Abstract
Research into the pathophysiological mechanisms of human disease and the development of targeted therapies have been hindered by a lack of predictive disease models that can be experimentally manipulated in vitro. This review describes the current state of modelling human diseases with the use of human induced pluripotent stem (iPS) cell lines. To date, a variety of neurodegenerative diseases, haematopoietic disorders, metabolic conditions and cardiovascular pathologies have been captured in a Petri dish through reprogramming of patient cells into iPS cells followed by directed differentiation of disease-relevant cells and tissues. However, realizing the true promise of iPS cells for advancing our basic understanding of disease and ultimately providing novel cell-based therapies will require more refined protocols for generating the highly specialized cells affected by disease, coupled with strategies for drug discovery and cell transplantation.
Keywords: induced pluripotent stem cells, disease modelling, reprogramming, embryonic stem cells
1. Introduction
Progress in understanding and treating many diseases has been impeded by the lack of in vitro models, especially for conditions for which affected cell types are inaccessible. Many cells affected by disease have defied attempts at in vitro culture, or their phenotype has been altered by accommodation to tissue culture. An abundant source of a pathological cell type of choice could be used for studies of cellular physiology and molecular mechanisms of disease, and provides a platform for drug, siRNA and other screens for potential therapeutic agents (summarized in figure 1). Pluripotent cells are an attractive source of cells when primary cells are difficult to obtain in sufficient numbers for in vitro studies or screening. Disease-specific pluripotent cell lines can be isolated from embryos subjected to preimplantation genetic diagnosis [1], engineered by in vitro mutagenesis [2], or derived from affected individuals through somatic cell reprogramming [3]. This review describes the current catalogue of human disease-specific induced pluripotent stem (iPS) cells, summarized in table 1.
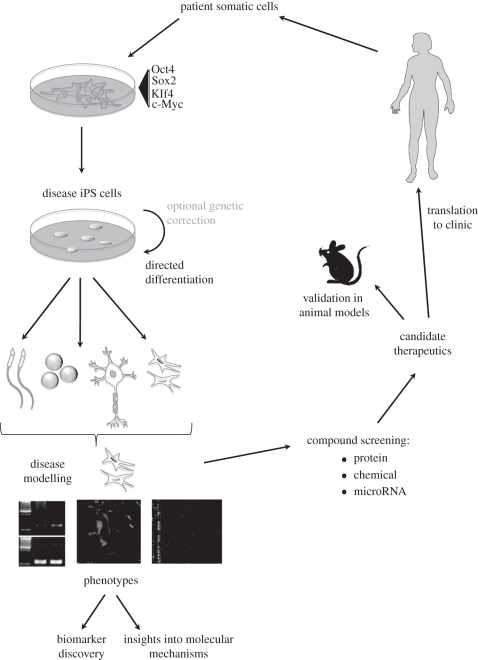
Figure 1.
Overview of the use of iPS cells for disease modelling. Tissue samples from patients are reprogrammed through exogenous expression of transcription factors, tested for pluripotency then differentiated to relevant cell types in vitro. Cells obtained are studied for observable phenotypes by a variety of laboratory procedures, and at this stage, biomarkers can be characterized or insights into molecular mechanisms of disease can be made. Differentiated cell populations are screened as appropriate, using protein, chemical or microRNA libraries, for possible therapeutic targets and/or insights into signalling pathways involved in the disease. Candidates can then be validated using animal models before progressing to clinical testing in humans.
Table 1.
Disease-specific cell lines. The table lists diseases from which iPS cells have been created by category, including information about genetic defect, if known, method of iPS cell generation, cells to which the iPS cells were differentiated, any reported disease phenotype, and drug or functional tests. Abbreviations: 2i, inhibition of MEK1, GSK3 with PD0325901 and CT99021; A1ATD or AAT, alpha 1-antitrypsin deficiency; ADA-SCID, adenosine deaminase severe combined immune deficiency; ALS, amyotrophic lateral sclerosis; FMR1, fragile X mental retardation 1; FH, familial hypercholesterolemia; GBA, acid beta-glucosidase; L, Lin28; M, c-Myc; MSC, mesenchymal stem cells; MStrC, mesenchymal stromal cell; MPS IH, mucopolysaccharidosis type I/Hurler syndrome; N, Nanog; n.a., not applicable; n.d., not determined; NK, not known; NS, nervous system; O, Oct4; S, Sox2; SMA, spinal muscular atrophy; VPA, valproic acid. Method abbreviations: AAV, adeno-associated virus; LV3, lentiviral OSK; LV4, lentiviral OSKM; RV3, retroviral OSK; RV4, retroviral OSKM; LVOSLN, lentiviral OSLN; iLV, inducible lentivirus; eLV, Cre-excisable lentivirus; x, xeno-free; 1eLV, single polycistronic vector excisable lentivirus; T, SV40 large T antigen.
