Abstract
There has been much recent interest and progress in the characterization of community structure and community assembly processes through the application of phylogenetic methods. To date most focus has been on groups of taxa for which some relevant detail of their ecology is known, for which community composition is reasonably easily quantified and where the temporal scale is such that speciation is not likely to feature. Here, we explore how we might apply a molecular genetic approach to investigate community structure and assembly at broad taxonomic and geographical scales, where we have little knowledge of species ecology, where community composition is not easily quantified, and where speciation is likely to be of some importance. We explore these ideas using the class Collembola as a focal group. Gathering molecular evidence for cryptic diversity suggests that the ubiquity of many species of Collembola across the landscape may belie greater community complexity than would otherwise be assumed. However, this morphologically cryptic species-level diversity poses a challenge for attempts to characterize diversity both within and among local species assemblages. Recent developments in high throughput parallel sequencing technology, combined with mtDNA barcoding, provide an advance that can bring together the fields of phylogenetic and phylogeographic analysis to bear on this problem. Such an approach could be standardized for analyses at any geographical scale for a range of taxonomic groups to quantify the formation and composition of species assemblages.
Keywords: high throughput sequencing, next generation sequencing, mesofauna, Collembola, cryptic species, DNA barcoding
1. Phylogeny and community analysis: a brief history
The past decade has witnessed an exciting coalescence of ecological investigation and evolutionary thinking. Webb's analysis of the phylogenetic structure of rainforest tree communities [1] can be seen as a catalyst for subsequent developments in the application of phylogenetic approaches both to characterize how communities of species are structured, and understand the processes that underlie structure (or lack thereof). Several recent complementary reviews provide readers with an understanding of both the state of the field, and useful future directions [2–5]. It is not our intention here to further review this field of research, but in the context of the focus of our manuscript, it is important to provide some backdrop for the ideas we wish to advocate. Hence what follows is a brief overview of the field of phylogenetic analysis of community structure.
The assembly and composition of communities of species is considered to be influenced by three processes, with some debate as to the relative importance of each of these [2,3]. The first of these three processes places importance on niche as a regulator of assembly, with two contrasting roles that niche may have. Given that closely related species are likely to have greater niche similarity than more distantly related species, one may expect closely related species to have a higher probability of co-occurrence than more distantly related species. Under such a scenario, the environment, or habitat, may function as a filter, selecting for sets of phylogenetically related species. However, given the competition that may ensue from the co-occurrence of phylogenetically related species, phylogenetic relatedness can equally be expected to limit coexistence, selecting for sets of phylogenetically less related species. Traits important for niche occupancy are considered to exhibit phylogenetic conservatism under both these scenarios. However, in cases where phylogenetic traits may be considered to exhibit evolutionary lability, the potential for strong competitive interactions to drive selection for divergent traits may facilitate the coexistence of closely related species, pushing community structure towards phylogenetically related groupings. The second process that is suggested to be of importance in the assembly of communities is neutrality, which gives greater emphasis to stochasticity over niche [6], where species within a trophic level are competitively equivalent and become persistent through the stochastic dynamics of dispersal, extinction and speciation. In this context, there are parallels with the theory of island biogeography [7], where species are examined in the absence of ecology, and patterns of phylogenetic relatedness are expected to conform to null expectations from stochastic assembly.
The niche-based and neutral processes just described correspond to the niche-based environmental filtering, niche-based species sorting, and neutral processes highlighted by Weiher et al. [8] as having much importance in ecological approaches to community assembly. The third process that is considered to be important in community assembly, but that receives less emphasis in ecological approaches, is history. In this context, Ricklefs [9] has argued that biogeographical and evolutionary processes cannot be ignored when one wants to understand community composition. This argument challenges the view that the time-scale over which ecological communities establish is sufficiently limited to consider the pool of species that may contribute membership to a community as fixed. Under this scenario the separation of the local species community and the regional pool from which it is sampled becomes blurred as interspecific interactions within communities can promote evolutionary change, contributing to speciation. This provides an interesting parallel to Emerson & Kolm's [10] proposition that increased membership of a local species community may promote evolutionary change within that community.
To take account of history, analyses of local diversity patterns must be placed in the context of regional distribution, together with evolutionary and environmental history [11]. Ricklefs [11] puts forward a number of suggestions for how this might be achieved, including: characterizing the distributions of species over the geographical and ecological gradients within which they interact; examining these within a phylogenetic context; examining local assemblages and distributions of species across important gradients of diversity; characterizing diversification rates across geographical or ecological gradients; incorporating phylogeographic data to characterize incipient speciation; embracing extinction as an important process in the establishment of diversity patterns. Phylogeny and phylogeography are implicit within the suggestions of Ricklefs. However, the incorporation of phylogeography within the analysis of community ecology has largely remained unexplored to date, although its potential has been noted [12].
2. Phylogeography and community analysis
Phylogeographic investigation seeks to infer the origin of geographical structuring of genetic variation within and among closely related species across the landscape by recourse to genealogical relationships of allelic variation within loci [13]. While other genetic data may be complementarily informative (e.g. microsatellites, single nucleotide polymorphisms, amplified fragment length polymorphisms (AFLPs)), it is the ability to construct genealogical relationships among DNA sequences that forms the core of phylogeographic analysis. As a discipline, the emergence of phylogeography coincided with increasing access to population level DNA sequence data afforded to biologists by the polymerase chain reaction (PCR). Early approaches to phylogeographic inference relied upon qualitative assessments of the correspondence between geography and genealogy, frequently relying on either mitochondrial (mt) or chloroplast (cp) DNA. Under this approach, phylogeographic history is inferred from a bifurcating tree topology with geographically coherent clade membership, and this may be calibrated to place an inferred demographic event into a temporal framework. While in some cases this may be acceptable, in many cases only limited information content will be extracted from the data, due to the vagaries of mutation and population-level processes (e.g. [14,15]). The field has subsequently developed with a greater emphasis on approaches that consider both mutational and coalescent variance, referred to as statistical phylogeography [16]. The availability of tools for statistical hypothesis testing, combined with the use of multiple loci for phylogeographic inference, means that one can undertake rigorous phylogeographic analyses either within or among species (see [12,17] for recent reviews). Thus, one can potentially undertake comparative phylogeographic analyses, with each focal species being sampled for multiple genetic loci, although this comprises a large task that has not been realized to date; so far studies have focused on one species and several genes (e.g. [14–17,18,19]) or several species and one gene (e.g. [20–23]).
It has previously been suggested that an informative approach to developing and understanding the general principles underlying community assembly may be to step back and look at the combined results of multiple phylogeographic studies for regional synthesis [24]. More recently, Hickerson et al. [12] have suggested the same, but noted that achieving this has been handicapped because comparative phylogeographic analyses typically only involve a handful of co-distributed taxa, and statistical tools for comparative phylogeography are in their infancy. An additional issue is that taxon sampling for comparative phylogeographic analysis is not necessarily community focused. If one considers that the temporal scale of community assembly may be sufficient to include speciation [3], then it must be acknowledged that statistical tools will need to incorporate models of both the coalescent and stochastic lineage/species emergence. This will be particularly relevant if species sampling for a comparative analysis is likely to include species harbouring cryptic diversity. One useful approach may be to incorporate the general mixed Yule coalescent (GMYC) model of Pons et al. [25] to first quantify probable species boundaries that will define the units of analysis for more detailed phylogeographic analyses within these.
