Abstract
Small molecules have been playing important roles in elucidating basic biology and treatment of a vast number of diseases for nearly a century, making their use in the field of stem cell biology a comparatively recent phenomenon. Nonetheless, the power of biology-oriented chemical design and synthesis, coupled with significant advances in screening technology, has enabled the discovery of a growing number of small molecules that have improved our understanding of stem cell biology and allowed us to manipulate stem cells in unprecedented ways. This review focuses on recent small molecule studies of (i) the key pathways governing stem cell homeostasis, (ii) the pluripotent stem cell niche, (iii) the directed differentiation of stem cells, (iv) the biology of adult stem cells, and (v) somatic cell reprogramming. In a very short period of time, small molecules have defined a perhaps universally attainable naive ground state of pluripotency, and are facilitating the precise, rapid and efficient differentiation of stem cells into somatic cell populations relevant to the clinic. Finally, following the publication of numerous groundbreaking studies at a pace and consistency unusual for a young field, we are closer than ever to completely eliminating the need for genetic modification in reprogramming.
Keywords: small molecules, stem cells, signalling, metabolites, reprogramming, transdifferentiation
1. Introduction
It took much of the second half of the twentieth century for the monumental discoveries in molecular biology and genetics to usher in the era of modern biology. But even as these fields of inquiry were in their infancy, the design and the synthesis of small-molecule compounds to elicit desired biological outcomes were a decades-old practice [1]. Over the past century, in both academic and industrial settings, an extraordinarily wide range of compounds—based on naturally occurring substances or entirely man-made—have been produced and screened for activity in diverse organisms, cell culture systems and molecular pathways. Among the earlier and most noteworthy fruits of such efforts were classic small-molecule antibiotics, which were so effective in curing previously life-threatening infections that they single-handedly and dramatically extended lifespans around the globe. Small-molecule drugs to treat a myriad of ailments from heart disease to depression eventually followed. To this day, the pharmaceutical industry as a whole still displays a strong preference for small molecules over macromolecular biologics in the drug-development process [2].
The development and the use of small molecules as the therapeutic method of choice are of course not coincidental. Their advantages are compelling: first, the current regulatory environment remains more-or-less aligned with the discovery and development of small molecules as therapeutics. Second, logistically, they are easily manufactured, stored and administered. Third, and perhaps most importantly, from a biochemical standpoint their effects are specific, dose-dependent, rapid and reversible. This set of attributes allows for very precise temporal and functional control in vivo. Small molecules also offer a distinct advantage in development; chemical synthesis of compounds based on biologically active molecular ‘scaffolds’ is a very effective means to quickly generate large ‘libraries’ of potentially effective small molecules [3,4]. However, since it is impossible to saturate chemical space and compound libraries are very large by necessity, the actual identification of a highly active and specific compound remains a herculean task [5]. Compounding this difficulty is the fact that many effective compounds are ultimately discovered to be unsuitable for clinical use, e.g. because of the toxicity or other dangerous off-target effects. Remarkably, over the years, screening technology has advanced to the point that millions of compounds can be rapidly and reliably tested in numerous biological contexts [6]. Such ‘ultra-high-throughput’ screening and related technological advances in design and manipulation have allowed us to overcome truly daunting odds on a fairly regular basis.
However, it is becoming increasingly obvious that effective development of drugs to combat diseases with complex biological underpinnings (e.g. many cancers and ageing-related disorders of the nervous system such as Parkinson's disease) will require a very detailed understanding of the molecular pathways involved, and no single small molecule is likely to represent a ‘magic bullet’ in our efforts to cure such diseases. Nonetheless, small molecules as a class will continue to be one of the most effective tools in biomedical research. At this juncture, it is clear that the signal transduction field will continue to use small molecules in the mapping of molecular pathways relevant to the development and progression of multi-factorial diseases. Importantly, small molecules will probably be key players in the quest to develop novel cell-based therapies for regenerative medicine, i.e. for repair and/or replacement of diseased tissues. Stem cells are currently at the forefront of such efforts, and small molecules—thanks in part to a large body of literature detailing their effects on cellular signalling pathways—are playing critical roles in stem cell derivation, maintenance and manipulation. As such, they offer perhaps the best hope for expeditious, effective and safe implementation of novel stem cell-based technologies in a clinical setting.
How exactly will small molecules help us accomplish these goals? Our review will address this question by covering, with an emphasis on the recent advances, a diverse array of small molecule-driven studies and their implications for the future. Uniquely positioned at the intersection of signal transduction, development and cellular plasticity, these studies offer a glimpse into one of the fastest developing areas of modern biology.
2. Stem cells and small molecules: an overview
Embryonic stem cells (ESCs), isolated from the inner cell mass (ICM) of the preimplantation embryo [7,8], are pluripotent: they possess theoretically boundless self-renewal capacity as well as the potential to generate nearly all cells of the organism from which they were derived. Other naturally occurring pluripotent cell types include epiblast stem cells (EpiSCs; derived from post-implantation embryos) and germline cells [9,10]. As these pluripotent cells progressively differentiate, they give rise to developmentally more restricted multi-potent progenitor cell populations that may persist into adulthood. While the existence and/or exact developmental potential of these ‘adult stem cells’ is not equally well documented for all tissues, there is a significant body of literature demonstrating their critical roles in tissue homeostasis and regeneration [11].
Since the first derivation of ESCs from mice (mESCs)—but especially after the successful isolation of human ESCs (hESCs)—their culture and directed differentiation have presented a unique set of challenges. The loss of pluripotency (i.e. spontaneous differentiation) and cell death under prolonged culture conditions, as well as the inability to generate homogeneous populations of differentiated cells, has been frustrating. Isolating and growing tissue-specific multipotent progenitors in culture have proved still more difficult.
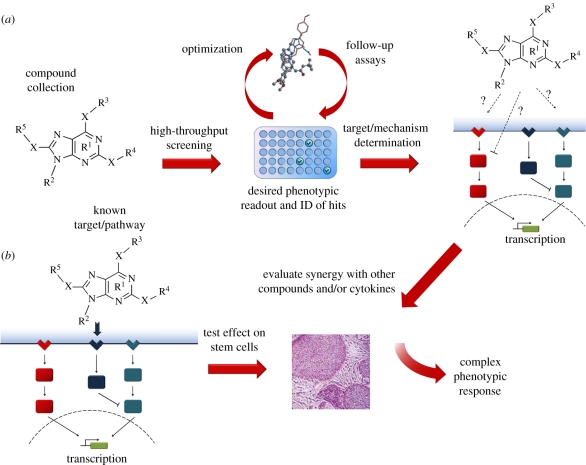
In using small molecules to study and address problems in stem cell biology, two distinct approaches can be taken: in the hypothesis-driven (i.e. target-based) approach, prior knowledge of signalling pathways and their small-molecule modulators is used to implicate them in key regulatory areas. Conversely, the discovery-driven (i.e. phenotype-based) approach assumes no prior knowledge of regulatory pathways, and instead uses unbiased high-throughput screening of small-molecule libraries to elicit a certain phenotype. The cellular targets of effective molecules or ‘hits’ can then be determined using follow-up assays, such as affinity chromatography. Eventually, a sufficiently large number of hits could conceivably allow for the extensive annotation of chemical libraries and the a priori prediction of small-molecule targets and/or activity. Both methods have their own distinct advantages, and have been quite effective in the past (reviewed in [6]).The following studies highlight how these approaches—individually, in tandem, or in combination with other cell-biological and genomics techniques—continue to improve our understanding of the complex biology of stem cells (figure 1). We identify five key categories: (i) elucidating key pathways of stem cell homeostasis, (ii) defining the stem cell niche, (iii) directed differentiation of stem cells, (iv) harnessing the potential of adult stem cells, and (v) somatic cell de-differentiation and fate switching (reprogramming).
Figure 1.
(a) Depicts the outline of a phenotypic screen using small molecules, whereas (b) illustrates the target-based approach. Their combined application in stem cell biology has proved very useful, as there is often a need to elicit, and eventually understand the molecular underpinnings of complex phenotypes.
3. Defining and maintaining a pluripotent ‘ground state’
Traditionally, mESCs and hESCs have been cultured on a monolayer of mitotically inactivated mouse embryonic fibroblasts (MEFs) termed ‘feeders’, with an additional requirement for leukaemia inhibitory factor (LIF; for mESCs) or basic fibroblast growth factor (bFGF or FGF2; for hESCs). In the mouse system, the downstream effectors of LIF were eventually identified as signal transducer and activator of transcription 3 (STAT3) and the pluripotency-associated genes Klf4 and c-Myc [12–14]. The critical component in the undefined portion of the serum-containing media was found to be bone morphogenetic protein 4 (BMP4), which lies upstream of the inhibitor of differentiation(Id)genes [15]. Feeder-conditioned media supplemented with LIF can thus inhibit spontaneous differentiation and allow indefinite self-renewal, but downstream experimental results can be erratic owing to intrinsic variability in serum and MEF quality, as well as varied manipulation techniques. Furthermore, even though a combination of purified BMP4 and LIF does support growth of mESCs in chemically defined media without feeders, experimental variability in differentiation potential and propensity remains an issue.