| disease | reference | molecular defect | donor cell | age, sex of donor | method | iPS cells differentiated to: | disease pheno-copied in iPS cells | disease phenocopied in differentiated cells | drug or functional tests |
|---|---|---|---|---|---|---|---|---|---|
| neurological | |||||||||
| ALS | [4] | superoxide dismutase L144F dominant allele | dermal fibroblasts | 82, 89 F | RV4 | motor neurons (HB9 + , ISLET1/2 + , ChAT + ) | n.a. | n.d. | no |
| SMA | [5] | uncharacterized; decreased SMN1 expression | fibroblast | 3 M | LVOSLN | neurons and astrocytes, mature motor neurons | n.a. | yes: reduced size and number of motor neurons, defective synapse formation, with culture | VPA and tobramycin in-creased total SMN1 protein and gem formation |
| Parkinson disease | [3,6–8] | unknown, LRRK2, undefined | dermal fibroblast | 71,53, 53,57, 60,60,85, M,F | RV4, LV3,4, iLV, eLV; some factor free, xRV4 | dopaminergic neurons | n.a. | no | transplant into brains of rats with Parkinson model; rescue of one of three measures of disease |
| Huntington disease | [3] | 72 CAG repeats, huntingtin gene | fibroblast | 20 F | RV4 | none | n.a. | n.d. | no |
| Down syndrome | [3,9] | trisomy 21 | fibroblast | 1 M | RV4 | teratoma | yes | yes; reduced microvessel density | no |
| Fragile X syndrome | [10] | CGG triplet repeat expansion resulting in silencing of FMR1 | dermal fibroblast (2 lines), foetal lung fibroblasts (1 line) | 4, 29 y/o, 22 week M | RV4 | none | yes; FMR1 silenced | n.d. | no |
| Familial dysautonomia | [11] | partial skipping of exon 20 of IKBKAP, reduced IKAP protein | dermal fibroblast | 10 F | LV4 | central NS, peripheral NS, haematopoietic, endothelial, endodermal | higher ratio normal: mutant transcripts in iPS than fibroblast | yes; decreased expression of genes involved in neurogenesis and neuronal differentiation | decreased migration in wound healing assay; kinetin-reduced mutant splice form, and in-creased per cent of differentiating neurons; no improvement in migration |
| Rett syndrome | [12,13] | heterozygous mutation in MECP2: C916T, 1155del32, C730T, C473T | fibroblast | 3,5,8 F | LV4 EOS, RV4 | none, neural progenitor cells | n.d., no | n.d., yes; reduced number of spines and density of glutamatergic synapse formation | yes; IGF1, high dose gentamicin treatment led to more glutamatergic synapses; decreased frequency/intensity of spontaneous currents |
| haematological | |||||||||
| ADA-SCID | [3] | GGG > AGG, exon 7 and Del(GAAGA) exon 10, ADA gene | fibroblast | 3 M | RV4 | none | n.a. | n.d. | no |
| Fanconi anaemia | [14] | FA-A, FA-D2 corrected | dermal fibroblasts | unknown M | RV4, 2 rounds with 2i | haematopoietic | no (corrected) | no (corrected) | no |
| Schwachman–Bodian–Diamond syndrome | [3] | multi-factorial | bone marrow mesenchymal cells | 4 M | RV4 | none | n.a. | n.d. | no |
| sickle cell anaemia | [15,16] | undefined? SS | fibroblast | 20 foetal week F | 1eLV4; eLV4 + T | none | n.d. | n.d. | no |
| beta thalassemia | [17] | homozygous for codon 41/42 4-bp (CTTT) deletion in beta-globin | dermal fibroblast | unknown | RV4 | haematopoietic | n.a. | n.d. | no |
| polycythemia vera | [18] | heterozygous JAK2 1849G > T | fibroblast | unknown | RV4 | haematopoietic | n.a. | yes; enhanced erythropoiesis | no |
| primary myelofibrosis | [18] | heterozygous JAK2 1849G > T | fibroblast | unknown | RV4 | none | n.d. | n.d. | no |
| metabolic | |||||||||
| Lesch-Nyhan syndrome (carrier) | [3,6,19] | heterozygosity of HPRT1; A > G mutation in exon 3 of HPRT1 | fibroblast | 34,11 F | iLV4 + N, iLV3; AAV | none | n.a. | n.d. | no |
| diabetes type I | [3,20] | multi-factorial; unknown | fibroblast | 42,32,30 F,M | RV3,4 | beta-like cells: somatostatin, glucagon, insulin + , glucose-responsive | n.a. | n.d. | no |
| Gaucher disease type III | [3] | AAC > AGC, exon 9, G-insertion, nucleotide 84 of cDNA, GBA gene | fibroblast | 20 M | RV4 | none | n.a. | n.d. | no |
| A1ATD | [15,21] | a1-antitrypsin deficiency: G342K | dermal or liver fibroblasts | 65, 55, 47, 57, 61, 64, 0.3 yr F,M | RV4, 1eLV4 | hepatocyte-like cells (foetal) | n.d. | yes; polymer accumulation | no |
| GSD1a | [21,22] | hepatic glucose-6-phosphate deficiency | dermal fibroblasts | 25, 7 M | RV4 | hepatocyte-like cells (foetal) | n.d. | yes; hyperaccumulation of glycogen | no |
| FH | [21] | familial hypercholesterolemia, autosomal dominant LDLR mutation | fibroblast | NK | RV4 | hepatocyte-like cells (foetal) | n.d. | yes; impaired LDL incorporation | no |
| Crigler–Najjar syndrome | [21,22] | 13 bp deletion, exon 2 of UGT1A1; or L413P | dermal fibroblasts | 2 month old, 19, 21 y/o M,F | RV4 | hepatocyte-like cells (foetal) | n.d. | n.d. | no |
| hereditary tyrosinemia type 1 | [21,22] | 553T > G V166G in fumarylaceto-acetate hydrolase | dermal fibroblasts | 2 month old, 6 y/o M,F | RV4 | hepatocyte-like cells (foetal) | n.d. | n.d. | no |
| progressive familial hereditary cholestasis | [22] | multi-factorial | dermal fibroblasts | 17 F | RV4 | hepatocyte-like cells (foetal) | n.d. | n.d. | no |
| Hurler syndrome (MPS IH) | [23] | IDUA deficiency; Y167X, W402X; W402X, W402X | keratinocyte, MStrC | M 1 | RV4 | haematopoietic | yes; higher GAG accumulation | no difference in CD34 or CD45+ cells or colony formation | no |
| cardiovascular | |||||||||
| LEOPARD syndrome | [24] | heterozygous T468M in PTPN11 | fibroblast | 25, 34 F,M | RV4 | cardiomyocytes | n.d. | yes; cardiomyocyte hypertrophy | antibody microarray: increase EGFR, MEK1 phosphorylation, no pERK response to bFGF |
| long QT syndrome | [25] | dominant R190Q in KCNQ1 | dermal fibroblast | 8,42 M | RV4 | cardio-myocytes | n.a. | yes; longer QT in ventricular and atrial myocytes; impaired cell membrane targeting of KCNQ1 protein | decreased IKs current density |
| other categories | |||||||||
| Duchenne muscular dystrophy | [3,26] | deletion of exon 45–52 or 46–50, dystrophin gene; | dermal fibroblast | 47, 13, 6 F carrier, M affected | RV4, 1LV4 | none | n.a. | n.d. | no |
| Becker muscular dystrophy | [3] | unidentified mutation in dystrophin | fibroblast | 38 M | RV4 | none | n.a. | n.d. | no |
| dyskeratosis congenita (DC) | [6,27] | del37L in DKC1 | fibroblast | 7,30 M | RV4, iLV3 | none | no | n.d. | no |
| cystic fibrosis | [15,28] | homozygous delta F508 in CFTR | dermal fibroblast | 8,21,29,31,33 F,M | 1eLV4 | none | n.d. | n.d. | no |
| scleroderma | [15] | unknown | dermal fibroblast | 47 F | 1eLV4 | none | n.d. | n.d. | no |
| osteogenesis imperfecta | [19] | G > A in exon 34 of COL1A2 | MSC | NK | AAV OSLN | none | n.a. | n.d. | no |
2. Diseases modelled