The rise of approximate Bayesian computation (ABC) [26] offers potential for intraspecific phylogeographic analysis. This potential has yet to be realized [12], but that is likely to change as sufficiently large and detailed comparative phylogeographic datasets come to hand. Hierarchical ABC is one promising development in this direction, and has recently been used to test competing hypotheses to explain the distributions of multiple taxon pairs [27]. Another promising investigative approach is that of Lemey et al. [28], who have introduced Bayesian modelling of character evolution for the inference of ancestral states. Using geographical locations as character states, Lemey et al. [28] are able to infer the posterior probabilities for the geographical location of ancestral nodes and migration events, while at the same time taking into account genealogical uncertainty. The implementation of Lemey et al. [28] is limited to populations conforming to panmixia, and it is not clear to what extent this would be robust to the structuring of populations [29], a feature that has been known to negatively influence other parameter estimates [30]. However, this is an exciting development, as the ability to infer ancestral geographical ranges and refugial areas is a central goal of phylogeography. It has previously been recognized that there is a relationship between the genealogical associations of alleles and their ancestry. Coalescent theory predicts that ancestral haplotypes will occur at high frequency, be represented in the greatest number of populations, have multiple connections to low frequency haplotypes, and be located at the interior of a network [31,32]. Within species that deviate from panmixia, such as those having a history involving geographical structure chequered by regional population extinction and recolonization, there may no longer be an obvious ancestral sequence [33] (figure 1). However, lower order ancestral sequences (ancestral sequences within a subsection of a network or gene tree) may be identifiable if the root location of a network can be identified [33–35] (figure 1). The spatial relationships among ancestral and descendant haplotypes may then be used to infer the geographical origin of a particular lineage of sequences [33–35]. Although this has not been done within a rigorous statistical framework, such a development would seem like a plausible future possibility.
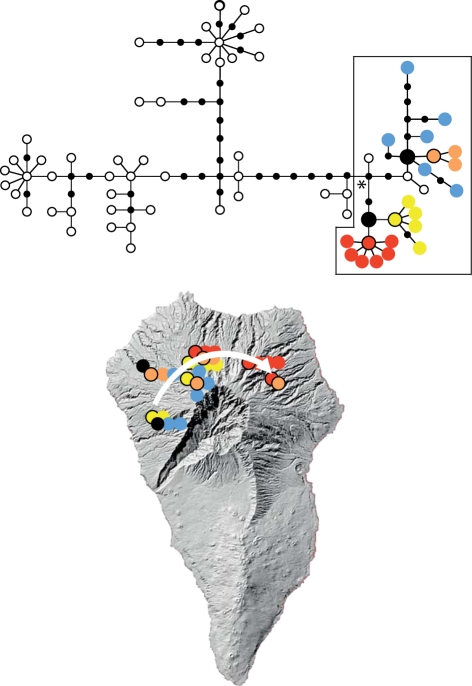
Figure 1.
A network of mtDNA haplotype relationships for a species of weevil, Brachyderes rugatus inhabiting the pine forest of La Palma, Canary Islands (from [33]). The most recent common ancestral (MRCA) sequence (denoted by an asterisk) of the framed lineage of haplotypes is either unsampled or extinct. However, the temporal sequence of mutations can be determined by progressing from the MRCA to tip haplotypes, indicating two lower order ancestral sequences with multiple descendant sequences derived from these. Plotting the geographical locations of ancestral and derived haplotypes indicates a western origin for the ancestral haplotypes with an eastern colonization of descendant haplotypes.
3. Phylobetadiversity and community analysis
Recently, Graham & Fine [36] have advocated an evolutionary genetic adaptation of beta diversity, a measure of how species composition of communities changes across space. Phylogenetic beta diversity (phylobetadiversity) differs from beta diversity by measuring how phylogenetic relatedness of communities changes across a landscape, and in this way it can be seen as a bridge between phylogeography, historical biogeography and phylogenetic analysis of community ecology. All three disciplines are reliant on a molecular approach, with the fundamental differences being that historical biogeography is typically concerned with relationships among species, while phylogeography is typically concerned with relationships within species or species complexes, and community phylogenetic analyses are limited to spatially defined assemblages of species. Communities are frequently difficult to demarcate because of the way they may interact with ecological gradients or change with geographical distance. Graham & Fine [36] argue that the analysis of phylobetadiversity can more easily accommodate this variation than analyses of community structure, allowing for the simultaneous assessment of how phenomena such as biotic interactions, phylogenetic constraints, current and past geographical isolation and environmental gradients might interact to structure diversity. Investigating phylobetadiversity along a continuous spatial scale will remove the subjectivity associated with investigator defined communities [36]. Thus, the characterization of regional and local species pools could be directly informed from sampling, rather than assumed.
Communities that either (i) lack clear demarcation of community boundaries or (ii) whose members are difficult to either sample or demarcate have posed a challenge for a phylogenetic approach to study community ecology, and there is a bias against these. Vamosi et al. [5] note that the difficulty of demarcating a community has contributed to a limited selection of case studies in community phylogenetics, dominated by plant communities or communities with discretely bounded habitats. This bias must be addressed if we are to understand what general principles may (or may not) underpin the assembly and structure of ecological communities. In terms of difficult-to-delineate communities, adopting an approach that incorporates phylobetadiversity will help to address the current bias. The second issue of difficulties associated with either sampling, or species circumscription, or a combination of these, requires an examination of recent efforts at the microbial scale to see how this issue might be best resolved. Microbial studies have addressed this issue by adopting a pyrosequencing approach to community sampling (e.g. [37–41]). While there have been some efforts to apply this approach to cryptic and complex eukaryote systems (e.g. [42,43]), the potential utility of this approach has not been fully explored. What follows is an examination of recent molecular insights into the mesofaunal component of one of the most complex and poorly studied habitats of terrestrial ecosystems—soils [44]. In the light of these results, we then suggest how progress might best be made to characterize community structure, assembly and ecology within this ecosystem, and other ecosystems.
4. Soil mesofaunal communities, with a focus on collembola
Soil has been referred to as the poor man's tropical rainforest [45,46] due to its seemingly relatively high biodiversity, within which only a proportion of all species have been described, and for which very little is known about their community structure and dynamics. For convenience, soil communities can be divided into arbitrary size classes, and here we use the scheme presented by Decaëns [44] that divides soil biota into three size classes: microflora/microfauna (up to 100 µm), mesofauna (100 µm—2 mm) and macrofauna (above 2 mm). Although, in general, soil is itself one of the most poorly studied habitats of terrestrial ecosystems, like any ecosystem the poverty of understanding is negatively associated with the size class of its constituent elements. Here, we focus attention at the mesofaunal scale of soil communities, where recent molecular analyses within the class Collembola strongly urge that we rethink about their diversity and community structure.
The known Collembola species are few in number, with approximately 8000 described. While the ‘true’ number of Collembola is estimated to be perhaps 50 000, this is attributed to geographical under-sampling rather than cryptic diversity within already described species [47]. Thus, the unusually broad geographical ranges characterizing many species are not viewed as a taxonomic artefact. Collembola are small (typically less than 2 mm), wingless, often profoundly associated with the soil, and have the broadest global distribution of any hexapod group. They occur throughout the world, including the Antarctic continent, and are probably the most abundant hexapods on Earth. Collembolans are a major component of terrestrial ecosystems (and particularly significant members of soil communities), constituting a significant proportion of animal biomass and are thus frequently and easily found. They are a key element for ecosystem functioning, and in forest soils they can reach densities of up to 600 000 000 individuals per hectare, or 60 000 per m2, densities only surpassed by the acarian soil population [47]. They are primarily found in soil and leaf litter, typically preferring wet or damp environments, from coastal regions to the highest of alpine environments.
