Abstract
The pattern of individual variation in brain component structure in pigs, minks and laboratory mice is very similar to variation across species in the same components, at a reduced scale. This conserved pattern of allometric scaling resembles robotic architectures designed to be robust to changes in computing power and task demands, and may reflect the mechanism by which both growing and evolving brains defend basic sensory, motor and homeostatic functions at multiple scales. Conserved scaling rules also have implications for species-specific sensory and social communication systems, motor competencies and cognitive abilities. The role of relative changes in neuron number in the central nervous system in producing species-specific behaviour is thus highly constrained, while changes in the sensory and motor periphery, and in motivational and attentional systems increase in probability as the principal loci producing important changes in functional neuroanatomy between species. By their nature, these loci require renewed attention to development and life history in the initial organization and production of species-specific behavioural abilities.
Keywords: brain evolution, allometry, domestication, individual variation, neocortex
1. Introduction: frameworks to relate natural variation in behaviour, brain structure and genes
Individual variation is the foundation of natural selection, and the idea of inheritance of adaptive characteristics is fundamentally a simple one. Yet, understanding individual behavioural variability and its inheritance is one of the most complex tasks facing current researchers. A good theory of the organization of every level of analysis from behaviour to gene is necessary: individual animals in populations, the computational structure of the brain, the mechanisms of development of the organism and the translation from genome to organism. How do we begin to break into the complex chain from gene to brain to behaviour to population? For example, a fairly recent review of the evolutionary biology of animal cognition gives an ambitious catalogue of variations in behaviour ranging from very specific differences in sensory systems to very complex changes in communication systems and cognition, some with known fitness consequences [1]. The associated brain ‘phenotypes’ range from photoreceptor opsins, to neurotransmitters and to whole brain organization and size, and the species range from humans to Drosophila, but in no case is the explanatory chain ever complete. For perfectly understandable reasons, researchers appreciate the complexity of the level of analysis of their primary area of expertise, and underestimate the complexity of other levels, producing predictable and recurring errors in explanations that attempt to bridge levels.
In the following paper, we will present some new data about individual variation in brain parts and relate it to phylogenetic variability. Relating the size of brain parts to species-typical adaptive behaviours has been the subject of studies of brain allometry for many years [2–6]. The nature of individual variation in the size of brain parts offered to selection informs this work directly. For this journal issue, however, we will place these results in the context of three links in the behaviour-to-gene chain, where new information has appeared. First, while the behavioural significance of changes in absolute and relative brain size in phylogeny has been the subject of analysis and speculation for many years, causal or even correlational links between relative brain size and fitness had never been demonstrated. Newer studies now allow a better understanding of what behavioural advantage an increase in relative brain size permits animals in natural contexts. Second, new work in computer science in robotics, the design of machines that must function in the real world, are beginning to yield insights on what kinds of computational architectures are robust to change, damage and growth. Finally, developmental neurobiology increasingly demonstrates the importance of epigenetic and developmental factors in aligning initially unspecified brain structures with their particular physical and social environments.
(a). Some insights from the evolution of colour vision
Of all the work in evolution of perceptual and motor systems, vertebrate colour vision comes closest to having a full description at every level, including genetic, developmental, physiological, computational and ecological. The history of this investigation can inform current work on other aspects of brain and behavioural evolution directly. Vertebrate colour vision contrasts the output of two to five very broadly tuned photoreceptors that cover the visible spectrum to code the reflectance characteristics of objects and environments with relative independence from the spectrum of light illuminating them. A typical mammal has two photopigments for diurnal vision (opsins), and most anthropoid primates have three [7–9]. The opsin molecule, composed of amino acids, is a rough cylinder to which a short-chain molecule, 11-cis retinal, is attached, like a tab on a cola can. Light absorbed by the 11-cis retinal changes its conformation to all-trans, initiating the chain of phototransduction. Of the several 100 amino acids comprising the opsin molecule (using the red–green opsin as a typical example), only a small fraction are placed in the opsin cylinder such that an amino acid substitution will change the best wavelength for reconformation of the 11-cis retinal molecule [10]. The probability of this class of mutations with direct functional consequences is reasonably well understood in terms of the ongoing ‘jitter’ in base-pair substitution that the genome continuously undergoes. Overall, the steps from gene to opsin to phototransduction are exceptionally well worked out.
From basic receptor photosensitivity to adaptive function in the real world, that is, to perception and behaviour, the path to understanding has been rockier. What does a changed best frequency of a photopigment signify for behavioural adaptation? A first guess was that there might be a direct relationship between the most sensitive frequency of the photoreceptors and a specific aspect of the adaptive environment. Perhaps the best frequencies of photoreceptors evolved to facilitate foraging or conspecific recognition—that is, detecting yellow preferentially to find bananas, or discriminating the redness of faces (e.g. [11]). However, no case of direct matching of best opsin frequency and any particular feature of the environment have ever been established as a basis for species-typical preferences (not negating the fact of species-typical preferences, only where in the brain they may be situated!). Instead, the opsins appear to have been selected to adequately cover the range of available light in each species' typical environment. In the case of signalling, the signaller (whether fruit or conspecific) typically evolves to be optimally detectable, while the receiver remains ‘generic’ [12,13].
In the special case of primate trichromacy, a single amino acid change in the original opsin may produce two slightly different opsin forms whose output can be compared in the central nervous system [14]. This produces better ‘colour acuity’ in the green–brown regions of colour space, with many plausible adaptive advantages [15–18]. Startlingly, the identical modification may be ‘knocked in’ to the retinas of mice or dichromatic monkeys, who can in short order learn to make behavioural use of the new colour information, with no other genetic changes in their nervous systems [19,20].
The lessons to be learned here are important, as they tend to recur over and over in brain–behaviour mapping questions. First, researchers equated prominent adaptive features of the behavioural environment (such as a coloured fruit) with features of the eye and brain (such as opsin best frequency, a particular cell type or a brain nucleus) far too quickly: the categories of the world are not the categories of the brain. Second, when investigators were interested in species-typical specializations in sensorimotor capacities, such as ability to identify a particular fruit, they tended to ignore the complex abilities in scene recognition, navigation and locomotion upon which such specializations ultimately depended. These complex, species-general abilities may dominate species-specific specializations in any currently measurable form of ‘neural commitment’ like neural volume or energetic expenditure. Finally, the perceptual, cognitive and motor systems of any extant vertebrate are the end result of continual compromise, having proved competent to deal with both species-typical and species-general problems.
(b). Evolution shapes development
New work in evo-devo adds a further dimension to our understanding of behavioural variability. The central researchers and theorists in evo-devo argue persuasively that the developmental programmes of existing creatures are as much a product of evolution as their mature phenotypes, and that these developmental programmes come to have the features of ‘evolvability’ and robustness. Considering the various aspects of ‘evolvability’, which have been discussed at length elsewhere [21–24], the one of central interest here is ‘facilitated variation’. Readers interested in a discussion of basic issues in brain development, including organization of the body plan, neural proliferation and segmentation, and activity-dependent organization of the central nervous systems are directed to any one of these sources.
Consider an evolutionary case of unfacilitated variation. Suppose we know that an animal would enjoy greater reproductive success if it had longer forelimbs, but we also know that there are no coordinating mechanisms between the developmental programmes producing its various organ systems. Production of longer forelimbs in this animal would require simultaneous random changes in its unlinked programmes for bone growth, vascular supply, muscle volume, muscle attachments, the length of other attached bones, the neural programme executing movement and so on. While gradual accretion of such changes is certainly not impossible, in principle, no such unlikely lineups of random events are needed to produce differences in relative and absolute amounts of limb growth in existing animals. A mutation changing only the rate or duration of bone production can be seen in adult limb morphology because of the epigenetic programmes controlling somatic growth in which bone growth is embedded.
In the colour vision example, a random change in the coding of a single amino acid in a much larger opsin molecule can directly change its best wavelength. If the neural system supporting wavelength selectivity required new committed processors from retina to brain (‘banana detectors’; ‘command neurons for banana grasping’), such changes at the photoreceptor level would rarely have consequences other than blindness. If the brain mechanism that looks at retinal information is a generalized wavelength comparator, however, as seems to be the case, prepared to analyse a new contrast and learn its relationship to environmental structure, the mutation can succeed, as has recently been demonstrated [19,20]. On the other hand, ‘nonsense’ genetic changes unsupported by epigenetic embedding or robust neural programmes will likely be invisible or be deleterious. Normal development, via evolution, acts as a filter of genetic mutations to promote meaningful variability that it has previously and successfully experienced, and opsin variations appear to be in that class.