Our laboratory produced the first large repository of disease-specific iPS cells, which provided a proof of principle for the robust application of transcription-factor-based reprogramming as a facile means of production of iPS cells [3]. We reprogrammed fibroblasts or bone marrow mesenchymal cells from individuals with 10 different disorders, and confirmed the causative genetic lesions in all seven of the cases in which the underlying gene defect was known. We reprogrammed fibroblasts from two individuals with Down syndrome and confirmed trisomy 21 by karyotype analysis. The immune deficiency in adenosine deaminase deficiency-related severe combined immunodeficiency (ADA-SCID) entails the absence of T, B and NK cells. iPS cells from an ADA-SCID patient's fibroblasts harboured two genetic defects: GGG to AGG in exon 7 (G215R mutation) of the adenosine deaminase gene, and on the other allele a frameshift deletion (–GAAGA) in exon 10. Shwachman–Bodian–Diamond syndrome (SBDS) is characterized by congenital exocrine pancreas insufficiency, skeletal abnormalities and bone marrow failure. In iPS cells derived from bone marrow mesenchymal cells from an SBDS patient, point mutations at the IV2 + 2T > C intron 2 splice donor site and IVS3–1G > A mutations were documented in the SBDS gene. Gaucher disease (GD) type III patients display pancytopenia and progressive neurological deterioration in this lysosomal storage disease caused by acid beta-glucosidase (GBA) gene mutations. iPS cells from fibroblasts from a GD patient revealed a 1226A > G point mutation (N370S mutation) as well as the frameshift insertion 84GG on the other allele. iPS cells were also derived from fibroblasts from a heterozygous carrier of Lesch–Nyhan syndrome, involving accumulation of uric acid owing to hypoxanthine–guanine phosphoribosyltransferase (HPRT) mutations, resulting in neurological deficits. Fibroblasts from a patient with Duchenne muscular dystrophy (DMD) were reprogrammed to yield iPS cells with deletion of exons 45–52, but the unknown defect in the iPS cells of a case of Becker-type muscular dystrophy (BMD) was not detected by multiplex PCR. In fibroblast-derived iPS cells from a patient with Huntington's disease (HD), 72 (CAG)n polyglutamine triplet repeat sequences were seen in the huntingtin gene. Parkinson disease (PD) and juvenile onset diabetes (JDM) iPS cells were also generated from patient fibroblasts, but the genetic defects for these diseases have not been characterized.
In this first study, we confirmed the capacity to derive disease-specific iPS cells from a variety of different individuals of different ages and afflicted by different conditions, but did not endeavour to document cell-based phenotypes for this range of diseases. However, soon after developing this first group of disease models, we employed the Down syndrome iPS lines in a collaborative study that implicated the chromosome 21 gene Down syndrome critical region-1 (DSCR-1 gene), a putative negative regulator of angiogenesis, in the curious observation of reduced solid tumour incidence in affected individuals. Murine strains engineered to overexpress the human DSCR-1 gene showed reduced capacity to support human tumour xenografts, which correlated with reduced angiogenesis. Importantly, a comparison of teratomas formed from Down syndrome and normal iPS cells in immune-deficient mice revealed a reduced microvessel density in the Down's samples, thereby providing evidence that the reduced tumour incidence in Down syndrome may be due to a reduced capacity to sustain tumour angiogenesis [9].
Laboratories that have been among the first to explore disease phenotypes in vitro have focused on disorders traceable to dysfunction of a specific cell type for which an effective protocol for in vitro differentiation is available, often times founded upon prior studies of directed differentiation of human embryonic stem (ES) cells. Because of the sophistication of prior studies that have recapitulated neuronal development, several of the most effective disease models have reflected neurological and neurodegenerative diseases, and have included amyotrophic lateral sclerosis (ALS), spinal muscular atrophy, PD, familial dysautonomia (FD) and retinal degeneration.
(a). Models of neurological and neurodegenerative conditions
Given the elegant demonstration of directed motor neuron differentiation from ES cells [29], ALS has been modelled by reprogramming dermal fibroblasts from two individuals aged 82 and 89, both heterozygous for the L144F mutation in the superoxide dismutase gene [4]. Only one of these individuals was symptomatic, and further characterization focused on the individual with active ALS. Upon differentiation of the resulting iPS cells to motor neurons with a sonic hedgehog agonist and retinoic acid, 20 per cent expressed the motor neuron marker HB9. Subsets of the motor neurons also expressed markers for other neuronal cell types. These cells await further functional and anatomical characterization to identify whether the neurons manifest disease-relevant phenotypes in culture.
Spinal muscular atrophy (SMA) is caused by autosomal recessive mutation in the survival motor neuron 1 (SMN1) gene, and is associated with reduced expression of SMN1 and the loss of lower motor neurons. iPS cells have been derived and studied from a patient with SMA type 1, the most severe form, and his unaffected mother, who served as a related control [5]. The molecular nature of the gene defect was not elucidated, but lower levels of full-length SMN1 transcripts were seen in SMA patient fibroblasts and iPS cells than in the control. The authors then generated neural stem cells from the iPS cells, further specified the neural stem cells to motor neuron fate as marked by motor neuron transcription factors HOXB4, OLIG2, ISLET1 and HB9, and mature motor neuron markers SMI-32 and choline acetyltransferase. After another two weeks in culture, however, a significant decrease was observed in motor neuron number and size, but not the total neuron pool, relative to control. SMN1 protein distribution, absent in nuclear structures called gems in SMA-iPS cell-derived neurons, could be induced by valproic acid or tobramycin, a finding that confirmed feasibility for drug screening. This was the first iPS cell modelling study to show disease-specific changes, selective motor neuron death, in the cell population of interest.
PD-specific iPS cells were generated by reprogramming fibroblasts from seven individuals with idiopathic disease using either three (OSK) or four (OKSM) lentiviral factors [6,7]. From these, dopaminergic neurons were successfully generated with similar proportions of dopaminergic neurons (tyrosine hydroxylase+ TUJ1+) differentiated from PD as from non-PD iPS cells using two distinct differentiation protocols. The cells were further characterized by transplantation into rat brains, with or without previous treatment with 6-hydroxydopamine (6-OHDA), a dopamine analogue that is toxic to dopaminergic neurons. The authors analysed the survival and behaviour of this rat model of PD, and observed viable grafts up to 16 weeks after transfer. No significant differences were observed between control and PD populations in axonal outgrowth or density; in both cases, the outgrowth was reduced, and the authors interpret these results to mean that the in vitro differentiation system results in inefficient patterning. Functionally, transplanted rats showed improvement in one aspect of the severe motor asymmetry observed in this model, ipsilateral amphetamine-induced rotations; in two other measures of complex motor functions, no improvement in performance was seen after transplant. No tumour formation was observed in these transplants, in contrast to an earlier study by Wernig et al. [30], which reprogrammed mouse fibroblasts, differentiated them to dopaminergic neurons, and transferred them into the rat model of PD. Although they did observe improvement of disease symptoms, they also reported tumour formation. More recently another study has demonstrated improved differentiation of dopaminergic neurons from iPS cells in a xenogenic-free defined medium [8]. This group also derived iPS cells from a PD patient with a LRRK2 mutation, the details of which have not yet been published.