In an effort to eliminate such variability and simplify media requirements for mESC culture, we carried out a phenotype-based screen of 50 000 small molecules to identify compounds that could support robust mESC culture in the absence of feeders, serum and LIF. For facile high-content imaging-based screening, we used a transgenic mESC line expressing green fluorescent protein (GFP) under control of the promoter of Oct4, the pluripotency master regulator. Using compact, domed colony morphology as a secondary phenotype, pluripotin (also known as SC1) was identified as a potent and specific small molecule supporting mESC expansion in the undifferentiated state [16]. Cells cultured without feeders, serum and cytokines in pluripotin-supplemented chemically defined media maintained homogeneous self-renewal and retained their ability for in vivo germline transmission over many passages, a standard requirement for the demonstration of pluripotency. Using affinity chromatography experiments, the targets of pluripotin were identified as extracellular-signal-regulated kinase 1 (ERK1) and Ras GTPase activating protein (RasGAP), both of which are known positive regulators of differentiation.
These findings have two very important implications: first, from a technical standpoint, they demonstrate that a phenotypic screen can identify a single small molecule capable of eliciting a very complex response/phenotype—in this case, maintenance of pluripotency, by virtue of pluripotin having specific and synergistic polypharmacological activity. This is a remarkable feat, and one that bodes well for future such efforts. Second, in terms of the underlying biology, it is now obvious that inhibition of targets/mechanisms promoting differentiation is sufficient to keep mESCs in a pluripotent ‘ground state’. This finding has been corroborated by others' subsequent demonstration that concurrent inhibition of the FGF4–ERK1/2 mitogen-activated protein kinase (MAPK) pathway and glycogen synthase kinase 3 (GSK3) also supports long-term mESC propagation [17]. By using LIF and BMP4, which negatively regulate each other's differentiation-inducing activities, traditional culture methods appear to accomplish the same; however, such continuous input can have the undesirable side effect of activating certain differentiation-inducing mechanisms at detectable levels (e.g. the ERK pathway mediates ectoderm induction, while BMP signalling mainly induces mesoderm) [18–20]. These secondary effects, in turn, lead to high experimental variability by creating a heterogeneous and unstable culture of cells possessing different, and continuously fluctuating, degrees of pluripotency. The more stable and homogeneous nature of cultures maintained in the presence of inhibitors indicates that the core pluripotency network is inherently stable, as long as it is not perturbed by external signalling.
Most recently, Wagner et al. [21] used a combination of pharmacological and genetic methods to implicate liver receptor homolog-1 (Lrh-1) as a novel β-catenin target gene required for maintaining proper levels of Oct4 and Nanog in mESCs. They have postulated that this signalling axis might represent the in vivo counterpart of the LIF–STAT3 pathway (which is only required in vitro) [21].
In the interim, our improved understanding of the pluripotent ground state has been successfully applied to the derivation of ESCs from mouse strains that had hitherto proven refractory to ESC isolation. We have shown that germline-competent ESCs can be derived from non-obese diabetic (NOD) mice through combined treatment with LIF and pluripotin [22]. Subsequently, Hanna et al. [23] published a study showing that the same could be accomplished by constitutive expression of Klf4 or c-Myc, or small molecules that can replace these factors. Finally, Nichols et al. [24] used LIF in combination with the small molecules PD0325901 and CHIR99021 (CHIR) to derive NOD mouse ESCs by inhibiting GSK3 and ERK pathway signalling, respectively.
In summary, small-molecule discoveries have enabled the stable long-term culture of homogeneous mESC cultures, and the derivation of ESCs from refractory mouse strains. Perhaps most importantly, insights gleaned from the small-molecule studies outlined above are currently being applied to hESCs. For example, Burton et al. [25] have very recently succeeded in maintaining hESCs in the absence of both feeders and cytokines—including bFGF—by using the compound erythro-9-(2-hydroxy-3-nonyl)adenine (EHNA) and its analogues. It is tempting to speculate that we may soon be able to use chemically defined conditions to derive and maintain more robust hESCs with greatly improved therapeutic potential.
4. Small molecules help elucidate the stem cell signalling niche
One of the striking differences between mESCs and hESCs is that the latter are very sensitive to single cell dissociation (e.g. trypsin treatment). The inability to dissociate colonies into single cells drastically limits the scope of experiments that can be carried out with them; for example, genetic manipulation or any study requiring clonal analysis becomes quite difficult. We sought to find a small-molecule solution to this problem by performing a high-throughput phenotypic screen of 50 000 synthetic compounds, looking for molecules that could promote survival following dissociation by trypsin. We ultimately found two: thiazovivin (Tzv; a 2,4-disubstituted thiazole) and pyrintegrin (Ptn; a 2,4-disubstituted pyrimidine). Both compounds enhance survival following dissociation more than 30-fold, without adversely affecting pluripotency over long-term culture in chemically defined medium [26].
Neither compound has an appreciable effect on cell proliferation; rather, both substantially increase adhesion to extracellular matrix (ECM, e.g. Matrigel or laminin), but not gelatin. The increased cell–ECM interaction, in turn, promotes cell survival and growth by activating integrin signalling and the downstream phosphatidylinositol-3-kinase (PI3K) and MAPK pathways [27,28]. In short, integrin-mediated cell–ECM interactions form the basis of an essential survival niche for hESCs.
We found that survival in the niche is regulated by a positive feedback loop involving E-cadherin-mediated cell–cell interactions and the Rho–ROCK signalling pathway: in a healthy, growing hESC colony, cell–cell interactions inhibit Rho–ROCK signalling, which then further enhances both cell–cell and cell–ECM adhesion. However, when cells are fully dissociated, e.g. by trypsinization, Rho–ROCK signalling is upregulated, resulting in destabilization of E-cadherin-mediated cell–cell adhesion. The loss of cell–cell adhesion, in turn, increases Rho–ROCK activity even further. This positive feedback loop culminates in the hyperactivation of ROCK signalling and the complete, irreversible disruption of cell–cell and cell–ECM adhesion. The integrin-dependent survival niche for hESCs is thus disturbed, leading to cell death. Conversely, this irreversible disruption of cell–cell adhesion and upregulation of ROCK signalling are not observed after trypsinization of mESCs. Upon closer investigation, we found that slower endocytosis of newly synthesized E-cadherin was the main reason that cell–cell and cell–ECM interactions are not adversely affected following single-cell dissociation of mESCs. This finding also raised the possibility that the signalling environment of the stem cell niche might be a determining factor in the post-dissociation rate of endocytosis. Indeed, when hESCs were grown under mESC-like culture and signalling conditions using the small molecules PD0325901 (for ERK inhibition) and SB203580 (a p38 kinase inhibitor) [29], cell-surface E-cadherin became much more stable and cell death upon trypsin treatment was greatly reduced.
The above example is an elegant, multi-faceted small-molecule study. While the first part of the paper takes a phenotype-based approach, using a screen to identify two compounds that implicate a known pathway, the second part makes use of a different set of small molecules to conduct a target-based follow-up. Together, they demonstrate a novel way in which the signalling environment of the stem cell niche can specify multiple pluripotent states, each characterized by different modes of cell survival and self-renewal.
Another, perhaps underappreciated, component of the stem cell niche comprises metabolites, the multitude of endogenous small molecules generated as by-products of intracellular reactions. Given how potent synthetic small molecules' effects on stem cell signalling and homeostasis can be, it is hard to imagine that metabolites would not in some fashion affect stem cell homeostasis. For example, might there be a connection between the characteristics of stem cells' metabolomes and the fate decisions they make?
To answer this question, we collaborated with Siuzdak and co-workers to take an untargeted metabolomics approach: using liquid chromatography–electrospray ionization–time of flight mass spectrometry (LC-ESI-TOF-MS), we quantitatively characterized and compared the metabolomes of mESCs and mESC-derived neurons or cardiomyocytes [30]. The chemical formulae for over 150 differentially regulated (difference of greater than 2 ×) metabolites and their degree of unsaturation were determined. Interestingly, the pluripotent metabolome was found to have a significantly higher degree of unsaturation (more than fivefold higher relative abundance) compared with that of either terminally differentiated cell type.
