Abstract
Induced pluripotent stem (iPS) cells have considerable promise as a novel tool for modelling human disease and for drug discovery. While the generation of disease-specific iPS cells has become routine, realizing the potential of iPS cells in disease modelling poses challenges at multiple fronts. Such challenges include selecting a suitable disease target, directing the fate of iPS cells into symptom-relevant cell populations, identifying disease-related phenotypes and showing reversibility of such phenotypes using genetic or pharmacological approaches. Finally, the system needs to be scalable for use in modern drug discovery. Here, we will discuss these points in the context of modelling familial dysautonomia (FD, Riley–Day syndrome, hereditary sensory and autonomic neuropathy III (HSAN-III)), a rare genetic disorder in the peripheral nervous system. We have demonstrated three disease-specific phenotypes in FD-iPS-derived cells that can be partially rescued by treating cells with the plant hormone kinetin. Here, we will discuss how to use FD-iPS cells further in high throughput drug discovery assays, in modelling disease severity and in performing mechanistic studies aimed at understanding disease pathogenesis. FD is a rare disease but represents an important testing ground for exploring the potential of iPS cell technology in modelling and treating human disease.
Keywords: human induced pluripotent stem cells, disease modelling, peripheral nervous system, human genetic disease, stem cells, drug discovery
1. Introduction
The ability to generate patient-tailored stem cells has been a long standing dream in the field of regenerative medicine. For many years the main strategy to achieve this goal was the development of efficient procedures for somatic cell nuclear transfer (SCNT) based on the remarkable studies by Wilmut et al. demonstrating that the nucleus of an adult somatic cell can be reprogrammed upon transplantation into an enucleated egg gaining the potential to form a complete organism, such as Dolly the sheep [1]. Over the years, the feasibility of SCNT has been demonstrated for more than a dozen mammalian species, including mouse [2], cattle [3], horse [4], cat [5], pig [6] and dog [7]. Nuclear transfer embryonic stem (ES) cells have been derived from both mouse [8,9] and non-human primate [10] hosts. However, the success of SCNT in humans has remained elusive and using current technologies, with the associated technical and ethical concerns, it appears unlikely that SCNT will become a viable option for routine generation of patient-specific cells. In contrast, the discovery of induced pluripotent stem (iPS) cells by Yamanaka and colleagues [11,12] has made the routine generation of patient-specific cells not just feasible but a routine technology used in many laboratories worldwide (for review [13]).
The ease of generating patient-specific iPS cells raises the question of what may represent the most important applications of the technology. The original idea of using patient-specific cells for generating immunocompatible tissue cell types remains an important goal. However, there are considerable challenges to safely implement cell therapy approaches for the treatment of human disease, and only a limited subset of disorders may be amenable to this approach with a well-defined pathology. A more immediate application may be the use of patient-specific iPS cells in modelling human disease and as a novel tool to generate tissue-specific cell types for drug discovery. A number of studies have made important steps towards this goal. Early work focused on generating iPS cells from a range of diseases [14] such as adenosine deaminase deficiency-related severe combined immunodeficiency, Schwachman–Bodian–Diamond syndrome, Gaucher disease type III, Duchenne and Becker muscular dystrophy, Parkinson disease (PD), Huntington disease (HD), juvenile-onset, type 1 diabetes mellitus, Down syndrome/trisomy 21, and Lesch–Nyhan syndrome. Several early studies were also able to demonstrate that patient-specific cells can be differentiated into symptom-relevant cell types such as spinal motoneurons in the case of ALS [15] or midbrain dopamine neuron-like cells in the case of PD [16]. A small number of subsequent studies went beyond generating symptom-relevant cells towards demonstrating in vitro disease phenotypes such as decreased survival of cholinergic motoneurons and changes in the number of ‘gems’ (deposits of SMN (survival of motor neuron) protein) in an iPS cell model of spinal muscular atrophy (SMA) [17]. Other examples include evidence of in vitro hypertrophy of cardiac cells derived from iPS cells of patients suffering from Leopard syndrome [18], demonstrating long QT syndrome in vitro as measured by electrophysiology in iPS-derived cardiomyocytes obtained from families with long QT syndrome [19], or measuring metabolic changes in iPS cell-derived hepatocytes from various metabolic disorders such as α1-antitrypsin deficiency, familial hypercholesterolaemia and glycogen storage disease type 1a [20]. Most recently, two studies using patient-specific iPS cells from girls suffering from an autism-related disorder (Rett syndrome) showed that Rett-iPS-derived neural precursors exhibit increased rates of L1 retrotransposition [21], a robust and interesting result though of unclear disease-relevance for Rett syndrome [22], and demonstrated that Rett-iPS cell-derived neurons have reduced numbers and densities of dendritic spines associated with reduced electrophysiological maturation [23].
In the current review, we will describe our work on familial dysautonomia (FD) [24], an example of human iPS cell-based disease modelling that is quite advanced in moving from iPS cell generation, the derivation of symptom-relevant cell types and discovery of multiple disease-associated phenotypes, to the successful use of candidate drugs to reverse in vitro phenotypes and the use of this system for both mechanistic studies and for applications in drug discovery. The goal is to present both the promise and the challenges involved in making iPS cell-based disease modelling a reality.
2. Familial dysautonomia
FD was originally described as Riley–Day syndrome [25], a rare autosomal recessive disorder [26] characterized by extensive autonomics nervous system deficits and dysfunction of small-fibre sensory neurons [27]. FD belongs to the category of hereditary sensory and autonomic neuropathy (HSAN) and is classified as HSAN-III. FD, unlike other types of HSANs, occurs nearly exclusively within people of Ashkenazi Jewish heritage, who have an estimated carrier frequency of 1 : 32 [28]. Worldwide about 650 registered cases of FD are known [29] and the incidence of the disease appears to have further decreased in recent years owing to systematic prenatal screening of the risk population [30]. A major milestone in the field was the identification of a single point mutation in the I-κ-B kinase complex-associated protein (IKBKAP) gene. This point mutation in intron 20 of IKBKAP (IKBKAP IVS20+6T→C) is responsible for 99.5 per cent of all FD cases [31,32] and results in a tissue-specific splicing defect with various levels of exon 20 skipping and reduced levels of IKAP protein [32]. Other very rare forms of FD include patients heterozygous for the IKBKAP IVS20+6T→C mutation but with additional missense mutations in the IKBKAP gene at R696P [31,32] or P914L [33].
Many of the clinical symptoms of FD can be linked to the autonomic nervous system dysfunction [27]. Gastrointestinal (GI) problems include poor oropharyngeal coordination leading to aspiration, frequent vomiting and reflux disease. Respiratory problems are caused by frequent aspiration from the GI tract and by a primary insensitivity to low O2 (hypoxia) and high CO2 levels (hypercapnia). Cardiovascular problems are characterized by positional hypotension as well as reactive hypertension following autonomic crises, particularly in older patients. Ophthalmological problems are also very common and are typically related to reduced tear production and an insensitivity of the cornea resulting in low blinking rates and indifference to corneal damage. Furthermore, patients frequently suffer from postural problems and commonly develop juvenile forms of scoliosis. Cognitive function remains generally intact and most patients display a normal IQ [34]. However, there is a distinct subset of patients with mild to severe central nervous system (CNS) deficits that appear unrelated to the severity of the peripheral symptoms. Using improved symptomatic treatments, the last decades have shown a dramatic increase in life expectancy from 50 per cent of patients reaching age 5 (at around 1960) to approximately 50 per cent of patients reaching 20, with some patients reaching age 40 [27].