Yet another neurological condition to be modelled by reprogramming fibroblasts from patients and non-affected controls is FD, an autosomal recessive degenerative sensory and autonomic neural disease in which there is a tissue-specific splicing defect in the IKBKAP gene [11]. In a tour de force of complex protocols, differentiation along central nervous system (neural crest), peripheral nervous system, haematopoietic, endothelial and endodermal lines were similar for disease and control iPS cells, but normal versus mutant transcript ratios varied, with the lowest ratio seen in endodermal, haematopoietic and neural crest precursors. The authors focused on the neural crest lineages, in keeping with the disease pathophysiology. Microarray transcriptome analysis was performed, comparing FACS-sorted neural crest precursors from FD- and control iPS cell-derived cells. This revealed 35 transcripts significantly higher and 54 lower in FD versus control iPS cell-derived cells; many of the genes with the most significant expression deficit were those involved in peripheral neurogenesis and neuronal differentiation. A wound-healing assay revealed a decrease in the migration of the FD-iPS cell-derived neural crest precursors compared with controls, again consistent with disease pathophysiology. A panel of assays was performed on multiple FD-iPS cell lines, with similar results. In an important proof of principle, the authors tested several drugs previously shown to have an effect on splicing, and demonstrated that kinetin but not two other drugs reduced the proportion of the mutant splice form, and specifically in the disease-specific iPS cell-derived cells. While short-term kinetin treatment did not affect the expression of neurogenic markers or migratory behaviour, treatment during all 28 days of culture resulted in significant improvements in the percentage of differentiating neurons and in the expression of two peripheral neuron markers, ASCL1 and SCG10; the migratory phenotype remained unchanged. These studies present encouraging evidence that the use of iPS cells to explore disease mechanisms and treatment will lead to improved understanding of pathology and strategies for therapy.
At least one study has defined a marked difference between the fidelity of disease modelling when pluripotent cells derived from embryos subjected to preimplantation diagnosis (PGD) were compared with iPS cells generated from the fibroblasts of living affected individuals. The Benvenisty group has previously isolated a human ES cell line from an embryo affected by fragile X (FX) syndrome, a common inherited cause of mental retardation [1]. FX is caused by a CGG triplet repeat expansion in the 5′ UTR of the FX mental retardation 1 (FMR1) gene. The FMR1 protein is expressed in human ES cells, but is transcriptionally silenced following in vitro differentiation, which reflects the dynamic silencing that is observed during foetal development in FX individuals. Working together with the Benvenisty group, we reprogrammed fibroblasts from affected individuals, but observed that the FMR1 gene remained silenced in the pluripotent iPS cells, and consistent with its transcriptionally silent state, there was H3K9 methylation and no histone acetylation or H3K4 methylation. The FMR1 locus thus appears to be resistant to reprogramming, even with iPS cells achieving pluripotency by stringent criteria, demonstrating that human ES (hES) cells and iPS cells are not equivalent, and suggesting that in some circumstances, ES cells may more faithfully reflect the disease process [10].
Retinal degeneration, as a consequence of gene defects or degeneration of retinal pigment epithelia or photoreceptors themselves, causes many blinding disorders, and given that these two tissues can be readily differentiated from hES cells, the appeal of modelling disease and treatment with iPS cells have compelled the direct comparison of retinal development in hES cells and iPS lines [31]. Similar frequencies of retinal cell types were obtained from both, demonstrating that iPS cells are equivalent to hES cells in their ability to generate these lineages. However, the cell populations achieved only a primitive stage of eye development, and the authors noted considerable variability in the ability of individual iPS lines to undergo neurectodermal differentiation towards retinogenesis; thus they chose to analyse a single clone in depth. While this paper did not model a specific retinal disease, with the systems established here, studies of disease-specific retinal cells should soon follow. Expectations for these cells are high [32] and it is thought that retinal degeneration is an example of a disease most amenable to treatment with iPS cells.
Rett syndrome, an X-linked neurodevelopmental disorder characterized by the loss of verbal and other skills in early childhood and mental retardation, was modelled as a proof of principle in the use of early transposon promoter and Oct-4 and Sox2 enhancers (EOS) lentiviral vector system to derive iPS cells; in this initial study, characterization or differentiation of these cells was not reported beyond confirmation of genotype and embryonic body (EB) formation [12]. However, a more complete study recently published documented a remarkable set of abnormalities in neuronal populations, including reduced synapse and dendritic spine density, smaller size, altered calcium signalling and electrophysiological defects in the diseased cells [13]. Such highly sophisticated cell phenotyping, which is possible for neuronal populations in vitro, represents a major new opportunity for exploring disease mechanisms and strategies for drug screening.
(b). Models of haematopoietic disorders
Haematopoietic conditions represent a second broad class of disorders that has generated interest among modellers, owing to the relative ease of differentiating pluripotent stem cells into the haematopoietic lineages. Beta-thalassemia, which results from defective synthesis of beta globin leading to progressive anaemia requiring blood transfusions and iron chelation therapy, has been modelled by reprogramming patient fibroblasts, amniotic fluid cells or chorionic villus sample cells [17]. The iPS cells were differentiated into haematopoietic colonies and haemoglobinized erythroid cells were observed, although no specific disease phenotype was reported. Because protocols for directed haematopoietic differentiation most faithfully recapitulate the embryonic and foetal stages of blood development, observation of phenotypes in adult blood is limited and must await improved differentiation protocols. This is particularly true for lymphoid lineages, which remain challenging to achieve in culture.