Hypothesizing that the activation of oxidative pathways might be linked to the induction of differentiation, we inhibited the pro-oxidative eicosanoid signalling pathway. Indeed, this inhibition promoted a pluripotent state, even under differentiation conditions. Conversely, individual metabolites downstream of oxidative pathways improved cardiac or neural differentiation three to 15-fold. The same effect was observed to a somewhat lesser degree (50–100% improvement) in hESC differentiation, implying that the role of these metabolites is conserved. Our results are corroborated by the observations that hypoxia sustains the pluripotent state [31,32] and that ESCs maintain lower levels of reactive oxygen species by mostly using non-oxidative glycolysis (rather than oxidative phosphorylation) [33]. Intriguingly, inflammation-resolving metabolites appear to accelerate differentiation, potentially implicating metabolite regulation in the regenerative process during wound healing.
Prior to our study, these abundant, naturally-occurring endogenous small molecules had been almost completely overlooked in the context of stem cell biology. In another study, Garcia-Gonzalo & Izpisua Belmonte [34] demonstrated that albumin-associated lipids contained in knock-out serum replacer (KSR) are responsible for its promotion of hESC self-renewal. In short, it is now clear that metabolites have active role(s) in balancing self-renewal and differentiation.
5. Directed differentiation of embryonic stem cells
To realize the true potential of pluripotent stem cells, significant improvements will be required in the methods used to differentiate them into functional cells relevant from both a basic research and clinical standpoint. Traditionally, the first step in the process has been the formation of embryoid bodies (EBs) in suspension and spontaneous differentiation. The cells of interest must then be isolated from this very heterogeneous mass comprising numerous cell types from all three germ layers. This method leaves much to be desired in terms of efficiency, speed and reproducibility. The resulting cells are not therapeutically useful, and do not lend themselves to detailed analysis of particular differentiation pathways. Likewise, the use of co-culture set-ups or conditioned media brings with it many of the same issues. Ideally, directed differentiation protocols would use only chemically defined media to avoid these problems. Tremendous headway has no doubt been made over the years (reviewed in [35]), but much remains unknown, and small molecule approaches can make critical, lasting contributions. Specifically, they can play inductive roles in differentiation by synergizing with growth factors and cytokines to incrementally recapitulate in vivo development in a precise and efficient fashion. In addition, they may allow the controlled expansion of desired precursor populations. Finally, they can inhibit certain processes, such as the self-renewal of stem cells or progression along an unwanted developmental route at the intermediate stages of differentiation.
For example, the compound stauprimide can promote the induction of Sox17+ endoderm by acting synergistically with the cytokine Activin A. Follow-up biochemical studies indicated that it inhibits the nuclear localization of NME2, a transcription factor that lies upstream of c-Myc. Stauprimide is thus a general destabilizer of the pluripotent state, and was shown to facilitate mesodermal and endodermal differentiation as well [36]. In a different study, the molecules IDE1 and IDE2 were identified as potent inducers of definitive endoderm—even in the absence of Activin A treatment. While both compounds' direct targets remain unknown, they were shown to induce Smad2 phosphorylation in mESCs [37]. Further, the cells generated using IDE1 and 2 can be subsequently differentiated into pancreatic cells when treated with the small molecule indolactam V, which is known to activate protein kinase C (PKC). Provided the relevant developmental pathways are relatively well characterized, the stepwise nature of developmental processes lends itself well to multi-step screens that can produce highly synergistic and specific effects. Such efforts have led to the development of protocols that successfully generate mature neurons [38,39], cardiomyocytes [40,41] and pancreatic cell types [42] from hESCs.
On the other hand, a phenotype-based screening approach can be taken to identify the contributions of one or more pathways to a specific developmental outcome. For example, our earlier efforts in this realm include a cell-based screen that identified TWS119, a neurogenic compound that was found to inhibit GSK3 [43]. We also identified the compounds cardiogenol (A–D) and purmorphamine as effective inducers of the cardiogenic and oesteogenic programmes, respectively [44,45]. More recently, we have discovered a synthetic compound dubbed neuropathiazol in a high-content image-based screen for inducers of neurons from hippocampal progenitor cells. Neuropathiazol is highly neurogenic, even under unfavourable gliogenic conditions [46].
These advances underscore the relevance of small molecule-driven approaches to the elucidation of basic developmental biology as well as the eventual clinical use of ESC-derived differentiated cells. From a clinical perspective, small molecules will impact not only technical feasibility, but also cost-effectiveness (e.g. by replacing expensive growth factors, etc.) In any case, as we make progress on the clinical front, it will be imperative that questions regarding cellular equivalence (i.e. small molecule versus naturally generated) and the in vivo relevance of pathways characterized in vitro be answered clearly and definitively.
6. Small molecules can modulate the behaviour of tissue-specific stem cells
Many mature tissues harbour rare populations of multi- and oligopotent adult stem/progenitor cells. These lineage-restricted precursors can play critical roles in tissue homeostasis and, more importantly, the regenerative process following injury and disease. These qualities make adult stem cells excellent candidates for cell-based therapy approaches, provided their potential can be harnessed as required. This approach was pioneered decades ago with the advent of bone marrow transplants, of which haematopoietic stem cells (HSCs) are the critical component. However, subsequent attempts using other cell types in various organ/tissue systems have typically not met with the same level of success. Aside from issues relating to the histocompatibility of allogeneic cell transplants, difficulties in determining the cell type-specific requirements for the ex vivo survival and proliferation of adult stem cells have universally impeded progress.
Fortunately, phenotypic screens for compounds mediating survival, proliferation and differentiation of these cells have yielded promising results. Two studies representing successful implementation of small-molecule approaches are outlined below. Ultimately, such research should lead to the development of drugs that either facilitate ex vivo manipulation of adult stem cells or enable/enhance the regenerative process by exerting precise control over these cells in vivo. We have also provided examples of current drug-development efforts to underscore the immediate clinical relevance of adult stem cell studies that use small molecules.
Three years ago, our laboratory did a collaborative study on the small-molecule control of cardiac precursor cell proliferation and differentiation [47]. At this time, it was known that multi-potent Isl1+ cardiac precursors could be expanded in co-culture with cardiac mesenchymal cells (CMCs) following their isolation from post-natal hearts. In order to gain a better understanding of the extracellular signals regulating this expansion, we used genetically-marked (LacZ) Isl1+ precursors in a screen of approximately 15 000 compounds. We identified three small molecules that could dramatically expand this small population, among which 6-bromoindirubin-3′-oxime (BIO) stood out as a known inhibitor of GSK3 and hence a positive regulator of the canonical Wnt pathway [48]. Treatment with BIO reproducibly induced a greater than sevenfold increase in Isl1+ cell number. Follow-up experiments revealed that the Wnt/β-catenin pathway has a multi-phasic role in cardiac development. Importantly, the proliferation-inducing effects of BIO, and hence of the canonical Wnt pathway, appear to be conserved in humans.
Around the same time, North et al. [49] published the results of a phenotypic screen for modulators of HSC numbers in vivo using zebrafish embryos. They identified small-molecule regulators of prostaglandin E2 (PGE2) synthesis as potent regulators of HSC number, and went on to show that 16,16-dimethyl PGE2 (a stable PGE2 analogue) improved kidney marrow recovery following irradiation injury in adult zebrafish. Furthermore, this effect was found to be conserved in mice, where PGE2 treatment enhanced progenitor cell proliferation in ESC differentiation assays. Finally, ex vivo treatment of bone marrow cells with 16,16-dimethyl PGE2 was observed to increase successful homing of HSCs in the body.
This study and others (reviewed in [50]) indicated that small molecules may have important role(s) to play in the ex vivo manipulation of HSCs prior to transplantation, a possibility borne out only 2 years later when Fate Therapeutics, Inc. announced promising results using a compound named FT1050 in clinical trials [51]. Other companies have been pursuing similar strategies: for example, Brain Cells, Inc. has found that its compound BCI-540 specifically activates neurogenesis in the hippocampus, and has potential for treatment of anxiety and depression. The company Nuvelo is developing a drug called R-spondin 1 that treats inflammatory bowel disease by inducing stem cell proliferation in the intestinal crypt and a corresponding increase in epithelial cells. Interestingly, like BIO, both BCI-540 and R-spondin 1 exert their effects through modulation of the Wnt pathway [52–54].
7. Facilitating somatic cell reprogramming
In late 2006, Shinya Yamanaka et al. [55] demonstrated that it was possible to revert murine fibroblasts in vitro to a pluripotent mESC-like state by retrovirally overexpressing just four genes: Oct4, Sox2, Klf4 and c-Myc. Given the dogma-shattering implications of this finding for cell and developmental biology—and an astonishing simplicity that could eventually lend itself to the development of autologous cell-based therapies—reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) quickly became an intensely studied area of cell biology. Over the past 4 years, iPSCs have been generated from human fibroblasts and a multitude of other cell types, some using fewer or different sets of genes [7,56–58]. A number of non-viral methods (reviewed in [59]) have also been developed, including the use of recombinant proteins [60].