Consistent with the strong autonomic neuron dysfunction, there are pathological studies performed more than 30 years ago that demonstrated dramatically reduced neuron numbers in the superior cervical sympathetic ganglia or the sphenopalatine ganglia and a near complete loss of autonomic neuron terminals at peripheral blood vessels [35]. The few remaining sympathetic neurons were shown to upregulate the expression of tyrosine hydroxylase potentially as a strategy to compensate for sympathetic neuron loss [36]. Sensory neurons are also impaired in FD with decreased neuron numbers in the dorsal root ganglia [37] and a particular loss of non-myelinated neurons and small-fibre myelinated neurons [38]. In contrast, several parasympathetic ganglia such as the ciliary ganglion appear to be relatively spared by the disease [39]. For a summary of the clinical presentation of FD see table 1.
Table 1.
Familial dysautonomia. Overview of the key facts about FD at the level of genetics, epidemiology, pathology and physiology, and clinical presentation.
| FD (OMIM no. 223900) | |
|---|---|
| Riley–Day syndrome, HSAN III | |
| genetics | IKBKAP gene (encoding IKAP protein that is a functional subunit of elongator complex) |
| majority of patients (greater than 95%) have IVS20 +6T → C mutation (protein truncation) | |
| less common mutation G → C transversion in exon 19 (missense translation, R696P) | |
| epidemiology | around 650 registered patients across the world (1 in 3703 live births in Israel) |
| carrier rate ranged from 1 in 27 to 1 in 32 | |
| mostly Ashkenazi Jewish/Eastern European background | |
| 59% of patients die before reaching the age of 20 | |
| pathology and physiology | reduced size of peripheral sensory ganglia |
| decrease of neuronal number and size of sympathetic ganglia | |
| chemoreceptor and baroreceptor dysfunction | |
| catecholamine hypersensitivity | |
| abnormal release of renin and aldosterone | |
| clinical presentation | dysautonomic/hypertensive |
| dyscoordination of the GI tract | |
| respiratory dysfunction | |
| altered sensitivity to nociception and proprioception | |
| cardiovascular dysfunction | |
| spinal curvature | |
| renal dysfunction | |
| ophthalmological disorders | |
The discovery that a single point mutation in IKBKAP causes FD has stimulated numerous studies aimed at disease pathogenesis. At the molecular level, it has been shown that the point mutation results in decreased splicing efficiency and variable levels of exon 20 exclusion resulting in the production of both wild-type (WT) and an unstable mutant IKBKAP transcript. There is no evidence that the mutant transcript yields IKAP protein, and therefore, FD can be considered as a partial loss of IKBKAP function disease. Interestingly, the extent of IKBKAP missplicing in FD patients shows considerable tissue-specific differences [32,40] that may underlie the tissue-specific clinical symptoms of the disease.
IKBKAP, also called ELP-1, is a component of the transcription elongation complex that has histone acetyltransferase activity, primarily directed towards histone H3 [41]. However, a substantial fraction of the elongator complex is located in the cytoplasm and acetylation of non-histone proteins has also been suggested to play an important role in IKBKAP function. In fact, recent studies [42] have demonstrated that loss-of-function of elongator activity results in acetylation defects of α-tubulin with subsequent deficits in cortical migration and neurite extension. Other proposed functions for the elongator complex include roles in exocytosis [43], in c-Jun N-terminal kinase-signalling, demethylation of zygotic paternal genomic DNA [44] and in t-RNA modification [45], although there is no common agreement about the contribution of each of those mechanisms to elongator function (for discussion, see [46]).
With the identification of the genetic defect responsible for FD, it was important to test whether loss of IKBKAP function in the mouse can be used to model the human disease. It has been shown that mice null for IKBKAP are developmentally arrested at mid-gastrulation resulting in early embryonic lethality [47]. The generation of bacterial artificial chromosome (BAC) transgenic mice carrying the human BAC with the FD disease-causing mutation has been used to model tissue-specific splicing of the IKBKAP gene [40]. However, introducing the human disease-specific BAC onto an IKBKAP-null mouse background leads to a complete rescue of embryonic lethality without any signs of disease in the resulting mouse [47]. Therefore, there are currently no mouse models of FD that can recapitulate the human disease phenotype.
3. Towards an induced pluripotent stem cell model of familial dysautonomia
(a). Traditional cell-based models of familial dysautonomia
Several cellular models of FD have been established over many years to study the human disease, including patient-specific fibroblast or lymphoblast cell lines. Studies in FD fibroblasts have been used to pursue a broad range of hypotheses on FD pathogenesis, such as changes in mitochondrial function [48] or neurotrophic factor secretion [49]. The more widely accepted differences in cell motility [41] and splicing [32] have also been first characterized in patient-specific fibroblasts. Traditional cell-based models have also been used to screen for candidate drugs that may reverse the splicing phenotype observed in FD. In particular, human FD lymphoblast cell lines were screened against a panel of 1040 bioactive compounds (National Institute of Neurological Disorders and Stroke (NINDS) Custom Collection, MicroSource Discovery Systems). This study monitored expression of both WT and mutant IKBKAP transcript using conventional and quantitative RT-PCR assays [50]. Kinetin showed a dose-dependent decreased in mutant transcript with a concomitant increase in WT transcript. Overall levels of IKBKAP mRNA were unchanged except at very high kinetin concentrations. While these studies demonstrated the power of cell-based models in FD, they remained limited by the fact that neither fibroblasts nor lymphoblasts represent symptom-relevant cell types. The availability of the cell types affected in FD such as neural crest and possibly CNS neural lineages is critical for modelling disease pathogenesis and to understand the tissue-specific manifestation of the disease. Given the obvious challenges in harvesting neural cells directly from patients, it became critical to identify a renewable human cell source that could be used for such studies. One of the most obvious strategies was the isolation of neural cells from human pluripotent stem cells, such as human ES or iPS cells. Over the last few years, multiple protocols have been established for the directed differentiation and purification of CNS and peripheral nervous system (PNS) lineages from human pluripotent stem cells such as human ES cells [51,52]. The derivation of neural crest lineages including multipotent neural crest stem cells [53] may be of particular relevance for generating disease-relevant cell types in FD.
(b). Strategies to generate patient-specific pluripotent stem cells
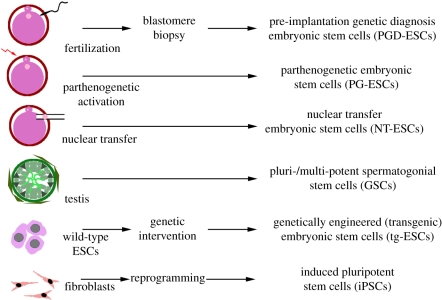
In addition to iPS cell technology, there are a number of potential strategies for deriving patient-specific cells relevant to modelling FD (figure 1). The classic paradigm of using SCNT to generate genetically matched cells has not become a viable approach owing to the lack of success with human SCNT and the difficulties in procuring sufficient numbers of human donor oocytes. The use of parthenogenetic [54] (from unfertilized oocyte) or various types of testis-derived [55–57] (from male germ-cell precursors) pluripotent stem cells are potential alternatives for generating matched human stem cell populations. However, either strategy is limited to male or female donors at the appropriate age, respectively, and the resulting stem cell lines typically carry genetic and imprinting-related differences that make those cells an imperfect source for disease modelling.