(c). From individual variation to species variation in morphology
Links of natural phenotypic variation to evolutionary dimensions of fitness, speciation and genotype have been made in non-behavioural contexts. Linking within-species individual phenotypic variability to patterns of speciation has been evaluated in wide-ranging natural contexts and phyla, the most well-known instances involving readily observable aspects of morphology, for example, plant ecology [25], skeletal alterations in three-spine sticklebacks [26] and the beaks of Darwin's finches [27]. In a particularly relevant study, Schluter [28] related details of stickleback morphological individual variation in skeletal structure to speciation. For a founder species, the largest set of co-varying features was termed ‘Gmax’, ‘the phylogenetic path of least resistance’. For closely related species, it was predicted that this factor should be highly represented, as it is the corpus of accessible variation on which selection can easily act. For distantly related species, it was expected that the variation observed would diverge systematically. Instead, Gmax remained as high in the distant taxa as in the immediately related ones. ‘Developmental constraint’ was offered to account for these results, operating over both short and long time spans. Alternatively, however, the co-varying dimensions could represent the operation of conserved developmental programmes, facilitating variability along the dimensions associated with viable outcomes in the past and filtering out others.
In natural instances of variation and speciation, going to the second step of linking phenotypic changes directly to genotypic changes has been attempted in only a few contexts, such as bird plumage [29], the explosive radiation of the Lake Malawi cichlids [30] and, recently, adaptive specialization of mouse fur colour to the environment [31]. These studies are ‘existence proofs’ that the links from fitness as measured in natural settings to genetic mechanisms can be made, when we understand the structure of individual and species variability in features with demonstrated fitness relationships.
2. Conservation and variation in brain size and behaviour, in an evo-devo context
(a). Relative brain size can be directly linked to aspects of fitness
The largest possible focus for brain–behaviour relationships, the whole brain, is a surprisingly informative place to begin. Pervasive increases in relative brain size with respect to body size (‘encephalization’) in multiple lineages over evolutionary time and its association with behavioural complexity (without any direct measure of fitness) were first described systematically by Jerison [2]. More recently, the same simple feature of relative brain size (at the species level) has been shown to have direct links to fitness, in innovative studies of multiple species of both birds and mammals in natural ecology [32]. These fitness indicators include annual mortality rate, probability of an individual species' survival when introduced into a new niche, and behavioural innovation in diet. These ecological indicators can be further correlated with several measures of learning flexibility in laboratory contexts in a smaller number of species. Although entirely different in emphasis, similar studies actually exist for individual differences in humans: individual variations in absolute brain size have a small, but statistically significant link to intelligence quotient (e.g. [33]), and thus to multiple measures of socioeconomic status, if not ‘fitness’. The relative sizes and developmental pattern of enlargement of the parts of the brain most disproportionately large in the largest individual brains, the frontal and parietal cortex, vary directly with individual differences in cognitive flexibility [34].
(b). Allometric variability in brains, between species
To make the links from behavioural flexibility to brain size to initial genetic specification of brain size, via development, we have empirical work at every step. Phylogenetic variation between mammalian species' brains is well described [3,35]. Developmental alterations associated with species variations in brain size and sensory systems are beginning to be understood [36–40]. A growing body of knowledge exists on those genes or gene loci involved in brain size regulation including studies in mouse models [41–44], in development [45–48] and in phylogeny [49].
(c). Theories of brain function underlying the first hypotheses of brain evolution
The history of our understanding the relationship of complex behavioural functions to phylogenetic brain variation is one of bootstrapping back and forth from the most general structure–function mappings to progressively more sophisticated versions, in much the same way that the understanding of the evolution of colour vision changed over time. Here, we must digress somewhat to consider contrasting theories of brain function in a general evolutionary context. As mentioned, Jerison [2] first proposed a relationship between encephalization, the possession of a relatively large brain with respect to body size, and behavioural complexity. He also hypothesized the strategic allocation of neural volume according to niche, which is the principle of ‘proper mass’. Particularly, he argued more volume came to be allocated to neocortex in highly visual, diurnal primates and more to olfactory bulb and the limbic system in early nocturnal insectivores. This axis of variation in relative olfactory bulb and limbic brain size proves to be a recurring ‘principal component’ of vertebrate brain variability, not only of primates [37,50,51], whose significance we will explore later.
In parallel with Jerison's description, the growth of neuroethology on the one hand, with its attention to species-specific adaptations, and the early discoveries of localization or ‘modularization’ of mature neural function in cognitive neuroscience led the field to concentrate on the relationship of lifestyle and niche to specific brain parts. The basic hypothesis appeared that brains are collections of special-purpose devices or ‘modules’ that can be the objects of selection, a point of view that retains strong adherents in evolutionary psychology (e.g. [52]), and some fields of cognitive neuroscience [53]. It is often assumed without question in literature about brain evolution [3,5].
To compare species, the immediate corollary of the ‘collection of devices’ hypothesis was that the relative size of a brain part should reflect the importance or complexity of the behaviour dependent upon it, ‘proper mass’. When investigated widely across species and brain structures, relating non-shared residual variation in the relative size of brain parts to niche or behavioural specializations typically met with only modest success, capturing statistically significant but small amounts of variation (e.g. [3,5]). Many reasonable predictions based on the idea of ‘proper mass’ fail to find any relationship of relative size and utility at all, for example whether there is a correlation between a mammal's nocturnality or diurnality and the volume of its visual cortex [54]. Any behavioural or fitness advantage directly owing to change in the relative size of a brain part has yet to be demonstrated.
Several apparent exceptions to this generality occur, which we will introduce now and discuss fully later in terms of the component structure of brain variability. Dunbar [4] posited that the relative size of the neocortex in primates, compared with the rest of the brain, was related to species' complexity in social structure, the ‘social brain’ hypothesis, though more recent analyses target whole brain size rather than cortex alone [55]. Species and individual variation in the hippocampus also stands out in its consistent relationship to requirements of memory in foraging and navigation, often dependent on experience [56–58]. The hippocampus associates with the olfactory bulb and olfactory cortex and dissociates from neocortex scaling across mammalian species [35], and is part of the neocortex/limbic contrast originally laid out by Jerison [2].
Simply because species differences in sensory capabilities, motor adaptations and social structure do not link readily to relative size of anatomically defined chunks of the brain in mammals (for example, equating ‘motor skill’ and ‘cerebellar volume’), this does not signify that species-specific adaptations do not exist, or that they are not important. It simply suggests that species-typical specializations must arise through other mechanisms. Some plausible, demonstrated mechanisms producing species diversity are: (i) the sensory or motor periphery imposing its order on the central nervous system, (ii) changed distributions of neuromodulators and receptors altering social motivation and attention, and (iii) dynamic reallocation of neural tissue to the most active channels by instruction through the sensory channels and motivational preferences of each animal. We have finally set the stage for looking at the relationship between phylogenetic variability in brain structure and scaling, and individual differences in brain organization.
3. Phylogenetic variations in brain structure
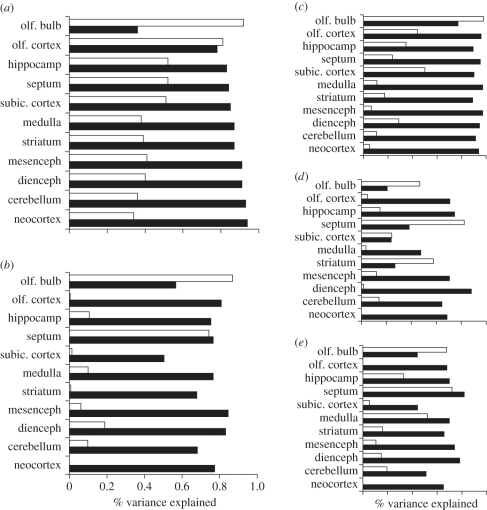
Three features predominate in mammalian brain scaling: high intercorrelation of structure volumes, distinct allometric scaling for each structure and relative independence of the olfactory–limbic system from the rest of the brain. At this point, we can consider nine major taxonomic groups [35], using the brain divisions employed in the original analyses of Stephan & Manolescu [59]. The first two principal components of volume variation account for 99.01 per cent of the total volume variation in this set of 160 mammals. The first factor is highly loaded on the cortex, cerebellum, diencephalon and mesencephalon, accounting for 96.47 per cent of the total variance (figure 1a, black bars). The second principal component loads most highly on the olfactory bulb, and next on olfactory cortex, hippocampus, septum and subicular cortices, accounting for 2.61 per cent of the variance (figure 1a, white bars). In addition, a third component relates body size independently to the sizes of the medulla and cerebellum—this factor is small, but significant, and has been reported in other kinds of analyses (e.g. [60]). Finally, each brain division has a distinct allometry, with each brain subdivision increasing at a different slope as brain volume increases overall. In particular, the neocortex and cerebellum scale with absolute brain volume at high slopes so that very large brains become disproportionately composed of these two structures.
Figure 1.
In all graphs, the percentage variance in each structure described by the first principal component (PC1) is graphed by the black bars, and the second principal component (PC2) by the white bars, the total percentage variance differing in each case. (a) ‘Phylogenetic variability’, based on a sample of 131 species of bats, primates and insectivores. PC1 accounts in total for 96.47 and PC2, 2.61%. (b) ‘Individual variability, Composite’ includes 47 individuals whose scores were entered as deviations from cell means so as to exclude species and sex differences, where the cells were six male wild mink, six female wild mink, six male domestic mink, six female domestic mink, six wild pigs of unknown sex, six domestic pigs of unknown sex and 11 mouse strains. PC1 accounts for 72.48% of the variance, and PC2 for 7.9%. For the individual species, (c) pig, (d) mink and (e) mouse, data are plotted so that their overall pattern might be examined, but no statistical claims about factor loadings on individual structures are made at the individual species level.
