Abstract
Epigenetic reprogramming in the germline provides a developmental model to study the erasure of epigenetic memory as it occurs naturally in vivo in the course of normal embryonic development. Our data show that germline reprogramming comprises both active DNA demethylation and extensive chromatin remodelling that are mechanistically linked through the activation of the base excision DNA repair pathway involved in the DNA demethylation process. The observed molecular hallmarks of the germline reprogramming exhibit intriguing similarities to other dedifferentiation or regeneration systems, pointing towards the existence of unifying molecular pathways underlying cell fate reversal. Elucidation of molecular processes involved in the resetting of epigenetic information in vivo will thus add to our ability to manipulate cell fate and to restore pluripotency in in vitro settings.
Keywords: reprogramming, germ cells, chromatin, DNA demethylation
1. Introduction
Embryonic development begins in the zygote where maternal and paternal genomes combine following fertilization (figure 1). The epigenetic information carried by mature gametes to the embryo has been shown to be essential for further embryonic development—a phenomenon that gave basis to the postulation of genomic imprinting [1,2]. Soon after fertilization (4–6 h post-fertilization in the mouse), the paternal genome undergoes a wave of genome-wide DNA demethylation that affects single copy genes as well as repetitive elements [3,4]. Interestingly, the genomic imprints sustain through this process and are propagated during the whole embryonic development. The zygotic DNA demethylation is targeted specifically to the paternal pronucleus while the maternal genome seems to be unaffected [3,4]. The epigenetic asymmetry between the parental genomes is further exemplified by the asymmetry in chromatin configuration—the paternal chromatin is preferentially associated with histone variant H3.3, while the maternal pronucleus is generally enriched in repressive histone modification marks such as H3K27me3 and H3K9me2 [5–7]. While some of the parent-of-origin-specific chromatin differences persist through the syngamy and first one to two cleavage divisions, the differences in DNA methylation between the parental genomes seem to disappear during the course of preimplantation development owing to the passive loss of DNA methylation [5,6,8].
Figure 1.
The life cycle of epigenome. Scheme representing major changes in epigenetic information as they occur during the mouse development. DNA demethylation proceeds in the male pronucleus of the zygote followed later by the erasure of polycomb (Pc) marks in the ICM of blastocyst. PGCs undergo erasure of both DNA methylation and polycomb marks at E11.5. 5mc, 5-methyl-cytosine.
The appearance of pluripotent cells in the inner cell mass (ICM) of the mouse blastocyst is connected with another wave of major epigenetic changes—the Nanog expressing ICM cells of the female mouse embryos undergo re-activation of the inactive X chromosome (Xi) that is observable by the loss of association of Xi with the non-coding Xist RNA and by the loss of enrichment of Ezh2 and Eed as well as of polycomb H3K27me3 marks localized to the Xi [6]. This erasure of imprinted X inactivation is potentially a readout of underlying changes in the chromatin configuration, which may occur in a gender non-specific manner. It should be noted that the reactivation of Xi has been well documented in the case of mouse embryos; the clear evidence for existence of this phenomenon in other species is however still missing. The formation of post-implantation pluripotent epiblast is further connected with the induced expression of the DNA methyltransferase Dnmt3b followed by a wave of de novo DNA methylation [9].
The last wave of major epigenetic reprogramming occurs in the developing germline; the post-migratory primordial germ cells (PGCs) residing in the genital ridges undergo genome-wide DNA demethylation, which includes erasure of genomic imprints and extensive chromatin remodelling (see below) as well as re-activation of Xi in female embryos [5,10,11]. It is at this point that the epigenome reaches its most ‘naive’ state during the development and sets the scene for the acquisition of new epigenetic information and genomic imprints that will be transmitted to the next generation through mature gametes (figure 1).
2. Towards the epigenetic ‘ground state’
The key biological function of every organism is to reproduce. In mammals, the basis of the future generation is formed with the appearance of PGCs (future gametes) in the gastrulating embryo [5]. These cells will create functional gametes and eventually restore totipotency upon fertilization in the zygote. In the mouse, the germ cell precursors are formed from the pluripotent epiblast cells through the interaction of signalling pathways and transcriptional networks [12]. The specification of early PGCs is dependent on the expression of several key germline factors such as Blimp1 and Prdm14, and the absence of either of these proteins causes early loss of germ cells [13,14].
