Abstract
Reprogramming differentiated cells towards pluripotency can be achieved by different experimental strategies including the forced expression of specific ‘inducers’ and nuclear transfer. While these offer unparalleled opportunities to generate stem cells and advance disease modelling, the relatively low levels of successful reprogramming achieved (1–2%) makes a direct analysis of the molecular events associated with productive reprogramming very challenging. The generation of transient heterokaryons between human differentiated cells (such as lymphocytes or fibroblasts) and mouse pluripotent stem cell lines results in a much higher frequency of successful conversion (15% SSEA4 expressing cells) and provides an alternative approach to study early events during reprogramming. Under these conditions, differentiated nuclei undergo a series of remodelling events before initiating human pluripotent gene expression and silencing differentiation-associated genes. When combined with genetic or RNAi-based approaches and high-throughput screens, heterokaryon studies can provide important new insights into the factors and mechanisms required to reprogramme unipotent cells towards pluripotency.
Keywords: reprogramming, embryonic stem cell, pluripotency, cell fusion, heterokaryon, nuclear organization
1. Introduction
The early 1970s saw the remarkable and scientifically important discovery that cells from different origins can be fused to generate experimental heterokaryons and hybrid cells. Researchers found that they could combine cells representing distinct stages of differentiation, normal and malignant cells and even cells from different species, to trace whether specific cellular traits were dominant or recessive [1,2]. Experimental heterokaryons and hybrid cells have shaped our understanding of malignancy [3] and tumour-suppressor activity [4]. They were forerunners to modern genetics, enabling specific biological properties to be traced to particular chromosomes and domains [5]. The possibility of combining the properties of different parental cells within heterokaryon and stable hybrid cells has also provided the means to produce monoclonal antibodies [6], to interrogate the mechanisms underlying cellular plasticity [7], gene activation and gene silencing [8,9], and to recapitulate sequential stages of tissue-specific differentiation [10,11].
2. Heterokaryon-mediated resetting of lineage potential
Recently, heterokaryon and hybrid approaches have been used to investigate reprogramming—the restoration of multi-lineage potential to cells with a previously restricted cellular fate—by fusing unipotent somatic cells with pluripotent embryonic stem (ES) cells. Cell fusion results in the formation of heterokaryons in which the two nuclei initially remain discrete. Nuclear fusion occurs later and gives rise to hybrid cells with a single tetraploid nucleus. A proportion of heterokaryons and resulting hybrid cells undergo successful reprogramming, acquiring similar growth and expression characteristics as pluripotent cells [12,13]. By generating interspecies heterokaryons between mouse ES cells and human lymphocytes, where the parental nuclei can easily be distinguished and their activity monitored, it is possible to trace the immediate sequence of events that occur within each cell type as reprogramming is achieved [14,15].
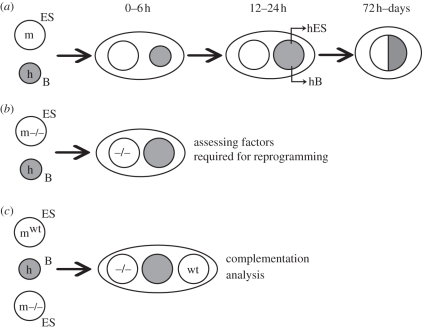
In our studies, mouse ES cells and human B cells were pre-labelled with different cell-surface dyes, mixed together and fused with polyethylene glycol. Heterokaryons containing a human and a mouse nucleus were isolated by fluorescence-activated cell sorting of double-labelled cells. Subsequent changes in gene expression and nuclear morphology were assessed by RT–PCR (using primers that distinguish human and mouse genes) and microscopy (using DAPI (4′,6-diamidino-2-phenylindole) staining patterns or DNA probes to identify human and mouse nuclei), respectively. Using this approach, we showed that human B cells initiate the expression of many pluripotency genes within 24–48 h, including human OCT4, NANOG, CRIPTO, TLE1 and REX1 as well as genes encoding ES-associated enzymes such as DNMT3b. Coordinated expression of these human ES-associated genes defines successful reprogramming [15]. Expression of differentiation-associated genes such as human CD45, CD19 and PAX5 declines within the first 48 h and is progressively extinguished. Hybrid cells, in which parental nuclei are fused, first become evident at 72 h and cellular division can be detected shortly thereafter (figure 1a shows a schematic representation of these events).
Figure 1.