The regularity of allometric brain relationships across mammals is echoed in a corresponding conservation of patterns of neurogenesis [36]. In a pattern that we termed ‘late equals large’, the latest-generated neuronal populations in a conserved order of mammalian development are the ones that show hyperallometry. Extending the duration of development to produce a larger brain has the greatest effect on the relative numbers of cells in precursor pools generating cells for the longest duration, such as those producing the cortex or cerebellum. Furthermore, the duration of neurogenesis maps onto an axis in the conserved embryonic brain plan in vertebrates such that the medially located, ‘basal’ cell groups in brain segments (‘segments’ here are spinal cord segments, rhombomeres and prosomeres) cease precursor generation early. Laterally located, ‘alar’ groups stop last, or in the case of hippocampus and olfactory bulb continue into adulthood [38].
Taxa included in the phylogenetic analysis of brain variability are very diverse, including species of mega- and microbats, shrews, armadillos, polar bears, llamas, humans and manatees, with brain sizes ranging over 20 000-fold [35]. Within any particular species, brain sizes will only range over a tiny fraction of this amount, but as phylogenetic variability must arise from the heritable components of individual variability, it is entirely reasonable to ask what aspects of phylogenetic variability in brains are mirrored in individual variability. One excellent source of brain volume measurements of multiple individual members of single species exists, measured in a way comparable with the original Stephan dataset and the Reep extended dataset. Dieter Kruska measured a variety of individual animals to study the effects of domestication. He compared a sample of six wild boars with six domestic swine (brain sizes range from 92 to 204 g, ratio 2.21 [61]) and 12 adult wild mink brains with 12 adult ranch mink brains (brain sizes range from 7.2 to 10.4 g, ratio 1.44 [62]; the electronic supplementary material, table S1). He compared a number of other domestic and wild species with fewer individuals (reviewed and discussed in [63–65]). In the present analysis, our interest was not domestication per se, but the availability of measurements of a number of individuals of the same species with the bonus of the added variation produced by domestication. In addition, morphometric analyses of a wide variety of mouse brains used for genetic analyses are now available from an online database ([66]; Box 1 includes individual strain descriptions). In this case, single individual examples of 11 different strains were chosen (ranging from 0.30 to 0.53 g, ratio 1.8), using the neuroanatomic delineations identical to those described in Reep et al. [35]. These individuals were examined for the same principal component structure examined previously, considering as covariates species, sex and domestication, as described in Reep et al. [35] and Finlay & Darlington [36] (brain measurements, table 1).
Box 1. Mouse strain details.
In general, most strains are wild-derived inbred strains with, at most, inducible non-neural diseases. Only WSB/Ei and CAST/Ei are referred to as wild strains.
CAST/Ei—is referred to as a wild strain (not inbred) and is often used as a control line. In a study characterizing behavioural phenotypes, CAST/Ei always came out somewhere between inbred strains, with no significant behavioural phenotype evident from the approximately 13 tests described [67,68].
CASA/Rk—wild-derived inbred strain, no abnormal phenotype.
Molf/Ei—wild-derived inbred strain, which has no abnormal behavioural phenotype but is extremely susceptible to infection with Salmonella typhimurium [69].
SWR/J—wild-derived inbred strain, which is susceptible to chemically induced colorectal cancer, but responds well to the anti-tumour drug lentinan [70,71]. The SWR/J strain also exhibits more extensive corneal clouding after UV exposure than other inbred strains do, and control SWR/J mice exhibits a low activity variant phenotype for the major ocular aldehyde dehydrogenase (ALDH) AHD-4, and decreased levels of soluble protein in corneal extracts. Theory: ALDH assists the cornea in protecting the eye against ultraviolet radiation-induced tissue damage [72].
WSB/Ei—wild strain very rarely used.
SM/J—non-diabetic [73], small inbred strain. ‘In the mouse the naturally occurring inbred strain SM/J presents with a number of phenotypic abnormalities that have been attributed to reduced neuraminidase activity. SM/J mice were originally characterized by their altered sialylation of several lysosomal glycoproteins. This defect was linked to a single gene, neu-1, on chromosome 17, which was mapped by linkage analysis to the H-2 locus. In addition, these mice have an altered immune response that has also been coupled to a deficiency of the Neu-1 neuraminidase. Here, we report the identification in SM/J mice of a single amino acid substitution (L209I) in the Neu-1 protein that is responsible for the partial deficiency of lysosomal neuraminidase.’ [74]. Also, ‘Compared with other inbred strains, SM/J mice have both abnormally high responses to B cell mitogens and hyper NK cell and K cell activity.’ [75].
RIIIS/J—inbred strain—‘highly susceptible to collagen-induced arthritis’ [76] and ‘produce low antibody responses to several polysaccharide Ag of bacterial origin.’ [77].
SJL/J—inbred strain—not much information, no abnormal phenotype described.
Molc/Rk—inbred strain—a light bellied-agouti.
PL/J—inbred strain with susceptibility to skin disease—‘Psoriasis is a frequently occurring inflammatory skin disease characterized by thickened erythematous skin that is covered with silvery scales. It is a complex genetic disease with both heritable and environmental factors contributing to onset and severity. The CD18 hypomorphic PL/J mouse reveals reduced expression of the common chain of β2 integrins (CD11/CD18) and spontaneously develops a skin disease that closely resembles human psoriasis. In contrast, CD18 hypomorphic C57BL/6J mice do not demonstrate this phenotype.' [78].
Thr/ft—wild-derived inbred strain—no abnormal phenotype was described.
Table 1.
Volumes of brain structures for eleven mouse strains. Definitions of the inclusions of each structure, which together comprise the entire brain, can be found in Reep et al. [35].
| mouse strains |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAST/Ei | CASA/RK | Molf/Ei | SWR/J | WSB/Ei | Thr/ft | SM/J | RIIIS/J | CAST/Ei 433a | SJL/J | MOLC/Rk | PL/J | |
| structure volume (mm3) | ||||||||||||
| mes | 32.23 | 28.19 | 25.38 | 32.88 | 34.51 | 48.8 | 41.5 | 39.06 | 47.2 | 34.6 | 38.6 | 35.9 |
| di | 34.75 | 32.58 | 25.24 | 29.51 | 46.45 | 50.07 | 47.28 | 42.61 | 34.2 | 28.5 | 32 | 37.5 |
| cer | 51.87 | 54.19 | 45.47 | 49.69 | 61.06 | 80.86 | 67.1 | 77.69 | 53.1 | 54.9 | 59.7 | 58.8 |
| med | 41.8 | 43.24 | 36.09 | 43.91 | 39.54 | 55.7 | 47.8 | 55.88 | 33.1 | 35.64 | 42.5 | 45.3 |
| tel | 208.469 | 218.98 | 163.48 | 230.29 | 251.218 | 295.57 | 253.5 | 255.1 | 235.1 | 224.21 | 218.8 | 230.5 |
| sept | 4.28 | 3.76 | 2.53 | 3.89 | 8.628 | 6.4 | 6.4 | 6.7 | 5.9 | 4.85 | 4.9 | 4.4 |
| schiz | 6.04 | 8.15 | 6.99 | 8.08 | 9.67 | 11.9 | 11.4 | 8 | 11.5 | 7.2 | 6.4 | 6.8 |
| striat | 23.42 | 24.12 | 20.9 | 28.34 | 29.4 | 40.4 | 28.3 | 32.6 | 25.1 | 27.3 | 21.9 | 27.9 |
| obulb | 20.14 | 17.86 | 16.1 | 20.34 | 14.09 | 32.8 | 27.6 | 23.4 | 23.7 | 22.36 | 25.9 | 18.5 |
| hippo | 25.43 | 21.3 | 14.7 | 20.83 | 31.14 | 30.9 | 26.1 | 26.8 | 33.2 | 21.2 | 26.8 | 20.8 |
| neo | 88.37 | 103.69 | 77.78 | 103.3 | 114.86 | 122.1 | 104.8 | 110.3 | 93.5 | 97.7 | 95.8 | 111.9 |
| palaeo | 40.789 | 40.1 | 24.48 | 45.51 | 43.43 | 51.07 | 48.9 | 47.3 | 42.2 | 43.6 | 37.1 | 40.2 |
4. Principal component structure and allometric scaling in individual brain variability
(a). First principal component
The factor loadings for phylogenetic variability already described are shown in figure 1a, and for individual variability in figure 1b. In each case, the per cent factor loading on each brain structure within the total variance of each component is plotted. The first principal component (PC1; black bars) accounts for much more variance (phylogenetic variability, 96%; individual variability, 72%) than the second component (white bars; 3 and 8%, respectively). The principal component analysis is also broken down by individual species in figure 1c–e. Although the number of individuals is too small for statistical comparison between species, these graphs are included so that the relationship of individual species' data to massed data can be examined. Comparing the individual analysis with the phylogenetic analysis, the PC1 loads on a similar range of brain parts. Though the total amount of variance explained is less than in the phylogenetic analysis, 72.48 per cent is remarkable considering the 20 000-fold range in the absolute brain sizes of the phylogenetic dataset versus the approximately twofold range of within-species variability. The similarity of the structure of the variance is the more striking in that this dataset includes the peculiar effects of directed selection for different aspects of domestication, and various indeterminate effects of laboratory rearing on the mouse strains, and not ‘natural’ selection.