The transcriptional network operating in specified PGCs shows clear similarities to the described core pluripotency transcriptional network [5]. While Oct4 is continuously expressed from epiblast through to the nascent Blimp1-positive PGCs, Sox2 expression is transiently downregulated in early Blimp1-positive cells [15]. Nanog, another core transcriptional regulator of pluripotency, starts to be expressed in the germline only around embryonic day (E) 7.75 [16,17]. The importance of the pluripotency-associated transcriptional network for the maintenance of the embryonic germline is documented by the experiments using germline-specific deletion of Oct4 and Nanog—deletion of either of the factors results in apoptosis of germ cells [17–19]. Initial observations on chimeras produced from Nanog null embryonic stem (ES) cells showed disappearance of PGCs around the time of epigenetic reprogramming [19]. These experiments hence suggested a potential role of Nanog in the reprogramming process. However, an independent study using the in vivo knockdown approach of Nanog revealed a gradual loss of PGCs prior to the reprogramming step, supporting the notion that Nanog is involved in the maintenance of the germline [17].
From the epigenetic perspective, the nascent PGCs are successors of epiblast cells and as such they have acquired layers of epigenetic information including DNA methylation and numerous histone modifications in a manner similar to other epiblast cells that will give rise to the soma. However, following the specification, the chromatin of germ cells starts to change. The arginine methyltransferase Prmt5 translocates to the nucleus of PGCs, where it forms a complex with the transcriptional regulator Blimp1 and drives repressive modification of histones H2A and H4 (H2A/H4R3me2s) [20]. Additionally, repressive H3K9me2 disappears from the chromatin of germ cells owing to the transcriptional downregulation of Glp1 which is a key component of the G9a/Glp1 histone methyltransferase complex [21–23]. Loss of H3K9me2, a hallmark of facultative heterochromatin, is counteracted by the upregulation of Ezh2 and H3K27me3 modification deposited by the core component of the PRC2 polycomb complex [21–23]. The importance of chromatin re-configuration in the early germline is documented by the experiments using the Prdm14 knockout model—PGCs lacking this factor fail to downregulate H3K9me2 and to upregulate the polycomb system which leads to their failure to give rise to pluripotent embryonic germ (EG) cells in culture [14]. The early chromatin changes set the germline apart from the surrounding soma and are potentially crucial for the successful reprogramming step that occurs later when PGCs reside in the genital ridges.
In order to give rise to functional gametes, PGCs need to reset epigenetic information including the genomic imprints. The existence of the erasure step in the germline was first documented by Monk and colleagues [24], however the timing and kinetics of the process have been further refined only in 2002 [10]. The studies revealed that the erasure of imprinted DNA methylation occurs in post-migratory PGCs between E11.5 and E12.5 in a distinct step. Given the doubling time of PGCs (approx. 16 h), these findings clearly hinted towards an active DNA demethylation mechanism being a part of the reprogramming process [10]. Additionally, bisulphite analysis of various imprinted as well as non-imprinted genes showed that the DNA demethylation affects all examined regions (imprinted as well as non-imprinted). This data suggested an unprecedented scale of DNA demethylation, which has been further documented by recent studies [10,25].
It is interesting to note that among various cell types, it is only the developing PGCs that have the ability to erase all layers of epigenetic information. This includes not only the reversal of genomic imprints and erasure of most of the DNA methylation, but also major changes in histone modifications and histone variant composition (see below). Thus, in the context of mammalian development, the epigenome reaches the lowest point (contains the lowest amount of information) in the developing germ cells following the reprogramming step. PGCs are, however, highly specialized cells that will give rise only to gametes and thus it is important to note that the erasure of epigenetic information that occurs in PGCs and the occurrence of pluripotency and developmental naivety in the cells of the ICM of blastocyst are spatially and temporally separated during the embryonic development.
3. Role of base excision dna repair in active dna demethylation
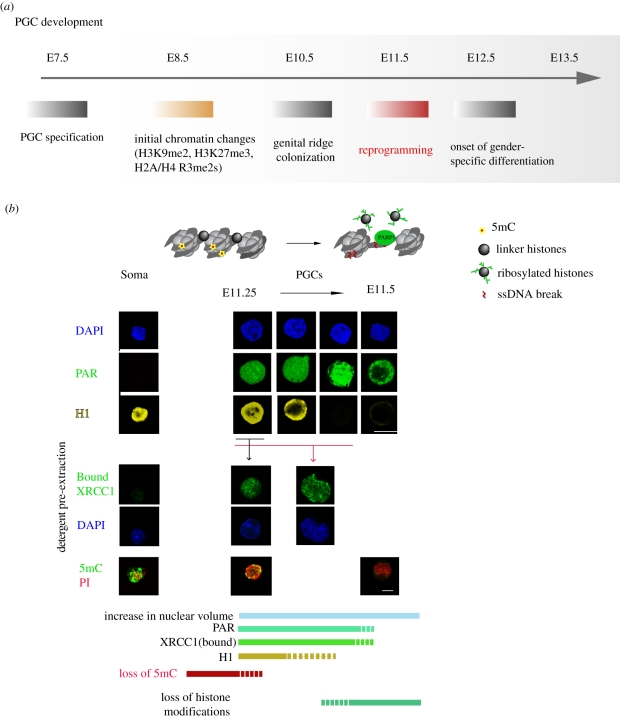
Following PGC specification (completed by E7.5), the nascent PGCs start to migrate along the developing hindgut (from E8.5) until they reach the genital ridges at E10.5 (figure 2).