Evolutionarily, Collembola are closely related to insects, and together with Diplura these comprise the Hexapoda [48]. Collembolan fossils of Rhyniella praecursor from the Devonian (ca 400 million years ago (Mya)) are among the oldest known records of terrestrial arthropods. The virtual ubiquity of modern Collembola in terrestrial systems, and their ancient origin, renders them one of the more successful arthropod lineages. For a group that is so old, so widely distributed, and with such limited dispersal ability, it is odd that their biodiversity is characterized by few species with frequently broad distributions. To put this into context, among the nine species of the genus Entomobrya occurring in the UK, six have distributions that extend across the Palearctic into eastern Russia, with several of these also extending into continental Africa. This is not atypical for the Collembola. Thirty per cent of Collembola species considered to be native to Tenerife can also be found in the UK. To put this into context, we are not aware of any native flightless insects shared between these two areas, despite the much larger number of native insects on Tenerife (4293 species, of which 89 are Collembola).
Fossil Collembola are scarce, and are mostly associated with amber deposits. Baltic amber fossils from 50 to 45 Mya are dominated by Collembola affiliated to contemporary genera, and in some cases these have been assigned to modern species names [49]. Morphological identity between fossil and modern assemblages is also reported in early Miocene (23–16 Mya) amber from Chiapas, Mexico, where seven specimens could be placed into extant species [50]. Miocene amber from the Dominican Republic (23–20 Mya) also illustrates this pattern where, with the exception of one species, all other specimens are believed to belong to species that are alive today [51]. Given this demonstrated propensity for morphological stasis over long periods of time, it is a plausible hypothesis that classically defined morphospecies are underestimates of true collembolan biodiversity. Further, Collembola lack genitalia which seriously limits traditional taxonomic approaches to discriminate species and quantify diversity within this Class [47]. Recent molecular genetic analyses reviewed below lend compelling support to this hypothesis.
(a). Molecular genetic insights
Increasing evidence from molecular genetic studies suggests that species diversity within the Collembola is vastly underestimated. A recent study has revealed deep and extensive lineage diversity among seven traditionally described morphospecies from the genus Lepidocyrtus in the northwestern Mediterranean basin [52]. A combined analysis of highly concordant mtDNA and nuclear EF1a gene sequences, with a conservative rate calibration, revealed an Oligocenic or pre-Oligocenic origin more than 23 Mya, with 35 evolutionary lineages in the region that were already distinct more than 5.8 Mya, indicating the survival and persistence of these lineages through the Messinian Salinity Crisis (figure 2). The Pleistocene is characterized by 52 evolutionary lineages prior to its onset that survived through this 1.8 Myr period. While it is clear that these genetically distinct and geographically discrete evolutionary lineages represent a fundamental component of uncharacterized diversity within the Collembola, their formal interpretation as species understandably depends upon the species concept one wishes to employ. The application of a GMYC model [25] identifies 83–91 branches of the tree to be consistent with a speciation branching pattern, and no instances of lineage sympatry reveal evidence for gene exchange between lineages [52]. That is to say, patterns of mtDNA and nuclear gene linkage are not disrupted in sympatry, consistent with the biological species concept [53]. While these results alone are remarkable, the implications for cryptic diversity within the Collembola are more profound when one considers the broader geographical ranges of these morphospecies. The distributions of all seven morphospecies extend far beyond the northwestern Mediterranean basin, with five of these spanning the Palearctic and Nearctic regions. Data for other Lepidocyrtus morphospecies in Iberia (M. A. Arnedo, unpublished data) and Central America (F. Cicconardi, F. Fanciulli & B. C. Emerson, in preparation) reveal similarly high levels of lineage diversity consistent with biological species.
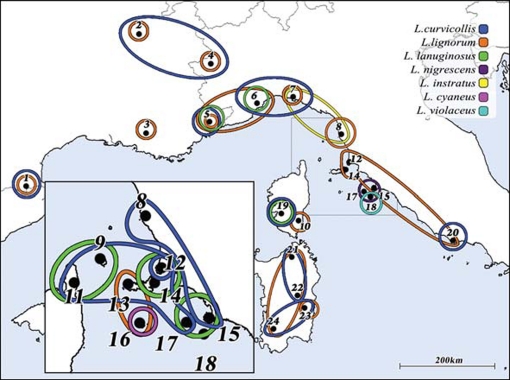
Figure 2.
The present distributions of 35 evolutionary lineages of Collembola sampled from within seven morphologically defined species of the genus Lepidocyrtus. Lineages are colour coded according to species. All 35 lineages are estimated to have originated sometime prior to the Messinian Salinity Crisis that is estimated to have occurred between 5.49 and 5.45 Mya. Numbers represent sampling locations (see ref. [52] for details). Reprinted with permission from [52].
Other collembolan genera investigated to date demonstrate a similar propensity for high levels of cryptic lineage diversity within traditionally described morphospecies. Timmermans et al. [54] found deeply divergent mtDNA lineages, estimated to have originated more than 3 Mya, with concordantly structured AFLP variation within the European range of Orchesella cincta. Sampling of the Cryptopygus antarcticus species complex across its continental and maritime circumpolar Antarctic distribution has revealed divergent lineages within this morphospecies, estimated to have originated more than 12 Mya [55]. Torricelli et al. [56] chose two individuals of the Antarctic Collembola species Friesea grisea from South Shetland Islands and Victoria Land for a whole mitochondrial genome sequencing project. MtDNA genome divergence between these two individuals is saturated and comparable to family level divergences in other arthropods, and it is estimated that the two lineages predate the Miocene (more than 23 Mya). Mitochondrial and nuclear DNA phylogeographic analyses of a newly recognized morphospecies in the genus Acanthanura, occupying a narrow southeast Australian forest belt stretching for approximately 100 km, have revealed a very fine scale phylogeographic structuring, with six geographically distinct lineages with an estimated Pliocene origin between 3.5 and 5 Mya [57] (figure 3). Concordant geographical divisions were subsequently genetically characterized within a newly recognized and similarly distributed morphospecies in the family Pseudachorutinae [21]. Although limited in number, molecular analyses within Collembola species reveal much cryptic diversity, and a broader taxonomic assessment using publicly available sequence data supports this.
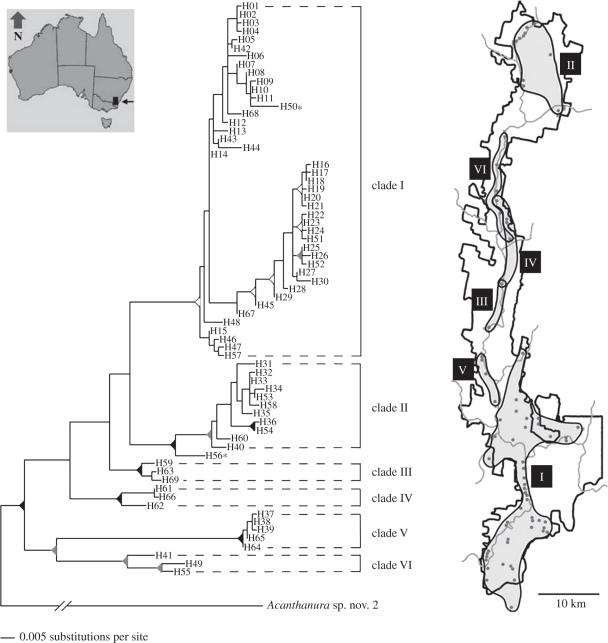
Figure 3.