However, despite such major progress in the field, at least two aspects of reprogramming remain problematic. First, a requirement for genetic modification greatly restricts the downstream utility of virally generated iPSCs, i.e. they are not therapeutically viable as long as insertional mutagenesis and reactivation of exogenous genes—especially oncogenes like c-Myc—remain a possibility. And while the aforementioned use of non-integrating vectors or purified proteins appears to circumvent these issues, these methods dramatically lower the efficiency and speed of iPSC generation. This trade-off raises a second concern, namely that the rarity of iPSC formation might indicate that cells with (epi)genetic abnormalities are being selected for in the process [61]. Alarmingly, there is substantial evidence that downregulation of tumour suppressor genes improves reprogramming outcomes [62–64].
Small molecules have the potential to alleviate both of these concerns by (i) functionally replacing the Yamanaka factors, and (ii) improving speed and efficiency to minimize the potential for acquisition of undesirable traits in culture. Importantly, the induction of pluripotency using small molecules represents a radically different (i.e. indirect) approach when compared with the derivation of iPSCs or somatic cell nuclear transfer (SCNT), which undoubtedly overlap considerably from a mechanistic standpoint. Finding such compounds is an admittedly difficult task, but the combined power of modern synthetic chemistry and high-throughput screening technology should not be underestimated. Moreover, certain cell types, e.g. neural precursor cells (NPCs) and keratinocytes, naturally express high levels of one or more of the Yamanaka factors, a feature that can be taken advantage of in the generation of iPSCs [56,65]. Such cells are especially good candidates for chemical reprogramming, as they may require as few as one factor for reprogramming (Oct4 is the only pluripotency factor that is expressed exclusively in stem cells). Finally, reprogramming appears to involve a mesenchymal-to-epithelial transition (MET) step, suggesting that the use of cells with epithelial phenotypes might be preferable [57].
Small molecules that improve reprogramming generally fall into one of two categories. Some compounds have a global effect on cellular plasticity; these include global epigenetic modifiers like valproic acid (VPA; a histone deacetylase (HDAC) inhibitor), RG108 (a DNA methyltransferase inhibitor) and parnate (an H3K4 histone demethylase inhibitor). In human cells, low levels of the naturally occurring HDAC inhibitor butyrate have been recently shown to non-specifically improve reprogramming efficiency as much as 50-fold [66]. Other compounds exert their effects by modulating a specific signalling pathway to replace one or more of the reprogramming factors. Our work has identified compounds in both categories. For example, we have shown that inhibition of the histone methyltransferase G9a with the compound BIX01294, when combined with the L-calcium channel agonist BayK8644, enables the reprogramming of fibroblasts transduced with only Oct4 and Klf4 [67]. Interestingly, BIX01294 is also capable of replacing Oct4 in NPCs; however, this requires exogenous overexpression of all three remaining Yamanaka factors [68]. Strikingly, Huangfu et al. [69] have demonstrated that VPA treatment is sufficient to replace both Klf4 and c-Myc in neonatal human fibroblasts.
In the context of directed pathway modulation, we have shown that the GSK3 inhibitor CHIR can replace Sox2 in the reprogramming of mouse fibroblasts transduced only with Oct4 and Klf4. When combined with parnate, CHIR permits the generation of iPSCs from human fibroblasts overexpressing the same two factors [70]. Ichida et al. [71] were also able to replace Sox2 in the reprogramming of mouse fibroblasts with the compound E-616452 (renamed RepSox), which inhibits the transforming growth factor-beta (TGF-β) receptor to upregulate the expression of Nanog. Another likely way in which the inhibition of TGF-β signalling contributes to reprogramming success is by facilitating MET [72]. It is thus not surprising that Maherali & Hochedlinger [73] could replace both Sox2 and c-Myc by using an inhibitor of TGF-β receptor I kinase/activin-like kinase 5 (Alk5). Lyssiotis et al. [74] have shown that kenpaullone, a GSK3 inhibitor, can replace Klf4 in the reprogramming of MEFs overexpressing Oct4, Sox2 and c-Myc—albeit in an as-of-yet unknown, GSK3-independent manner. Finally, in the human system, we have shown that combined treatment of fibroblasts with the Alk5 inhibitor SB431542, the MEK inhibitor PD0325901 and thiazovivin improves four-factor reprograming efficiency greater than 200-fold [75].
Aside from re-establishing the pluripotency programme in somatic cells, reprogramming has also been used to transition between different pluripotent states, specifically those of ESCs and EpiSCs. The former functionally represent the ICM of the pre-implantation blastocyst, and can generate chimeric or even entirely mESC-derived animals when transplanted back into the blastocyst using the appropriate methods [76]. EpiSCs, on the other hand, are obtained from post-implantation egg cylinder-stage epiblasts of mice and rats [10]. Just like mESCs, EpiSCs appear to be pluripotent, in that they express the same core pluripotency transcription factors (Oct4, Sox2 and Nanog) and can give rise to tissues of all three germ layers in vitro or in vivo (only in teratoma assays). However, they exhibit a number of striking differences in cell morphology, culture traits and signalling mechanisms [77,78]: mESCs form domed colonies, have a doubling time of approximately 12 h, and are not appreciably affected by single-cell dissociation, whereas EpiSCs grow in flattened sheets, double roughly every 36 h, and do not tolerate single-cell dissociation well. Moreover, treatment with LIF (JAK–STAT3 pathway) and BMP4 (SMAD–Id pathway) have traditionally been used to promote mESC self-renewal, while EpiSCs depend on FGF2 and Activin A for long-term maintenance in culture [12,79]. Most importantly, though, EpiSCs' poor contribution to chimerism suggested that they represent a different and less pluripotent state than mESCs.
This hypothesis has recently been confirmed by the finding that overexpression of Klf4, Nr5a and Nanog can revert EpiSCs to an mESC-like state [80,81]. Furthermore, we have identified a combination of four small molecules that induce the same reversion: concomitant treatment of EpiSCs with inhibitors of the histone demethylase LSD1, ALK5, GSK3 and the MEK pathway produces chimerism-competent mESC-like cells [82].
Interestingly, hESCs very closely resemble EpiSCs in all aspects of their phenotype and culture requirements. This observation raises the possibility that, despite also being derived from the ICM, hESCs are not equivalent to mESCs in terms of their pluripotency state. While mESCs have been successfully cultured in a naive ground state, one might speculate that hESCs have hitherto eluded capture at this stage, undergoing limited differentiation—equivalent to mouse EpiSCs—prior to establishment in culture. Given the field's recent success in reverting EpiSCs back to a more naive mESC-like pluripotent state, we and others have asked whether the same conversion might work for hESCs. This question can be addressed in two ways: by starting with somatic cells and using reprogramming to artificially induce the ground state, or by attempting to derive naive pluripotent cells directly from human blastocysts. Taking the former approach, we found that four-factor transduced human fibroblasts gave rise to mESC-like pluripotent cells when cultured in mESC medium containing human LIF. The nascent colonies could be reliably expanded in the presence of a small-molecule cocktail containing PD0325901, A-83-01 and CHIR99021—inhibitors of MEK, ALK5 and GSK3, respectively [83]. In a related study, Hanna et al. [84] were able to isolate the same type of naive cells by ectopically expressing Oct4 in hESCs and simultaneously treating the cells with LIF, inhibitors of GSK3 and ERK1/2, and forskolin, a protein kinase A (PKA) pathway agonist that can induce Klf2 and Klf4 expression. The same conditions were also permissive for the derivation of human iPSCs.
Unfortunately, despite these important advances, attempts to derive mESC-like hESCs from embryos have thus far not succeeded. Lengner et al. [85] have shown that it is possible to derive female hESCs that have not inactivated one of their X chromosomes, a hallmark of mESCs and fully reprogrammed mouse iPSCs. However, in every other way, these cells still resemble regular hESCs. The fact that relatively few mouse strains have spontaneously given rise to stable mESC lines suggests that their naive pluripotent state is a metastable one. This metastability is at least in part owing to genetic and epigenetic differences between various mouse strains, which have yet to be fully characterized. A more detailed understanding of the impact of genetic and epigenetic background on pluripotency may be needed before we can derive and maintain the mESC-like naive pluripotent stem cells from humans and other mammalian species. A recent study elegantly demonstrates the dramatic impact a slight difference in genetic makeup can have on the way different species regulate the stability of the pluripotent state [86]: mESCs, with their uniquely stunted cell cycle, were found to be critically dependent on high levels of threonine dehydrogenase activity as a source of one-carbon metabolism for purine synthesis. Intriguingly, humans do not have a functional threonine dehydrogenase gene, a deficiency that would probably put hESCs at a significant disadvantage for self-renewal under mESC-like conditions.