(b). Second and third components
The second principal component of variation (figure 1; white bars) loads most strongly on the olfactory bulb in both cross-species and individual cases. This component contributes more highly to total variance in the individual than in the phylogenetic comparison, 7.9 per cent versus 3 per cent, explaining 86 per cent of the residual variance not accounted for by the first factor. In each of the individual species, (i.e. pig, mink and mouse), the second principal component loads most highly on olfactory bulb though loading on other brain subcomponents varies widely.
We further examined the relationship of body size to brain components, because a relationship of body mass to medulla and mesencephalon, partialing out brain size, has been noted previously. Because individual body weight was not available for the mice, a regression analysis controlling for the effects of species, sex and domestication was done on the remaining animals. Body weight correlated highest with medulla (r = 0.198) and second highest with mesencephalon (r = 0.1143). Since the correlation coefficient for medulla is 74 per cent higher than even the second highest value for the mesencephalon, we can be reasonably confident that individual variability also echoes cross-species variability, in both cases, a small effect.
(c). Mice follow the pattern, but some mouse strains are outliers
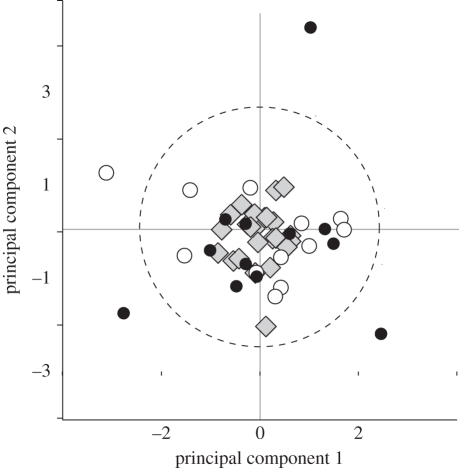
An interesting feature to the pattern of variability for the first and second principal components is plotted in figure 2. For each individual pig, mink and mouse, a point is plotted for their value of PC1 and PC2. Of the four individual animals lying outside the 90 per cent confidence circle, three are commonly used strains of laboratory mice. Two are aberrant on PC1, one loading abnormally high (WSB/ei) and one abnormally low (MOLF/ei). This means that for WSB/ei, the major brain parts associated with the PC1 (cortex, cerebellum and so forth) are unusually invariant with respect to each other (two standard deviations from the mean), while MOLF/ei is unusually variable. The third mouse strain, abnormally high on PC2[MOLC/ei], has large and highly correlated limbic system components, five deviations from the mean. What feature of the ‘ordinary’ domestication of minks and pigs retains the factorial structure in brain variation resembling phylogenetic variability, which appears to be lost in part in the form of selection exerted on individual mouse strains, is not clear. This observation suggests that unusual variation in brain organization should be considered as a factor in the research use of these strains.
Figure 2.
Values of PC1 and PC2, expressed as a z-score for each individual animal for the population of 47 individuals. Three of the mice and one pig fall outside the 90% delimiter (dashed line); these mouse strains are WSB/ei, MOLC/ei and MOLf/ei. Mouse strains may be intrinsically more variable because of greater genetic difference than the minks and pigs, whose lineages are unknown, but whatever the cause, their deviation from the phylogenetic and the other individuals is pronounced. Diamonds, minks; open circles, pigs; filled circles, mice.
A recent magnetic resonance imaging study of recombinant inbred individuals of two strains of mice for the purposes of identifying genetic influences on brain volumes and neuron numbers used different subdivisions of brain from those employed here, but retrieves a generally similar structure, though the range in brain sizes is reduced still further [44]. The PC1 in both strains loads highest on cortex, and the third highest on olfactory bulb; the second principal component loads highest on midbrain and medulla, reversing the order of the second and third components we observed. Earlier work of this same group [41] strongly linked overall control of brain neuron number to gene regions related to transcription factors and to overall somatic growth, consistent with the idea of the PC1 of the brain region related to duration or rate of development linking the proliferation of all brain regions. Another study of control of neuron number and size of the olfactory bulb noted that the bulb size (unlike the brain) was highly variable, was related to sex and experience, and also changed in volume late in maturation [79]. Four different genetic loci from the one controlling whole brain size were related to olfactory bulb variation.
(d). Cortex hyperallometry
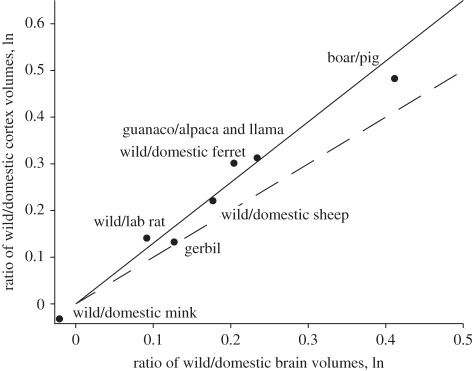
Finally, the hyperallometry of the cortex can be examined in this dataset by comparison of the various domesticated and wild animals studied by Kruska [62,64,65]. We confirm his observations of the relative greater reduction of the cortex compared with whole brain volume. We are able to further describe the relative reduction in cortex size of the domesticated species compared with our cross-species database (figure 3). In general, the effect of domestication is a decrease in relative brain size to body size or encephalization, ranging from a 26 per cent reduction from the wild guanaco to the domesticated alpaca or llama to equality between the wild and domesticated minks; no pair has the reverse relationship. The solid line plotted in figure 3 is the regression line of slope of 1.3 previously computed for the phylogenetic relationship of neocortex volume to the total brain volume [35], not the regression line for these data. For visual comparison, the dashed line plots the slope of 1.0, the line that would be observed if the cortex decreased isometrically with the total brain volume. Five of the seven comparisons fall on or above equal proportionality. So, as domesticated species decrease in relative brain volume, they decrease disproportionately in neocortex volume, in accord with the phylogenetic pattern.
Figure 3.
Comparison of the ratio of cortex to total brain volume in seven wild/domestic comparisons gathered by Kruska, compared with equal proportionality (dashed line) or to cortex hyperallometry determined phylogenetically (solid line). In five of the seven comparisons, cortex volume is reduced more than overall brain volume.
(e). Species differences and ‘principal component 2’
Both across and within mammalian taxa, the volume of the neocortex (the structure loading highest on PC1) versus the volume of PC2 ‘limbic’ structures (olfactory bulb and cortex, hippocampus, amygdala and septum), both compared with the rest of the brain, are negatively correlated [35]. Various multi-variate analyses of the initial Stephan dataset (e.g. [5,36,50] and others) capture this same contrast. The negative correlation of the volume of limbic structures and the cortex contributes to a large number of studies showing mosaic, part-by-part, or system-by-system covariation (e.g. [80,81]). The olfactory bulb and hippocampus have the further unusual feature of continued neurogenesis into adulthood in mammals, which is one clear substrate for their linkage to sex and experiential factors [79]. In the very large number of studies linking hippocampus size to niche and foraging strategy, in mammals and in birds, across and within species, the whole brain or some adjacent structure is typically used for calibration of hippocampus volume, so it is unclear if the hippocampus varies independently from the rest of ‘PC2’ in all cases [56,57]; see reviews in [58,82,83]. Of the various contrasts that have been made in the studies of hippocampus, avian versus mammalian, foraging range and the presence of hording, seasonality of foraging and gender, it would be interesting to see if some contrasts claim the entire set of ‘principal component 2’ structures, and if some are specific to the hippocampus. In any case, the two dominant components of mammalian brain variation, which we have recently shown also characterize chondrichthyans (sharks and rays; [51]), are an enduring ‘hot spot’ for species-typical variation.
5. A computational context for brain variation
The close resemblance of phylogenetic variation to individual variation was quite surprising, given the orders of magnitude differentiating the variation of phylogenetic and individual samples. Reiterating, we find consistent covariation in the relative size of brain parts with respect to each other. We find the same dissociation, the independent variation of olfactory bulb with respect to the rest of the brain. Finally, we find the predicted disproportionate reduction of the neocortex with respect to the total brain volume, in those domesticated species that have regressed in total brain volume. The similarity is the more impressive in that domestication and laboratory animal membership should minimally exert ‘atypical’ genetic pressure on the individuals of these species. Independent of genetics and any kind of selection, domestication itself should have large effects on rearing conditions, nutrition and general experience of individual animals likely to confound an analysis of brain variability, not enhance it. Of course, the seeds of phylogenetic variability must be found in individual variability, but it is surprising that the pattern of phylogenetic variability should conform in such detail. The morphological evolution of stickleback species, where individual variation matched species variation, over both short and long phylogenetic differences, closely resembles these results [28].