Fine scale phylogeographic structure within Collembola. Phylogenetic relationships of mtDNA COI haplotypes and the geographical distributions of major clades for the Collembolan Acanthanura sp. n., a species known to occur only in the Tallaganda National Park and State Forest of New South Wales, Australia. Inset: map of Australia showing location of the Tallaganda National Park and State Forest. Reprinted with permission from [57].
(b). The taxonomic extent of cryptic diversity
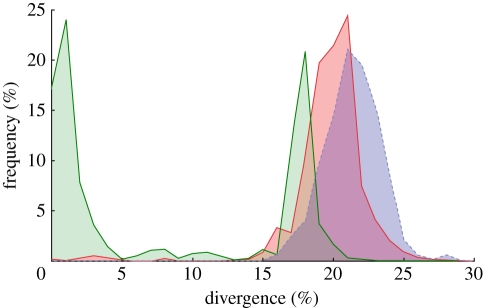
To more fully explore the taxonomic extent of cryptic diversity within the Collembola, we sequenced the 5′ region of the cytochrome oxidase subunit 1 (COI) of the mtDNA for 184 specimens from 61 species of Collembola using modifications of the primers designed by Folmer et al. [58]. Sequences were uploaded to BOLDSYSTEM database [59] and merged with the other publicly available homologous Collembola sequences yielding a dataset of 866 sequences belonging to 105 species. Using bioinformatics tools implemented within the BOLDSYSTEM database, we performed a distance summary analysis computing a pairwise distance matrix from an alignment of sequences longer then 420 bp (excluding sequences with contamination, stop codons or errors), and then plotted the distributions of the pairwise distances observed within species, genera and families (figure 4). This analysis reveals a clear bimodal distribution of pairwise distances within species, with one peak at 1 per cent divergence (representing 24.03% of all sequence comparisons) and another at 18 per cent (representing 20.89% of all sequence comparisons). The distribution of this cryptic diversity overlaps substantially with the distributions of divergences observed both within genera and within families of Collembola (figure 4), consistent with cryptic diversity within putative morphospecies.
Figure 4.
Pairwise genetic distances calculated among 866 individuals belonging to 105 species of Collembola for the mtDNA COI gene. Frequencies of pairwise distances calculated between individuals belonging to the same morphologically recognized species are shown in green. Frequencies of pairwise differences calculated between individuals belonging to the same genus (but not the same species) are shown in red. Frequencies of pairwise differences calculated between individuals belonging to the same family (but not the same genus) are shown in blue. Green, within species; red, within genera; blue, within families.
To exclude a potential bias that may arise due to a limited number of sequences (866 public sequences against the 9376 non-public sequences), we performed an additional analysis (data not shown). All GenBank sequences of COI associated to a Collembola species name (1065 sequences) were downloaded and clustered into groups with a threshold of 5% pairwise divergence. From each of the resulting 250 clusters a sequence was queried against the BOLDSYSTEM database to quantify similarity to unpublished sequences. The 99 most similar taxa were recorded and from these divergences within species, genera and family were calculated, revealing a pattern broadly similar to the previous analysis.
All these point to very high levels of cryptic species diversity, often associated with very deep genetic divergences, being a taxonomically and geographically pervasive feature within the Collembola. This reconciles the apparently broad distributions, and evolutionary long-term morphological stasis of many Collembolan morphospecies, with their limited dispersal potential. The most plausible explanation for the patterns emerging from molecular analyses is that Collembola are both morphologically conservative and locally persistent through time. Their extraordinarily high population densities (up to 60 000 m−2) confer a degree of local demographic continuity through time at spatial scales unlikely to confer such opportunity to other far less densely populated arthropod species [21]. A consequence of this is an increased probability, relative to the majority of arthropod species, of fragmented populations persisting and surviving during periods of climatic and/or ecological change. It now seems certain that the limited information content of Collembolan morphology has confounded traditional approaches to taxonomy and species delimitation. If the study of community ecology within soil was not hard enough already, it has just become harder, and there is no reason to suspect that this issue of cryptic diversity is limited to Collembola. Molecular genetic assessments of other soil mesofaunal groups such as Nematoda, Tardigrada and Acari all point to similar issues [60–66], although it should be noted that in [60] samples of nematodes were from beaches. The exciting implication is that there is more diversity than we thought, and it is structured over smaller geographical scales than has previously been assumed. The challenge for community ecologists and evolutionary biologists is to characterize this diversity and its spatial patterning.
5. Quantifying cryptic soil mesofaunal community structure with high throughput sequencing
Faced with a fauna where both community and species boundaries are not readily quantifiable, parallels between the challenges for quantifying microbial and soil mesofaunal community structure become apparent. Recent efforts in the microbial domain have seen amplicon high throughput parallel (HTP) sequencing brought to bear on the task (e.g. [37,38,40,41,67–69]). To date the gene of choice for amplicon HTP sequencing of bacteria has been the 16S small-subunit ribosomal gene, due to its ubiquitous presence in microbes and the existence of conserved sequence motifs facilitating primer design for cross species amplification (e.g. [37,38,40,41]. Similarly in fungi (e.g. [41]), protists (e.g. [70,71]) and marine meiofaunal elements [42,43] coding genes for nuclear ribosomal RNA (rRNA) have been employed for amplicon HTP sequencing. Again, it is their ubiquity and the fact that rRNA contains highly conserved regions that span more variable regions that have made them the marker of choice for amplicon HTP sequencing. Another useful feature of these markers is that they are well represented on the public sequence databases.
(a). Nuclear ribosomal versus mitochondrial protein-coding amplicons
Given the similar difficulties for the quantification of microbial communities and soil mesofaunal communities, it would appear that there is a good case for exploring amplicon HTP sequencing as an approach to quantify spatial structuring of soil mesofaunal phylogenetic diversity. The idea of pooled DNA analysis of soil faunal communities is not entirely new. Hamilton et al. [72] extracted DNA directly from soil samples for the amplification and subsequent cloning and sequencing of a fragment of the 18S rRNA gene. However, rather than continuing with a nuclear rRNA marker approach, it is important to consider the relative merits of more traditionally employed mtDNA genes that have been used for the assessment of intraspecific and interspecific analyses of mesofaunal taxa. Important criteria for any amplicon HTP sequencing project are the following: (i) the amplicon should effectively be single copy, (ii) the amplicon should be present in all taxa of interest, (iii) primers should be capable of amplifying the amplicon across all taxa, (iv) taxonomic diversity within the focal group should be captured by sequence diversity within the amplicon. Beyond these four criteria, additional desirable criteria are (v) a taxonomic reference library for assigning species names to amplicon HTP sequences, (vi) amplicons should minimize the generation of, or facilitate the identification of, artefact sequences from the HTP parallel sequencing process, a phenomenon that if uncorrected may result in spurious estimates of diversity [73,74].