Heterokaryon-based approaches for somatic cell reprogramming. (a) A human B (hB) cell can be reprogrammed following fusion with a murine ES cell. A heterokaryon containing two spatially discrete nuclei, but shared cytoplasm, is formed. After remodelling of the hB cell-derived nucleus, human pluripotency-associated genes are transcribed, whereas human lymphocyte-associated genes are progressively silenced. Tetraploid hybrid cells arise subsequently following nuclear fusion. (b) Factors required for the successful reprogramming of hB cells can be assayed by depleting candidate factors from mES cells (m−/−) prior to (during or after) cell fusion. (c) Complementation studies test whether successful reprogramming can be restored (or mutant ES cells are functionally dominant) using trikaryons formed between different mES cells (wt and −/− shown here).
In fusions performed between mouse ES cells and human B cells, only a proportion of cells (approx. 15%) express SSEA4, a surface glycoprotein that together with TRA-1-60 and TRA-181 characterizes human ES cells [16]. As the enrichment of these SSEA4-positive heterokaryons co-purifies all cells induced to express human OCT4, CRIPTO and NANOG [14,15], SSEA4 provides a useful surrogate marker to estimate the frequency of productive reprogramming. This is particularly relevant since long-term interspecies hybrids are notoriously unstable (they frequently succumb to non-random chromosomal loss), and this can impair their functional characterization.
3. Successful reprogramming requires oct4 and polycomb repression
Using the approach described here, we have begun to dissect the factors and processes required for the pluripotent reprogramming of human lymphocytes. These studies employ mouse ES cells in which specific genes are absent or mutated, conditionally deleted (figure 1b) or where expression is knocked down using interfering RNA strategies. For example, we demonstrated that ES cells deprived of Oct4 can no longer successfully reprogramme human B cells, whereas Sox2, a factor believed to be critical for generating induced pluripotent stem (iPS) cells from fibroblasts [17], was dispensable for reprogramming human B cells in heterokaryons [15]. Similar studies in which short interfering RNAs were used to knock down activation-induced cytidine deaminase (AID) in heterokaryons formed between mouse ES cells and human fibroblasts [18] have argued that AID may be essential for active DNA demethylation required for efficient reprogramming. Although the case for replication-independent DNA demethylation remains controversial [19,20], the future application of sophisticated molecular and genetic approaches in heterokaryon studies will allow the stepwise processes contributing to reprogramming to be systematically investigated.
Recent comparisons of the reprogramming activity of mouse ES cells lacking individual chromatin remodelling factors showed that ES cells require Polycomb repressive complex (PRC) activity to successfully reprogramme human B cells. In complementation assays where trikaryons were generated that contained a wild-type and a PRC-deficient mouse ES nucleus (in addition to the human B cell target), mouse ES cells lacking PRC1/2 dominantly suppressed the reprogramming ability of wild-type ES cells [21] (figure 1c illustrates the approach). This surprising result implied that PRCs were important for silencing factors that if inappropriately expressed by ES cells actively interfered with reprogramming. Although the identity of such factors is currently unknown, the observation that lymphocyte reprogramming by iPS cells is enhanced when Pax5 (a PRC1/2 target that is important for maintaining B cell identity) is depleted [22], or when DNA methyltransferase activity is inhibited [23], suggests that reprogramming of lineage potential and plasticity can be enhanced when factors maintaining cell identity are ablated.