Figure 1.
Multiple potential strategies to generate disease-specific pluripotent stem cell lines. There are multiple ways to establish disease-specific pluripotent stem cells. These include the isolation of pluripotent cells from (i) embryos carrying a genetic defect as defined by pre-implantation genetic diagnosis (PGD); (ii) embryos from parthenogenetically activated oocytes obtained from female patients; (iii) embryos reconstructed following nuclear transfer from a somatic cell of the patient; (iv) spermatogonial tissues induced towards pluripotency from male patients; (v) normal embryonic stem cells genetically engineered to model human disease; (vi) following reprogramming of somatic cells such as skin fibroblasts using defined factors (introduction of transcription factors and/or exposure to small molecule compounds promoting reprogramming).
A more practical strategy is the generation of ES cells derived from embryos that underwent pre-implantation genetic diagnosis (PGD). PGD lines have been generated for a broad range of genetic diseases, such as HD [58,59], cystic fibrosis [58,59], myotonic dystrophy [58], fragile X syndrome [59,60], Gaucher's syndrome [61], Duchenne dystrophy [61], Saethre–Chotzen syndrome [61], torsion dystonia [61], Alport syndrome [61], multiple endocrine neoplasia [59], spino-cerebellar ataxia [59], Fabry syndrome [59], Charcot Marie Tooth disease [62], osteogenesis imperfecta [62] and facioscapulohumeral muscular dystrophy [62]. Despite the availability of those lines there are very few examples of successful disease modelling using PGD lines [60,63]. Furthermore, we are not aware of any available PGD human ES cell line for FD.
Another powerful strategy to model human diseases is the use of genetic engineering to introduce or delete disease-related genes. An early successful example of this approach is knockout human ES cell lines for hypoxanthine–guanine phosphoribosyl transferase (HPRT), modelling Lesch–Nyhan disease [64]. In the case of FD, gene targeting may be a particularly interesting to determine sufficiency of the IKBKAP point mutation to cause disease phenotypes. Nearly, all patients with FD are of Ashkenazi Jewish origin and 99.5 per cent of the patients carry the identical homozygous point mutation. The obvious question therefore is whether introducing the same point mutation into unrelated human ES or iPS cell lines of non-Ashkenazi Jewish background can mimic the disease phenotype or whether other contributing genes in the donor background are required. Such studies remain technically challenging as they involve successful targeting of both IKBKAP alleles. However, novel gene-targeting approaches in human ES and iPS cells such as the use of zinc finger nucleases [65,66] or adeno-associated virus-mediated gene targeting [67] should help in making such approaches feasible in the near future.
Clearly, the currently most promising strategy to generate human disease-specific stem cell lines is the use of iPS cell technology to reprogramme FD fibroblasts or other somatic cell types. A broad number of vectors and delivery systems have been used to successfully reprogramme somatic cells including classic retroviral [11] or lentiviral [68] vectors, vectors allowing for inducible expression [69] or transgene excision [16], piggyBAC transposons [70], episomal expression vectors [71], protein-based reprogramming [72] and, most recently, RNA-based reprogramming methods using RNA viruses [73] or synthetic modified RNAs [74]. Furthermore, there is intense work on the use of small molecules-based reprogramming strategies [75]. The advantages of iPS cell technology as compared with all the alternative approaches is the relative ease of use, the ability to generate multiple independent clones and the possibility of generating iPS cell lines from a large number of individuals. Potential disadvantages include technical concerns related to any given vector system such as insertional mutagenesis for integrating vectors systems, the risk of generating partially reprogrammed clones, uncertainties about whether iPS cells are truly equivalent to the ES cell counterparts [76,77], questions related to incomplete reprogramming of epigenetic memory [78,79] and the lack of complete reactivation of the X-chromosome in female lines commonly seen in human iPS cells [80].
In the case of generating FD-iPS cells suitable for disease modelling, we have primarily used a lentiviral vector approach that enables real-time monitoring of transgene expression using a P2A element linked to a unique colour for each of the four reprogramming factors [24,81]. Using this system, we can ascertain silencing of transgenes both at the undifferentiated iPS cell stage and upon differentiation into symptom-relevant cell types. However, reliance on spontaneous silencing of transgenes in pluripotent cells remains a suboptimal approach, and future studies should include the use of transgene-free FD-iPS cell lines in an effort to further reduce technical variability for mechanistic studies and for drug discovery.
(c). Familial dysautonomia-induced pluripotent stem-based disease model
A critical issue for iPS cell-based human disease models is the identification and derivation of the symptom-relevant cells suitable for in vitro studies. In the case of modelling FD, we had to resolve two major questions related to disease pathogenesis. On the one hand, there was evidence for tissue-specific splicing as the potential culprit for the specific-disease symptoms in the neural crest and other neural lineages [32]. For the development of a relevant disease model that reproduces tissue-specific splicing in vitro, it was critical to derive multiple independent tissue-specific cell types such as CNS and PNS precursors but also haematopoietic, endothelial and endoderm cell types [24,81]. While our study was able to detect tissue-specific differences in IKBKAP splicing the differences were not dramatic and quantitative analyses for the expression of WT transcript were required to demonstrate the particularly low IKBKAP levels in the neural crest lineage. Future studies should determine whether specific cell types within the neural crest lineage such as autonomic neurons or sensory neurons show distinct IKBKAP splicing compared with other neural crest lineages and whether such subtype-specific results may be more predictive for modelling the tissue specificity of the human disease. While the splicing study required the derivation of a broad set of cell types across all three germ-layers, the studies related to pathogenesis within the neural crest lineage were focused on the use of neural crest precursor cells [24]. For these studies, we made use of the neural crest precursor cell isolation strategies based on the generation of neural plate stage neuroepithelial precursors followed by prospective isolation of p75+ and/or HNK1+ fractions as previously described in detail [52,53]. Those cells are of cranial neural crest identity [53] and it will be interesting to see whether novel differentiation protocols under development—for the generation of trunk and other anterior–posterior regions of neural crest—will show identical or distinct disease patterns.
Beyond IKBKAP splicing, the most robust phenotype observed across all FD-iPSC lines and subclones tested was the reduction in ASCL1 both at the mRNA and protein level [24]. ASCL1 is a particularly interesting gene as the knockout of the mouse orthologue (Mash1) resulted in a complete loss of autonomic neuron generation [82], the neuron type most dramatically affected in FD [27]. Our work demonstrated that the generation of ASCL1+ neurons from purified FD-iPS cell-derived neural crest precursors is dramatically reduced and delayed, an in vitro disease phenotype compatible with the clinical symptoms and the pathological observations in FD patients. However, we have not yet further characterized whether the reduction is because of a problem in specification or early maintenance of autonomic neuron precursors. It would be interesting to determine whether there are specific subtypes of autonomic neurons that are more or less affected in our model and that may inform about differential sensitivity in FD. Furthermore, the iPS cell system should enable studies to determine whether the autonomic neuron defects is a cell autonomous or non-autonomous defect by mixing FD- and control-iPS precursors at defined differentiation stages and cell numbers. Defects in cell motility as observed in our FD-iPS cell-derived neural crest precursors have been reported previously in fibroblast genetically depleted of IKBKAP [41]. Similar to the splicing defect, it will be interesting to define the cell-type specificity of the defect and to determine whether reduced motility can be linked to changes in acetylation of alpha-tubulin as demonstrated for the migration of IKBKAP depleted cortical neurons [42].