Consistent with our hypothesis, we found that our above cocktail of compounds (PD0325901, A-83-01 and CHIR99021), when supplemented with LIF, did allow for the generation of naive, mESC-like rat iPSCs that were capable of contributing to chimerism [83]. Similarly, others have found that rat ESCs can be derived from blastocysts under chemically defined conditions that include PD0325901, CHIR99021 and LIF [87,88]. More recently, another study generated rat ESCs using four small molecules, including PD0325901, A-83-01, CHIR99021 and Y27632 [89]. Y27632 is a Rho-associated protein kinase inhibitor that can stabilize E-cadherin [26]. These derived cells successfully contributed to the generation of transgenic rats via germline transmission, proving their occupancy of the naive ground state. These advances have paved the way for the use of homologous recombination in rat ESCs to generate knock-in strains or loss-of-function mutants. Recently, a p53 knockout rat has been successfully generated using this strategy [90]. Such strains represent invaluable genetic tools, as rats are potentially better suited as a model to study human biology and diseases, especially multi-factorial ones. Furthermore, the successful establishment of rat ESC cultures may provide the technical framework for the derivation of naive ESCs from other model organisms (e.g. primates) and livestock.
In summary, significant progress has been made towards ‘chemical reprogramming’, insofar as all four Yamanaka factors have now been individually replaced by small molecules. The real challenge will be to find a combination that can replace all four simultaneously, and do so rapidly and efficiently. Mechanistically, this process would undoubtedly represent a completely novel path to reprogramming compared with SCNT or defined transcription factor-based induction of pluripotency. These two established methods of reprogramming both rely on maternally stored or ectopically expressed transcription factors, the highly specific functions of which have evolved over millions of years. We unfortunately cannot escape the fact that man-made small molecules, as effective as they can be, do not possess any functional entitie(s) analogous to the highly specific DNA recognition and transcription domains that render transcription factors so extraordinarily powerful. Attaining the same degree of potency and specificity with small molecules, which by necessity must indirectly bring about the desired outcome, represents both a fundamental challenge and an opportunity to achieve truly groundbreaking progress.
Small molecules have also helped define distinct metastable states of pluripotency and self-renewal, thereby facilitating transitions from one state to another. A related future role for small molecules in reprogramming might be to ensure that the epigenomes of iPSCs are fully reset, as it was very recently shown that certain iPSCs may retain an epigenetic ‘memory’ of their origins, developmentally restricting or biasing them towards these particular lineages [91].
8. Small molecules help establish a new transdifferentiation paradigm
Most recently, our laboratory has been focusing on a hitherto unexplored aspect of somatic cell reprogramming. We and others have observed that reprogramming using the Yamanaka factors often gives rise to rare side populations of cells that do not yield any iPSCs. While some of these clusters consist of nondescript granular cells reported to be unproductive iPSC intermediates [92,93], others exhibit more complex morphological traits. Intriguingly, this latter group may express marker genes characteristic of partially or fully differentiated cells entirely unrelated to the starting population—the hallmark of transdifferentiation. We have, for example, observed spontaneously contracting foci of cells and/or the presence of neurons in a very small numbers.
This unexpected observation led us to ask the following question: might it be possible to more specifically instruct reprogramming towards one or more of these side populations by altering reprogramming conditions? Implicit in this question is the idea that iPSCs may represent but one of many possible outcomes of the Yamanaka factor-based reprogramming process. We speculated that overexpression of the Yamanaka factors—especially early on—could be priming the cells for rapid lineage switching by transiently creating an epigenetically very unstable and plastic state. To test this hypothesis, we modified standard reprogramming procedures in ways we thought might ‘unmask’ alternative outcomes. First, we employed a doxycycline-inducible system to limit Yamanaka factor expression to the bare minimum required for reprogramming to take place, as indicated by various morphological changes culminating in colony formation. We simultaneously tested conditions conducive to the generation and/or survival of lineage-restricted cells, e.g. cardiomyocytes, rather thaniPSCs: among other things, this entailed omitting LIF from the media, and switching to chemically defined conditions (including growth factors/cytokines) early on.
Importantly, we again enlisted the help of small molecules in the process. Not only did we use an inhibitor of the JAK/STAT pathway to suppress signalling critical to the establishment of pluripotency, but we also employed a Wnt pathway modulator to promote cardiogenesis during an empirically determined optimal time window. This strategy has been very successful in generating spontaneously contracting cardiomyocytes from embryonic and adult fibroblasts in 12–13 days [94]. A recent study has accomplished much the same using cardiac lineage-specific transcription factor overexpression [95]; however, it is important to note that the time required for the development of contraction is considerably longer in this case.
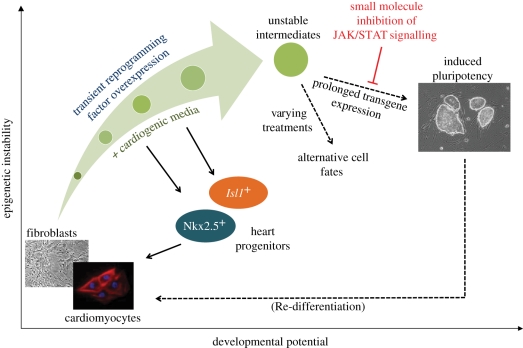
Since we have achieved transdifferentiation using only the Yamanaka factors, it has broad ramifications and applicability. Accordingly, we were able to successfully adapt the process to the rapid and efficient generation of NPCs from fibroblasts [96]. We believe that we have thus established a new transdifferentiation paradigm, and small molecules have once again played a prominent role in the process (figure 2).
Figure 2.
A new transdifferentiation paradigm. In our model, transient high-level reprogramming factor expression leads to the formation of epigenetically unstable (i.e. more naive and ‘activated’) intermediates, which subsequently ‘relax’ back into more stable state(s). In this scheme, the generation of iPSCs is unlikely, as complete de-differentiation requires prolonged overexpression of pluripotency factors. On the other hand, direct reprogramming to various progenitor cells—and perhaps terminally differentiated cells as well—is a feasible outcome, especially if (i) culture conditions allow and/or promote their formation and (ii) establishment of pluripotency is prevented by using a small-molecule inhibitor of JAK/STAT signalling.
9. Conclusions and perspectives
Our understanding of the biology of stem cells, especially with respect to hESCs and somatic cell reprogramming, remains relatively superficial. Likewise, our knowledge of small-molecule mechanisms of action is also incomplete; some compounds that appear to specifically modulate enzymatic action or receptor signalling may have as-of-yet undetected secondary effects. It is important that dogma does not become embedded around such compounds as this may cloud understanding of their true mechanism of action, a sine qua non for their deployment clinically. As has been the case in the past few years, the coming decade will no doubt see numerous advances in these areas, which should lead to the development of effective novel drugs.
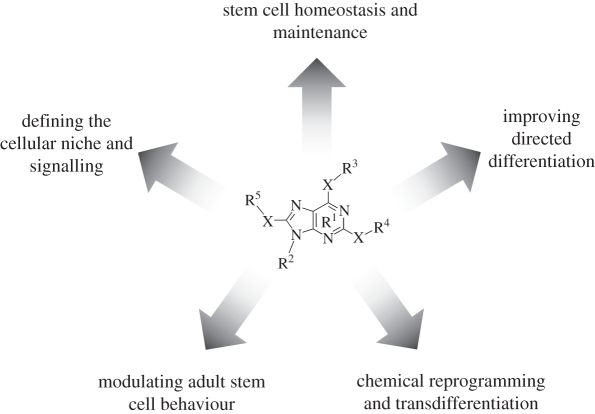
In this review, we have summarized the ways in which small molecules have contributed to the advancement of stem cell research (figure 3 and table 1), and how they will continue to shape these efforts. By allowing us to systematically probe known signalling pathways, small-molecule studies are defining very specific requirements for the robust propagation of pluripotent cells. Such optimization and standardization of growth conditions, in turn, are greatly facilitating the effective and reproducible induction of directed differentiation (one of the key goals being large/industrial-scale generation of desired populations). Small molecules have also come to play prominent roles in this next step, e.g. by mimicking niche signalling to more precisely recapitulate in vivo development and/or by inhibiting unwanted signalling input that invariably reduces efficiency. At the level of basic biology, small molecules have proved useful in defining how particular elements of the niche influence stem cell survival and proliferation. Consequently, their utility in activating or potentiating adult stem cells is already being tested in clinical trials. Finally, small molecules represent a highly desirable alternative to genetic modification in reprogramming and transdifferentiation; true ‘chemical reprogramming’ would be easier, more effective and much safer from a disease treatment perspective. Many, if not all, of the above small molecule approaches could conceivably lend themselves to both in vivo and ex vivo clinical implementation. Which route—or combination of routes—is ultimately chosen will largely depend on empirical success, and is very difficult to ascertain at this point. It is almost certain, however, that the interest in these efforts will continue to grow at its current rapid pace for the foreseeable future.