In terms of the above six criteria, like nuclear rRNA, mtDNA typically satisfies criteria 1 and 2. Criterion 3 is also satisfied, most easily for the 12S and 16S mtDNA genes that share the similar property of nuclear rRNA genes of containing highly conserved regions that span more variable regions. However, evolutionary properties of the protein-coding genes of the mtDNA genome also provide the potential for universal cross species amplification [58], and recent bioinformatic tools greatly facilitate the design of degenerate primer pairs for this purpose [75,76]. The possibility to use mtDNA genes for amplicon HTP sequencing of soil mesofaunal communities provides for enhanced optimization of criteria 4, 5 and 6, particularly so for protein-coding genes, and especially so for the COI gene. Regarding criterion 4, the faster evolutionary substitution rate of the mtDNA genome over the nuclear genome provides for greatly enhanced taxonomic resolution for any taxonomic group. Regarding criterion 5, mtDNA gene sequences are well represented on public databases, and in particular the 5′ end of the COI gene is the focal region for the BOLDSYSTEM database [59], with an extensive representation of the Collembola, Acari, Nematoda and Tardigrada. Thus there are several clear advantages to a mtDNA COI amplicon approach to mesofaunal HTP sequencing, and an additional advantage exists for criterion 6. The current platform of choice for amplicon HTP parallel sequencing is 454 Roche's GS series Titanium sequencer, and it is recognized that there is a significant noise component for sequence generation, particularly associated with miscalling of homopolymer runs [73], but also due to noise associated with PCR. It is this noise component that confounds accurate diversity assessments (e.g. 43]), and it is here that protein-coding genes offer a key advantage over rRNA genes, due to their differing functional constraints. While rRNA genes may naturally exhibit insertions or deletions (indels), the 5′ region of the mtDNA COI gene does not, with the very rare exception of amino acid insertions or deletions. Thus, while the distinction between a genuine indel, and an indel generated by a homopolymer read error may complicate the denoising of 454 sequence data for a rRNA gene, the expected absence of indels for mtDNA COI nullifies this issue. Additionally, the conservative evolutionary nature of many COI amino acid residues [58] provides an additional resource for sequence alignment and homopolymer error correction, and the identification of probable PCR error (mutations associated with improbable amino acid residues).
(b). Dealing with nuclear copies
While mtDNA COI offers many advantages over nuclear rRNA genes for the analysis of soil mesofaunal communities, there is one disadvantage. Nuclear copies of mitochondrial DNA (Numts) are widely reported in the animal kingdom, and their complications for evolutionary analyses have been well documented [77]. Numts will have two negative consequences for amplicon HTP sequencing. First, from a practical point of view, their amplification will reduce the number of beads available for the sequencing of genuine mitochondrial copies. Second, from an analytical point of view, if left unchecked in a dataset Numts will confound downstream estimates of intra- and inter-species diversity within and among sampling sites. While the first issue is not controllable, the second is. As Numts accumulate mutations in the nuclear genome, they become increasingly recognizable from genuine mitochondrial copies by the accumulation of indels and point mutations associated with improbable amino acid residues. Additionally, with primer equivalency, Numts will amplify at a lower frequency than the genuine mtDNA sequences from which they are descended. Thus, sequences that consistently co-occur at a low frequency with related sequences, and/or exhibit atypical mtDNA COI mutations, can be identified and excluded from downstream analysis.
(c). Sampling strategies
In terms of sampling the mesofauna at a given sampling site, one can take the strategy of Hamilton et al. [72] and simply extract all DNA from a given quantity of soil (in their case a 1 g subsample of 20 g of ground soil). This will have the constraint of placing an upper limit on the number of mesofaunal individuals sampled, which depends on the volume of soil that can be extracted from. A fundamental difference between the eukaryote mesofauna and the microbial microfauna is that the mesofauna is more amenable to working at the level of the individual. This offers a degree of flexibility for how one may sample for amplicon HTP sequencing. Extraction tools such as Tullgren and Baermann funnels can be employed both to increase and standardize the mesofaunal biomass for subsequent DNA extraction. Further to this, a sample can be divided into major taxonomic groups (e.g. Acari, Nematoda, Collembola, Tardigrada, Diplura, Protura), enabling one either to focus on one particular taxonomic group or normalize DNA concentrations for their joint analysis. Normalization will alleviate potential problems that may arise from different proportional biomass representation of different taxonomic groups. As an example, in the study of Hamilton et al. [72], DNA sequences were dominated by Nematoda and Acari by up to 100 per cent, reflective of their typically greater biomass in soil relative to other groups. Although Nematoda and Acari may be the more abundant, it does not follow that they are also the more taxonomically diverse, and the under-sampling of sequences from other groups may result in a downward bias in diversity of other groups under-sampled for their DNA sequences.
The ability to work at the level of the individual has an additional advantage for the quantification of soil mesofaunal communities. For the microfauna, bacterial DNA may be sequenced on an individual basis, but this may only be achieved for the subset of laboratory-culturable bacteria. Mesofaunal eukaryotes, on the other hand, are sufficiently sized for individual based Sanger sequencing of DNA. This greatly enhances the possibility of linking sequences derived from amplicon HTP sequencing to single specimen Sanger sequence data, and such an approach has recently been applied with good effect in a survey of alveolate diversity (Ciliophora and Dinophlaceae) in a freshwater lake [70]. The ability to individually sequence taxa that are the subject of amplicon HTP sequencing surveys increases the reference library available for assigning taxonomic identity to a given sequence derived from amplicon HTP sequencing, that would otherwise rely on publicly available DNA sequence databases. In effect, one can rapidly sequence thousands of individuals from multiple locations, and then work backwards from there with the public databases and Sanger sequencing to assign morphospecies to sequences. Even when faced with cryptic diversity within morphospecies, this still allows one to associate a given sequence with a particular ecological role associated with a morphospecies where it may be known (as an example, Collembola form ecologically differentiated feeding guilds, with bacterial and yeast feeders, fungivores, predators, phytophages, detritivores, omniphages and other specialists [47]).
6. Conclusions
Our ability to understand the structure, origins and dynamics of many communities is complicated by difficulties defining the boundaries of a community, quantifying membership, sampling taxonomically densely or sampling geographically broadly. Such issues are likely frequently to coincide with a lack of niche-informative trait data for member species, which is important for a fine-grained understanding of community assembly processes [8]. However, although confronted with an absence of trait data, molecular methodologies are opening the door to quantifying the structure of communities that exhibit some or all of the remaining difficulties. An approach that harnesses HTP sequencing technology together with analytical developments in community phylogenetics, phylogeography and the analysis of phylobetadiversity offers promise in this direction. Such an approach may allow a researcher to simultaneously quantify both the geographical and ecological vagaries of community composition together with community membership. There are of course limitations to what can be extrapolated from a single gene sequence, but as long as due care is taken not to extend inferences beyond the limits of the data, much can be achieved. A shift away from nuclear rRNA genes to the mtDNA COI gene region will yield resolution on more recent temporal and finer geographical scales than could otherwise be achieved. However, the temporal resolution of mtDNA COI does not allow for the estimation of deeper phylogenetic relationships among taxa. Recognizing this limitation, such analyses can be complemented with individual-based Sanger sequencing to increase gene coverage and phylogenetic resolution among mtDNA COI defined lineages where this may be needed. We have chosen Collembola as an example group from soil mesofauna where existing molecular data argue for the need for such an approach to address a gap in our understanding of how diversity is structured across the landscape. Other ecosystems where similar challenges are faced by community ecologists, such as meso- and macrofaunal assemblages of ocean floors and forest canopies, could also be approached in an analogous way.
Acknowledgements
This work was supported by a Research Fellowship awarded to Brent Emerson from The Leverhulme Trust. The authors are grateful to the International Biogeography Society (IBS) for the opportunity to present this work at the 2011 meeting in Heraklion, Crete. Brent Emerson wishes to express thanks to the School of Biological Sciences of the University of East Anglia and the International Biogeography Society for financial support to present this work at the 2011 IBS meeting. Authors are grateful to Dave Jenkins, Bob Ricklefs and Evan Weiher, for comments on an earlier version of this manuscript.