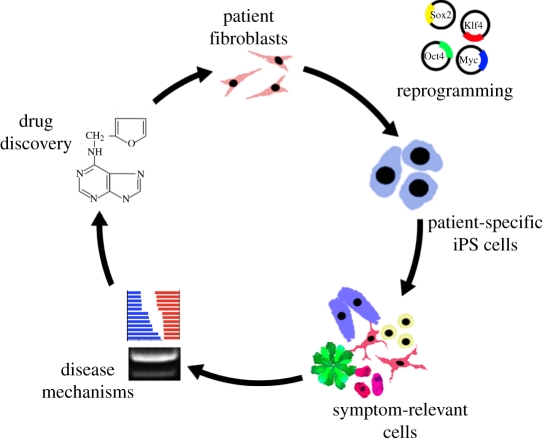
One of the major goals of establishing iPS cell-based disease models is the promise of a novel area of drug discovery based on access to unlimited numbers of patient-specific, symptom-relevant cell types. To date, there have been no published reports on using iPS cell-based disease models for large-scale drug discovery efforts, but there are clearly a number of groups that are currently pursuing such strategies. Our own published work, as well as similar efforts in SMA [17], has focused on testing the iPS-based disease model against a small set of candidate compounds. We were able to test most of the key compounds previously proposed to be of potential value for treating FD such as the vitamin E derivative tocotrienols [83], the polyphenol epigallocatechin gallate [84] and the plant hormone kinetin [50]. We only observed robust rescue of FD-iPS cell phenotypes such as splicing and the generation of autonomic neurons with kinetin but not any of the other compounds. However, most of those assays were limited to testing kinetin as the other compounds did not show robust phenotypes at the fibroblast stage and therefore were not further pursued. However, we cannot rule out that under different conditions and treatment regimens those other compounds may also beneficially affect splicing or overall expression levels of IKBKAP or impact other FD-related phenotypes. Our kinetin studies indicate that long-term and likely early-stage treatment is required to rescue disease phenotypes. One interpretation of those data is FD patients would need to be treated with kinetin already at very early developmental stages, potentially even during in utero development. However, it may also be possible that prolonged treatment at a later stage could still be effective by inducing neurogenesis over time in the small set of remaining endogenous neural crest precursor populations or by arresting the progressive degenerative nature of the disease. It will be interesting to test whether our iPS cell model will allow us to distinguish among those various alternatives by running multiple treatment regimens as a sort of in vitro clinical trial across a large set of independent FD-iPS cell lines. For a summary of the steps involved in FD-iPS cell-based disease modelling see figure 2.
Figure 2.
iPS cell-based disease modelling in FD. Patient fibroblasts were reprogrammed using lenvitral vectors expressing the four reprogramming factors in combination with a unique colour. The resulting patient-specific iPS cells were validated for transgene silencing and other pluripotent properties and differentiated into multiple disease-relevant cell types in order to assess disease mechanisms, such as tissue-specific splicing and defects in neurogenesis. Finally, the in vitro assays were used to test for candidate drugs that could rescue FD-iPS cell disease phenotypes. One such compound, the plant hormone kinetin, was capable of partially reversing both the splicing and neurogenesis defects.
4. What are the next steps?
(a). Genetic rescue
A critical point for modelling any type of monogenic disease is the ability to genetically rescue disease phenotypes. In the case of FD, one could think of a number of possible strategies. FD can be considered a partial loss-of-function disease by the fact that the mutant IKAP transcript does not get translated into protein, and that only patients homozygous for the disease appear to show symptoms. However, precise levels of IKBKAP expression may be critical for its function in transcriptional elongation, and it is clearly preferable to have a system that maintains tissue specificity of expression. If both levels and tissue-specific expression are critical, there may be two main strategies for genetic rescue in FD-iPSCs: gene targeting and BAC transgenesis. The use of BAC transgenes has been shown to completely rescue IKBKAP deficiency in a null mutant mice [47]. In this case, the IKBKAP null mouse is not a representative model of the disease per se, since it results in early embryonic lethality. However, embryonic lethality can be rescued when using the human BAC containing the FD mutation [47] and the resulting mouse can be used for modelling tissue-specific IKBKAP splicing though it does not exhibit any FD-related disease symptoms [47].
The use of BAC transgenesis in the human FD-iPS model should retain tissue specificity of IKBKAP expression and enable transgene dosing by isolating independent BAC rescue clones with various copy number. Similar approaches have been used previously to model the role of Zfx in the pluripotency in mouse [85] and human ES cells (M. Tomishima 2011, personal communication). In addition to BAC-based rescue, the most obvious strategy is homologous recombination to knock-in the WT allele into the endogenous disease locus of FD-iPS cells. Currently, there have been no published reports at performing homologous recombination for the rescue of an iPS cell disease phenotype but several groups are working on making this approach a reality. One critical question in the context of FD is whether repair of a single allele will lead to a full rescue of the iPS cell in vitro phenotypes. Our original studies did not include the analysis of iPS cells from heterozygous FD carriers that may represent a good control for these rescue studies. In fact, it will be very interesting to compare in vitro disease phenotypes of iPS cells derived from a normal carrier with those of FD-iPS cells rescued to a carrier status after gene targeting and those induced to a carrier status by introducing the disease-causing allele into WT hES and iPS cells. However, a key issue remains the question of genetic sufficiency and whether inducing a homozygous mutation into WT human ES or iPS cell is capable of reproducing the full in vitro phenotype independent of ethnic background.
A related question is whether the iPS cell-based model can predict differences in clinical manifestation of the disease. As it turns out, there are considerable differences in disease severity among FD patients and most of the FD-iPS cell lines generated to date are from patients with severe disease. Furthermore, most FD patients are of normal intelligence while a subset of patients shows reduced cognitive function [34,86]. There has been a lot of discussion of iPS cell technology and personalized medicine. The case of FD could provide again a testing ground to put these ideas into practice. For example, will our iPS cell model enable the identification of severe versus more mild cases of FD purely based on in vitro disease manifestation? Will it be possible to predict which patient are prone to CNS complications by in vitro behaviour of neural crest and CNS-derived neural cells across patients? If it is indeed feasible to model disease severity and differential CNS involvement across patients, the FD-iPS cell could turn into a powerful system to identify the genetic factors responsible for patient-specific disease symptoms. The ability to reliably detect differences among various FD patients would also indicate that iPS cell-based modelling can discriminate phenotypic changes associated with complex genetic background factors independent of the key monogenic defect shared among all FD patients. The identification of complex genetic phenotypes will be a key requirement for the use of iPS cell technology in modelling disorders such as Alzheimer's disease or PD. Those much more common human diseases generally involve a complex array of contributing genetic and non-genetic factors and occur late in life, all factors that will make iPS cell-based disease modelling considerably more challenging than in the case of FD [87].