(a). Brain architectures that scale gracefully
The hyperallometry of cortex (and cerebellum), which allocates the profits of extended brain production preferentially to multi-functional brain components, may be useful both at the individual and species level. We can take some hints from current computing research, where it is of obvious practical use to develop computing structures in which more elements can be added as necessary, or where an architecture is desired which can lose components and remain at least partially functional (as in warfare). These scalable computer architectures, or ‘subsumption architectures’, allow for the addition and subtraction of components gracefully without interference in fundamental operations [84,85]. One basic insight arising from this literature is that locating new computational circuits directly in command lines executing central functions impairs processing speed and prohibits scaling, but locating new computational power as ancillary loops modifying basic functions improves speed and enables scaling. Restating, if more computing power is located between sensory input and motor output, it slows the entire device and makes it vulnerable to damage at any point. If, however, a ‘model brain’ is produced beside the basic command lines but able to intercept and modify commands as they are made, a more robust architecture with more computing power and no loss in speed is the result. This, of course, is a good description of the computational position of most of the cortex and cerebellum. Since this computational claim is a property about brain scaling in general, and not taxonomic levels of brain organization, it should thus hold true for variations in brain growth, adult variations in brain size and species level. To draw an analogy with our original colour vision example, while it might be extremely useful for a monkey to rapidly recognize bananas, placing a banana-recognition device in the retina may never be computationally feasible if the design requirements of the whole brain are considered. Testing this hypothesis about the ‘evolvability’ of particular types of circuit organization will eventually require examination of circuits known to range in size in diverse vertebrate and invertebrate groups, and comparison of their properties.
Considering the partial dissociation of the olfactory bulb and limbic structures, we speculate that the pattern of variation at the individual level may be either or both the source and product of evolution. Evolution must certainly serve as a filter against deleterious variations, but might pass on neutral ones. We speculate that the independent variation of olfactory bulb from the rest of the brain may be not so much selection for olfactory variability, but rather selection for tighter coupling of the other sensory systems that must share thalamic projections and neocortical representations. Modelling an independent enlargement of a single defined ‘module’ in a neural net consumed disproportionate amounts of the neural resources of the other modules sharing the same net resources as its excessive input–output requirements propagated through the net [86]. Sequestering the variation of the single mammalian brain part that can vary its size by generation of neurons throughout life may aid this computational requirement.
6. Two large classes of behavioural variation?
Returning to our initial discussion of brain variability and its behavioural correlates, we suggest there are two very different kinds of change typical in brain evolution. The first is the general increase in computational power and flexibility afforded by a large brain, which can be measured in the very most general ways: surviving longer by avoiding predation, exploiting new food sources and learning new strategies to find shelter, attract mates and protect offspring [32]. These classes of behaviour are only incidentally niche- and modality dependent, and would be ill served by strong modular commitments. Variation in the relative size of cortex among humans has a small but significant relationship to ‘general intelligence’. As yet, no such relationship between brain size and general behavioural capacity has been shown at the individual level for any other species; the work by Lefebvre et al. [32] compares species, but the relationship should hold for individuals as well. Overall, for general problems, the demands of general-purpose architectures should predominate, and should be seen both in individual and species variations, the kind we describe here.
On the other hand, species-typical behaviours of every possible sort have evolved. On the sensory side, we have everything from variants of colour vision, to whole new sensory systems like electroreception in mammals, to sensorimotor combinations like echolocation. On the motor side, we have crawling, flying and competitive gymnastics. Every variation of social preference and aggression exists, generating from individual preferences lifelong monogamy, cooperative predation, solitary individuals and herds. The data here do not bear directly on the mechanisms of these changes. However, evolutionary changes must occur largely within the numeric confines established by general brain scaling. Specializations in the sensory periphery impose their structure on the brain, causing the representation of the whole body surface to disproportionately represent the specialization, from the palpating nose of the star-nosed mole, to hands and tongues, to whiskers and tusks. Many such cases of redirection of generic brain structure are well described (e.g. [87–90]). These allocations may develop in infancy, or may dynamically relocate as required, as in the reallocation of the visual cortex for Braille reading even in the late blind [91].
A second source of rich variability is just beginning to be investigated, the churning of the nonapepetide modulator and receptor distribution in basal forebrain and midbrain networks, where small changes in expression patterns give rise to large changes in the motivational and attentional structure of individual animals, likely to result in the emergence of new social organization in populations [92,93]. The third source of evolutionary behavioural variation is intrinsically developmental: over and over again, although the cortex comes equipped with initially highly specified input connectivity, it has been shown that computational space is allocated in cortex on the basis of activity, from micro- to macro-scales (reviewed in [94]). Since the source of our information on this has often come from developmental accidents of deafness or blindness [91,95,96] or deliberate experimental subtractions [97], the plasticity observed is often seated in the context of recovery from pathology. Dynamic reallocation of function according to relative activity, however, both during development and at adulthood (e.g. [98]) is probably the normal state of affairs, and should be studied more systematically in non-manipulated brain, and as an emergent property in species adaptations. The massive re-use of brain tissue implied by imaging studies, showing the same tissue lighting up repeatedly in diverse contexts and tasks (e.g. [99,100]), is probably no artefact of experimental design or poor task distinctions, but a basic operating principle of a general purpose device put to use in multiple adaptive contexts.
7. Evo-devo, the brain and behaviour
We can now return to highlight how some features of brain scaling may be examples of the facilitated variability and evolvability that have been much discussed in the evo-devo literature. In general, in the mammals discussed here, simple duration of brain growth is tightly linked to brain size [101]. Changes in the duration of growth (of genetic, but possibly environmental origin as well) do not produce isometric changes in all brain parts, but allocate volume changes preferentially to the structures produced over the longest embryonic duration. In neural development, the basal-to-alar axis of repeating brain segments (the embryonic medial-to-lateral direction) is the axis in which increasing duration of neurogenesis is roughly represented [35,38]. While this conserved axis could be argued to be a ‘developmental constraint’ on brain structure, we have presented arguments here that a brain axis in which duration of neuron production systematically varies may be essential to a useful brain architecture, one that scales gracefully and is robust to damage.
Note also that most of the mechanisms that transform initial, ‘generic’ brain architecture into a species-specific instantiation are developmental or epigenetic mechanisms, which take key changes, often in the sensory and motor periphery, and amplify their effects throughout the nervous system. We return to our visual system example. Rather than producing a ‘more visual’ primate with trichromatic colour vision and fovea by adding parts—generating more cells in the fovea, a new set of photoreceptors, more stages of processing, a larger-than-expected lateral geniculate, larger primary visual cortex, genetically specified ‘colour vision modules’ and so forth—alterations are made within the overall scaling architecture [102,103]. Nevertheless, the restructuring and processing changes are profound. To produce the high visual acuity of the primate fovea, the retina is differentially stretched, a topological rather than additive change, to compact cells in the fovea and spread out the cells of the periphery, conserving the ‘expected’ number of cells in the retina. Consequently, processing resources of the cortex are concentrated automatically on the central few degrees of the visual field. A new opsin is expressed, but adds no total photoreceptors to the retina, subdividing the initial set. The new chromatic information is then analysed by the generalized comparator mechanism for wavelength already present in the animal [20]. More interest and attention to visual features, perhaps mediated by subcortical changes in motivation, will cause activity-dependent processes to allocate more and more ‘brain space’ to visual information. Aspects of modularity may emerge in cortex, for example, for colour processing, or ‘face areas’ from the same Hebbian ‘fire together, wire together’ processes. Some of these changes arise from the immediate epigenetic effects of connecting up the nervous system, but many more depend on later interactions with the environment, guided by the animal's attentional and motivational preferences. The study of brain evolution and special behavioural adaptations may essentially become the study of guided brain development.
Acknowledgements
Supported by NSF DBI0848612 to B. Finlay. We thank Christine Charvet for her help with the production of this manuscript, and also the three anonymous reviewers for their clarifying suggestions.
Footnotes
One contribution of 10 to a Theme Issue ‘Evolutionary developmental biology (evo-devo) and behaviour’.