References
- 1.Webb C. O. 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 156, 145–155 10.1086/303378 (doi:10.1086/303378) [DOI] [PubMed] [Google Scholar]
- 2.Cavender-Bares J., Kozak K. H., Fine P. V. A., Kembel S. W. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715 10.1111/j.1461-0248.2009.01314.x (doi:10.1111/j.1461-0248.2009.01314.x) [DOI] [PubMed] [Google Scholar]
- 3.Emerson B. C., Gillespie R. G. 2008. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. Evol. 23, 619–630 10.1016/j.tree.2008.07.005 (doi:10.1016/j.tree.2008.07.005) [DOI] [PubMed] [Google Scholar]
- 4.Johnson M. T. J., Stinchcombe J. R. 2007. An emerging synthensis between community ecology and evolutionary biology. Trends Ecol. Evol. 22, 250–257 10.1016/j.tree.2007.01.014 (doi:10.1016/j.tree.2007.01.014) [DOI] [PubMed] [Google Scholar]
- 5.Vamosi S. M., Heard S. B., Vamosi J. C., Webb C. O. 2009. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 18, 572–592 10.1111/j.1365-294X.2008.04001.x (doi:10.1111/j.1365-294X.2008.04001.x) [DOI] [PubMed] [Google Scholar]
- 6.Hubbell S. P. 2001. The unified neutral theory of biodiversity and biogeography. Princeton, NJ: Princeton University Press [Google Scholar]
- 7.MacArthur R. H., Wilson E. O. 1967. The theory of island biogeography. Princeton, NJ: Princeton University Press [Google Scholar]
- 8.Weiher E., Freund D., Bunton T., Stefanski A., Lee T., Bentivenga S. 2011. Advances, challenges, and a developing synthesis of ecological community assembly theory. Phil. Trans. R. Soc. B. 366, 2403–2413 10.1098/rstb.2011.0056 (doi:10.1098/rstb.2011.0056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ricklefs R. E. 1987. Community diversity: relative roles of local and regional processes. Science 235, 167–171 10.1126/science.235.4785.167 (doi:10.1126/science.235.4785.167) [DOI] [PubMed] [Google Scholar]
- 10.Emerson B. C., Kolm N. 2005. Species diversity can drive speciation. Nature 434, 1015–1017 10.1038/nature03450 (doi:10.1038/nature03450) [DOI] [PubMed] [Google Scholar]
- 11.Ricklefs R. E. 2006. Evolutionary diversification and the origin of the diversity–environment relationship. Ecology 87, S3–S13 10.1890/0012-9658(2006)87[3:EDATOO]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[3:EDATOO]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 12.Hickerson M. J., Carstens B. C., Cavender-Bares J., Crandall K. A., Graham C. H., Johnson J. B., Rissler L., Victoriano P. F., Yoder A. D. 2010. Phylogeography's past, present, and future: 10 years after Avise, 2000. Mol. Phylogenet. Evol. 54, 291–301 10.1016/j.ympev.2009.09.016 (doi:10.1016/j.ympev.2009.09.016) [DOI] [PubMed] [Google Scholar]
- 13.Avise J. C. 2000. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press [Google Scholar]
- 14.Carstens B. C., Knowles L. L. 2007. Shifting distributions and speciation: species divergence during rapid climate change. Mol. Ecol. 16, 619–627 10.1111/j.1365-294X.2006.03167.x (doi:10.1111/j.1365-294X.2006.03167.x) [DOI] [PubMed] [Google Scholar]
- 15.Knowles L. L., Carstens B. C. 2006. Estimating a geographically explicit model of population divergence. Evolution 61, 477–493 10.1111/j.1558-5646.2007.00043.x (doi:10.1111/j.1558-5646.2007.00043.x) [DOI] [PubMed] [Google Scholar]
- 16.Knowles L. L., Maddison W. P. 2002. Statistical phylogeography. Mol. Phylogeogr. 11, 2623–2635 [DOI] [PubMed] [Google Scholar]
- 17.Knowles L. L. 2009. Statistical phylogeography. Annu. Rev. Ecol. Evol. Syst. 40, 593–612 10.1146/annurev.ecolsys.38.091206.095702 (doi:10.1146/annurev.ecolsys.38.091206.095702) [DOI] [Google Scholar]
- 18.Galbreath K. E., Hafner D. J., Zamudio K. R., Agnew K. 2010. Isolation and introgression in the intermountain west: contrasting gene genalogies reveal the complex biogeographic history of the American pike (Ochotona princeps). J. Biogeogr. 37, 344–362 10.1111/j.1365-2699.2009.02201.x (doi:10.1111/j.1365-2699.2009.02201.x) [DOI] [Google Scholar]
- 19.Peters J. L., Zhuravlev Y. N., Fefelov I., Humphries E. M., Omland K. E. 2008. Multilocus phylogeography of a holarctic duck: colonization of North America from Eurasia by Gadwall (Anas strepera). Evolution 62, 1469–1483 10.1111/j.1558-5646.2008.00372.x (doi:10.1111/j.1558-5646.2008.00372.x) [DOI] [PubMed] [Google Scholar]
- 20.Carnaval A. C., Hickerson M. J., Haddad C. F. B., Rodrigues M. T., Moritz C. 2009. Stability predicts genetic diversity in a Brazilian Atlantic forest hotspot. Science 323, 785–789 10.1126/science.1166955 (doi:10.1126/science.1166955) [DOI] [PubMed] [Google Scholar]
- 21.Garrick R. C., Rowell D. M., Simmons C. S., Hillis D. M., Sunnucks P. 2008. Fine-scale phylogeographic congruence despite demographic incongruence in two low-mobility saproxylic springtails. Evolution 62, 1103–1118 10.1111/j.1558-5646.2008.00349.x (doi:10.1111/j.1558-5646.2008.00349.x) [DOI] [PubMed] [Google Scholar]
- 22.Ilves K. L., Huang W., Wares J. P., Hickerson M. J. 2010. Colonization and/or mitochondrial selective sweeps across the North Atlantic intertidal assemblage revealed by multi-taxa Bayesian computation. Mol. Ecol. 19, 4505–4519 10.1111/j.1365-294X.2010.04790.x (doi:10.1111/j.1365-294X.2010.04790.x) [DOI] [PubMed] [Google Scholar]
- 23.McCulloch G. A., Wallis G. P., Waters J. M. 2010. Onset of glaciation drove simultaneous vicariant isolation of alpine insects in New Zealand. Evolution 64, 2033–2043 10.1111/j.1558-5646.2010.00980.x (doi:10.1111/j.1558-5646.2010.00980.x) [DOI] [PubMed] [Google Scholar]
- 24.Emerson B. C., Hewitt G. M. 2005. Phylogeography. Curr. Biol. 15, R367–R371 10.1016/j.cub.2005.05.016 (doi:10.1016/j.cub.2005.05.016) [DOI] [PubMed] [Google Scholar]
- 25.Pons J., Barraclough T. G., Gomez-Zurita J., Cardoso A., Duran D. P., Hazell S., Kamoun S., Sumlin W., Vogler A. 2006. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609 10.1080/10635150600852011 (doi:10.1080/10635150600852011) [DOI] [PubMed] [Google Scholar]
- 26.Beaumont M. A., Zhang W., Balding D. J. 2002. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickerson M. J., Meyer C. P. 2008. Testing comparative phylogeographic models of marine vicariance and dispersal using a hirearchical Bayesian approach. BMC Evol. Biol. 8, 322. 10.1186/1471-2148-8-322 (doi:10.1186/1471-2148-8-322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemey P., Rambaut A., Drummond A., Suchard M. A. 2009. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5, e1000520. 10.1371/journal.pcbi.1000520 (doi:10.1371/journal.pcbi.1000520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navascués M., Depaulis F., Emerson B. C. 2010. Combining contemporary and ancient DNA in population genetic and phylogeographical studies. Mol. Ecol. Resour. 10, 760–772 10.1111/j.1755-0998.2010.02895.x (doi:10.1111/j.1755-0998.2010.02895.x) [DOI] [PubMed] [Google Scholar]