Figure 2.
Mouse germline development and the kinetics of reprogramming. (a) Overview of germline development in the mouse. (b) Immunofluorescence detection of markers of active BER response in PGCs at the time of DNA demethylation. Bottom panel shows the kinetics of molecular events observed during the epigenetic reprogramming in PGCs.
At E11.5, soon after their entry into the genital ridges, PGCs start to undergo pronounced changes in their nuclear architecture connected with the decondensation of heterochromatin, loss of chromocentres and increase in nuclear size [23]. Interestingly, this correlates with the upregulation of key factors involved in the base excision DNA repair (BER) pathway at the transcriptional as well as protein level [11] (figure 2). Induction of BER response in PGCs at this time is further documented by the presence of active poly(ADP)-ribose polymerase 1 (PARP1) as judged by the appearance of poly-ADP-ribose (PAR) polymers in the PGC nuclei as well as chromatin-bound XRCC1, indicating the presence of single-strand DNA (ssDNA) breaks. Importantly, these processes coincide with the loss of 5-methyl-cytosine (5mC), supporting the idea that, as observed in plants, active DNA demethylation in mammals proceeds through BER [11] (figure 2).
There are two waves of active DNA demethylation in mammalian development. The germline demethylation proceeds at E11.5 in post-migratory PGCs. Another wave of demethylation occurs in the zygote where the paternal pronucleus undergoes genome-wide loss of 5mC just a few hours following fertilization [6] (figure 1 and table 1). Interestingly, both developmental models of active DNA demethylation display similar hallmarks of BER activation. PARP1 as well as chromatin-bound XRCC1, signalling the presence of ssDNA breaks, are detected specifically in the paternal pronucleus of the zygote at the time of DNA demethylation. Additionally, small molecule inhibitors of BER inhibit the progression of DNA demethylation, further supporting the mechanistic link between those two processes [11]. While some of the previous studies have suggested a possible link between the nucleotide excision repair (NER) pathway and active DNA demethylation [26,27], it is noteworthy that no evidence has been found for NER involvement either in the germline or in zygotic DNA demethylation [11]. The cumulative evidence thus points towards at least some level of mechanistic conservation between DNA demethylation processes in mammals and plants.
Table 1.
Comparison of epigenetic reprogramming in mouse zygote and in the developing germline.
| zygotic reprogramming | germline reprogramming |
|---|---|
| active DNA demethylation (paternal genome) linked to the activation of BER | active DNA demethylation linked to the activation of BER |
| maintenance of genomic imprints | erasure of genomic imprints |
| lack of ability to re-activate the Xi (SCNT experiments) | re-activation of inactive X chromosome (Xi) in female embryos |
| maintenance of polycomb histone modifications (H3K27me3) | loss of polycomb histone modifications (H3K27me3) |
| presence of key pluripotency factors (Oct4, Sox2, Nanog missing) | presence of key pluripotency factors (Oct4, Sox2 and Nanog) |
Investigation of the molecular mechanism implicated in active DNA demethylation in flowering plants revealed the existence of the 5mC-specific glycosylases, Dme1 and Ros1 [28–30]. As proteins homologous to Dme1 or Ros1 have not been found in higher vertebrates, the molecular events leading to the activation of BER in the course of active DNA demethylation in mammals remain to be elucidated. 5mC can be either targeted and excised directly by a yet unknown DNA glycosylase or, alternatively, BER response can be triggered by further chemical modification of 5mC (figure 3). To this end, DNA deaminases Aid and Apobec1 have been shown to deaminate 5mC to T in vitro [31]. In vivo, in the context of a double-stranded DNA template, the resulting T–G mismatch could be recognized and further processed by the T–G mismatch glycosylases Tdg1 or Mbd4. Alternatively, the modification of 5mC to 5-hydroxymethyl-cytosine (5hmC) has recently been reported [32,33] and the presence of this modified base could potentially trigger the BER response (figure 3).
