Abstract
Apoptosis is a tightly regulated cell suicide program that plays an essential role in the development and maintenance of tissue homeostasis by eliminating unnecessary or harmful cells. Impairment of this native defense mechanism promotes aberrant cellular proliferation and the accumulation of genetic defects, ultimately resulting in tumorigenesis, and frequently confers drug resistance to cancer cells. The regulation of apoptosis at several levels is essential to maintain the delicate balance between cellular survival and death signaling that is required to prevent disease. Complex networks of signaling pathways act to promote or inhibit apoptosis in response to various cues. Apoptosis can be triggered by signals from within the cell, such as genotoxic stress, or by extrinsic signals, such as the binding of ligands to cell surface death receptors. Various upstream signaling pathways can modulate apoptosis by converging on, and thereby altering the activity of, common central control points within the apoptotic signaling pathways, which involve the BCL-2 family proteins, inhibitor of apoptosis (IAP) proteins, and FLICE-inhibitory protein (c-FLIP). This review highlights the role of these fundamental regulators of apoptosis in the context of both normal apoptotic signaling mechanisms and dysregulated apoptotic pathways that can render cancer cells resistant to cell death. In addition, therapeutic strategies aimed at modulating the activity of BCL-2 family proteins, IAPs, and c-FLIP for the targeted induction of apoptosis are briefly discussed.
Introduction
Apoptosis is a tightly regulated form of cell death that plays a critical role in normal development and tissue homeostasis by eliminating unnecessary and unwanted cells.1,2 Proper apoptotic signaling is vitally important in maintaining a healthy balance between cell survival and cell death and in safeguarding the integrity of the genome, as highlighted by the establishment of the evasion of apoptosis as a prominent hallmark of cancer.3 In response to severe DNA damage, such as that found in precancerous lesions, induction of apoptosis via activation of the DNA damage checkpoint pathway can serve to remove potentially harmful DNA-damaged cells and thereby block cancer development.4,5 In line with apoptosis acting as a barrier to tumorigenesis, cancer cells typically harbor alterations that result in impaired apoptotic signaling, which facilitates tumor development and metastasis.3,6,7
Importantly, dysregulation of the apoptotic pathways can not only promote tumorigenesis, but can also render cancer cells resistant to conventional anti-cancer agents, since chemotherapy- and radiotherapy-induced killing of cancer cells is mainly mediated through activation of apoptosis.8–10 Furthermore, TRAIL, a candidate for targeted therapy that has gained much attention for its ability to preferentially kill cancer cells, is frequently hampered by various mechanisms of resistance of cancer cells to apoptosis.11,12 TRAIL is a cytokine that induces apoptosis by binding one of its cognate death receptors and is known to play an important role in immune surveillance against tumors.11–13 Nevertheless, in many types of cancer cells, effective anti-tumor activity of TRAIL requires the suppression of aberrantly expressed negative regulators of apoptosis.11,12 To enhance cancer cell sensitivity to apoptosis, and thus overcome treatment failure, therapeutic strategies targeting molecules implicated in apoptosis resistance have been developed.8–10
As regulation of apoptosis relies on multiple cell signaling mechanisms, cancer cells can employ a number of different strategies to suppress a protective apoptotic response.3,6,7 Numerous signaling pathways, including survival signaling pathways14,15 and stress-induced signaling pathways,16,17 are linked to the apoptotic machinery, directly modulating components of the apoptotic machinery itself or key regulatory molecules within the core apoptotic signaling pathways. Notably, various upstream signaling pathways often impinge on the same few central apoptosis control points, which involve the critical apoptosis regulators in the BCL-2 family of proteins; inhibitor of apoptosis (IAP) proteins; and FLICE-inhibitory protein (c-FLIP).6,7,10,18,19 In this review, we provide an overview of mechanisms by which these regulatory molecules govern apoptosis in normal cells and describe modes of apoptotic dysregulation based on alterations in their function that facilitate the evasion of apoptosis in cancer cells. We will also briefly discuss the development of some promising cancer treatment strategies aimed at restoring the apoptotic response.
Insight, innovation, integration.
Apoptosis plays an essential role in the development and maintenance of tissue homeostasis and its deregulation results in a variety of diseases including tumorigenesis. Moreover, evasion of apoptosis is one of the hallmarks of cancer and also confers drug resistance to cancer cells. Here, we will highlight the role and therapeutic values of various regulators of apoptosis. We will also briefly discuss the importance of preclinical models utilizing three dimensional systems and genetically engineered mouse models in validation of novel apoptosis-based cancer therapeutics.
