1993;234:826–-36. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- 34.Mannherz HG, Goody RS, Konrad M, Nowak E. The interaction of bovine pancreatic deoxyribonuclease I and skeletal muscle actin. Eur. J. Biochem. 1980;104:367–-79. doi: 10.1111/j.1432-1033.1980.tb04437.x. [DOI] [PubMed] [Google Scholar]
- 35.Miralles F, Visa N. Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 2006;18:261–-66. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–-45. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 37.Oda T, Maeda Y. Multiple conformations of F-actin. Structure. 2010;18:761–-67. doi: 10.1016/j.str.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010;11:353–-65. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–-94. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 40.Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–-11. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 41.Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 2008;182:51–-59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paunola E, Mattila PK, Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 2002;513:92–-97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- 43.Polka JK, Kollman JM, Agard DA, Mullins RD. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J. Bacteriol. 2009;191:6219–-30. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:451–-77. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 45.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–-65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 46.Popp D, Lednev VV, Jahn W. Methods of preparing well-orientated sols of F-actin containing filaments suitable for X-ray diffraction. J. Mol. Biol. 1987;197:679–-84. doi: 10.1016/0022-2836(87)90474-8. [DOI] [PubMed] [Google Scholar]
- 47.Popp D, Narita A, Ghoshdastider U, Maeda K, Maeda Y, et al. Polymeric structures and dynamic properties of the bacterial actin AlfA. J. Mol. Biol. 2010;397:1031–-41. doi: 10.1016/j.jmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Popp D, Narita A, Oda T, Fujisawa T, Matsuo H, et al. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 2008;27:570–-79. doi: 10.1038/sj.emboj.7601978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–-84. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 50.Rohr G, Mannherz HG. Isolation and characterization of secretory actin. DNAase I complex from rat pancreatic juice. Eur. J. Biochem. 1978;89:151–-57. doi: 10.1111/j.1432-1033.1978.tb20907.x. [DOI] [PubMed] [Google Scholar]
- 51.Sawaya MR, Kudryashov DS, Pashkov I, Adisetiyo H, Reisler E, Yeates TO. Multiple crystal structures of actin dimers and their implications for interactions in the actin filament. Acta Crystallogr. D. 2008;64:454–-65. doi: 10.1107/S0907444908003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol. Life Sci. 2004;61:2614–-23. doi: 10.1007/s00018-004-4225-6. [DOI] [PubMed] [Google Scholar]
- 53.Tirion MM, ben-Avraham D, Lorenz M, Holmes KC. Normal modes as refinement parameters for the F-actin model. Biophys. J. 1995;68:5–-12. doi: 10.1016/S0006-3495(95)80156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–-44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 55.Deleted in proof [AU: Renumbered thruout as Ref. 54.]
- 56.van den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–-43. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wegner A, Isenberg G. 12-fold difference between the critical monomer concentrations of the two ends of actin filaments in physiological salt conditions. Proc. Natl. Acad. Sci. USA. 1983;80:4922–-25. doi: 10.1073/pnas.80.16.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, et al. Crystal structures of a Formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–-23. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 59.Yarmola EG, Somasundaram T, Boring TA, Spector I, Bubb MR. Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J. Biol. Chem. 2000;275:28120–-27. doi: 10.1074/jbc.M004253200. [DOI] [PubMed] [Google Scholar]
- 60.Yeung KS, Paterson I. Actin-binding marine macrolides: total synthesis and biological importance. Angew. Chem. Int. Ed. Engl. 2002;41:4632–-53. doi: 10.1002/anie.200290057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.