Figure 3.
Epigenetic reprogramming—connection between DNA demethylation and chromatin remodelling. The model depicts possible routes leading to the activation of BER in the course of DNA demethylation in mouse development and the connection between observed DNA demethylation and chromatin remodelling.
Possible involvement of DNA deaminases Aid or Apobec1 in the active DNA demethylation in vivo has also been recently suggested [34,35], including a mechanistic link between Aid and the active DNA demethylation in the mouse germline [25]. The genetic ablation of Aid has, however, only limited impact on the 5mC levels in PGCs and the expression of Aid is detectable only after the wave of DNA demethylation [11,25]. Taken together with the fact that T–G mismatch glycosylases are either not detectable (Tdg) or that their loss has no impact on germline development or fertility (Mbd4), clear evidence for the involvement of DNA deamination in the context of mouse germline reprogramming is still missing [11].
In contrast, there is a detectable upregulation of Tet1 responsible for the conversion of 5mC to 5hmC, specifically in the PGCs at the time of DNA demethylation [11]. While high levels of Tet1 and 5hmC are also seen in the zygote at the time of DNA demethylation (Rachel Amouroux, Aditya Sankar and Petra Hajkova 2010, unpublished data), further experiments are needed to elucidate the exact role of 5hmC in the process of active DNA demethylation during mouse development.
4. Chromatin remodelling in the process of epigenetic reprogramming
Besides DNA demethylation, the reprogramming process in PGCs is also associated with the loss of numerous histone modifications [23]. The loss of repressive histone marks (H3K9me3, H3K27me3 and H2A/H4R3me2s) as well as marks associated with active chromatin (H3K9ac) has been described and while some of the modifications disappear only for a few hours, a number of changes persist [23]. As the observed changes affect modifications of all four core histones, it is unlikely that the process is a result of a concerted action of numerous histone de-modification enzymes. Instead, we favour the explanation of nucleosome disassembly and histone replacement. This idea is also supported by observable changes in the composition of histone variants as well as relocalizaton of Nap-1, a histone chaperone involved in chromatin disassembly in vitro, to the nucleus of PGCs at the time of epigenetic reprogramming [23].
The experimental evidence shows that in the course of germline epigenetic reprogramming, the onset of chromatin remodelling connected with the change of histone modification profile follows the onset of DNA demethylation [23]. The possible mechanistic link between those processes is provided by the activation of PARP1 observed in the course of BER response (figure 2). One of the hallmarks of the PGCs undergoing reprogramming is a transient loss of linker H1 histones [23]. In this context, PARP1 has been shown not only to replace H1 in the chromatin, but experimental evidence also shows that H1 is one of the preferred targets of PARP1 ribosylation activity and intensive ribosylation can potentially contribute to the removal/eviction of histones from the chromatin [36–38]. The connection between pronounced changes in chromatin compaction and the activation of PARP1 has been previously reported during the heat shock-driven transcriptional activation or in the course of DNA repair. It is also found in PGCs where the detection of PAR polymers in the germ cell nuclei undergoing reprogramming shows direct correlation with the level of chromatin decondensation [11,39–41].
Our data show that active DNA demethylation in PGCs is connected with the activation of BER involving active Parp1, which in turn could be a driving force behind the large-scale chromatin remodelling that follows the loss of 5mC [11] (figure 3). However, it is also possible that the activation of BER and Parp1 are only partially responsible for the observed chromatin remodelling and changes in histone modifications, and hence other independent molecular mechanisms could additionally contribute to the reprogramming process.
5. What information needs to be erased?
Two major repressive systems, DNA methylation and polycomb silencing, contribute mechanistically to the propagation of epigenetic memory through cell divisions. In order to reset genomic imprinting, to erase the epigenetic modifications imposed during the embryonic development as well as to prevent the accumulation of epimutations induced by the environment, the germline has to intervene with both the DNA methylation and the polycomb silencing. It is interesting to note that this feature is a unique attribute of germline reprogramming as the reprogramming in the zygote and ICM/early post-implantation epiblast involves only the DNA demethylation and the erasure of polycomb marks, respectively (see above, figure 1 and table 1).
The cumulative evidence shows that the epigenetic reprogramming involves erasure of DNA methylation as well as numerous histone modifications (including H3K27me3 polycomb histone marks). Moreover, the reprogramming process in the mouse germline is also connected with the change in the histone variant composition as well as major changes in nuclear architecture. This suggests that the epigenetic information in PGCs is erased at numerous distinct levels possibly including the erasure of structural information embedded in the higher order chromatin folding [23].