Caspases: the central effectors of apoptosis
Apoptosis is primarily executed by a family of proteases known as the caspases (cysteinyl, aspartate-specific proteases).20 As fundamental players in death-inducing signaling pathways, the regulation of caspase activation must be subject to various layers of control to ensure apoptotic cell death is triggered only under appropriate conditions. Accordingly, caspases are synthesized in the cell as inactive zymogens with an N-terminal prodomain in addition to a p20 large subunit and a p10 small subunit that make up the catalytic domain.20,21 In response to an apoptosis-inducing signal, the initiator caspases are recruited to oligomeric complexes by activating adaptor proteins; the consequent induced proximity of two caspase monomers then promotes dimerization and subsequent activation.21,22 Activated initiator caspases, in turn, activate a second class of caspases, the executioner or effector caspases with short prodomains that occur as dimers, by proteolytic cleavage of the linker separating the two subunits of the catalytic domain.21
While the identification of apoptotic cells cannot be based on caspase activation alone,23 and several studies have shown that caspases can be involved in biological processes other than cell death, including cell proliferation and differentiation,20,23 the importance of caspases in executing the classical apoptotic cell death programs has been firmly established.24 Once activated, caspases can dismantle the cell by cleaving a number of key proteins. Indeed, caspase activity may be required to induce the biochemical and morphological changes specific to apoptotic cells.25 These characteristic alterations include blebbing of the plasma membrane, exposure of phosphatidylserine at the external surface of the cell membrane, shrinkage of the cytoplasm, chromatin condensation, and DNA fragmentation.25,26 The morphological transformation of the apoptotic cell concludes with the formation of apoptotic bodies that are recognized and eliminated by specialized phagocytes and neighboring cells via phagocytosis.26,27
Significant advances toward the identification of the full set of caspase substrates, driven by the creation of a compiled list of reported caspase substrates (accessible via the CASBAH online database http://bioinf.gen.tcd.ie/casbah/)25 as well as large-scale proteomic studies of proteolytic events following the induction of apoptosis,28 have revealed that hundreds of proteins can be subject to caspase-mediated cleavage. Given the multitude of identified caspase substrates, the true challenge has become the ability to distinguish those cleavage events that are important for the apoptosis process and the accompanying biochemical and morphological changes from the cleavage of “innocent bystander” caspase substrates.25 Evidence is lacking as to the functional importance of most of the hundreds of identified caspase substrates, with the cleavage of only a small number of these caspase substrates linked to the signature alterations of apoptotic cells.25
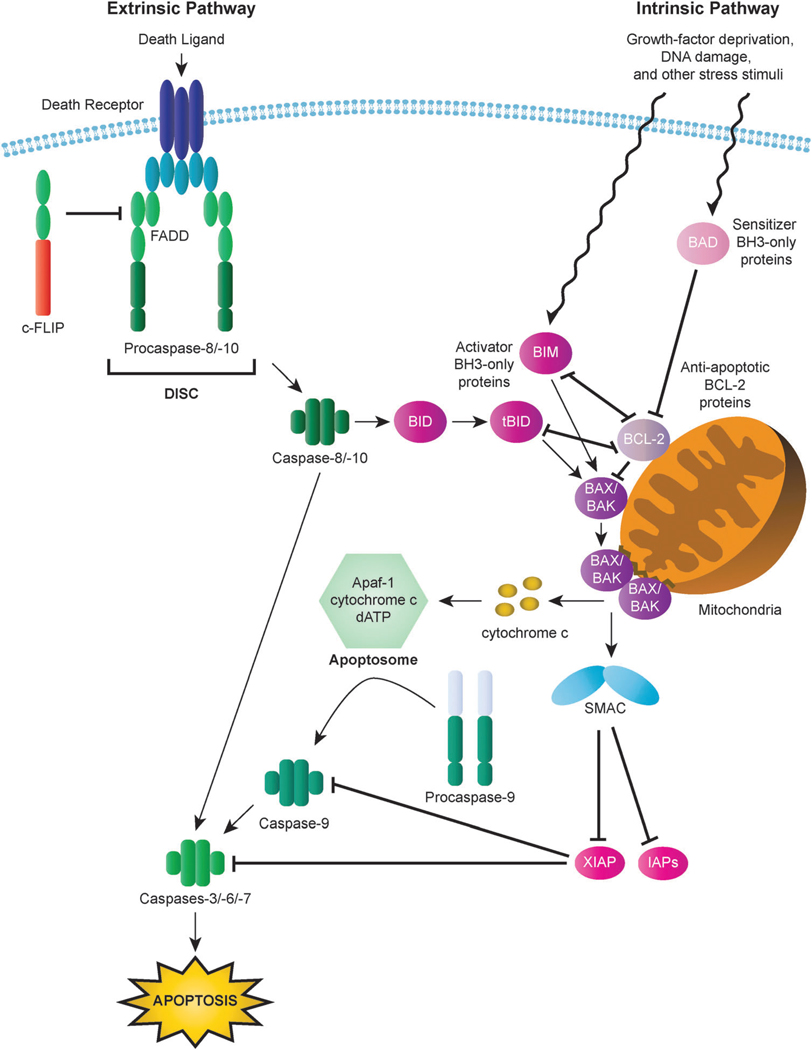
The irreversible and extensive proteolysis resulting from the activation of the executioner caspases, and the consequent induction of cell death, can be triggered by death receptor- or mitochondria-dependent apoptotic pathways, known as the intrinsic and extrinsic apoptotic pathways, respectively (Fig. 1).29
Fig. 1.