4. Early molecular events that define pluripotent conversion
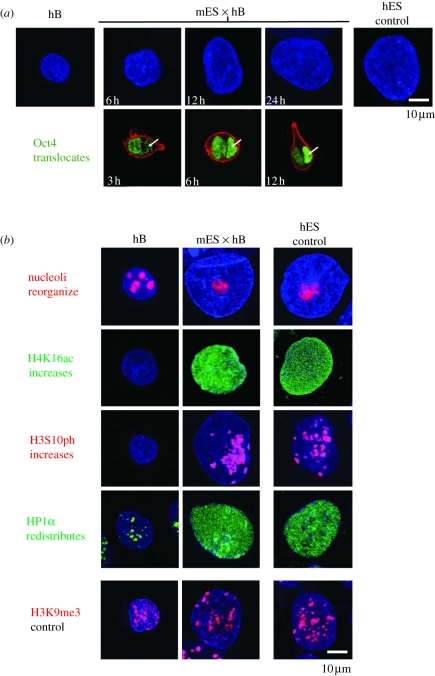
Since mouse ES cells depleted of Oct4 do not initiate pluripotent conversion and mouse ES cells depleted of PRC1/PRC2 fail to successfully complete B cell reprogramming in heterokaryons, both models are useful for discriminating events associated with genuine conversion towards pluripotency from effects associated with heterokaryon formation per se. For example, ourselves and others have shown that somatic cells (such as human B lymphocytes) undergo a dramatic increase in the nuclear volume when incorporated within stable muscle heterokaryons [10] or in transient heterokaryons formed with mouse ES cells (mES × hB, illustrated in figure 2a, upper panels) [21]. We estimated that the nucleus of human B cells undergoes a 2.5-fold increase in volume within 6–24 h of mES × hB heterokaryon formation and this is preceded (3 h) by an influx and steady accumulation of mouse (ES-derived) Oct4 protein within the human nucleus (arrowed, figure 2a, lower panel, Oct4 protein shown in green). Accumulation of Oct4 and changes in the nuclear volume are not predictive of productive conversion to pluripotency because they are still induced by non-reprogramming PRC1/2-deficient ES cells [21]. Likewise, the reorganization of human nucleoli and the increase in acetylated histone H4 at lysine 16 (H4K16ac) that characterizes mES × hB heterokaryon formation (figure 2b) are also induced during non-productive reprogramming with PRC1/2-deficient ES cells [21]. Some changes were, however, selectively induced in heterokaryons containing wild-type but not PRC1/2-deficient ES cells. These include global increase in the level of phosphorylated histone H3 at serine 10 (H3S10ph) and the redistribution of heterochromatin protein 1α (HP1α) in human B nuclei (figure 2b) [21]. Phosphorylation of H3S10 is catalysed by the activity of Aurora B kinase [24] and was shown by Gurdon and co-workers [25] to be induced by reprogramming during nuclear transfer. To understand whether the phosphorylation of H3S10 is indeed required for pluripotent induction, we performed cell fusion experiments in the presence of kinase inhibitors. Although inhibition of Aurora B kinase dramatically reduced H3S10ph levels in human B nuclei, and prevented HP1α redistribution, it did not abolish successful reprogramming (data not shown). This result suggested that although changes in H3S10ph and HP1α mark reprogrammed cells, neither appears to be essential for pluripotent conversion.
Figure 2.
Remodelling of somatic nuclei during pluripotent reprogramming. (a) DAPI-labelled hB cells before and at sequential times (in hours) after fusion and heterokaryon formation with mouse ES cells (mES × hB) reveal an increased nuclear size (blue) and influx of Oct4 protein (green). Oct4 is detected in the hB (arrowed) nucleus within 3 h (and peaks at 12 h), ahead of human Oct4 transcription [15]. Representative confocal images are shown. (b) Human nuclei before (hB) and 24 h after fusion (mES × hB), labelled to reveal specific nuclear changes that occur early during reprogramming. Human ES cells are shown as the control. Antibodies used detect nucleolar marker B23 (nucleophosmin, red), acetylated H4K16 (H4K16ac, green), phosphorylated H3S10 (H3S10ph, red), HP1α (green) and tri-methylated H3K9 (H3K9me3, red). H3K9me3, which does not appreciably change, is provided as a control. Representative confocal images are shown.
It is also worth noting that in heterokaryons formed between human B cells and mouse ES cells lacking PRC1/2 (or conditionally depleted of Oct4), expression of human lymphocyte-associated genes was efficiently extinguished—even though pluripotent genes were not properly induced. This indicates that reprogramming is a multi-step and multi-factorial process rather than an ‘all-or-nothing’ phenomenon as suggested by Tada and co-workers [26].