Upon completion of epigenetic reprogramming in PGCs at E11.5, the epigenome reaches the most ‘naive’ state in the development—the PGCs are devoid of most DNA methylation, they lack genomic imprints, have very low levels of H3K9me2 and transiently also have very low levels of H3K27me3. This state of the epigenome, which has no parallel in any other cell during development, creates a baseline for setting up of new information that will eventually be transmitted to the next generation through mature gametes. Notably, this epigenetically ‘naive’ state is acquired by highly specialized unipotent germ cells in a tightly regulated fashion (potentially through strong transcriptional networks) as erasure of epigenetic memory could lead to the reversal of developmental decision and possible change of cell fate.
6. The role of epigenetic reprogramming
The need for epigenetic reprogramming in the germline is possibly driven by the inductive type of germ cell specification in mammals [12]. The absence of the inherited germ plasm and the fact that the germ cell precursors originate from the post-implantation epiblast that has accumulated layers of epigenetic marks during early development may drive the need for the reprogramming step in the germline that ensures the erasure of this information. Alternatively, the necessity of the germline reprogramming is connected with the existence of genomic imprinting—the parent-of-origin marks carried by mature gametes are combined in the zygote and inherited by nascent PGCs from their precursors in post-implantation epiblast. These marks need to be erased and reset in a sex-specific manner in order to create future functional gametes.
Notably, epigenetic reprogramming occurs in the germline prior to the sex differentiation (E12.5) and the onset of meiosis (E13.5 in female germline). It is thus also possible that the reprogramming process is one of the requirements for the successful execution of meiosis. However, further, in-depth studies are needed to address this hypothesis.
7. Mechanistic links to other reprogramming systems
Resetting of epigenetic information in the mammalian germline is a highly efficient process that is a key part of a normal developmental programme in the germline of every individual. Resetting of epigenetic information mechanistically underlies also the processes of cell fate switch, regeneration and regaining of pluripotency, and hence it is intriguing to investigate the possible parallels between these processes and the molecular hallmarks of the germline reprogramming.
Extensive chromatin remodelling and heterochromatin decondensation have been observed in somatic cell nuclear transfer as well as during protoplast formation in plants and is one of the first observable features of the ongoing reprogramming process in both systems [42–44]. Changes in H3K9me as well as release of heterochromatin protein 1 (HP1) from chromatin observed during germline reprogramming have also been associated with the protoplast formation [43,44]. Loss of linker histones and chromatin decondensation have been observed upon incubation of chicken erythrocytes with Xenopus egg extracts or upon transplantation of somatic nuclei into Xenopus eggs [45,46]. Last but not least, a histone demethylase removing H3K27me3 polycomb marks which are also lost during germline reprogramming has been implicated in the regeneration process in zebrafish [47].
Reversal of the differentiation programme and regaining of pluripotency followed by directed differentiation has been one of the major goals in regenerative medicine, and several in vitro systems have been designed and used in order to pursue the reversal of cell fate. Transfer of somatic cell nucleus into the enucleated oocyte (SCNT—somatic cell nuclear transfer) can lead, albeit with low frequency, to the reversal of somatic cell fate and re-acquisition of totipotency [48]. Reversal of the somatic cell programme is also observed upon transplantation of somatic nuclei into the Xenopus egg or upon fusion of a somatic cell with a pluripotent ES or EG cell [49–51]. The latest advances in the field showed that reversal of somatic fate and regaining of pluripotency can be achieved through the delivery of four stem cell-related transcription factors into somatic cells [52]. However, even using this method, the efficiency of generating induced pluripotent stem (iPS) cells is relatively low. The efficiency can be increased by addition of drugs modulating chromatin structure, such as histone deacetylase (HDAC) inhibitors or inhibitors of H3K9 methyltransferase [53,54], adding to the notion that disruption of epigenetic memory is one of the key steps underlying cell fate reversal. Increase of reprogramming efficiency is also induced by drugs interfering with the maintenance of DNA methylation [55] and, interestingly, inefficient erasure of this epigenetic mark and persistence of the original somatic type of DNA methylation signature has been linked to the low efficiency of iPS generation [56,57].
8. Outlook
Epigenetic reprogramming in the mouse germline provides a unique model to study molecular mechanisms underlying erasure of epigenetic memory as it occurs naturally in vivo. Detailed mechanistic understanding of this process and the connections between active DNA demethylation and chromatin remodelling will shed light on the processes connected with the cell fate reversal and regaining of pluripotency and will eventually help to design new, more efficient in vitro reprogramming protocols.