Figure 3.
A schematic of stem cell-related small molecule efforts as presented in this review.
Table 1.
Small molecules used in the studies reviewed and their relevance to stem cell biology.
| name of compound | target(s) | effect(s) | reference no. |
|---|---|---|---|
| (−) indolactam V | PKC | pancreatic differentiation of ESC-derived endoderm | [37] |
| (+) Bayk 8644 | L-type Ca2+ channel | enhances reprogramming of MEFs | [67] |
| 16,16-dimethyl prostaglandin E2 | EP receptor(s) | increases frequency of long-term HSCs; improved kidney regeneration | [49] |
| 6-bromoindirubin-3′-oxime (BIO) | GSK3 | self-renewal of mESCs and proliferation of Isl1+ cardiac precursors | [47,48] |
| A-83-01 | ALK4, ALK5 and ALK7 | rat iPSC self-renewal, in combination with CHIR99021 and PD0325901 | [83,89] |
| BCI-540 | Wnt pathway | neurogenesis in the hippocampus | [51] |
| BIX-01294 | G9a HMTase | reprogramming of NPCs and MEFs transduced with only Oct4 and Klf4 | [67,68] |
| cardiogenols (A–D) | unknown | efficient induction of cardiogenesis in mESCs | [44] |
| CHIRON99021 (CHIR) | GSK3 | activates canonical Wnt signalling, promotes mESC self-renewal | [24,70,83,87–89] |
| erythro-9-(2-hydroxy-3- nonyl)adenine (EHNA) | phosphodiesterase (PDE2) | allows hESC maintenance in chemically defined media | [25] |
| forskolin | PKA | promotes neurogenesis along with retinoic acid | [84] |
| FT1050 | Wnt and prostaglandin E2 | improves HSC function and engraftment | [51] |
| IDE1/2 | unknown | Smad2 phosphorylation in ES cells; differentiation into endoderm | [37] |
| kenpaullone | GSK3 and CDKs | replaces Klf4 in the reprogramming of MEFs | [74] |
| neuropathiazol | unknown | induces neuronal differentiation of multi-potent adult hippocampal NPCs | [46] |
| parnate | lysine-specific demethylase I | reprogramming of human keratinocytes transduced with Oct4/Klf4 | [70] |
| PD0325901 | MEK | inhibits differentiation of mESCs | [24,29,75,83,87–89] |
| pluripotin (SC1) | RasGAP and ERK1 | promotes mESC self-renewal | [16,22] |
| purmorphamine | Hedgehog | induction of osteogenesis | [45] |
| RG108 | DNA MTase | facilitates reprogramming of MEFs | [66] |
| R-spondin 1 | Wnt pathway | stem cell proliferation in the intestinal crypt | [54] |
| SB203580 | p38-MAPK | used in conjunction with PD0325901 to maintain hESCs in an mESC-like signalling niche | [29] |
| SB431542 | ALK4, ALK5 and ALK7 | efficient neural differentiation of hES cells in combination with Noggin | [75] |
| stauprimide | NME2 | enhanced differentiation of ES cells | [36] |
| thiazovivin (Tzv) | ROCK | improved survival of hESCs upon dissociation | [26,75] |
| TWS119 | GSK3 | neurogenesis enhancer | [43] |
| pyrintegrin (Pyr) | unknown | improved survival of hESCs upon dissociation | [26] |
| valproic acid (VPA) | histone deacetylase | enables reprogramming of human fibroblasts transduced with only Oct4 and Sox2; globally acting factor | [66,69] |
| Y27632 | ROCK | improved survival of hESCs upon dissociation | [26,89] |
As it becomes increasingly clear that advances in stem cell biology have the potential to radically transform the way diseases are treated in the not-so-distant future, small molecules may be poised to once again take centre stage in a profound paradigm shift, just as they did decades ago with the discovery of antibiotics. They might be considered small in the molecular realm, but their impact on humanity has been, and continues to be, enormous.
Acknowledgements
We thank all members of the Ding laboratory for their tireless work and stimulating discussions. S.D. is supported by funding from NICHD, NHLBI and NIMH/NIH, California Institute for Regenerative Medicine, Prostate Cancer Foundation, Fate Therapeutics, Esther B. O'Keeffe Foundation and The Scripps Research Institute. J.A.E. is a Lowe Family Foundation fellow.
References
- 1.Drews J. 2000. Drug discovery: a historical perspective. Science 287, 1960–1964 10.1126/science.287.5460.1960 (doi:10.1126/science.287.5460.1960) [DOI] [PubMed] [Google Scholar]
- 2.Cayen M. N. 2010. Early drug development: strategies and routes to first-in-human trials. Hoboken, NJ: Wiley [Google Scholar]
- 3.Dandapani S., Marcaurelle L. A. 2010. Current strategies for diversity-oriented synthesis. Curr. Opin. Chem. Biol. 14, 362–370 10.1016/j.cbpa.2010.03.018 (doi:10.1016/j.cbpa.2010.03.018) [DOI] [PubMed] [Google Scholar]
- 4.Wilk W., Zimmermann T. J., Kaiser M., Waldmann H. 2010. Principles, implementation, and application of biology-oriented synthesis (BIOS). Biol Chem. 391, 491–497 10.1515/BC.2010.013 (doi:10.1515/BC.2010.013) [DOI] [PubMed] [Google Scholar]
- 5.Dobson C. M. 2004. Chemical space and biology. Nature 432, 824–828 10.1038/nature03192 (doi:10.1038/nature03192) [DOI] [PubMed] [Google Scholar]
- 6.Emre N., Coleman R., Ding S. 2007. A chemical approach to stem cell biology. Curr. Opin. Chem. Biol. 11, 252–258 10.1016/j.cbpa.2007.04.024 (doi:10.1016/j.cbpa.2007.04.024) [DOI] [PubMed] [Google Scholar]
- 7.Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 10.1126/science.282.5391.1145 (doi:10.1126/science.282.5391.1145) [DOI] [PubMed] [Google Scholar]
- 8.Evans M. J., Kaufman M. H. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 10.1038/292154a0 (doi:10.1038/292154a0) [DOI] [PubMed] [Google Scholar]
- 9.Kanatsu-Shinohara M., et al. 2004. Generation of pluripotent stem cells from neonatal mouse testis. Cell 119, 1001–1012 10.1016/j.cell.2004.11.011 (doi:10.1016/j.cell.2004.11.011) [DOI] [PubMed] [Google Scholar]
- 10.Brons I. G., et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 10.1038/nature05950 (doi:10.1038/nature05950) [DOI] [PubMed] [Google Scholar]
- 11.Wagers A. J., Weissman I. L. 2004. Plasticity of adult stem cells. Cell 116, 639–648 10.1016/S0092-8674(04)00208-9 (doi:10.1016/S0092-8674(04)00208-9) [DOI] [PubMed] [Google Scholar]
- 12.Niwa H., Burdon T., Chambers I., Smith A. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048–2060 10.1101/gad.12.13.2048 (doi:10.1101/gad.12.13.2048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichols J., Chambers I., Taga T., Smith A. 2001. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development 128, 2333–2339 [DOI] [PubMed] [Google Scholar]
- 14.Li Y., McClintick J., Zhong L., Edenberg H. J., Yoder M. C., Chan R. J. 2005. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood 105, 635–637 10.1182/blood-2004-07-2681 (doi:10.1182/blood-2004-07-2681) [DOI] [PubMed] [Google Scholar]
- 15.Ying Q. L., Smith A. G. 2003. Defined conditions for neural commitment and differentiation. Methods Enzymol. 365, 327–341 10.1016/S0076-6879(03)65023-8 (doi:10.1016/S0076-6879(03)65023-8) [DOI] [PubMed] [Google Scholar]
- 16.Chen S., Do J. T., Zhang Q., Yao S., Yan F., Peters E. C., Scholer H. R., Schultz P. G., Ding S. 2006. Self-renewal of embryonic stem cells by a small molecule. Proc. Natl Acad. Sci. USA 103, 17 266–17 271 10.1073/pnas.0608156103 (doi:10.1073/pnas.0608156103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 10.1038/nature06968 (doi:10.1038/nature06968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan H., Corbi N., Basilico C., Dailey L. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9, 2635–2645 10.1101/gad.9.21.2635 (doi:10.1101/gad.9.21.2635) [DOI] [PubMed] [Google Scholar]
- 19.Stefanovic S., Abboud N., Desilets S., Nury D., Cowan C., Puceat M. 2009. Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. J. Cell Biol. 186, 665–673 10.1083/jcb.200901040 (doi:10.1083/jcb.200901040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder M., Huang X. Y., Zhang J. J. 2010. Stat3 directly controls the expression of Tbx5, Nkx2.5, and GATA4 and is essential for cardiomyocyte differentiation of P19CL6 cells. J. Biol. Chem. 285, 23 639–23 646 10.1074/jbc.M110.101063 (doi:10.1074/jbc.M110.101063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner R. T., Xu X., Yi F., Merrill B. J., Cooney A. J. 2010. Canonical Wnt/β-catenin regulation of liver receptor homolog-1 mediates pluripotency gene expression. Stem Cells. 28, 1794–1804 10.1002/stem.502 (doi:10.1002/stem.502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W., et al. 2009. Pluripotin combined with leukemia inhibitory factor greatly promotes the derivation of embryonic stem cell lines from refractory strains. Stem Cells 27, 383–389 10.1634/stemcells.2008-0974 (doi:10.1634/stemcells.2008-0974) [DOI] [PubMed] [Google Scholar]