Fanconi anaemia (FA), the most common genetic bone marrow failure syndrome, is caused by recessive autosomal or X-linked mutation in one of 13 genes in the FA pathway. Fibroblasts from patients with this condition show enhanced sensitivity to agents that break DNA strands, and the FA pathway has been documented to be important for the repair of DNA double-strand breaks. Interestingly, attempts to reprogramme FA fibroblasts or keratinocytes were unsuccessful unless the gene defect was first corrected via lentiviral gene delivery [14]. In only a single patient with a defect in FANCD2 could fibroblasts be reprogrammed before gene correction, but the reprogrammed cells could not be maintained for more than three passages. Patients with defects in FANCD2 tend to express some residual protein, suggesting that only hypomorphic alleles of the FANCD2 gene are compatible with life. This overall resistance of FA fibroblasts to reprogramming implies that the FA pathway is necessary for reprogramming. The FA pathway was functional in the gene-corrected iPS cells and their derivatives, as demonstrated by relocation of FANCD2 to damaged subnuclear areas upon UV irradiation or hydroxyurea treatment. In vitro differentiation towards haematopoietic lineages resulted in similar proportions of both erythroid and myeloid colony-forming cells in both gene-corrected iPS cells relative to normal iPS controls. Another strategy employed gene knockdown in hES cells to demonstrate that the FA pathway is dispensable for self-renewal [33]; these cells were then differentiated to haematopoietic lineages via EB culture with haematopoietic cytokines, followed by methylcellulose haematopoietic colony formation. FANCD2-knockdown hES cells demonstrated significant reduction in total colony-forming units, both FANCD2- and FANCA-knockdown hES cells were hypersensitive to DNA damage induced by mitomycin C, and both showed a reduction in the ratio between gamma- and epsilon-globin in comparison with controls, rescued by overexpression. Thus in this case, owing to the apparent role of FA pathway members in reprogramming, iPS cells do not present a feasible platform for the study of FA, while more can be learned about the pathophysiology of the disease by knocking the protein down in wild-type hES (or presumably iPS) cells.
Sometimes an attempt at disease modelling teaches an unanticipated lesson about the mechanisms of the pluripotent state itself. We endeavoured to explore the role of telomerase in reprogramming by studying cells from patients with dyskeratosis congenita (DC), a disorder of telomerase deficiency that results in shortened telomeres, premature senescence and fibrosis of lungs and liver, skin abnormalities and marrow failure. We began by attempting to reprogramme cells from individuals with an X-linked form of the disorder caused by mutations in the dyskerin gene, an RNA-binding protein that stabilizes the telomerase RNA component (TERC). Although reprogramming efficiency was poor, fully pluripotent iPS cells were obtained and could be indefinitely passaged, in contrast to parental fibroblasts, which senesced after three to four passages. Telomere length, which was shortened in parental fibroblasts, decreased immediately after reprogramming, but increased with serial passage of iPS cells until it was comparable to the parental line; TERC expression, which was decreased to 10–15% of normal in parental fibroblasts, increased six to eightfold in the iPS cells. A similar increase in TERC levels was observed in normal pluripotent iPS and ES cells, therefore demonstrating that high levels of TERC are a hallmark of the pluripotent state. Upon differentiation of DC-iPS cells to fibroblasts, TERC expression and telomere length reverted to the pre-reprogrammed state, recapitulating the disease phenotype [27]; it will be interesting to test differentiation to lineages most severely affected in DC, such as haematopoietic precursors, where early senescence is expected.
Attempts to capture disease-specific mutations that arise in somatic tissues were first achieved by reprogramming peripheral blood CD34+ cells of patients with the myeloproliferative disorder (MPD) caused by JAK2-V617F mutation, which is present in most polycythaemia vera (PV) patients and half of essential thrombocytosis and primary myelofibrosis cases [18]. Haematopoietic differentiation by modified EB formation, feeder- and serum-free culture produced myeloid and erythroid colonies in multiple lineages. As predicted, consistent with disease phenotype, the PV-iPS cells produced more erythroid colonies than did controls, and culture conditions favouring erythropoiesis resulted in significantly more erythroid cells in the PV-iPS cell-derived case than in controls. Comparing iPS cells and their derivatives from the other MPDs (besides PV) could shed light on the variety of diseases caused by a single mutation. iPS cells have also been created for sickle cell disease [15,16], but these await disease modelling and characterization.
(c). Models of cardiovascular conditions
Cardiovascular disease is a third class that can be modelled because of the availability of protocols for directed differentiation into cardiomyocytes. Long QT syndrome type I, a condition that predisposes to potentially life-threatening cardiac arrhythmias that can be exacerbated by drugs, has now been modelled by deriving iPS cells from skin fibroblasts of two family members with the dominant mis-sense mutation in KCNQ1 (R190Q), a component of a potassium channel [25]. The iPS cells were then differentiated into cardiomyocytes that demonstrated the classical electrocardiographic derangement of a long QT interval, reflecting a slower repolarization velocity, in the atrial and ventricular populations, but normal QT intervals in the nodal cells. The QT intervals in cardiomyocytes differentiated from iPS cells from unaffected individuals were normal. Furthermore, the authors demonstrated impaired trafficking of mutant KCNQ1 from endoplasmic reticulum to plasma membrane, as previously shown, and electrophysiological analyses showed the expected decrease in the delayed rectifier IKs current density when compared with control cells. Response to adrenergic stimuli was significantly impaired, and as anticipated from clinical experience, protected by beta adrenergic receptor blockade. The iPS cells provided a significant improvement over available animal models, in which changes non-specific to the disease could have been due to transgenic expression of dominant-negative mutants or to limitations of the animal host [34]. This study realizes a long-standing ambition of stem cell biology to employ human cells of a specific genotype for characterizing potential adverse and toxic side effects of medications, and thus to advance the prospects for drug development.
LEOPARD syndrome, an autosomal dominant disorder including hypertrophic cardiomyopathy, facial dysmorphisms, lentigines or liver spots, growth retardation, abnormal genitalia and deafness, has been modelled by deriving iPS cells from two adult patients, and a non-affected sibling as control [24]. As in the majority of cases, a mis-sense mutation, T468M, in the PTPN11 gene is the cause of the disease. The iPS cells met criteria for pluripotency, and DNA fingerprinting verified that the iPS cell source was patient-derived fibroblasts. Two lines from each patient were differentiated into haematopoietic and cardiac lineages. Beating EBs were observed as expected, and cells from LEOPARD lines had significantly increased median surface area, increased sarcomere assembly and increased proportion of cells with nuclear NFATC4, consistent with cardiomyocyte hypertrophy. As acknowledged by the authors, several of the relevant cellular phenotypes could not be adequately assessed because the relevant cell population could not be generated through in vitro culture at this time; thus none of the non-cardiac disease phenotypes were characterized in this report. However, signalling pathways and molecular targets involved in this syndrome were queried by an antibody microarray, and verified in several cases by Western blot. Epidermal growth factor receptor (EGFR) and MEK1 phosphorylation were increased to a variable degree in the disease lines, and basic fibroblast growth factor (bFGF) stimulation failed to elicit a phosphorylated ERK (pERK) response in the LEOPARD iPS cell lines, as it did in the control lines. The analyses performed in this paper provide examples of the productive use of disease-specific iPS cell lines for elucidation of pathology and affected signalling pathways.