Acknowledgements
I would like to thank members of my laboratory: R. Amouroux, A. Turp and A. Sankar, as well as Prof. A. Surani and past and present members of his laboratory for their contribution, help and discussions. I would also like to thank T. Carroll for helpful comments to the manuscript. The research in my laboratory is funded by the Medical Research Council.
References
- 1.McGrath J., Solter D. 1984. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183 10.1016/0092-8674(84)90313-1 (doi:10.1016/0092-8674(84)90313-1) [DOI] [PubMed] [Google Scholar]
- 2.Barton S. C., Surani M. A., Norris M. L. 1984. Role of paternal and maternal genomes in mouse development. Nature 311, 374–376 10.1038/311374a0 (doi:10.1038/311374a0) [DOI] [PubMed] [Google Scholar]
- 3.Mayer W., Niveleau A., Walter J., Fundele R., Haaf T. 2000. Demethylation of the zygotic paternal genome. Nature 403, 501–502 10.1038/35000656 (doi:10.1038/35000656) [DOI] [PubMed] [Google Scholar]
- 4.Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J. 2000. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10, 475–478 10.1016/S0960-9822(00)00448-6 (doi:10.1016/S0960-9822(00)00448-6) [DOI] [PubMed] [Google Scholar]
- 5.Surani M. A., Hayashi K., Hajkova P. 2007. Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762 10.1016/j.cell.2007.02.010 (doi:10.1016/j.cell.2007.02.010) [DOI] [PubMed] [Google Scholar]
- 6.Hajkova P. 2010. Epigenetic reprogramming–taking a lesson from the embryo. Curr. Opin. Cell. Biol. 22, 342–350 10.1016/j.ceb.2010.04.011 (doi:10.1016/j.ceb.2010.04.011) [DOI] [PubMed] [Google Scholar]
- 7.Santos F., Peters A. H., Otte A. P., Reik W., Dean W. 2005. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 280, 225–236 10.1016/j.ydbio.2005.01.025 (doi:10.1016/j.ydbio.2005.01.025) [DOI] [PubMed] [Google Scholar]
- 8.Puschendorf M., et al. 2008. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40, 411–420 10.1038/ng.99 (doi:10.1038/ng.99) [DOI] [PubMed] [Google Scholar]
- 9.Hirasawa R., Sasaki H. 2009. Dynamic transition of Dnmt3b expression in mouse pre- and early post-implantation embryos. Gene Expr. Patterns 9, 27–30 10.1016/j.gep.2008.09.002 (doi:10.1016/j.gep.2008.09.002) [DOI] [PubMed] [Google Scholar]
- 10.Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., et al. 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117, 15–23 10.1016/S0925-4773(02)00181-8 (doi:10.1016/S0925-4773(02)00181-8) [DOI] [PubMed] [Google Scholar]
- 11.Hajkova P., Jeffries S. J., Lee C., Miller N., Jackson S. P., Surani M. A. 2010. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329, 78–82 10.1126/science.1187945 (doi:10.1126/science.1187945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi K., de Sousa Lopes S. M., Surani M. A. 2007. Germ cell specification in mice. Science 316, 394–396 10.1126/science.1137545 (doi:10.1126/science.1137545) [DOI] [PubMed] [Google Scholar]
- 13.Ohinata Y., et al. 2005. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213 10.1038/nature03813 (doi:10.1038/nature03813) [DOI] [PubMed] [Google Scholar]
- 14.Yamaji M., Seki Y., Kurimoto K., Yabuta Y., Yuasa M., Shigeta M., Yamanaka K., Ohinata Y., Saitou M. 2008. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 40, 1016–1022 10.1038/ng.186 (doi:10.1038/ng.186) [DOI] [PubMed] [Google Scholar]
- 15.Kurimoto K., Yabuta Y., Ohinata Y., Shigeta M., Yamanaka K., Saitou M. 2008. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 22, 1617–1635 10.1101/gad.1649908 (doi:10.1101/gad.1649908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi S., Kimura H., Tada M., Nakatsuji N., Tada T. 2005. Nanog expression in mouse germ cell development. Gene Expr. Patterns 5, 639–646 10.1016/j.modgep.2005.03.001 (doi:10.1016/j.modgep.2005.03.001) [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi S., Kurimoto K., Yabuta Y., Sasaki H., Nakatsuji N., Saitou M., Tada T. 2009. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development 136, 4011–4020 10.1242/dev.041160 (doi:10.1242/dev.041160) [DOI] [PubMed] [Google Scholar]