References
- 1.Dukas R. 2004. Evolutionary biology of animal cognition. Annu. Rev. Ecol. Evol. Syst. 35, 347–374 10.1146/annurev.ecolsys.35.112202.130152 (doi:10.1146/annurev.ecolsys.35.112202.130152) [DOI] [Google Scholar]
- 2.Jerison H. J. 1973. Evolution of the brain and intelligence. New York, NY: Academic Press [Google Scholar]
- 3.Stephan H., Baron G., Frahm H. D. 1988. Comparative size of brain and brain components. In Comparative primate biology, pp. 1–38 New York, NY: Alan R. Liss [Google Scholar]
- 4.Dunbar R. I. M. 1993. Coevolution of neocortical size, group size and language in humans. Behav. Brain Sci. 16, 681–694 10.1017/S0140525X00032325 (doi:10.1017/S0140525X00032325) [DOI] [Google Scholar]
- 5.Barton R. A., Harvey P. H. 2000. Mosaic evolution of brain structure in mammals. Nature 405, 1055–1058 10.1038/35016580 (doi:10.1038/35016580) [DOI] [PubMed] [Google Scholar]
- 6.Striedter G. 2005. Principles of brain evolution. New York, NY: Sinauer [Google Scholar]
- 7.Jacobs G. H. 1993. The distribution and nature of colour vision among the mammals. Biol. Rev. 68, 413–471 [DOI] [PubMed] [Google Scholar]
- 8.Jacobs G. H. 2008. Primate color vision: a comparative perspective. Vis. Neurosci. 25, 619–633 10.1017/S0952523808080760 (doi:10.1017/S0952523808080760) [DOI] [PubMed] [Google Scholar]
- 9.Hunt D. M., Carvalho L. S., Cowing J. A., Davies W. L. 2009. Evolution and spectral tuning of visual pigments in birds and mammals. Phil. Trans. R. Soc. B 364, 2941–2955 10.1098/rstb.2009.0044 (doi:10.1098/rstb.2009.0044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernald R. D. 1997. The evolution of eyes. Brain Behav. Evol. 50, 253–259 10.1159/000113339 (doi:10.1159/000113339) [DOI] [PubMed] [Google Scholar]
- 11.Changizi M. A., Zhang Q., Shimojo S. 2006. Bare skin, blood and the evolution of primate colour vision. Biol. Lett. 2, 217–221 10.1098/rsbl.2006.0440 (doi:10.1098/rsbl.2006.0440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persons M. H., Fleishman L. J., Frye M. A., Stimphil M. E. 1999. Sensory response patterns and the evolution of visual signal design in anoline lizards. J. Comp. Physiol. A 184, 585–607 10.1007/s003590050358 (doi:10.1007/s003590050358) [DOI] [Google Scholar]
- 13.Osorio D., Vorobyev M. 2008. A review of the evolution of animal colour vision and visual communication signals. Vis. Res. 48, 2042–2051 10.1016/j.visres.2008.06.018 (doi:10.1016/j.visres.2008.06.018) [DOI] [PubMed] [Google Scholar]
- 14.Mollon J. D. 1989. ‘Tho she kneeled at the place they grew…’ The uses and origins of primate color vision. In Principles of sensory coding and processing (ed. Laughlin S. B.), pp. 21–38 Cambridge, UK: The Company of Biologists, Ltd [Google Scholar]
- 15.Dominy N. J., Lucas P. W. 2001. Ecological importance of trichromatic vision to primates. Nature 410, 363–366 10.1038/35066567 (doi:10.1038/35066567) [DOI] [PubMed] [Google Scholar]
- 16.Kingdom F. A. A. 2003. Color brings relief to human vision. Nat. Neurosci. 6, 641–644 10.1038/nn1060 (doi:10.1038/nn1060) [DOI] [PubMed] [Google Scholar]
- 17.Hansen T., Gegenfurtner K. R. 2009. Independence of color and luminance edges in natural scenes. Vis. Neurosci. 26, 35–49 10.1017/S0952523808080796 (doi:10.1017/S0952523808080796) [DOI] [PubMed] [Google Scholar]
- 18.Caine N. G., Mundy N. I. 2000. Demonstration of a foraging advantage for trichromatic marmosets (Callithrix geoffroyi) dependent on food colour. Proc. R. Soc. Lond. B 267, 439–444 10.1098/rspb.2000.1019 (doi:10.1098/rspb.2000.1019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs G. H., Williams G. A., Cahill H., Nathans J. 2007. Emergence of novel color vision in mice engineered to express a human cone photopigment. Science 315, 1723–1725 10.1126/science.1138838 (doi:10.1126/science.1138838) [DOI] [PubMed] [Google Scholar]
- 20.Mancuso K., Hauswirth W. W., Li Q., Connor T. B., Kuchenbecker J. A., Mauck M. C., Neitz J., Neitz M. 2009. Gene therapy for red-green colour blindness in adult primates. Nature 461, 784–787 10.1038/nature08401 (doi:10.1038/nature08401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerhart J., Kirschner M. 1997. Cells, embryos and evolution. Malden, MA: Blackwell Science [Google Scholar]
- 22.Jablonka E., Lamb M. 2005. Evolution in four dimensions. Cambridge, MA: MIT Press [Google Scholar]
- 23.Kirschner M. W., Gerhart J. C. 2005. The plausibility of life: resolving Darwin's dilemma. New Haven, CT: Yale University Press [Google Scholar]
- 24.Wagner A. D. 2005. Robustness and evolvability in living systems. Princeton, NJ: Princeton University Press [Google Scholar]
- 25.Westoby N., Falster D. S., Moles A. T., Vesk P. A., Wright I. J. 2002. Plant ecological strategies: some leading dimensions of variation between species. Annu. Rev. Ecol. Evol. Syst. 33, 125–129 10.1146/annurev.ecolsys.33.010802.150452 (doi:10.1146/annurev.ecolsys.33.010802.150452) [DOI] [Google Scholar]
- 26.Shapiro M. D., Marks M. E., Peichel C. L., Blackman B. K., Nereng K. S., Junsson B., Schluter D., Kingsley D. M. 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428, 717–723 10.1038/nature02415 (doi:10.1038/nature02415) [DOI] [PubMed] [Google Scholar]
- 27.Schluter D. 1984. Morphological and phylogenetic relations among the Darwin's finches. Evolution 38, 921–930 10.2307/2408428 (doi:10.2307/2408428) [DOI] [PubMed] [Google Scholar]
- 28.Schluter D. 1996. Adaptive radiation along the lines of least resistance. Evolution 50, 1766–1774 10.2307/2410734 (doi:10.2307/2410734) [DOI] [PubMed] [Google Scholar]
- 29.Mundy N. I., Badcock N. S., Hart T., Scribner K., Janssen K., Nadeau N. J. 2004. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science 303, 1870–1873 10.1126/science.1093834 (doi:10.1126/science.1093834) [DOI] [PubMed] [Google Scholar]
- 30.Allender C. J., Seehausen O., Knight M. E., Turner G. F. 2003. Divergent selection during speciation of Lake Malawi cichlids inferred from parallel radiations in nuptial coloration. Proc. Natl Acad. Sci. USA 24, 14 074–14 079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steiner C. C., Rompler H., Boettger L. M., Schoneberg T., Hoekstra H. E. 2009. The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol. Biol. Evol. 26, 35–45 10.1093/molbev/msn218 (doi:10.1093/molbev/msn218) [DOI] [PubMed] [Google Scholar]
- 32.Lefebvre L., Reader S. M., Sol D. 2004. Brains, innovations and evolution in birds and primates. Brain Behav. Evol. 63, 233–246 10.1159/000076784 (doi:10.1159/000076784) [DOI] [PubMed] [Google Scholar]
- 33.tramo M. J., Loftus W. C., Stukel T. A., Green R. L., Weaver J. B., Gazzaniga M. S. 1998. Brain size, head size, and intelligence quotient in monozygotic twins. Neurology 50, 1246–1252 [DOI] [PubMed] [Google Scholar]
- 34.Shaw P., Greenstein D., Lerch J., Clasen L., Lenroot R., Gogtay N., Evans A., Rapoport J., Giedd J. 2006. Intellectual ability and cortical development in children and adolescents. Nature 440, 676–679 10.1038/nature04513 (doi:10.1038/nature04513) [DOI] [PubMed] [Google Scholar]
- 35.Reep R., Darlington R. B., Finlay B. L. 2007. The limbic system in mammalian brain evolution. Brain Behav. Evol. 70, 57–70 10.1159/000101491 (doi:10.1159/000101491) [DOI] [PubMed] [Google Scholar]
- 36.Finlay B. L., Darlington R. B. 1995. Linked regularities in the development and evolution of mammalian brains. Science 268, 1578–1584 10.1126/science.7777856 (doi:10.1126/science.7777856) [DOI] [PubMed] [Google Scholar]
- 37.Finlay B. L., Darlington R. B., Nicastro N. 2001. Developmental structure in brain evolution. Behav. Brain Sci. 24, 263–307 [PubMed] [Google Scholar]
- 38.Finlay B. L., Hersman M. N., Darlington R. B. 1998. Patterns of vertebrate neurogenesis and the paths of vertebrate evolution. Brain Behav. Evol. 52, 232–242 10.1159/000006566 (doi:10.1159/000006566) [DOI] [PubMed] [Google Scholar]