(b). A novel system for mechanistic studies of familial dysautonomia pathogenesis
The use of cell-based systems to study mechanism of disease is one of the great promises of iPS cell research. It is now possible to generate nearly unlimited numbers of iPS-derived disease-relevant cell types for biochemical or genetic studies. In the case of FD, those cells could be used to address mechanisms underlying the splicing defect and its tissue specificity. Previous work in HEK293 cells has attempted to address the mechanisms for IKBKAP missplicing in FD [88]. Exclusion of exon 20 of the IKBKAP gene was modelled in silico and validated using a mini-gene in vitro splicing assay. The in silico modelling data suggested that the missplicing event cannot be explained alone by the disease-causing point mutation in the intron at position 6, a position that does not affect splicing of most other introns. Based on these studies there is a weak upstream 3′ splicing site that contributes to exon 20 exclusion in combination with the FD mutated 5′ splicing site [88]. The use of iPS cells for such studies should enable addressing the tissue-specific differences in alternative splicing by comparing cell types that are most differentially affected by the FD mutation such as cardiomyoctyes (low levels of missplicing) versus central and peripheral neurons (thought to exhibit highest levels of mutant transcript). Access to unlimited numbers of tissue-specific FD-iPS-derived cell types may allow the isolation of tissue-specific co-factors that control the extent of missplicing in the disease cells. There is also is considerable interest in using FD-iPS cells to access CNS neurons and to test the hypothesis derived from IKBKAP loss-of-function studies in mouse cortical neurons [41,42]. These data suggest that the elongator complex is critical for acetylation of non-histone proteins including α-tubulin, and that the resulting cellular migration defect in IKBKAP depleted neurons may underlie FD pathogenesis. Human iPS-derived tissues should provide a powerful source of material for the systematic analysis of non-histone acetylation events across the proteome using modern mass spectrometry technologies [89,90].
It is currently unclear whether reduced ASCL1 expression levels observed in neural crest precursors across all FD-iPS cell clones is a primary or secondary event during disease pathogenesis. Loss-of-function data for Mash1 (orthologoue of human ASCL1 gene) in the mouse are striking, with a loss of autonomic neurons, the cell type mostly affected in FD. However, low ASCL1 levels in FD-iPS cell-derived neural crest precursors could also be simply a secondary consequence rather than the cause of the disease, as both ASCL1 and SCG10 represent classic autonomic markers [91] that were similarly affected in our system.
(c). Platform for drug screening and future therapies
FD may be one of the first disorders to yield therapeutically relevant compounds from iPS cell-based drug screening efforts. Key advantages for FD-iPS cell-based drug discovery are the relatively robust phenotypes such as the splicing and neurogenesis defect, the proof-of-concept for successful high throughput screening (HTS) approaches in FD lymphoblast cell lines [50], and the ability to generate symptom-relevant cell types in a scalable, reproducible and fairly homogeneous manner. We observed that human iPS cell-derived neural crest precursors can be proliferated after initial isolation [53] in the presence of fibrobalst growth factor (FGF)2/epidermal growth factor (EGF). Under those in vitro expansion conditions neural crest precursors retain basic marker expression profiles and differentiation behaviour for multiple passages. This enables the routine generation of up to 1 × 109 cells within about three to four weeks of expansion. Some of the critical steps in making FD-iPS cell-derived neural crest precursors compatible for HTS assays include optimizing the conditions for cell adhesion during initial plating, determining suitable cell densities, defining the length of incubation prior to and post compound addition and implementing the use of kinetin as a positive control. While any of the three assays described initially (splicing, neurogenesis and cell motility) could be developed into a screening platform, measuring transcript levels of WT and mutant IKBKAP may be the most direct assay for a primary screen. Furthermore, our data suggest that rescuing neurogenesis or cell motility is much more challenging and requires lengthy exposure to bioactive compounds such as kinetin. The protracted culture periods (up to 28 days) necessary for such assays are not readily suitable for translation to HTS platform [24]. A critical question is whether the use of symptom-relevant cell types such as neural crest precursor cells will yield a set of molecules distinct from those obtained in patient-specific fibroblast or lymphoblast cell lines. Given the intricacies of the splicing defect and the highly distinct physiology of neural crest precursors as opposed to fibroblast of lymphoblast cell lines it is reasonable to assume more relevant screening results, but this issue remains to be proven.
The identification of kinetin or any other interesting compound from FD-iPSC-based drug discovery raises the question of how to translate in vitro results towards ultimate clinical trials. We have proposed to focus our initial HTS assays on the set of US Food and Drug Administration (FDA)-approved drugs and natural product libraries that may facilitate clinical translation (approx. 6000 compound screen). In fact, the initial discovery of kinetin emerged from a screen of 1040 compounds of FDA-approved drugs from the NINDS collection [50]. FDA-approved compounds have a known safety profile that will facilitate translation and the key question is whether the effective dose range for the approved indication is comparable to the doses required for affecting IKBKAP levels. One major impediment in the FD field is the lack of a mouse model that appropriately reflects human disease. The existing models are suitable for testing impact on IKBKAP splicing but not on any FD-related functional parameters in vivo. Preliminary testing of kinetin in humans has been performed in a set of 29 non-affected heterozygous carriers. Subjects were exposed to kinetin at various doses for up to 8 days of treatment to determine whether blood levels of the drug are sufficient to impact IKBKAP splicing in peripheral blood cells and to address side effects [92]. These preliminary data suggest that blood levels achieved through oral administration are sufficient to affect in vivo IKBKAP splicing [92].
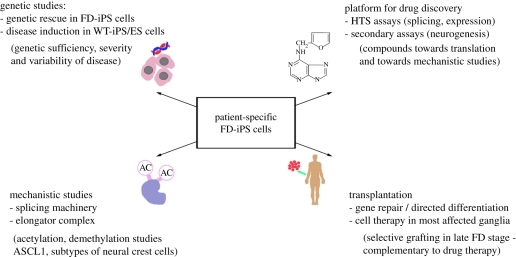
While the main focus of our work is directed towards the discovery of novel bioactive compounds that may be developed into a useful treatment, there is also the possibility of developing cell therapy-based approaches to the disease. For example it may be possible to use FD-iPS cell-derived neural populations as an autologous source of neurons for targeted transplantation into those peripheral ganglia most affected by the disease. Such an approach would probably require gene correction prior to transplantation and improved techniques to direct and purify specific iPS cell-derived autonomic neuron subtypes. Given the progressive, degenerative nature of late-stage disease and the difficulties of treating patients during the earliest stages of neuron development, cell-based approaches may offer a valuable complementary approach to exploit the full potential of iPS cell technology towards a treatment and eventually a cure of this debilitating disease. For a summary of the next steps in FD-iPS cell-based disease modelling see figure 3.
Figure 3.
Next steps in FD-iPS cell-based disease modelling (see text for details).
Acknowledgements
We would like to thank members of the Studer laboratory for valuable discussions on the manuscript. Our own work described in this review was supported by grants from NYSTEM, the Starr Foundation and a Druckenmiller fellowship from NYSCF to G.L.