- 18.Kehler J., et al. 2004. Oct4 is required for primordial germ cell survival. EMBO Rep. 5, 1078–1083 10.1038/sj.embor.7400279 (doi:10.1038/sj.embor.7400279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers I., et al. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 10.1038/nature06403 (doi:10.1038/nature06403) [DOI] [PubMed] [Google Scholar]
- 20.Ancelin K., Lange U. C., Hajkova P., Schneider R., Bannister A. J., Kouzarides T., Surani M. A. 2006. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell. Biol. 8, 623–630 10.1038/ncb1413 (doi:10.1038/ncb1413) [DOI] [PubMed] [Google Scholar]
- 21.Seki Y., et al. 2007. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134, 2627–2638 10.1242/dev.005611 (doi:10.1242/dev.005611) [DOI] [PubMed] [Google Scholar]
- 22.Seki Y., Hayashi K., Itoh K., Mizugaki M., Saitou M., Matsui Y. 2005. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 278, 440–458 10.1016/j.ydbio.2004.11.025 (doi:10.1016/j.ydbio.2004.11.025) [DOI] [PubMed] [Google Scholar]
- 23.Hajkova P., et al. 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452, 877–881 10.1038/nature06714 (doi:10.1038/nature06714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monk M., Boubelik M., Lehnert S. 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99, 371–382 [DOI] [PubMed] [Google Scholar]
- 25.Popp C., Dean W., Feng S., Cokus S. J., Andrews S., Pellegrini M., Jacobsen S. E., Reik W. 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463, 1101–1105 10.1038/nature08829 (doi:10.1038/nature08829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barreto G., et al. 2007. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445, 671–675 10.1038/nature05515 (doi:10.1038/nature05515) [DOI] [PubMed] [Google Scholar]
- 27.Schmitz K. M., Schmitt N., Hoffmann-Rohrer U., Schafer A., Grummt I., Mayer C. 2009. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol. Cell. 33, 344–353 10.1016/j.molcel.2009.01.015 (doi:10.1016/j.molcel.2009.01.015) [DOI] [PubMed] [Google Scholar]
- 28.Gehring M., Reik W., Henikoff S. 2009. DNA demethylation by DNA repair. Trends Genet. 25, 82–90 10.1016/j.tig.2008.12.001 (doi:10.1016/j.tig.2008.12.001) [DOI] [PubMed] [Google Scholar]
- 29.Choi Y., Gehring M., Johnson L., Hannon M., Harada J. J., Goldberg R. B., Jacobsen S. E., Fischer R. L. 2002. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 110, 33–42 10.1016/S0092-8674(02)00807-3 (doi:10.1016/S0092-8674(02)00807-3) [DOI] [PubMed] [Google Scholar]
- 30.Gong Z., Morales-Ruiz T., Ariza R. R., Roldan-Arjona T., David L., Zhu J. K. 2002. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 10.1016/S0092-8674(02)01133-9 (doi:10.1016/S0092-8674(02)01133-9) [DOI] [PubMed] [Google Scholar]
- 31.Morgan H. D., Dean W., Coker H. A., Reik W., Petersen-Mahrt S. K. 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 279, 52 353–52 360 10.1074/jbc.M407695200 (doi:10.1074/jbc.M407695200) [DOI] [PubMed] [Google Scholar]
- 32.Tahiliani M., et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 10.1126/science.1170116 (doi:10.1126/science.1170116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kriaucionis S., Heintz N. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 10.1126/science.1169786 (doi:10.1126/science.1169786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rai K., Huggins I. J., James S. R., Karpf A. R., Jones D. A., Cairns B. R. 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135, 1201–1212 10.1016/j.cell.2008.11.042 (doi:10.1016/j.cell.2008.11.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhutani N., Brady J. J., Damian M., Sacco A., Corbel S. Y., Blau H. M. 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463, 1042–1047 10.1038/nature08752 (doi:10.1038/nature08752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnakumar R., Gamble M. J., Frizzell K. M., Berrocal J. G., Kininis M., Kraus W. L. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 10.1126/science.1149250 (doi:10.1126/science.1149250) [DOI] [PubMed] [Google Scholar]
- 37.Althaus F. R., Hofferer L., Kleczkowska H. E., Malanga M., Naegeli H., Panzeter P. L., Realini C. A. 1994. Histone shuttling by poly ADP-ribosylation. Mol. Cell. Biochem. 138, 53–59 10.1007/BF00928443 (doi:10.1007/BF00928443) [DOI] [PubMed] [Google Scholar]