(d). Models of metabolic disorders
Inherited metabolic disorders of the liver have been modelled by reprogramming patient fibroblasts, followed by directed differentiation using chemically defined medium with polyvinyl alcohol, Activin A, FGF2, bone morphogenetic protein (BMP)4 and phosphoinositide 3-kinase (PI3K) inhibitor to derive endoderm cells, followed by Activin-A and B27 for hepatic progenitors, and hepatocyte growth factor (HGF)/Oncostatin-M to derive cells with functional properties of hepatocytes [21]. The resulting foetal-like liver cells demonstrated glycogen and low density lipoprotein (LDL) storage, albumin secretion, drug metabolism and glucagon response. Curiously, 10 per cent of the iPS cell lines did not support differentiation to the hepatocyte lineage. Alpha1-antitrypsin deficiency (A1ATD), the most common pathology requiring liver transplantation, involves hepatotoxicity owing to accumulation of the poorly secreted, mutant α1-antitrypsin Z molecule [35]. Hepatocytes from patients with A1ATD, but not control iPS cells, were demonstrated to accumulate α1-antitrypsin polymers in the endoplasmic reticulum, phenocopying an important feature of the disease. Familial hypercholesterolaemia (FH) is caused by mutation of the LDL receptor, and is characterized by premature atherosclerotic cardiovascular disease [36]. Hepatocytes from FH-iPS cells could not incorporate LDL, as expected, while those from normal iPS cells could. Glycogen storage disease type 1a (GSD1a), a metabolic disorder characterized by inability to maintain glucose homeostasis and sequelae including growth retardation, hepatomegaly, lactic acidaemia and hyperlipidaemia, is caused by deficiency of glucose-6-phosphatase [37]. In a GSD1a model, patient iPS cell-derived hepatocytes accumulated greater amounts of intracellular glycogen and lipid, and produced more lactic acid, than those from normal individuals, as expected in glucose-6-phosphatase deficiency. Another panel of liver diseases including tyrosinemia type I, GSD1, progressive familial hereditary cholestasis and Crigler–Najjar syndrome have been modelled by creating iPS cell lines from patient fibroblasts, differentiating into hepatocyte-like lineages by generating EBs, then supplementing with Activin A (definitive endoderm), then varying concentrations of knockout serum replacement, followed by culture in media containing HGF then dexamethasone, yielding cells with epithelial morphology positive for hepatic markers that secrete albumin and take up LDL, among other characteristics [22].
Since mucopolysaccharidosis type I (also known as Hurler syndrome; MPS IH) is an attractive candidate for gene-corrected autologous cell transplant, iPS cells were raised from patient samples and their phenotype characterized. In this disease, deficiency in α-l-iduronidase (IDUA) causes build-up of glycosaminoglycans (GAGs), leading to progressive dysfunction in various organ systems, and death within the first decade. iPS cells derived from patient keratinocytes and mesenchymal stromal cells were found to contain elevated levels of GAGs, with partial rescue upon exogenous (lentiviral) expression of IDUA. Wild-type (WT) and mutant iPS differentiated equally well into haematopoietic lineages [23]. These results suggest that disease aetiology may be from a very early stage in development, providing an example of a condition in which much can be learned from disease modelling by iPS cells.
As facility with reprogramming and directed differentiation protocols becomes more widely applied, a host of diseases that are not defined by single genes with Mendelian penetrance, and for which specific cellular phenotypes may not be obvious, will nonetheless lend themselves to exploration using iPS cells as tools. iPS cells have been produced from patients with type I diabetes, which has unknown genetic and molecular aetiology but results from the autoimmune destruction of insulin-secreting pancreatic beta cells [20]. The iPS cells could be differentiated towards pancreatic lineage by a stepwise protocol through definitive endoderm (using Wnt3A and Activin A), gut tube endoderm (FGF10 and cyclopamine), pancreatic progenitors (FGF10, cyclopamine, RA, (−)-Indolactam V) and beta-like cells (EX-4, DAPT, HGF and IGF1), albeit at low efficiency. This regime produced cells positive for somatostatin, glucagon and insulin, which released C-peptide, indicative of insulin release, upon glucose stimulation, at levels similar to controls. Given that type I diabetes appears to reflect an interplay of autoimmune dysregulation among T cells, thymic epithelia and beta cells, identifying cellular phenotypes that reflect the disease process may require coordinated differentiation and analysis of multiple tissue types, transplanted into an immune-deficient mouse host, so that the autoimmune process can be recapitulated and studied.
(e). An expanding potpourri of disease models
Several forms of lung disease including emphysema (α-1 antitrypsin deficient), cystic fibrosis (homozygous delta F508 in CFTR) and scleroderma (unknown genetic component) have been represented in iPS cells via reprogramming dermal or liver fibroblasts from patients using a single excisable lentiviral vector with four factors [15]. iPS cells were then differentiated to definitive endoderm (from which lung and liver epithelium were derived) by culture with BMP4, then Activin A, FGF2 and BMP4. No comparison was made with WT-iPS cell-derived definitive endoderm, and no disease correlations were made.
Reprogramming fibroblasts from a carrier and an affected individual with Duchenne muscular dystrophy, an X-linked disease, yielded insights into X chromosome inactivation [26], as some clones from the carrier expressed mutant and some wild-type dystrophin. The authors demonstrated that, while X-inactivation is random in the starting cell population, iPS cells maintain the status of the X chromosome present in the starting fibroblast in a clonal manner: the X chromosome is not re-inactivated in human iPS cells, as in the mouse. These lines (and similar ones derived from individuals who are carriers for other X-linked diseases) will provide well-matched experimental and control iPS cell populations for disease modelling and drug screening.
3. Progress and challenges
While considerable progress has been made in deriving iPS cells from patients, and differentiating them into tissues of interest, the use of those cells as platforms for understanding disease pathogenesis and the development of therapies is just beginning. Of the 25 modelling studies cited, 12 could successfully demonstrate a phenotype of any kind in the cell type of interest, and seven revealed a specific functional deficit. Further studies are expected to reveal aspects of pathophysiology not yet understood. Chemical screens, none of which have to date been published, represent an area of exceptional promise in the iPS modelling field: the ability to generate large quantities of pluripotent cells, and therefore differentiated cells, will enable large-scale studies of small-molecule libraries with the potential for both biological insights and therapeutics.