References
- 1.Wilmut I., Schnieke A. E., McWhir J., Kind A. J., Campbell K. H. 1997. Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 10.1038/385810a0 (doi:10.1038/385810a0) [DOI] [PubMed] [Google Scholar]
- 2.Wakayama T., Perry A. C., Zuccotti M., Johnson K. R., Yanagimachi R. 1998. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394, 369–374 10.1038/28615 (doi:10.1038/28615) [DOI] [PubMed] [Google Scholar]
- 3.Cibelli J. B., Stice S. L., Golueke P. J., Kane J. J., Jerry J., Blackwell C., de León F. A. P., Robl J. M. 1998. Transgenic bovine chimeric offspring produced from somatic cell-derived stem-like cells. Nat. Biotechnol. 16, 642–646 10.1038/nbt0798-642 (doi:10.1038/nbt0798-642) [DOI] [PubMed] [Google Scholar]
- 4.Galli C., Lagutina I., Crotti G., Colleoni S., Turini P., Ponderato N., Duchi R., Lazzari G. 2003. Pregnancy: a cloned horse born to its dam twin. Nature 424, 635. 10.1038/424635a (doi:10.1038/424635a) [DOI] [PubMed] [Google Scholar]
- 5.Shin T., et al. 2002. A cat cloned by nuclear transplantation. Nature 415, 859. 10.1038/nature723 (doi:10.1038/nature723) [DOI] [PubMed] [Google Scholar]
- 6.Lee G. S., et al. 2005. Production of transgenic cloned piglets from genetically transformed fetal fibroblasts selected by green fluorescent protein. Theriogenology 63, 973–991 10.1016/j.theriogenology.2004.04.017 (doi:10.1016/j.theriogenology.2004.04.017) [DOI] [PubMed] [Google Scholar]
- 7.Lee B. C., et al. 2005. Dogs cloned from adult somatic cells. Nature 436, 641. 10.1038/436641a (doi:10.1038/436641a) [DOI] [PubMed] [Google Scholar]
- 8.Wakayama T., Tabar V., Rodriguez I., Perry A. C. F., Studer L., Mombaerts P. 2001. Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer. Science 292, 740–743 10.1126/science.1059399 (doi:10.1126/science.1059399) [DOI] [PubMed] [Google Scholar]
- 9.Munsie M. J., Michalska A. E., O'Brien C. l M., Trounson A. O., Pera M. F., Mountford P. S. 2000. Isolation of pluripotent embryonic stem cells from reprogrammed adult mouse somatic cell nuclei. Curr. Biol. 10, 989–992 10.1016/S0960-9822(00)00648-5 (doi:10.1016/S0960-9822(00)00648-5) [DOI] [PubMed] [Google Scholar]
- 10.Byrne J. A., Pedersen D. A., Clepper L. L., Nelson M., Sanger W. G., Gokhale S., Wolf D. P., Mitalipov S. M. 2007. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature 450, 497–502 10.1038/nature06357 (doi:10.1038/nature06357) [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131, 861–872 10.1016/j.cell.2007.11.019 (doi:10.1016/j.cell.2007.11.019) [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 (doi:10.1016/j.cell.2006.07.024) [DOI] [PubMed] [Google Scholar]
- 13.Stadtfeld M., Hochedlinger K. 2010. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 24, 2239–2263 10.1101/gad.1963910 (doi:10.1101/gad.1963910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park I. H., et al. 2008. Disease-specific induced pluripotent stem cells. Cell 134, 877–886 10.1016/j.cell.2008.07.041 (doi:10.1016/j.cell.2008.07.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimos J. T., et al. 2008. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 321, 1218–1221 10.1126/science.1158799 (doi:10.1126/science.1158799) [DOI] [PubMed] [Google Scholar]
- 16.Soldner F., et al. 2009. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136, 964–977 10.1016/j.cell.2009.02.013 (doi:10.1016/j.cell.2009.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebert A. D., Yu J., Rose F. F., Mattis V. B., Lorson C. L., Thomson J. A., Svendsen C. N. 2009. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457, 277–280 10.1038/nature07677 (doi:10.1038/nature07677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvajal-Vergara X., et al. 2010. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature 465, 808–812 10.1038/nature09005 (doi:10.1038/nature09005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moretti A., et al. 2010. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 363, 1397–1409 10.1056/NEJMoa0908679 (doi:10.1056/NEJMoa0908679) [DOI] [PubMed] [Google Scholar]
- 20.Rashid S. T., et al. 2010. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Invest. 120, 3127–3136 10.1172/JCI43122 (doi:10.1172/JCI43122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muotri A. R., Marchetto M. C. N., Coufal N. G., Oefner R., Yeo G., Nakashima K., Gage F. H. 2010. L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446 10.1038/nature09544 (doi:10.1038/nature09544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Studer L. 2010. Neuroscience: excessive mobility interrupted. Nature 468, 383–384 10.1038/468383a (doi:10.1038/468383a) [DOI] [PubMed] [Google Scholar]
- 23.Marchetto M. C., Carromeu C., Acab A., Yu D., Yeo G. W., Mu Y., Chen G., Gage F. H., Muotri A. R. 2010. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143, 527–539 10.1016/j.cell.2010.10.016 (doi:10.1016/j.cell.2010.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee G., et al. 2009. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461, 402–406 10.1038/nature08320 (doi:10.1038/nature08320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley C. M., Day R. L., Greeley D. M., Langford W. S. 1949. Central autonomic dysfunction with defective lacrimation. I. Report of 5 cases. Pediatrics 3, 468–478 [PubMed] [Google Scholar]
- 26.Brunt P. W., McKusick V. A. 1970. Familial dysautonomia. A report of genetic and clinical studies, with a review of the literature. Medicine (Baltimore) 49, 343–374 [PubMed] [Google Scholar]
- 27.Axelrod F. B. 2004. Familial dysautonomia. Muscle Nerve 29, 352–363 10.1002/mus.10499 (doi:10.1002/mus.10499) [DOI] [PubMed] [Google Scholar]
- 28.Dong J., Edelmann L., Bajwa A. M., Kornreich R., Desnick R. J. 2002. Familial dysautonomia: detection of the IKBKAP IVS20(+6T −> C) and R696P mutations and frequencies among Ashkenazi Jews. Am. J. Med. Genet. 110, 253–257 10.1002/ajmg.10450 (doi:10.1002/ajmg.10450) [DOI] [PubMed] [Google Scholar]
- 29.Axelrod F. B. 2005. Familial dysautonomia: a review of the current pharmacological treatments. Expert. Opin. Pharmacother. 6, 561–567 10.1517/14656566.6.4.561 (doi:10.1517/14656566.6.4.561) [DOI] [PubMed] [Google Scholar]
- 30.Lerner B. H. 2009. When diseases disappear—the case of familial dysautonomia. N. Engl. J. Med. 361, 1622–1625 10.1056/NEJMp0809587 (doi:10.1056/NEJMp0809587) [DOI] [PubMed] [Google Scholar]