- 38.Aubin R. J., Dam V. T., Miclette J., Brousseau Y., Huletsky A., Poirier G. G. 1982. Hyper(ADP-ribosyl)ation of histone H1. Can. J. Biochem. 60, 1085–1094 10.1139/o82-139 (doi:10.1139/o82-139) [DOI] [PubMed] [Google Scholar]
- 39.Tulin A., Spradling A. 2003. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299, 560–562 10.1126/science.1078764 (doi:10.1126/science.1078764) [DOI] [PubMed] [Google Scholar]
- 40.Kim M. Y., Mauro S., Gevry N., Lis J. T., Kraus W. L. 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119, 803–814 10.1016/j.cell.2004.11.002 (doi:10.1016/j.cell.2004.11.002) [DOI] [PubMed] [Google Scholar]
- 41.Caldecott K. W. 2007. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 6, 443–453 10.1016/j.dnarep.2006.10.006 (doi:10.1016/j.dnarep.2006.10.006) [DOI] [PubMed] [Google Scholar]
- 42.Campbell K. H., Loi P., Otaegui P. J., Wilmut I. 1996. Cell cycle co-ordination in embryo cloning by nuclear transfer. Rev. Reprod. 1, 40–46 10.1530/ror.0.0010040 (doi:10.1530/ror.0.0010040) [DOI] [PubMed] [Google Scholar]
- 43.Williams L., Zhao J., Morozova N., Li Y., Avivi Y., Grafi G. 2003. Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev. Dyn. 228, 113–120 10.1002/dvdy.10348 (doi:10.1002/dvdy.10348) [DOI] [PubMed] [Google Scholar]
- 44.Grafi G. 2004. How cells dedifferentiate: a lesson from plants. Dev. Biol. 268, 1–6 10.1016/j.ydbio.2003.12.027 (doi:10.1016/j.ydbio.2003.12.027) [DOI] [PubMed] [Google Scholar]
- 45.Blank T., Trendelenburg M., Kleinschmidt J. A. 1992. Reactivation of DNA replication in erythrocyte nuclei by Xenopus egg extract involves energy-dependent chromatin decondensation and changes in histone phosphorylation. Exp. Cell Res. 202, 224–232 10.1016/0014-4827(92)90069-K (doi:10.1016/0014-4827(92)90069-K) [DOI] [PubMed] [Google Scholar]
- 46.Kikyo N., Wade P. A., Guschin D., Ge H., Wolffe A. P. 2000. Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science 289, 2360–2362 10.1126/science.289.5488.2360 (doi:10.1126/science.289.5488.2360) [DOI] [PubMed] [Google Scholar]
- 47.Stewart S., Tsun Z. Y., Izpisua Belmonte J. C. 2009. A histone demethylase is necessary for regeneration in zebrafish. Proc. Natl Acad. Sci. USA 106, 19 889–19 894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilmut I., Schnieke A. E., McWhir J., Kind A. J., Campbell K. H. 1997. Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 10.1038/385810a0 (doi:10.1038/385810a0) [DOI] [PubMed] [Google Scholar]
- 49.Byrne J. A., Simonsson S., Western P. S., Gurdon J. B. 2003. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 13, 1206–1213 10.1016/S0960-9822(03)00462-7 (doi:10.1016/S0960-9822(03)00462-7) [DOI] [PubMed] [Google Scholar]
- 50.Tada M., Tada T., Lefebvre L., Barton S. C., Surani M. A. 1997. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 16, 6510–6520 10.1093/emboj/16.21.6510 (doi:10.1093/emboj/16.21.6510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tada M., Takahama Y., Abe K., Nakatsuji N., Tada T. 2001. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11, 1553–1558 10.1016/S0960-9822(01)00459-6 (doi:10.1016/S0960-9822(01)00459-6) [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 (doi:10.1016/j.cell.2006.07.024) [DOI] [PubMed] [Google Scholar]
- 53.Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. 2008. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 10.1038/nbt1418 (doi:10.1038/nbt1418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi Y., Desponts C., Do J. T., Hahm H. S., Scholer H. R., Ding S. 2008. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 3, 568–574 10.1016/j.stem.2008.10.004 (doi:10.1016/j.stem.2008.10.004) [DOI] [PubMed] [Google Scholar]
- 55.Mikkelsen T. S., et al. 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 10.1038/nature07056 (doi:10.1038/nature07056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polo J. M., et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855 10.1038/nbt.1667 (doi:10.1038/nbt.1667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim K., et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 10.1038/nature09342 (doi:10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]