- 39.Finlay B. L., Silveira L. C. L., Reichenbach A. 2005. Comparative aspects of visual system development. In The structure, function and evolution of the primate visual system (ed. Kremers J.), pp. 37–72 New York, NY: John Wiley and Sons [Google Scholar]
- 40.Dyer M. A., Martins R., da Silva Filho M., Muniz J. A., Silveira L. C. L., Cepko C., Finlay B. L. 2009. Developmental sources of conservation and variation in the evolution of the primate eye. Proc. Natl Acad. Sci. USA 106, 8963–8968 10.1073/pnas.0901484106 (doi:10.1073/pnas.0901484106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams R. W., Strom R. C., Goldowitz D. 1998. Natural variation in neuron number in mice is linked to a major quantitative trait locus on Chr 11. J. Neurosci. 18, 138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Airey D. C., Lu L., Williams R. W. 2001. Genetic control of the mouse cerebellum: identification of quantitative trait loci modulating size and architecture. J. Neurosci. 21, 5099–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu L., Airey D. C., Williams R. W. 2001. Complex trait analysis of the hippocampus: mapping and biometric analysis of two novel gene loci with specific effects on hippocampal structure in mice. J. Neurosci. 21, 3503–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Badea A., Johnson G. A., Williams R. W. 2009. Genetic dissection of the mouse brain using high-field magnetic resonance microscopy. NeuroImage 45, 1067–1079 10.1016/j.neuroimage.2009.01.021 (doi:10.1016/j.neuroimage.2009.01.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gleeson J. G., et al. 1998. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signalling protein. Cell 92, 63–72 10.1016/S0092-8674(00)80899-5 (doi:10.1016/S0092-8674(00)80899-5) [DOI] [PubMed] [Google Scholar]
- 46.Kingsbury M. A., Rehen S. K., Contos J. J. A., Higgins C. M., Chun J. 2003. Non-proliferative effects of lysophosphatidic acid enhance cortical growth and folding. Nat. Neurosci. 6, 1292–1299 10.1038/nn1157 (doi:10.1038/nn1157) [DOI] [PubMed] [Google Scholar]
- 47.Evans P. D., Gilbert S. L., Mekel-Bobrov N., Vallender E. J., Anderson J. R., Vaez-Azizi L. M., Tishkoff S. A., Hudson R. R., Lahn B. T. 2005. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science 309, 1717–1720 10.1126/science.1113722 (doi:10.1126/science.1113722) [DOI] [PubMed] [Google Scholar]
- 48.Hill R. S., Walsh C. A. 2005. Molecular insights into human brain evolution. Nature 437, 64–67 10.1038/nature04103 (doi:10.1038/nature04103) [DOI] [PubMed] [Google Scholar]
- 49.Vallender E. J. 2008. Exploring the origins of the human brain through molecular evolution. Brain Behav. Evol. 72, 168–177 10.1159/000151476 (doi:10.1159/000151476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gould S. J. 1975. Allometry in primates, with emphasis on scaling and the evolution of the brain. In Approaches to primate paleobiology (ed. Szalay F. S.), pp. 244–292 Basel, Switzerland: Karger; [PubMed] [Google Scholar]
- 51.Yopak K. E., Lisney T. J., Darlington R. B., Collin S. P., Montgomery J. C., Finlay B. L. 2010. A conserved pattern of brain scaling from sharks to primates. Proc. Natl Acad. Sci. USA 107, 12 946–12 951 10.1073/pnas.1002195107 (doi:10.1073/pnas.1002195107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duchaine B., Cosmides L., Tooby J. 2001. Evolutionary psychology and the brain. Curr. Opin. Neurobiol. 11, 225–230 10.1016/S0959-4388(00)00201-4 (doi:10.1016/S0959-4388(00)00201-4) [DOI] [PubMed] [Google Scholar]
- 53.Op de Beeck H. P., Baker C. I. 2010. The neural basis of visual object learning. Trends Cogn. Sci. 14, 22–30 10.1016/j.tics.2009.11.002 (doi:10.1016/j.tics.2009.11.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaskan P., Franco C., Yamada E., Silveira L. C. L., Darlington R., Finlay B. L. 2005. Peripheral variability and central constancy in mammalian visual system evolution. Proc. R. Soc B 272, 91–100 10.1098/rspb.2004.2925 (doi:10.1098/rspb.2004.2925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez-Barberia F. J., Shultz S., Dunbar R. I. M., Janis C. 2009. Evidence for coevolution of sociality and relative brain size in three orders of mammals. Evolution 61, 2811–2821 10.1111/j.1558-5646.2007.00229.x (doi:10.1111/j.1558-5646.2007.00229.x) [DOI] [PubMed] [Google Scholar]
- 56.Jacobs L. F., Spencer W. D. 1994. Natural space-use patterns and hippocampal size in kangaroo rats. Brain Behav. Evol. 44, 125–132 10.1159/000113584 (doi:10.1159/000113584) [DOI] [PubMed] [Google Scholar]
- 57.Clayton N. S. 1995. Development of memory and the hippocampus: comparison of food-storing and nonstoring birds on a one-trial associative memory task. J. Neurosci. 15, 2796–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherry D. F., Hoshooley J. S. 2010. Seasonal hippocampal plasticity in food-storing birds. Phil. Trans. R. Soc. B 365, 933–943 10.1098/rstb.2009.0220 (doi:10.1098/rstb.2009.0220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stephan H., Manolescu J. 1980. Comparative investigations on hippocampus in insectivores and primate. Z mikrosk-anat Forsch. 94, 1025–1050 [PubMed] [Google Scholar]
- 60.Fox J. H., Wilczynski W. 1986. Allometry of major CNS divisions: towards a reevaluation of somatic brain-body scaling. Brain Behav. Evol. 28, 157–169 10.1159/000118700 (doi:10.1159/000118700) [DOI] [PubMed] [Google Scholar]
- 61.Kruska D. 1970. Vergleichend cytoarchitektonische Untersuchungen an Gehirnen von Wild- und Hausschweinen. Z. Anat. Entwickl. Gesch. 131, 291–324 10.1007/BF00519973 (doi:10.1007/BF00519973) [DOI] [PubMed] [Google Scholar]
- 62.Kruska D. 1996. The effect of domestication on brain size and composition in the mink (Mustela vison). J. Zool. 239, 645–661 10.1111/j.1469-7998.1996.tb05468.x (doi:10.1111/j.1469-7998.1996.tb05468.x) [DOI] [Google Scholar]
- 63.Kruska D. 1980. Domestikations bedingte Hirngrosse nanderungen bei Saugetieren. J. Zool. Syst. Evol. Res. 18, 161–195 10.1111/j.1439-0469.1980.tb00738.x (doi:10.1111/j.1439-0469.1980.tb00738.x) [DOI] [Google Scholar]
- 64.Kruska D. 1988. Mammalian domestication and its effect on brain structure and behavior. In Intelligence and evolutionary biology (eds Jerison H. J., Jerison I.). Berlin, Germany: Springer [Google Scholar]
- 65.Kruska D. C. 2005. On the evolutionary significance of encephalization in some eutherian mammals: effects of adaptive radiation, domestication, and feralization. Brain Behav. Evol. 65, 73–108 10.1159/000082979 (doi:10.1159/000082979) [DOI] [PubMed] [Google Scholar]
- 66.Williams R. W. 2007. Mouse brain library. See http://www.mbl.org/
- 67.Koide T., Moriwaki K., Ikeda K., Niki H., Shiroishi T. 2000. Multi-phenotype behavioural characterization of inbred strains derived from wild stocks of Mus musculus. Mamm. Genome 11, 664–670 (doi:10.1007/s003350010129) [DOI] [PubMed] [Google Scholar]
- 68.Santos J., Cole Y., Pellicer A. 1993. Phylogenetic relationships among lab and wild-origin Mus musculus strains on the basis of genomic DNA RFLPs. Mamm. Genome 4, 485–492 (doi:10.1007/BF00364782) [DOI] [PubMed] [Google Scholar]
- 69.Sebastiani G., Olien L., Gauthier S., Skamene E., Morgan K., Gros P., Malo D. 1998. Mapping of genetic modulators of natural resistance to infection with Salmonella typhimurium in wild-derived mice. Genomics 47, 180–186 (doi:10.1006/geno.1997.5116) [DOI] [PubMed] [Google Scholar]
- 70.Maeda Y. Y., Takahama S., Kohara Y., Yonekawa H. 1996. Two genes controlling acute phase responses by the antitumor polysaccharide, lentinan. Immunogenetics 43, 215–219 (doi:10.1007/s002510050048) [DOI] [PubMed] [Google Scholar]