- 31.Anderson S. L., Coli R., Daly I. W., Kichula E. A., Rork M. J., Volpi S. A., Ekstein J., Rubin B. Y. 2001. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 68, 753–758 10.1086/318808 (doi:10.1086/318808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slaugenhaupt S. A., et al. 2001. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 68, 598–605 10.1086/318810 (doi:10.1086/318810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leyne M., et al. 2003. Identification of the first non-Jewish mutation in familial dysautonomia. Am. J. Med. Genet. A 118A, 305–308 10.1002/ajmg.a.20052 (doi:10.1002/ajmg.a.20052) [DOI] [PubMed] [Google Scholar]
- 34.Welton W., Clayson D., Axelrod F. B., Levine D. B. 1979. Intellectual development and familial dysautonomia. Pediatrics 63, 708–712 [PubMed] [Google Scholar]
- 35.Pearson J., Pytel B. A. 1978. Quantitative studies of sympathetic ganglia and spinal cord intermedio-lateral gray columns in familial dysautonomia. J. Neurol. Sci. 39, 47–59 10.1016/0022-510X(78)90187-9 (doi:10.1016/0022-510X(78)90187-9) [DOI] [PubMed] [Google Scholar]
- 36.Pearson J., Brandeis L., Goldstein M. 1979. Tyrosine hydroxylase immunoreactivity in familial dysautonomia. Science 206, 71–72 10.1126/science.39339 (doi:10.1126/science.39339) [DOI] [PubMed] [Google Scholar]
- 37.Pearson J., Pytel B. A., Grover-Johnson N., Axelrod F., Dancis J. 1978. Quantitative studies of dorsal root ganglia and neuropathologic observations on spinal cords in familial dysautonomia. J. Neurol. Sci. 35, 77–92 10.1016/0022-510X(78)90103-X (doi:10.1016/0022-510X(78)90103-X) [DOI] [PubMed] [Google Scholar]
- 38.Pearson J., Axelrod F., Dancis J. 1974. Trophic functions of the neuron. V. Familial dysautonomis. Current concepts of dysautonomia: neuropathological defects. Ann. N. Y. Acad. Sci. 228, 288–300 10.1111/j.1749-6632.1974.tb20517.x (doi:10.1111/j.1749-6632.1974.tb20517.x) [DOI] [PubMed] [Google Scholar]
- 39.Pearson J., Pytel B. 1978. Quantitative studies of ciliary and sphenopalatine ganglia in familial dysautonomia. J. Neurol. Sci. 39, 123–130 10.1016/0022-510X(78)90193-4 (doi:10.1016/0022-510X(78)90193-4) [DOI] [PubMed] [Google Scholar]
- 40.Hims M. M., Shetty R. S., Pickel J., Mull J., Leyne M., Liu L., Gusella J. F., Slaugenhaupt S. A. 2007. A humanized IKBKAP transgenic mouse models a tissue-specific human splicing defect. Genomics 90, 389–396 10.1016/j.ygeno.2007.05.012 (doi:10.1016/j.ygeno.2007.05.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Close P., et al. 2006. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell 22, 521–531 10.1016/j.molcel.2006.04.017 (doi:10.1016/j.molcel.2006.04.017) [DOI] [PubMed] [Google Scholar]
- 42.Creppe C., et al. 2009. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 136, 551–564 10.1016/j.cell.2008.11.043 (doi:10.1016/j.cell.2008.11.043) [DOI] [PubMed] [Google Scholar]
- 43.Rahl P. B., Chen C. Z., Collins R. N. 2005. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell 17, 841–853 10.1016/j.molcel.2005.02.018 (doi:10.1016/j.molcel.2005.02.018) [DOI] [PubMed] [Google Scholar]
- 44.Okada Y., Yamagata K., Hong K., Wakayama T., Zhang Y. 2010. A role for the elongator complex in zygotic paternal genome demethylation. Nature 463, 554–558 10.1038/nature08732 (doi:10.1038/nature08732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esberg A., Huang B., Johansson M. J., Bystrom A. S. 2006. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 24, 139–148 10.1016/j.molcel.2006.07.031 (doi:10.1016/j.molcel.2006.07.031) [DOI] [PubMed] [Google Scholar]
- 46.Svejstrup J. Q. 2007. Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19, 331–336 10.1016/j.ceb.2007.04.005 (doi:10.1016/j.ceb.2007.04.005) [DOI] [PubMed] [Google Scholar]
- 47.Chen Y. T., Hims M. M., Shetty R. S., Mull J., Liu L., Leyne M., Slaugenhaupt S. A. 2009. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell Biol. 29, 736–744 10.1128/MCB.01313-08 (doi:10.1128/MCB.01313-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strasberg P., Bridge P., Merante F., Yeger H., Pereira J. 1996. Normal mitochondrial DNA and respiratory chain activity in familial dysautonomia fibroblasts. Biochem. Mol. Med. 59, 20–27 10.1006/bmme.1996.0059 (doi:10.1006/bmme.1996.0059) [DOI] [PubMed] [Google Scholar]
- 49.Schwartz J. P., Breakefield X. O. 1980. Altered nerve growth factor in fibroblasts from patients with familial dysautonomia. Proc. Natl Acad. Sci. USA 77, 1154–1158 10.1073/pnas.77.2.1154 (doi:10.1073/pnas.77.2.1154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slaugenhaupt S. A., Mull J., Leyne M., Cuajungco M. P., Gill S. P., Hims M. M., Quintero F., Axelrod F. B., Gusella J. F. 2004. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 13, 429–436 10.1093/hmg/ddh046 (doi:10.1093/hmg/ddh046) [DOI] [PubMed] [Google Scholar]
- 51.Elkabetz Y., Studer L. 2008. Human ESC-derived neural rosettes and neural stem cell progression. Cold Spring Harb. Symp. Quant. Biol. 73, 377–387 10.1101/sqb.2008.73.052 (doi:10.1101/sqb.2008.73.052) [DOI] [PubMed] [Google Scholar]
- 52.Lee G., Chambers S. M., Tomishima M. J., Studer L. 2010. Derivation of neural crest cells from human pluripotent stem cells. Nat. Protoc. 5, 688–701 10.1038/nprot.2010.35 (doi:10.1038/nprot.2010.35) [DOI] [PubMed] [Google Scholar]
- 53.Lee G., Kim H., Elkabetz Y., Al Shamy G., Panagiotakos G., Barberi T., Tabar V., Studer L. 2007. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 25, 1468–1475 10.1038/nbt1365 (doi:10.1038/nbt1365) [DOI] [PubMed] [Google Scholar]
- 54.Cibelli J. B., et al. 2002. Parthenogenetic stem cells in nonhuman primates. Science 295, 819. 10.1126/science.1065637 (doi:10.1126/science.1065637) [DOI] [PubMed] [Google Scholar]
- 55.Kanatsu-Shinohara M., et al. 2004. Generation of pluripotent stem cells from neonatal mouse testis. Cell 119, 1001–1012 10.1016/j.cell.2004.11.011 (doi:10.1016/j.cell.2004.11.011) [DOI] [PubMed] [Google Scholar]
- 56.Conrad S., et al. 2008. Generation of pluripotent stem cells from adult human testis. Nature 456, 344–349 10.1038/nature07404 (doi:10.1038/nature07404) [DOI] [PubMed] [Google Scholar]
- 57.Seandel M., et al. 2007. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature 449, 346–350 10.1038/nature06129 (doi:10.1038/nature06129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mateizel I., et al. 2006. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Hum. Reprod. 21, 503–511 10.1093/humrep/dei345 (doi:10.1093/humrep/dei345) [DOI] [PubMed] [Google Scholar]
- 59.Tropel P., et al. 2010. High-efficiency derivation of human embryonic stem cell lines following pre-implantation genetic diagnosis. In Vitro Cell. Dev. Biol. Anim. 46, 376–385 10.1007/s11626-010-9300-8 (doi:10.1007/s11626-010-9300-8) [DOI] [PubMed] [Google Scholar]
- 60.Eiges R., et al. 2007. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell 1, 568–577 10.1016/j.stem.2007.09.001 (doi:10.1016/j.stem.2007.09.001) [DOI] [PubMed] [Google Scholar]