- 30.Navascués M., Emerson B. C. 2009. Elevated substitution rate estimates from ancient DNA: model violation and bias of Bayesian methods. Mol. Ecol. 18, 4390–4397 10.1111/j.1365-294X.2009.04333.x (doi:10.1111/j.1365-294X.2009.04333.x) [DOI] [PubMed] [Google Scholar]
- 31.Crandall K. A., Templeton A. R. 1993. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny construction. Genetics 134, 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posada D., Crandall K. A. 2001. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol. 16, 37–45 10.1016/S0169-5347(00)02026-7 (doi:10.1016/S0169-5347(00)02026-7) [DOI] [PubMed] [Google Scholar]
- 33.Emerson B. C., Forgie S., Goodacre S. L., Oromi P. 2006. Testing phylogeographic predictions on an active volcanic island: Brachyderes rugatus (Coleoptera: Curculionidae) on La Palma (Canary Islands). Mol. Ecol. 15, 449–458 10.1111/j.1365-294X.2005.02786.x (doi:10.1111/j.1365-294X.2005.02786.x) [DOI] [PubMed] [Google Scholar]
- 34.Miraldo A., Hewitt G. M., Paulo O. S., Emerson B. C. Submitted Phylogeography and demographic history of Lacerta lepida in the Iberian Peninsula: multiple refugia, range expansions and secondary contact zones. BMC Evol. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarza E., Reynoso V. H., Emerson B. C. 2008. Diversification in the northern neotropics: mitochondrial and nuclear DNA phylogeography of the iguana Ctenosaura pectinata and related species. Mol. Ecol. 17, 3259–3275 10.1111/j.1365-294X.2008.03826.x (doi:10.1111/j.1365-294X.2008.03826.x) [DOI] [PubMed] [Google Scholar]
- 36.Graham C. H., Fine P. V. A. 2008. Phylogenetic beta diversity: linking ecological and evolutionary processes over space and time. Ecol. Lett. 11, 1265–1277 10.1111/j.1461-0248.2008.01256.x (doi:10.1111/j.1461-0248.2008.01256.x) [DOI] [PubMed] [Google Scholar]
- 37.Chu H. Y., Fierer N., Lauber C. L., Caporaso J. G., Knight R., Grogan P. 2010. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12, 2998–3006 10.1111/j.1462-2920.2010.02277.x (doi:10.1111/j.1462-2920.2010.02277.x) [DOI] [PubMed] [Google Scholar]
- 38.Fulthorpe R. R., Roesch L. F. W., Riva A., Triplett E. W. 2008. Distantly sampled soils carry few species in common. ISME J. 2, 901–910 10.1038/ismej.2008.55 (doi:10.1038/ismej.2008.55) [DOI] [PubMed] [Google Scholar]
- 39.Kaspari M., Stevenson B. S., Shik J., Kerekes J. F. 2010. Scaling community structure: how bacteria, fungi, and ant taxocenes differentiate along a tropical forest floor. Ecology 91, 2221–2226 10.1890/09-2089.1 (doi:10.1890/09-2089.1) [DOI] [PubMed] [Google Scholar]
- 40.Lauber C. L., Hamady M., Knight R., Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 10.1128/AEM.00335-09 (doi:10.1128/AEM.00335-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rousk J., Baath E., Brookes P. C., Lauber C. L., Lozupone C., Caporaso J. G., Knight R., Fierer N. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351 10.1038/ismej.2010.58 (doi:10.1038/ismej.2010.58) [DOI] [PubMed] [Google Scholar]
- 42.Creer S., et al. 2010. Ultra-deep sequencing of the meiofaunal biosphere: practice, pitfalls and promises. Mol. Ecol. 19, 4–20 10.1111/j.1365-294X.2009.04473.x (doi:10.1111/j.1365-294X.2009.04473.x) [DOI] [PubMed] [Google Scholar]
- 43.Fonseca V. G., et al. 2010. Second-generation environmental sequencing unmasks marine metazoan biodiversity. Nat. Commun. 1, 98. 10.1038/ncomms1095 (doi:10.1038/ncomms1095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decaëns T. 2010. Macroecological patterns in soil communities. Global Ecol. Biogeogr. 19, 287–302 10.1111/j.1466-8238.2009.00517.x (doi:10.1111/j.1466-8238.2009.00517.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Usher M. B., Davis P., Harris J., Longstaff B. 1979. A profusion of species? Approaches towards understanding the dynamics of the populations of microarthropods in decomposer communities. In Population dynamics (eds Anderson A. M., Turner B. D., Taylor L. R.), pp. 359–384 Oxford, UK: Blackwell Scientific [Google Scholar]
- 46.Giller P. S. 1996. The diversity of soil communities, the ‘poor man's tropical rainforest’. Biodivers. Conserv. 5, 135–168 10.1007/BF00055827 (doi:10.1007/BF00055827) [DOI] [Google Scholar]
- 47.Hopkin S. P. 1997. Biology of the springtails (Insecta: Collembola). Oxford, UK: Oxford University Press [Google Scholar]
- 48.Regier J. C., Schultz J. W., Zwick A., Hussey A., Ball B., Wetzer R., Martin J. W., Cunningham C. W. 2010. Athropod relationships revealed by phylogenomic analysis of nuclear protein coding sequences. Nature 463, 1079–1083 10.1038/nature08742 (doi:10.1038/nature08742) [DOI] [PubMed] [Google Scholar]
- 49.Rapoport E. H. 1971. The geographical distribution of Neotropical and Antarctic Collembola. Pacific Insects Monogr. 25, 99–118 [Google Scholar]
- 50.Christiansen K. 1971. Notes on Miocene amber Collembola from Chiapas. Univ. California Publ. Entomol. 63, 45–48 [Google Scholar]
- 51.Mari Mutt J. A. 1983. Collembola in amber from the Dominican Republic. Proc. Entomol. Soc. Wash. 85, 575–587 [Google Scholar]
- 52.Cicconardi F., Nardi F., Emerson B. C., Frati F., Fanciulli P. P. 2010. Deep phylogeographic divisions and long-term persistence of forest invertebrates in the North-Western Mediterranean basin. Mol. Ecol. 19, 386–400 10.1111/j.1365-294X.2009.04457.x (doi:10.1111/j.1365-294X.2009.04457.x) [DOI] [PubMed] [Google Scholar]
- 53.Mayr E. 1942. Systematics and the origin of species from the viewpoint of a zoologist. Cambridge, MA: Harvard University Press [Google Scholar]
- 54.Timmermans M. J. T. N., Ellers J., Marien J., Verhoeff S. C., Ferwerda E. B. & Van Straalen, N. M. 2005 Genetic structure in Orchesella cincta (Collembola): strong subdivision of European populations inferred from mtDNA and AFLP markers. Mol. Ecol. 14, 2017–2024 10.1111/j.1365-294X.2005.02548.x (doi:10.1111/j.1365-294X.2005.02548.x) [DOI] [PubMed] [Google Scholar]