- 71.Rosenberg D. W. 1995. Non/homogeneous marking of distal colonic mucosa using Dolichos biflorus lectin. Cancer Lett. 98, 33–37 (doi:10.1016/S0304-3835(06)80007-8) [PubMed] [Google Scholar]
- 72.Downes J. E., Schwann P. G., Holmes R. S. 1994. Differential corneal sensitivity to ultraviolet light among inbred strains of mice: correlation of ultraviolet B sensitivity with aldehyde dehydrogenase deficiency. Cornea 13, 67–72 [DOI] [PubMed] [Google Scholar]
- 73.Kobayashi M., Lo F., Kawai T., Kumazawa M., Ikegami H., Nishimura M., Ohno T., Horio F. 2006. Major quantitative trait locus on chromosome 2 for glucose tolerance in diabetic SMXA-5 mouse established from non-diabetic SM/J and A/J strains. Diabetologia 49, 431–433 (doi:10.1007/s00125-005-0121-3) [DOI] [PubMed] [Google Scholar]
- 74.Rottier R. J., Bonten E., d'Azzo A. 1998. A point mutation in the neu-1 locus causes the neuraminidase defect in the SM/J mouse. Hum. Mol. Genet. 7, 313–321 (doi:10.1093/hmg/7.2.313) [DOI] [PubMed] [Google Scholar]
- 75.Clark E. A., Engel D., Windsor N. T. 1981. Immune responsiveness of SM/J mice: hyper NK cell activity mediated by NK 1+ Qa 5− cells. J. Immunol. 127, 2391–2395 [PubMed] [Google Scholar]
- 76.Nandakumar K. S., Holmdahl R. 2005. A genetic contamination in the MHC-congenic mouse strains reveals a locus on the chromosome 10 that determines autoimmunity and arthritis susceptibility. Eur. J. Immunol. 35, 1275–1282 (doi:10.1002/eji.200425925) [DOI] [PubMed] [Google Scholar]
- 77.Hiernaux J. R., Goidl E. A., McEvoy S. J., Stashak P. W., Baker P. J., Holmes K. L.1989. Characterization of the immunodeficiency of RIIIS/J mice. I. Association with the CD5 (LY-1) B cell lineage. J. Immunol. 142, 1813–1817 [PubMed] [Google Scholar]
- 78.Kess D., et al. 2006. Identification of susceptibility loci for skin disease in a murine psoriasis model. J. Immunol. 177, 4612–4619 [DOI] [PubMed] [Google Scholar]
- 79.Williams R., Airey D., Kulkarni A., Zhou G., Lu L. 2001. Genetic dissection of the olfactory bulbs of mice: QTLs on four chromosomes modulate bulb size. Behav. Genet. 31, 61–77 10.1023/A:1010209925783 (doi:10.1023/A:1010209925783) [DOI] [PubMed] [Google Scholar]
- 80.Barton R. A. 1998. Visual specialization and brain evolution in primates. Proc. R. Soc. Lond. B 265, 1933–1937 10.1098/rspb.1998.0523 (doi:10.1098/rspb.1998.0523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barton R. A., Aggleton J. P., Grenyer R. 2003. Evolutionary coherence of the mammalian amygdala. Proc. R. Soc. Lond. B 270, 539–543 10.1098/rspb.2002.2276 (doi:10.1098/rspb.2002.2276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brodin A. 2010. The history of scatter hoarding studies. Phil. Trans. R. Soc. B 365, 869–881 10.1098/rstb.2009.0217 (doi:10.1098/rstb.2009.0217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roth T. C., Brodin A., Smulders T. V., LaDage L. D., Pravosudov V. V. 2010. Is bigger always better? A critical appraisal of the use of volumetric analysis in the study of the hippocampus. Phil. Trans. R. Soc. B 365, 915–931 10.1098/rstb.2009.0208 (doi:10.1098/rstb.2009.0208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brooks R. 1986. A robust layered control system for a mobile robot. IEEE J. Robot. Autom. 2, 14–23 10.1109/JRA.1986.1087032 (doi:10.1109/JRA.1986.1087032) [DOI] [Google Scholar]
- 85.Hawes N., Sloman A., Wyatt J., Zillich M., Jacobsson A., Kruijff G., Brenner M., Berginc G., Skočaj D. 2007. Towards an integrated robot with multiple cognitive functions. Proc. Assoc. Adv. Artifical Intell. 7, 1–6 [Google Scholar]
- 86.Finlay B. L., Brodsky P. B. 2006. Cortical evolution as the expression of a program for disproportionate growth and the proliferation of areas. In Evolution of nervous systems (eds Kaas J. H., Krubitzer L. A.), pp. 73–96 Oxford, UK: Academic Press [Google Scholar]
- 87.Suga N., Kuzirai K., O'Neill W. E. 1981. How biosonar information is represented in the bat cerebral cortex. In Neuronal mechanisms of hearing (eds Syka J., Aitken L.). New York, NY: Plenum Press [Google Scholar]
- 88.Van der Loos H., Welker E. 1985. Development and plasticity of somatosensory brain maps. In Development, organization, and processing in somatosensory pathways; neurology and neurobiology (eds Rowe M., Willis W. D., Jr), pp. 53–67 New York, NY: Alan R. Liss [Google Scholar]
- 89.Silveira L. C. L., Pincanco-Diniz C. W., Sampaio L. F. S., Oswaldo-Cruz E. 1989. Retinal ganglion cell distribution in the Cebus monkey: a comparison with the cortical magnification factors. Vis. Res. 29, 1471–1483 10.1016/0042-6989(89)90131-4 (doi:10.1016/0042-6989(89)90131-4) [DOI] [PubMed] [Google Scholar]
- 90.Catania K. C. 2002. Barrels, stripes and fingerprints in the brain: implications for theories of cortical organization. J. Neurocytol. 31, 347–358 10.1023/A:1024186329012 (doi:10.1023/A:1024186329012) [DOI] [PubMed] [Google Scholar]
- 91.Burton H. 2003. Visual cortex activity in early and late blind people. J. Neurosci. 23, 4005–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Insel T. R., Young L. J. 2000. Neuropeptides and the evolution of social behavior. Curr. Opin. Neurobiol. 10, 784–789 10.1016/S0959-4388(00)00146-X (doi:10.1016/S0959-4388(00)00146-X) [DOI] [PubMed] [Google Scholar]
- 93.Goodson J. L., Evans A. K., Lindberg L., Allen C. D. 2005. Neuro-evolutionary patterning of sociality. Proc. R. Soc. B 272, 227–235 10.1098/rspb.2004.2892 (doi:10.1098/rspb.2004.2892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kingsbury M. A., Finlay B. L. 2001. The cortex in multidimensional space: where do cortical areas come from? Dev. Sci. 4, 125–156 10.1111/1467-7687.00158 (doi:10.1111/1467-7687.00158) [DOI] [Google Scholar]
- 95.Sadato N., Pascualleone A., Grafman J., Ibanez V., Deiber M. P., Dold G., Hallett M. 1996. Activation of the primary visual cortex by Braille reading in blind subjects. Nature 380, 526–528 10.1038/380526a0 (doi:10.1038/380526a0) [DOI] [PubMed] [Google Scholar]
- 96.Bavalier D., Tomann A., Hutton C., Mitchel T., Corina D., Liu G., Neville H. 2000. Visual attention in the periphery is enhanced in congenitally deaf individuals. J. Neurosci. 20, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pallas S. L. 2001. Cross-modal plasticity as a tool for understanding the ontogeny and phylogeny of cerebral cortex. Trends Neurosci. 24, 417–423 10.1016/S0166-2236(00)01853-1 (doi:10.1016/S0166-2236(00)01853-1) [DOI] [PubMed] [Google Scholar]
- 98.Gilbert C. D., Das A., Ito M., Kapadia M., Westheimer G. 1996. Spatial integration and cortical dynamics. Proc. Natl Acad. Sci. USA 93, 615–622 10.1073/pnas.93.2.615 (doi:10.1073/pnas.93.2.615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duncan J., Owen A. M. 2000. Common regions of the human frontal lobe recruited by diverse cognitive demands. Trends Neurosci. 23, 475–481 10.1016/S0166-2236(00)01633-7 (doi:10.1016/S0166-2236(00)01633-7) [DOI] [PubMed] [Google Scholar]
- 100.Anderson M. L. 2010. Neural reuse as a fundamental organizational principal of the brain. Behav. Brain Sci. 33, 245–266 10.1017/S0140525X10000853 (doi:10.1017/S0140525X10000853) [DOI] [PubMed] [Google Scholar]
- 101.Passingham R. E. 1985. Rates of brain development in mammals including man. Brain Behav. Evol. 26, 167–175 10.1159/000118773 (doi:10.1159/000118773) [DOI] [PubMed] [Google Scholar]
- 102.Finlay B. L., Franco E. C., Yamada E. S., Crowley J. C., Parsons M., Muniz J. A., Silveira L. C. 2008. Number and topography of cones, rods and optic nerve axons in New and Old World primates. Vis. Neurosci. 25, 289–299 10.1017/S0952523808080371 (doi:10.1017/S0952523808080371) [DOI] [PubMed] [Google Scholar]
- 103.Finlay B. L. 2008. The developing and evolving retina: using time to organize form. Brain Res. 1192, 5–16 10.1016/j.brainres.2007.07.005 (doi:10.1016/j.brainres.2007.07.005) [DOI] [PubMed] [Google Scholar]