5. Optimizing reprogramming: future directions and technologies
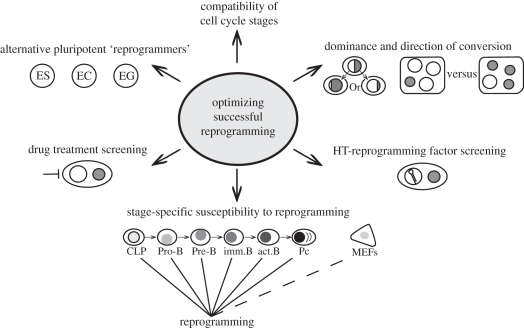
The ability of ES cells to induce the expression of pluripotency-associated genes by differentiated somatic nuclei in transient heterokaryons is a property shared by other embryo-derived cell types [27]. These include embryonic germ (EG) cells cultivated from primordial germ cells that colonize the genital ridge [28] and embryonic carcinoma (EC) cells derived from spontaneous or induced teratocarcinomas [29]. EG cells, unlike ES and EC cells, are also capable of inducing the erasure of DNA methylation at imprinted genes in somatic cell hybrids [12,30], although exactly how this is achieved is not fully understood. Trophoblast stem and extra-ES cell lines (XEN), derived from the developing trophectoderm and primitive endoderm, respectively, have also been shown to dominantly direct the conversion of human B cells towards appropriate extra-embryonic fates within transient heterokaryons [31]. Collectively, these data suggest that stem cell lines derived from primitive cells that emerge at early stages of development possess powerful and often dominant reprogramming capabilities. Whether reprogramming is necessarily uni- or bi-directional has been the subject of much recent [32,33] and past debate [8,29,34], although it seems reasonable to assume that the eventual outcome depends on the relative abundance of certain factors, for example, Nanog [35], as well as the ‘compatibility’ of the parental cell types and culture conditions used to select the hybrids. Future studies will need to address these uncertainties, for example, by examining the impact of nuclear ‘dosage’ on lineage conversion (figure 3) and by assessing the reprogramming outcomes using cell-cycle-stage-enriched ES and target cells.
Figure 3.
Approaches to advance the understanding of reprogramming. Future strategies aimed at improving our knowledge of the molecular events that define the successful conversion of unipotent differentiated cells to pluripotency. Diploid somatic nuclei, or the contribution of the somatic chromosomes within tetraploid cells, are indicated in grey. HT, high throughput; CLP, common lymphocyte precursor. MEFs, mouse embryonic fibroblasts; Pc, plasma cell.
Claims that the differentiation state of a somatic cell acts as a barrier to efficient reprogramming into iPS [36] can be addressed by measuring the susceptibility of progressive differentiation stages to pluripotent conversion in ES-somatic heterkaryons, for example by comparing the reprogramming success of purified human pro-B, pre-B, immature, mature B and plasma cell targets (figure 3), as well as the effects of chromatin-modifying drugs that may weaken epigenetic memory. Perhaps the most significant future advances are likely to come from technologies that enable high-throughput heterokaryon and hybrid analysis to be established. One approach, reported recently by Voldman and Jaenisch, uses a microfluidic device to trap and efficiently pair cells prior to fusion with electrical or chemical agents [37]. Platforms of this kind, which can be coupled to high-throughput screens using RNAi, or drug libraries, and automated single cell time lapse imaging [38], are set to massively expand our knowledge of the early events and mechanisms that direct lineage-restricted cells towards pluripotency. With this in mind, it is sobering to remember that while many current discoveries have their origins in work performed nearly 40 years ago, including evidence that reprogramming (by iPS or in hybrids) does not entirely erase epigenetic marking [39] or morphological characteristics [40], we are now entering a decade of research that will bring us far closer to understanding the molecular basis of cellular memory and reprogramming than previously imagined.
Acknowledgements
The authors would like to thank their colleagues for providing mouse ES cell lines and reagents used in these studies, the MRC for supporting the work and fellowship support from HFSO (T.T.), the EU Epigenome Network of Excellence (C.F.P.) and EMBO (I.C.).