- 23.Hanna J., et al. 2009. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell 4, 513–524 10.1016/j.stem.2009.04.015 (doi:10.1016/j.stem.2009.04.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols J., Jones K., Phillips J. M., Newland S. A., Roode M., Mansfield W., Smith A., Cooke A. 2009. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat. Med. 15, 814–818 10.1038/nm.1996 (doi:10.1038/nm.1996) [DOI] [PubMed] [Google Scholar]
- 25.Burton P., et al. 2010. Identification and characterization of small-molecule ligands that maintain pluripotency of human embryonic stem cells. Biochem. Soc. Trans. 38, 1058–1061 10.1042/BST0381058 (doi:10.1042/BST0381058) [DOI] [PubMed] [Google Scholar]
- 26.Xu Y., Zhu X., Hahm H. S., Wei W., Hao E., Hayek A., Ding S. 2010. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl Acad. Sci. USA 107, 8129–8134 10.1073/pnas.1002024107 (doi:10.1073/pnas.1002024107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armstrong L., et al. 2006. The role of PI3K/AKT, MAPK/ERK and NFkappabeta signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum. Mol. Genet. 15, 1894–1913 10.1093/hmg/ddl112 (doi:10.1093/hmg/ddl112) [DOI] [PubMed] [Google Scholar]
- 28.Paling N. R., Wheadon H., Bone H. K., Welham M. J. 2004. Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J. Biol. Chem. 279, 48 063–48 070 10.1074/jbc.M406467200 (doi:10.1074/jbc.M406467200) [DOI] [PubMed] [Google Scholar]
- 29.Burdon T., Stracey C., Chambers I., Nichols J., Smith A. 1999. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 210, 30–43 10.1006/dbio.1999.9265 (doi:10.1006/dbio.1999.9265) [DOI] [PubMed] [Google Scholar]
- 30.Yanes O., et al. 2010. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 6, 411–417 10.1038/nchembio.364 (doi:10.1038/nchembio.364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezashi T., Das P., Roberts R. M. 2005. Low O2 tensions and the prevention of differentiation of hES cells. Proc. Natl Acad. Sci. USA 102, 4783–4788 10.1073/pnas.0501283102 (doi:10.1073/pnas.0501283102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Q., Lee Y. J., Yun Z. 2006. Differentiation arrest by hypoxia. J. Biol. Chem. 281, 30 678–30 683 10.1074/jbc.C600120200 (doi:10.1074/jbc.C600120200) [DOI] [PubMed] [Google Scholar]
- 33.Tsatmali M., Walcott E. C., Crossin K. L. 2005. Newborn neurons acquire high levels of reactive oxygen species and increased mitochondrial proteins upon differentiation from progenitors. Brain Res. 1040, 137–150 10.1016/j.brainres.2005.01.087 (doi:10.1016/j.brainres.2005.01.087) [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Gonzalo F. R., Izpisua Belmonte J. C. 2008. Albumin-associated lipids regulate human embryonic stem cell self-renewal. PLoS ONE 3, e1384. 10.1371/journal.pone.0001384 (doi:10.1371/journal.pone.0001384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murry C. E., Keller G. 2008. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680 10.1016/j.cell.2008.02.008 (doi:10.1016/j.cell.2008.02.008) [DOI] [PubMed] [Google Scholar]
- 36.Zhu S., Wurdak H., Wang J., Lyssiotis C. A., Peters E. C., Cho C. Y., Wu X. u, Schultz P. G. 2009. A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell 4, 416–426 10.1016/j.stem.2009.04.001 (doi:10.1016/j.stem.2009.04.001) [DOI] [PubMed] [Google Scholar]
- 37.Borowiak M., Maehr R., Chen S., Chen A. E., Tang W., Fox J. L., Schreiber S., Melton D. A. 2009. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 4, 348–358 10.1016/j.stem.2009.01.014 (doi:10.1016/j.stem.2009.01.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying Q. L., Stavridis M., Griffiths D., Li M., Smith A. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 10.1038/nbt780 (doi:10.1038/nbt780) [DOI] [PubMed] [Google Scholar]
- 39.Li X. J., Du Z. W., Zarnowska E. D., Pankratz M., Hansen L. O., Pearce R. A., Zhang S.-C. 2005. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 23, 215–221 10.1038/nbt1063 (doi:10.1038/nbt1063) [DOI] [PubMed] [Google Scholar]
- 40.Kattman S. J., Huber T. L., Keller G. M. 2006. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev. Cell. 11, 723–732 10.1016/j.devcel.2006.10.002 (doi:10.1016/j.devcel.2006.10.002) [DOI] [PubMed] [Google Scholar]
- 41.Laflamme M. A., et al. 2007. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 25, 1015–1024 10.1038/nbt1327 (doi:10.1038/nbt1327) [DOI] [PubMed] [Google Scholar]
- 42.D'Amour K. A., Agulnick A. D., Eliazer S., Kelly O. G., Kroon E., Baetge E. E. 2005. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534–1541 10.1038/nbt1163 (doi:10.1038/nbt1163) [DOI] [PubMed] [Google Scholar]
- 43.Ding S., Wu T. Y., Brinker A., Peters E. C., Hur W., Gray N. S., Schultz P. G. 2003. Synthetic small molecules that control stem cell fate. Proc. Natl Acad. Sci. USA 100, 7632–7637 10.1073/pnas.0732087100 (doi:10.1073/pnas.0732087100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X., Ding S., Ding Q., Gray N. S., Schultz P. G. 2004. Small molecules that induce cardiomyogenesis in embryonic stem cells. J. Am. Chem. Soc. 126, 1590–1591 10.1021/ja038950i (doi:10.1021/ja038950i) [DOI] [PubMed] [Google Scholar]
- 45.Wu X., Ding S., Ding Q., Gray N. S., Schultz P. G. 2002. A small molecule with osteogenesis-inducing activity in multipotent mesenchymal progenitor cells. J. Am. Chem. Soc. 124, 14 520–14 521 10.1021/ja0283908 (doi:10.1021/ja0283908) [DOI] [PubMed] [Google Scholar]
- 46.Lu Q., Paredes M., Zhang J., Kosik K. S. 1998. Basal extracellular signal-regulated kinase activity modulates cell-cell and cell-matrix interactions. Mol. Cell Biol. 18, 3257–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qyang Y., et al. 2007. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell 1, 165–179 10.1016/j.stem.2007.05.018 (doi:10.1016/j.stem.2007.05.018) [DOI] [PubMed] [Google Scholar]
- 48.Meijer L., et al. 2003. GSK-3–selective inhibitors derived from Tyrian purple indirubins. Chem. Biol. 10, 1255–1266 10.1016/j.chembiol.2003.11.010 (doi:10.1016/j.chembiol.2003.11.010) [DOI] [PubMed] [Google Scholar]
- 49.North T. E., et al. 2007. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 10.1038/nature05883 (doi:10.1038/nature05883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Broxmeyer H. E. 2008. Chemokines in hematopoiesis. Curr. Opin. Hematol. 15, 49–58 10.1097/MOH.0b013e3282f29012 (doi:10.1097/MOH.0b013e3282f29012) [DOI] [PubMed] [Google Scholar]
- 51.Grayson P., Mendlein J., Thies S., Yingling J. 2009. Stem cell modulators (SCMs): a small molecule and biologic approach to stem cell modulators. Drug Discov. Today Ther. Strateg. 6, 141–145 10.1016/J.DDSTR.2009.06.005 (doi:10.1016/J.DDSTR.2009.06.005) [DOI] [Google Scholar]
- 52.Li X., Zhang Y., Kang H., Liu W., Liu P., Zhang J., Harris S. E., Wu D. 2005. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J. Biol. Chem. 280, 19 883–19 887 10.1074/jbc.M413274200 (doi:10.1074/jbc.M413274200) [DOI] [PubMed] [Google Scholar]
- 53.Semenov M., Tamai K., He X. 2005. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J. Biol. Chem. 280, 26770–26775 10.1074/jbc.M504308200 (doi:10.1074/jbc.M504308200) [DOI] [PubMed] [Google Scholar]
- 54.Binnerts M. E., et al. 2007. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc. Natl Acad. Sci. USA 104, 14 700–14 705 10.1073/pnas.0702305104 (doi:10.1073/pnas.0702305104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 (doi:10.1016/j.cell.2006.07.024) [DOI] [PubMed] [Google Scholar]
- 56.Kim J. B., et al. 2009. Oct4–induced pluripotency in adult neural stem cells. Cell 136, 411–419 10.1016/j.cell.2009.01.023 (doi:10.1016/j.cell.2009.01.023) [DOI] [PubMed] [Google Scholar]
- 57.Aasen T., et al. 2008. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26, 1276–1284 10.1038/nbt.1503 (doi:10.1038/nbt.1503) [DOI] [PubMed] [Google Scholar]