- 55.Stevens M. I., Greenslade P., Hogg I. D., Sunnucks P. 2006. Southern hemisphere springtails: could any have survived glaciation of Antarctica? Mol. Biol. Evol. 23, 874–882 10.1093/molbev/msj073 (doi:10.1093/molbev/msj073) [DOI] [PubMed] [Google Scholar]
- 56.Torricelli G., Carapelli A., Convey P., Nardi F., Boore J. L., Frati F. 2009. High divergence across the whole mitochondrial genome in the ‘pan-Antarctic’ springtail Friesea grisea: evidence for cryptic species? Gene 449, 30–40 10.1016/j.gene.2009.09.006 (doi:10.1016/j.gene.2009.09.006) [DOI] [PubMed] [Google Scholar]
- 57.Garrick R. C., Sands C. J., Rowell D. M., Hillis D. M., Sunnucks P. 2007. Catchments catch all: long-term population history of a giant springtail from the southeast Australian highlands—a multigene approach. Mol. Ecol. 16, 1865–1882 10.1111/j.1365-294X.2006.03165.x (doi:10.1111/j.1365-294X.2006.03165.x) [DOI] [PubMed] [Google Scholar]
- 58.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 [PubMed] [Google Scholar]
- 59.Ratnasingham S., Hebert P. D. N. 2007. BOLD: the barcode of life data system. Mol. Ecol. Notes 7, 355–364 10.1111/j.1471-8286.2007.01678.x (doi:10.1111/j.1471-8286.2007.01678.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blaxter M. L. 2004. The promise of a DNA taxonomy. Phil. Trans. R. Soc. B 359, 669–679 10.1098/rstb.2003.1447 (doi:10.1098/rstb.2003.1447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blaxter M. L., Elsworth B., Daub J. 2004. DNA taxonomy of a neglected anumal phylum: an unexpected diversity of tardigrades. Biol. Lett. 271, S189–S192 10.1098/rsbl.2003.0130 (doi:10.1098/rsbl.2003.0130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heethoff M., Domes K., Laumann M., Maraun M., Norton R. A., Scheu S. 2007. High genetic divergences indicate ancient separation of parthenogenetic lineages of the oribatid mite Platynothrus peltifer (Acari, Oribatida). J. Evol. Biol. 20, 392–402 10.1111/j.1420-9101.2006.01183.x (doi:10.1111/j.1420-9101.2006.01183.x) [DOI] [PubMed] [Google Scholar]
- 63.Mortimer E., van Vuuren B. J., Lee J. E., Marshall D. J., Convey P., Chwon S. L. 2010. Mite dispersal among the Southern Ocean Islands and Antarctica before the last glacial maximum. Proc. R. Soc. Lond. B 278, 1247–1255 10.1098/rspb.2010.1779 (doi:10.1098/rspb.2010.1779) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schäffer S., Pfingstl T., Koblmüller S., Winkler K. A., Sturmbauer C., Krisper G. 2010. Phylogenetic analysis of European Scutovertex mites (Acari, Oribatida, Scutoverticidae) reveals paraphyly and cryptic diversity: a molecular genetic and morphological approach. Mol. Phylogenet. Evol. 55, 677–688 10.1016/j.ympev.2009.11.025 (doi:10.1016/j.ympev.2009.11.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stevens M. I., Hogg I. D. 2006. Contrasting levels of mitochondrial DNA variability between mites (Penthalodidae) and springtails (Hypogastruridae) from Trans-Antarctic Mountains suggest long-term effects of glaciation and life history on substitution rates, and speciation process. Soil Biol. Biochem. 38, 3171–3180 10.1016/j.soilbio.2006.01.009 (doi:10.1016/j.soilbio.2006.01.009) [DOI] [Google Scholar]
- 66.Powers T. O., Neher D. A., Mullin P., Esquivel A., Giblin-Davis R. M., Kanzaki N., Stock S. P., Mora M. M., Uribe-Lorio L. 2009. Tropical nematode diversity: vertical stratification of nematode communities in a Costa Rican humid lowland rainforest. Mol. Ecol. 18, 985–996 10.1111/j.1365-294X.2008.04075.x (doi:10.1111/j.1365-294X.2008.04075.x) [DOI] [PubMed] [Google Scholar]
- 67.Huber J. A., Mark Welch D., Morrison H. G., Huse S. M., Neal P. R., Butterfield D. A., Sogin M. L. 2007. Microbial population structures in the deep marine biosphere. Science 318, 97–100 10.1126/science.1146689 (doi:10.1126/science.1146689) [DOI] [PubMed] [Google Scholar]
- 68.Roesch L. F., Fulthorpe R. R., Riva A., Casella G., Hadwin A. K. M., Kent A. D., et al. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1, 283–290 10.1038/ismej.2007.53 (doi:10.1038/ismej.2007.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sogin M. L., Morrison H. G., Huber J. A., Mark Welch D., Huse S. M., Neal P. R., Arrieta J. M., Herndl G. J. 2006. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc. Natl Acad. Sci. USA 103, 12 115–12 120 10.1073/pnas.0605127103 (doi:10.1073/pnas.0605127103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Medinger R., Nolte V., Pandey R. V., Jost S., Ottenwalder B., Schlotterer C., Boenigk J. 2010. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol. Ecol. 19, 32–40 10.1111/j.1365-294X.2009.04478.x (doi:10.1111/j.1365-294X.2009.04478.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nolte V., Pandey R. V., Jost S., Medinger R., Ottenwalder B., Boenigk J., Schlötterer C. 2010. Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol. Ecol. 19, 2908–2915 10.1111/j.1365-294X.2010.04669.x (doi:10.1111/j.1365-294X.2010.04669.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hamilton H. C., Strickland M. S., Wickings K., Bradford M. A., Fierer N. 2009. Surveying soil faunal communities using a direct molecular approach. Soil Biol. Biochem. 41, 1311–1314 10.1016/j.soilbio.2009.03.021 (doi:10.1016/j.soilbio.2009.03.021) [DOI] [Google Scholar]
- 73.Quince C., Lanzén A., Curtis T. P., Davenport R. J., Hall N., Head I. M., Read L. F., Sloan W. T. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Meth. 6, 639–641 10.1038/nmeth.1361 (doi:10.1038/nmeth.1361) [DOI] [PubMed] [Google Scholar]
- 74.Reeder J., Knight R. 2009. The ‘rare biosphere’: a reality check. Nat. Meth. 6, 636–637 10.1038/nmeth0909-636 (doi:10.1038/nmeth0909-636) [DOI] [PubMed] [Google Scholar]
- 75.Boyce R., Chilana P., Rose T. P. 2009. iCODEHOP: a new interactive program for designing COnsensus-DEgenerate Hybrid Oligonucleotide Primers from multiply aligned protein sequences. Nucleic Acids Res. 37, w222–w228 10.1093/nar/gkp379 (doi:10.1093/nar/gkp379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rose T. M., Henikoff J. G., Henikoff S. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31, 3763–3766 10.1093/nar/gkg524 (doi:10.1093/nar/gkg524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bensasson D., Zhang D.-X., Hartl D. L., Hewitt G. M. 2001. Mitochondrial pseudogenes: evolution's misplaced witnesses. Trends Ecol. Evol. 16, 314–321 10.1016/S0169-5347(01)02151-6 (doi:10.1016/S0169-5347(01)02151-6) [DOI] [PubMed] [Google Scholar]