References
- 1.Harris H. 1970. Cell fusion: the Dunham lectures. Cambridge, MA: Harvard University Press [Google Scholar]
- 2.Ephrussi B. 1972. Hybridisation of somatic cells. Princeton, NJ: Princeton University Press [Google Scholar]
- 3.Harris H. 1971. The Croonian Lecture, 1971: cell fusion and the analysis of malignancy. Proc. R. Soc. Lond. B 179, 1–20 10.1098/rspb.1971.0078 (doi:10.1098/rspb.1971.0078) [DOI] [PubMed] [Google Scholar]
- 4.Harris H., Miller O., Klein G., Worst P., Tachibana T. 1969. Suppression of malignancy by cell fusion. Nature 223, 363–368 10.1038/223363a0 (doi:10.1038/223363a0) [DOI] [PubMed] [Google Scholar]
- 5.Weiss M., Green H. 1967. Human–mouse hybrid cell lines containing partial complements of human chromosomes and functioning human genes. Proc Natl Acad. Sci. USA 58, 1104–1111 10.1073/pnas.58.3.1104 (doi:10.1073/pnas.58.3.1104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köhler G., Milstein C. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 (doi:10.1038/256495a0) [DOI] [PubMed] [Google Scholar]
- 7.Blau H. M., Pavlath G. K., Hardeman E. C., Chiu C. P., Silberstein L., Webster S. G., Miller S., Webster C. 1985. Plasticity of the differentiated state. Science 230, 758–766 10.1126/science.2414846 (doi:10.1126/science.2414846) [DOI] [PubMed] [Google Scholar]
- 8.McBurney M. W., Featherstone M. S., Kaplan H. 1978. Activation of teratocarcinoma-derived hemoglobin genes in teratocarcinoma-friend cell hybrids. Cell 15, 1323–1330 10.1016/0092-8674(78)90057-0 (doi:10.1016/0092-8674(78)90057-0) [DOI] [PubMed] [Google Scholar]
- 9.Junker S., Lamm M., Nielsen V., Matthias P. 1997. Extinction of immunoglobulin gene expression in B cells upon fusion with HeLa cells is preceded by rapid nuclear depletion of essential transcription factors and is accompanied by widespread inactivation of genes expressed in a B cell-specific manner. J. Cell Sci. 110, 2579–2587 [DOI] [PubMed] [Google Scholar]
- 10.Terranova R., Pereira C. F., Du Roure C., Merkenschlager M., Fisher A. G. 2006. Acquisition and extinction of gene expression programs are separable events in heterokaryon reprogramming. J. Cell Sci. 119, 2065–2072 10.1242/jcs.02945 (doi:10.1242/jcs.02945) [DOI] [PubMed] [Google Scholar]
- 11.Pomerantz J. H., Mukherjee S., Palermo A. T., Blau H. M. 2009. Reprogramming to a muscle fate by fusion recapitulates differentiation. J. Cell Sci. 122, 1045–1053 10.1242/jcs.041376 (doi:10.1242/jcs.041376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tada M., Takahama Y., Abe K., Nakatsuji N., Tada T. 2001. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11, 1553–1558 10.1016/S0960-9822(01)00459-6 (doi:10.1016/S0960-9822(01)00459-6) [DOI] [PubMed] [Google Scholar]
- 13.Cowan C. A., Atienza J., Melton D. A., Eggan K. 2005. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309, 1369–1373 10.1126/science.1116447 (doi:10.1126/science.1116447) [DOI] [PubMed] [Google Scholar]
- 14.Pereira C. F., Fisher A. G. 2009. Heterokaryon-based reprogramming for pluripotency: current protocols in stem cell biology. New York, NY: John Wiley & Sons, Inc; [DOI] [PubMed] [Google Scholar]
- 15.Pereira C. F., Terranova R. M., Ryan N. K., Santos J., Morris K. J., Cui W., Merkenschlager M., Fisher A. G., Roopenian D. C. 2008. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but Not Sox2. PLoS Genet. 4, e1000170. 10.1371/journal.pgen.1000170 (doi:10.1371/journal.pgen.1000170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 10.1126/science.282.5391.1145 (doi:10.1126/science.282.5391.1145) [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 10.1016/j.cell.2007.11.019 (doi:10.1016/j.cell.2007.11.019) [DOI] [PubMed] [Google Scholar]
- 18.Bhutani N., Brady J. J., Damian M., Sacco A., Corbel S. Y., Blau H. M. 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463, 1042–1047 10.1038/nature08752 (doi:10.1038/nature08752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han D., Do J., Gentile L., Stehling M., Lee H., Schöler H. 2008. Pluripotential reprogramming of the somatic genome in hybrid cells occurs with the first cell cycle. Stem Cells 26, 445–454 10.1634/stemcells.2007-0553 (doi:10.1634/stemcells.2007-0553) [DOI] [PubMed] [Google Scholar]
- 20.Hanna J., Saha K., Pando B., van Zon J., Lengner C. J., Creyghton M. P., van Oudenaarden A., Jaenisch R. 2009. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601 10.1038/nature08592 (doi:10.1038/nature08592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira C. F., et al. 2010. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6, 547–556 10.1016/j.stem.2010.04.013 (doi:10.1016/j.stem.2010.04.013) [DOI] [PubMed] [Google Scholar]