- 58.Loh Y. H., et al. 2009. Generation of induced pluripotent stem cells from human blood. Blood 113, 5476–5479 10.1182/blood-2009-02-204800 (doi:10.1182/blood-2009-02-204800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abujarour R., Ding S. 2009. Induced pluripotent stem cells free of exogenous reprogramming factors. Genome Biol. 10, 220. 10.1186/gb-2009-10-5-220 (doi:10.1186/gb-2009-10-5-220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou H., et al. 2009. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4, 381–384 10.1016/j.stem.2009.04.005 (doi:10.1016/j.stem.2009.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohm J. E., et al. 2010. Cancer-related epigenome changes associated with reprogramming to induced pluripotent stem cells. Cancer Res. 70, 7662–7673 10.1158/0008-5472.CAN-10-1361 (doi:10.1158/0008-5472.CAN-10-1361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marion R. M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M. A. 2009. A p53–mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460, 1149–1153 10.1038/nature08287 (doi:10.1038/nature08287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawamura T., Suzuki J., Wang Y. V., Menendez S., Morera L. B., Raya A., Wahl G. M., Belmonte J. C. I. 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144 10.1038/nature08311 (doi:10.1038/nature08311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H., Collado M., Villasante A., Strati K., Ortega S., Canamero M., Blasco M. A., Serrano M. l. 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–1139 10.1038/nature08290 (doi:10.1038/nature08290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hester M. E., Song S., Miranda C. J., Eagle A., Schwartz P. H., Kaspar B. K. 2009. Two factor reprogramming of human neural stem cells into pluripotency. PLoS ONE 4, e7044. 10.1371/journal.pone.0007044 (doi:10.1371/journal.pone.0007044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mali P., et al. 2010. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 28, 713–720 10.1002/stem.402 (doi:10.1002/stem.402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi Y., Desponts C., Do J. T., Hahm H. S., Scholer H. R., Ding S. 2008. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 3, 568–574 10.1016/j.stem.2008.10.004 (doi:10.1016/j.stem.2008.10.004) [DOI] [PubMed] [Google Scholar]
- 68.Shi Y., Do J. T., Desponts C., Hahm H. S., Scholer H. R., Ding S. 2008. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2, 525–528 10.1016/j.stem.2008.05.011 (doi:10.1016/j.stem.2008.05.011) [DOI] [PubMed] [Google Scholar]
- 69.Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D. A. 2008. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 26, 1269–1275 10.1038/nbt.1502 (doi:10.1038/nbt.1502) [DOI] [PubMed] [Google Scholar]
- 70.Li W., et al. 2009. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells 27, 2992–3000 10.1002/stem.240 (doi:10.1002/stem.240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ichida J. K., et al. 2009. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5, 491–503 10.1016/j.stem.2009.09.012 (doi:10.1016/j.stem.2009.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou H., Ding S. 2010. Evolution of induced pluripotent stem cell technology. Curr. Opin. Hematol. 17, 276–280 10.1097/MOH.0b013e328339f2ee (doi:10.1097/MOH.0b013e328339f2ee) [DOI] [PubMed] [Google Scholar]
- 73.Maherali N., Hochedlinger K. 2009. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 19, 1718–1723 10.1016/j.cub.2009.08.025 (doi:10.1016/j.cub.2009.08.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lyssiotis C. A., et al. 2009. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc. Natl Acad. Sci. USA 106, 8912–8917 10.1073/pnas.0903860106 (doi:10.1073/pnas.0903860106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin T., et al. 2009. A chemical platform for improved induction of human iPSCs. Nat. Methods 6, 805–808 10.1038/nmeth.1393 (doi:10.1038/nmeth.1393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nagy A., Gocza E., Diaz E. M., Prideaux V. R., Ivanyi E., Markkula M., Rossant J. 1990. Embryonic stem cells alone are able to support fetal development in the mouse. Development 110, 815–821 [DOI] [PubMed] [Google Scholar]
- 77.Wei C. L., et al. 2005. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells 23, 166–185 10.1634/stemcells.2004-0162 (doi:10.1634/stemcells.2004-0162) [DOI] [PubMed] [Google Scholar]
- 78.Ohtsuka S., Dalton S. 2008. Molecular and biological properties of pluripotent embryonic stem cells. Gene Ther. 15, 74–81 10.1038/sj.gt.3303065 (doi:10.1038/sj.gt.3303065) [DOI] [PubMed] [Google Scholar]
- 79.Qi X., Li T. G., Hao J., Hu J., Wang J., Simmons H., Miura S., Mishina Y., Zhao G.-Q. 2004. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc. Natl Acad. Sci. USA 101, 6027–6032 10.1073/pnas.0401367101 (doi:10.1073/pnas.0401367101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo G., Smith A. 2010. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 137, 3185–3192 10.1242/dev.052753 (doi:10.1242/dev.052753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W., Smith A. 2009. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 10.1242/dev.030957 (doi:10.1242/dev.030957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou H., et al. 2010. Conversion of mouse epiblast stem cells to an earlier pluripotency state by small molecules. J. Biol. Chem. 285, 29 676–29 680 10.1074/jbc.c110.150599 (doi:10.1074/jbc.c110.150599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li W., et al. 2009. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 4, 16–19 10.1016/j.stem.2008.11.014 (doi:10.1016/j.stem.2008.11.014) [DOI] [PubMed] [Google Scholar]
- 84.Hanna J., et al. 2010. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl Acad. Sci. USA 107, 9222–9227 10.1073/pnas.1004584107 (doi:10.1073/pnas.1004584107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lengner C. J., et al. 2010. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141, 872–883 10.1016/j.cell.2010.04.010 (doi:10.1016/j.cell.2010.04.010) [DOI] [PubMed] [Google Scholar]
- 86.Wang J., Alexander P., Wu L., Hammer R., Cleaver O., McKnight S. L. 2009. Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435–439 10.1126/science.1173288 (doi:10.1126/science.1173288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buehr M., et al. 2008. Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 10.1016/j.cell.2008.12.007 (doi:10.1016/j.cell.2008.12.007) [DOI] [PubMed] [Google Scholar]
- 88.Li P., et al. 2008. Germline competent embryonic stem cells derived from rat blastocysts. Cell 135, 1299–1310 10.1016/j.cell.2008.12.006 (doi:10.1016/j.cell.2008.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawamata M., Ochiya T. 2010. Generation of genetically modified rats from embryonic stem cells. Proc. Natl Acad. Sci. USA 107, 14 223–14 228 10.1073/pnas.1009582107 (doi:10.1073/pnas.1009582107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tong C., Li P., Wu N. L., Yan Y., Ying Q. L. 2010. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature 467, 211–213 10.1038/nature09368 (doi:10.1038/nature09368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim K., et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 10.1038/nature09342 (doi:10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mikkelsen T. S., et al. 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 10.1038/nature07056 (doi:10.1038/nature07056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T. W., Smith A. 2008. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, e253. 10.1371/journal.pbio.0060253 (doi:10.1371/journal.pbio.0060253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Efe J. A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G., Chen J., Ding S. 2011. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13, 215–222 10.1038/ncb2164 (doi:10.1038/ncb2164) [DOI] [PubMed] [Google Scholar]
- 95.Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 10.1016/j.cell.2010.07.002 (doi:10.1016/j.cell.2010.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim J., Efe J. A., Zhu S., Talantova M., Yuan X., Wang S., Lipton S. A., Zhang K., Ding S. 2011. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl Acad. Sci. USA 108, 7838–7843 10.1073/pnas.1103113108 (doi:10.1073/pnas.1103113108) [DOI] [PMC free article] [PubMed] [Google Scholar]