In some cases, traditional reprogramming techniques may not prove satisfactory. In the case of diseases in which the defect precludes generation of pluripotent cells, such as FA, a knockdown strategy has been shown to be an effective alternative. For those in which iPS cells do not provide an adequate model, such as FX syndrome, hES cells are the better option, unless future innovations prove to fully reprogramme such cells.
The main limiting factor for exploitation of iPS cells in modelling is availability of reliable and repeatable protocols for complete differentiation to a tissue type of choice. Great progress has been made towards this end, evidenced by papers reviewed here and elsewhere [38], but in most cases, differentiation to adult fate has not been accomplished, and the painstaking work of development of consistent protocols will continue.
While iPS cell technology offers unprecedented opportunities, investigators should be mindful of several cautions regarding their use. As noted by several of the authors, variability exists in the ability of iPS cell lines to differentiate to a population of choice [31,39]. This could reflect subtle differences in cell of origin, incomplete reprogramming, epigenetic memory [40,41] or native variability [42]. Testing multiple lines, preferably from multiple patients, is crucial. While iPS cells are a boon to the disease modelling field, they should be compared with hES cells whenever feasible, since there is ample evidence that iPS cells may retain different degrees of residual methylation owing to the technical limitations of the reprogramming process [10,40,43]. Aberrations occurring owing to prolonged culture can affect iPS cells, so culture periods should be minimized and lines should be tested frequently for genomic changes, at all stages including target somatic cells, iPS cell lines and differentiated tissues. Investigators should realize the potential for iPS cells, and modify their informed consent procedures for obtaining donated tissue for research, so that future investigators will not face limitations in the use of the cells for modelling purposes [44].
Diseases of ageing appear to present a particularly challenging barrier to iPS-based modelling, since the time frame for completing research projects, as well as a finite lifespan of differentiated cell types in culture, may preclude recapitulation of the disease process. For diseases such as PD, HD and retinal degeneration, alternative strategies will have to be developed, perhaps including in vitro treatment of cells with substances known to accelerate ageing, such as those that induce oxidative stress.
Many of the papers reviewed have shown promising results. In many cases, though, the reader is left with a sense that much more work is necessary in order to truly model the disease. Close collaboration between laboratories specializing in iPS cell technology, study of the disease modelled, pluripotent cell differentiation in specified directions and various screening methods will be essential in order to bring disease modelling to fruition.
4. Conclusions
The field of iPS cell disease modelling is gaining momentum. Investigators can increase the odds of successful modelling by carefully selecting diseases to model in iPS cells, based on genetics/epigenetics and clinical course of the disease, characteristics of cell types involved in pathology, and availability of patient tissues [45]. As tightly directed differentiation is accomplished in an expanding repertoire of cell types, iPS cell-derived cells can be used for studies of cellular function and for screening in ways that will further our understanding of disease and provide treatment options for patients.
Acknowledgements
G.Q.D. is supported by grants from the NIH (RO1-DK70055, RO1-DK59279, UO1-HL100001, and special funds from the ARRA stimulus package RC2-HL102815), the Roche Foundation for Anemia Research, and Alex's Lemonade Stand. G.Q.D. is a recipient of Clinical Scientist Awards in Translational Research from the Burroughs Wellcome Fund and the Leukaemia and Lymphoma Society, and is an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research. J.J.U. is supported by a grant from the NIH (T32-HL07623-23). We thank M. Willy Lensch for critical review of the manuscript, Patrick Cahan for contribution to the figure and Wolfgang Unternaehrer for assistance in preparing the table.
References
- 1.Eiges R., et al. 2007. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell 1, 568–577 10.1016/j.stem.2007.09.001 (doi:10.1016/j.stem.2007.09.001) [DOI] [PubMed] [Google Scholar]
- 2.Cova E., Ghiroldi A., Guareschi S., Mazzini G., Gagliardi S., Davin A., Bianchi M., Ceroni M., Cereda C. 2010. G93A SOD1 alters cell cycle in a cellular model of Amyotrophic Lateral Sclerosis. Cell. Signal. 22, 1477–1484 10.1016/j.cellsig.2010.05.016 (doi:10.1016/j.cellsig.2010.05.016) [DOI] [PubMed] [Google Scholar]
- 3.Park I. H., et al. 2008. Disease-specific induced pluripotent stem cells. Cell 134, 877–886 10.1016/j.cell.2008.07.041 (doi:10.1016/j.cell.2008.07.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimos J. T., et al. 2008. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321, 1218–1221 10.1126/science.1158799 (doi:10.1126/science.1158799) [DOI] [PubMed] [Google Scholar]
- 5.Ebert A. D., Yu J., Rose F. F., Jr, Mattis V. B., Lorson C. L., Thomson J. A., Svendsen C. N. 2009. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457, 277–280 10.1038/nature07677 (doi:10.1038/nature07677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soldner F., et al. 2009. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136, 964–977 10.1016/j.cell.2009.02.013 (doi:10.1016/j.cell.2009.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hargus G., et al. 2010. Differentiated Parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in Parkinsonian rats. Proc. Natl Acad. Sci. USA 107, 15 921–15 926 10.1073/pnas.1010209107 (doi:10.1073/pnas.1010209107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swistowski A., Peng J., Liu Q., Mali P., Rao M. S., Cheng L., Zeng X. 2010. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells 28, 1893–1904 10.1002/stem.499 (doi:10.1002/stem.499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baek K. H., et al. 2009. Down's syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature 459, 1126–1130 10.1038/nature08062 (doi:10.1038/nature08062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urbach A., Bar-Nur O., Daley G. Q., Benvenisty N. 2010. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell 6, 407–411 10.1016/j.stem.2010.04.005 (doi:10.1016/j.stem.2010.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee G., et al. 2009. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461, 402–406 10.1038/nature08320 (doi:10.1038/nature08320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotta A., et al. 2009. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods 6, 370–376 10.1038/nmeth.1325 (doi:10.1038/nmeth.1325) [DOI] [PubMed] [Google Scholar]