- 61.Frumkin T., Malcov M., Telias M., Gold V., Schwartz T., Azem F., Amit A., Yaron Y., Ben-Yosef D. 2010. Human embryonic stem cells carrying mutations for severe genetic disorders. In Vitro Cell. Dev. Biol. Anim. 46, 327–336 10.1007/s11626-010-9275-5 (doi:10.1007/s11626-010-9275-5) [DOI] [PubMed] [Google Scholar]
- 62.Mateizel I., Spits C., De R. M., Liebaers I., Sermon K. 2010. Derivation, culture, and characterization of VUB hESC lines. In Vitro Cell. Dev. Biol. Anim. 46, 300–308 10.1007/s11626-010-9284-4 (doi:10.1007/s11626-010-9284-4) [DOI] [PubMed] [Google Scholar]
- 63.Urbach A., Bar-Nur O., Daley G. Q., Benvenisty N. 2010. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell 6, 407–411 10.1016/j.stem.2010.04.005 (doi:10.1016/j.stem.2010.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urbach A., Schuldiner M., Benvenisty N. 2004. Modeling for Lesch–Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells 22, 635–641 10.1634/stemcells.22-4-635 (doi:10.1634/stemcells.22-4-635) [DOI] [PubMed] [Google Scholar]
- 65.Lombardo A., et al. 2007. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25, 1298–1306 10.1038/nbt1353 (doi:10.1038/nbt1353) [DOI] [PubMed] [Google Scholar]
- 66.Hockemeyer D., et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27, 851–857 10.1038/nbt.1562 (doi:10.1038/nbt.1562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khan I. F., et al. 2010. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 18, 1192–1199 10.1038/mt.2010.55 (doi:10.1038/mt.2010.55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu J., et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science. 318, 1917–1920 10.1126/science.1151526 (doi:10.1126/science.1151526) [DOI] [PubMed] [Google Scholar]
- 69.Maherali N., Ahfeldt T., Rigamonti A., Utikal J., Cowan C., Hochedlinger K. 2008. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3, 340–345 10.1016/j.stem.2008.08.003 (doi:10.1016/j.stem.2008.08.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woltjen K., et al. 2009. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 10.1038/nature07863 (doi:10.1038/nature07863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I. I., Thomson J. A. 2009. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 10.1126/science.1172482 (doi:10.1126/science.1172482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim D., et al. 2009. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4, 472–476 10.1016/j.stem.2009.05.005 (doi:10.1016/j.stem.2009.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. 2009. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. B 85, 348–362 10.2183/pjab.85.348 (doi:10.2183/pjab.85.348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Warren L., et al. 2010. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 10.1016/j.stem.2010.08.012 (doi:10.1016/j.stem.2010.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. 2010. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651–655 10.1016/j.stem.2010.11.015 (doi:10.1016/j.stem.2010.11.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. 2010. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181 10.1038/nature09017 (doi:10.1038/nature09017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chin M. H., Pellegrini M., Plath K., Lowry W. E. 2010. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell 7, 263–269 10.1016/j.stem.2010.06.019 (doi:10.1016/j.stem.2010.06.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim K., et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 10.1038/nature09342 (doi:10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Polo J. M., et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855 10.1038/nbt.1667 (doi:10.1038/nbt.1667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tchieu J., et al. 2010. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell 7, 329–342 10.1016/j.stem.2010.06.024 (doi:10.1016/j.stem.2010.06.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papapetrou E. P., et al. 2009. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl Acad. Sci. USA 106, 12 759–12 764 10.1073/pnas.0904825106 (doi:10.1073/pnas.0904825106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guillemot F., Lo L.-C., Johnson J. E., Auerbach A., Anderson D. J., Joyner A. L. 1993. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75, 463–476 10.1016/0092-8674(93)90381-Y (doi:10.1016/0092-8674(93)90381-Y) [DOI] [PubMed] [Google Scholar]
- 83.Anderson S. L., Qiu J., Rubin B. Y. 2003. Tocotrienols induce IKBKAP expression: a possible therapy for familial dysautonomia. Biochem. Biophys. Res. Commun. 306, 303–309 10.1016/S0006-291X(03)00971-9 (doi:10.1016/S0006-291X(03)00971-9) [DOI] [PubMed] [Google Scholar]
- 84.Anderson S. L., Qiu J., Rubin B. Y. 2003. EGCG corrects aberrant splicing of IKAP mRNA in cells from patients with familial dysautonomia. Biochem. Biophys. Res. Commun. 310, 627–633 10.1016/j.bbrc.2003.09.019 (doi:10.1016/j.bbrc.2003.09.019) [DOI] [PubMed] [Google Scholar]
- 85.Galan-Caridad J. M., Harel S., Arenzana T. L., Hou Z. E., Doetsch F. K., Mirny L. A., Reizis B. 2007. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell 129, 345–357 10.1016/j.cell.2007.03.014 (doi:10.1016/j.cell.2007.03.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clayson D., Welton W., Axelrod F. B. 1980. Personality development and familial dysautonomia. Pediatrics 65, 269–274 [PubMed] [Google Scholar]
- 87.Lee G., Studer L. 2010. Induced pluripotent stem cell technology for the study of human disease. Nat. Methods 7, 25–27 10.1038/nmeth.f.283 (doi:10.1038/nmeth.f.283) [DOI] [PubMed] [Google Scholar]
- 88.Ibrahim E. C., Hims M. M., Shomron N., Burge C. B., Slaugenhaupt S. A., Reed R. 2007. Weak definition of IKBKAP exon 20 leads to aberrant splicing in familial dysautonomia. Hum. Mutat. 28, 41–53 10.1002/humu.20401 (doi:10.1002/humu.20401) [DOI] [PubMed] [Google Scholar]
- 89.Kim S. C., et al. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 10.1016/j.molcel.2006.06.026 (doi:10.1016/j.molcel.2006.06.026) [DOI] [PubMed] [Google Scholar]
- 90.Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 10.1126/science.1175371 (doi:10.1126/science.1175371) [DOI] [PubMed] [Google Scholar]
- 91.Sommer L., Shah N., Rao M., Anderson D. J. 1995. The cellular function of MASH1 in autonomic neurogenesis. Neuron 15, 1245–1258 10.1016/0896-6273(95)90005-5 (doi:10.1016/0896-6273(95)90005-5) [DOI] [PubMed] [Google Scholar]
- 92.Gold-von S. G., et al. 2009. Kinetin in familial dysautonomia carriers: implications for a new therapeutic strategy targeting mRNA splicing. Pediatr. Res. 65, 341–346 10.1203/PDR.0b013e318194fd52 (doi:10.1203/PDR.0b013e318194fd52) [DOI] [PubMed] [Google Scholar]