- 22.Hanna J., et al. 2008. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133, 250–264 10.1016/j.cell.2008.03.028 (doi:10.1016/j.cell.2008.03.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikkelsen T. S., et al. 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 10.1038/nature07056 (doi:10.1038/nature07056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirota T., Lipp J. J., Toh B.-H., Peters J.-M. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438, 1176–1180 10.1038/nature04254 (doi:10.1038/nature04254) [DOI] [PubMed] [Google Scholar]
- 25.Murata K., Kouzarides T., Bannister A., Gurdon J. 2010. Histone H3 lysine 4 methylation is associated with the transcriptional reprogramming efficiency of somatic nuclei by oocytes. Epigenet. Chromatin 3, 4. 10.1186/1756-8935-3-4 (doi:10.1186/1756-8935-3-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura H., Tada M., Nakatsuji N., Tada T. 2004. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol. Cell Biol. 24, 5710–5720 10.1128/MCB.24.13.5710-5720.2004 (doi:10.1128/MCB.24.13.5710-5720.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Do J., Han D., Gentile L., Sobek-Klocke I., Stehling M., Lee H., Schöler H. R. 2007. Erasure of cellular memory by fusion with pluripotent cells. Stem Cells 25, 1013–1020 10.1634/stemcells.2006-0691 (doi:10.1634/stemcells.2006-0691) [DOI] [PubMed] [Google Scholar]
- 28.Matsui Y., Zsebo K., Hogan B. L. M. 1992. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70, 841–847 10.1016/0092-8674(92)90317-6 (doi:10.1016/0092-8674(92)90317-6) [DOI] [PubMed] [Google Scholar]
- 29.Andrews P., Goodfellow P. 1980. Antigen expression by somatic cell hybrids of a murine embryonal carcinoma cell with thymocytes and L cells. Somatic Cell Genet. 6, 271–284 10.1007/BF01538801 (doi:10.1007/BF01538801) [DOI] [PubMed] [Google Scholar]
- 30.Tada M., Tada T., Lefebvre L., Barton S. C., Surani M. A. 1997. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 16, 6510–6520 10.1093/emboj/16.21.6510 (doi:10.1093/emboj/16.21.6510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos J., Filipe Pereira C., Di-Gregorio A., Spruce T., Alder O., Rodriguez T., Azuara V., Merkenschlager M., Fisher A. G. 2010. Differences in the epigenetic and reprogramming properties of pluripotent and extra-embryonic stem cells implicate chromatin remodelling as an important early event in the developing mouse embryo. Epigenet. Chromatin 3, 1. 10.1186/1756-8935-3-1 (doi:10.1186/1756-8935-3-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Do J. T., Han D. W., Gentile L., Sobek-Klocke I., Wutz A., Scholer H. R. 2009. Reprogramming of Xist against the pluripotent state in fusion hybrids. J. Cell Sci. 122, 4122–4129 10.1242/jcs.056119 (doi:10.1242/jcs.056119) [DOI] [PubMed] [Google Scholar]
- 33.Palermo A., Doyonnas R., Bhutani N., Pomerantz J., Alkan O., Blau H. M. 2009. Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional. FASEB J. 23, 1431–1440 10.1096/fj.08-122903 (doi:10.1096/fj.08-122903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finch B. W., Ephrussi B. 1967. Retention of multiple developmental potentialities by cells of a mouse testicular teratocarcinoma during prolonged culture in vitro and their extinction upon hybridization with cells of permanent lines. Proc. Natl Acad. Sci. USA. 57, 615–621 10.1073/pnas.57.3.615 (doi:10.1073/pnas.57.3.615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva J., Chambers I., Pollard S., Smith A. 2006. Nanog promotes transfer of pluripotency after cell fusion. Nature 441, 997–1001 10.1038/nature04914 (doi:10.1038/nature04914) [DOI] [PubMed] [Google Scholar]
- 36.Eminli S., Foudi A., Stadtfeld M., Maherali N., Ahfeldt T., Mostoslavsky G., Hock H., Hochedlinger K. 2009. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat. Genet. 41, 968–976 10.1038/ng.428 (doi:10.1038/ng.428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skelley A. M., Kirak O., Suh H., Jaenisch R., Voldman J. 2009. Microfluidic control of cell pairing and fusion. Nat. Methods 6, 147–152 10.1038/nmeth.1290 (doi:10.1038/nmeth.1290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith Z. D., Nachman I., Regev A., Meissner A. 2010. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat. Biotech. 28, 521–526 10.1038/nbt.1632 (doi:10.1038/nbt.1632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim K., et al. 2010. Epigenetic memory in induced pluripotent stem cells Nature 467, 285–290 10.1038/nature09342 (doi:10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiener F., Cochran A., Klein G., Harris H. 1972. Genetic determinants of morphological differentiation in hybrid tumors. J. Natl Cancer Inst. 48, 465–486 [PubMed] [Google Scholar]