- 13.Marchetto M. C., Carromeu C., Acab A., Yu D., Yeo G. W., Mu Y., Chen G., Gage F. H., Muotri A. R. 2010. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143, 527–539 10.1016/j.cell.2010.10.016 (doi:10.1016/j.cell.2010.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raya A., et al. 2009. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 460, 53–59 10.1038/nature08129 (doi:10.1038/nature08129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somers A., et al. 2010. Generation of transgene-free lung disease-specific human iPS cells using a single excisable lentiviral stem cell cassette. Stem Cells 28, 1728–1740 10.1002/stem.495 (doi:10.1002/stem.495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P., Ye Z., Hommond H. H., Yu X., Lin J., Chen G., Zou J., Cheng L. 2008. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 26, 1998–2005 10.1634/stemcells.2008-0346 (doi:10.1634/stemcells.2008-0346) [DOI] [PubMed] [Google Scholar]
- 17.Ye L., Chang J. C., Lin C., Sun X., Yu J., Kan Y. W. 2009. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc. Natl Acad. Sci. USA 106, 9826–9830 10.1073/pnas.0904689106 (doi:10.1073/pnas.0904689106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Z., et al. 2009. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood 114, 5473–5480 10.1182/blood-2009-04-217406 (doi:10.1182/blood-2009-04-217406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan I. F., et al. 2010. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 18, 1192–1199 10.1038/mt.2010.55 (doi:10.1038/mt.2010.55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maehr R., Chen S., Snitow M., Ludwig T., Yagasaki L., Goland R., Leibel R. L., Melton D. A. 2009. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc. Natl Acad. Sci. USA 106, 15 768–15 773 10.1073/pnas.0906894106 (doi:10.1073/pnas.0906894106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashid S. T., et al. 2010. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Invest. 120, 3127–3136 10.1172/JCI43122 (doi:10.1172/JCI43122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghodsizadeh A., et al. 2010. Generation of liver disease-specific induced pluripotent stem cells along with efficient differentiation to functional hepatocyte-like cells. Stem Cell Rev 6, 622–632 10.1007/S12015-010-9189-3 (doi:10.1007/S12015-010-9189-3) [DOI] [PubMed] [Google Scholar]
- 23.Tolar J., et al. 2010. Hematopoietic differentiation of induced pluripotent stem cells from patients with mucopolysaccharidosis type I (Hurler syndrome). Blood 117, 839–847 10.1182/blood-2010-05-287607 (doi:10.1182/blood-2010-05-287607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvajal-Vergara X., et al. 2010. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature 465, 808–812 10.1038/nature09005 (doi:10.1038/nature09005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moretti A., et al. 2010. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med 363, 1397–1409 10.1056/NEJMoa0908679 (doi:10.1056/NEJMoa0908679) [DOI] [PubMed] [Google Scholar]
- 26.Tchieu J., et al. 2010. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell 7, 329–342 10.1016/j.stem.2010.06.024 (doi:10.1016/j.stem.2010.06.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agarwal S., et al. 2010. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature 464, 292–296 10.1038/nature08792 (doi:10.1038/nature08792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warren L., et al. 2010. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 10.1016/j.stem.2010.08.012 (doi:10.1016/j.stem.2010.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wichterle H., Lieberam I., Porter J. A., Jessell T. M. 2002. Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397 10.1016/S0092-8674(02)00835-8 (doi:10.1016/S0092-8674(02)00835-8) [DOI] [PubMed] [Google Scholar]
- 30.Wernig M., et al. 2008. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc. Natl Acad. Sci. USA 105, 5856–5861 10.1073/pnas.0801677105 (doi:10.1073/pnas.0801677105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer J. S., Shearer R. L., Capowski E. E., Wright L. S., Wallace K. A., McMillan E. L., Zhang S.-C., Gamm D. M. 2009. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA 106, 16 698–16 703 10.1073/pnas.0905245106 (doi:10.1073/pnas.0905245106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comyn O., Lee E., MacLaren R. E. 2010. Induced pluripotent stem cell therapies for retinal disease. Curr. Opin. Neurol. 23, 4–9 10.1097/WCO.0b013e3283352f96 (doi:10.1097/WCO.0b013e3283352f96) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tulpule A., Lensch M. W., Miller J. D., Austin K., D'Andrea A., Schlaeger T. M., Shimamura A., Daley G. Q. 2010. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood 115, 3453–3462 10.1182/blood-2009-10-246694 (doi:10.1182/blood-2009-10-246694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunner M., et al. 2008. Mechanisms of cardiac arrhythmias and sudden death in transgenic rabbits with long QT syndrome. J. Clin. Invest. 118, 2246–2259 10.1172/JCI33578 (doi:10.1172/JCI33578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlmutter D. H. 2004. Alpha-1-antitrypsin deficiency: diagnosis and treatment. Clin. Liver Dis. 8, 839–859(doi:10.1016/j.cld.2004.06.001) [DOI] [PubMed] [Google Scholar]
- 36.Rader D. J., Cohen J., Hobbs H. H. 2003. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J. Clin. Invest. 111, 1795–1803 10.1172/JCI18925 (doi:10.1172/JCI18925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou J. Y. 2001. The molecular basis of type 1 glycogen storage diseases. Curr. Mol. Med. 1, 25–44 10.2174/1566524013364112 (doi:10.2174/1566524013364112) [DOI] [PubMed] [Google Scholar]
- 38.Murry C. E., Keller G. 2008. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680 10.1016/j.cell.2008.02.008 (doi:10.1016/j.cell.2008.02.008) [DOI] [PubMed] [Google Scholar]
- 39.Hu B. Y., Weick J. P., Yu J., Ma L. X., Zhang X. Q., Thomson J. A., Zhang S. C. 2010. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl Acad. Sci. USA 107, 4335–4340 10.1073/pnas.0910012107 (doi:10.1073/pnas.0910012107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim K., et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 10.1038/nature09342 (doi:10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polo J. M., et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855 10.1038/nbt.1667 (doi:10.1038/nbt.1667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osafune K., Caron L., Borowiak M., Martinez R. J., Fitz-Gerald C. S., Sato Y., Cowan C. A., Chien K. R., Melton D. A. 2008. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26, 313–315 10.1038/nbt1383 (doi:10.1038/nbt1383) [DOI] [PubMed] [Google Scholar]
- 43.Chin M. H., et al. 2009. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5, 111–123 10.1016/j.stem.2009.06.008 (doi:10.1016/j.stem.2009.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aalto-Setala K., Conklin B. R., Lo B. 2009. Obtaining consent for future research with induced pluripotent cells: opportunities and challenges. PLoS Biol. 7, e42. 10.1371/journal.pbio.1000042 (doi:10.1371/journal.pbio.1000042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colman A., Dreesen O. 2009. Pluripotent stem cells and disease modeling. Cell Stem Cell 5, 244–247 10.1016/j.stem.2009.08.010 (doi:10.1016/j.stem.2009.08.010) [DOI] [PubMed] [Google Scholar]