Intrinsic and extrinsic apoptotic pathways. In the intrinsic (mitochondrial) pathway, mitochondrial outer membrane permeabilization (MOMP) results in the release of cytochrome c and other apoptogenic factors from mitochondria into the cytosol and the ensuing formation of the apoptosome, which triggers activation of the apoptosis-inducing caspase cascade via activation of caspase-9. Interactions among BCL-2 family proteins play a critical role in the mediating MOMP induction and consequent apoptosis. BH3-only proteins (activator BH3-only proteins are represented here by BIM and tBID and sensitizer BH3-only proteins are represented by BAD) can relay apoptotic signals to the mitochondria through activation of BAX or BAK, the principal effectors of the intrinsic apoptotic pathway. In contrast, anti-apoptotic BCL-2 proteins (represented by BCL-2) serve to inhibit apoptosis by blocking BAX/BAK activation. In the extrinsic (death receptor) pathway, binding of death receptors such as FAS or TRAIL receptors (TRAILR1, TRAILR2) by their cognate ligands triggers the recruitment of death domain (DD)-containing adaptor proteins (represented by FADD) and procaspases with a death effector domain (DED), specifically procaspase-8 and procaspase-10. The resulting complex is known as the death inducing signaling complex (DISC). High levels of active caspase-8 generated by large amounts of procaspase-8 processing at the DISC lead to the activation of executioner caspases, including caspase-3, and the induction of apoptosis. Activation of caspase-8 can also result in the cleavage of the BH3-only protein BID to generate the activated BID fragment tBID, which serves to transmit the death signal from the extrinsic to the intrinsic signaling pathway.
The intrinsic apoptotic pathway
Overview of the intrinsic apoptotic pathway
The intrinsic apoptotic pathway can be activated by a range of stress stimuli, including UV radiation, gamma irradiation, heat, viral virulence factors, growth-factor deprivation, most DNA-damaging agents, and the activation of some oncogenic factors (Fig. 1). These diverse stressors are recognized by multiple intracellular components that relay the signal to the mitochondria, resulting in mitochondrial outer membrane permeabilization (MOMP).26,30–32 MOMP prompts various proteins, which are normally confined to mitochondrial intermembrane space (IMS), to diffuse into the cytosol. Among the proteins that escape from the IMS, a set of proteins, known as apoptogenic factors, play a vital role in the mitochondria-dependent initiation of the death-inducing caspase cascade.33 In the cytosol, the apoptogenic factor cytochrome c binds to apoptosis protease activating factor-1 (Apaf-1) in a dATP-dependent manner to drive the formation of a complex known as the apoptosome that recruits procaspase-9, thus mediating the oligomerization and activation of caspase-9.34,35 Activation of caspase-9 leads to the processing and activation of executioner caspases-3, -6 and -7 which bring about the execution of the cell.36
The BCL-2 protein family
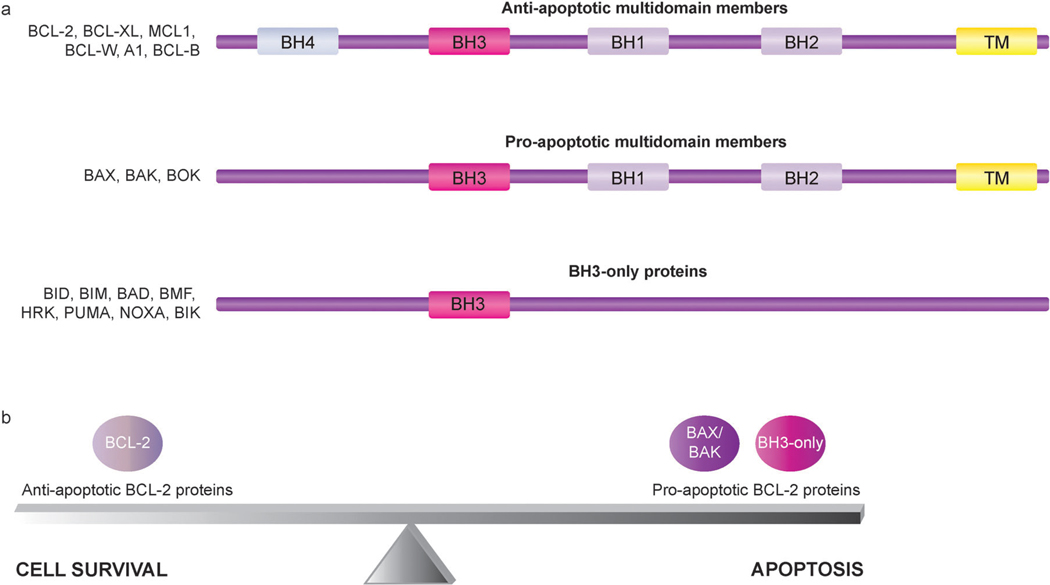
Molecules responsible for control of MOMP induction are central regulators of apoptosis, as MOMP is regarded as the critical event in the mitochondria-mediated apoptotic pathway that commits the cell to apoptosis.30 Accordingly, the BCL-2 family of proteins serves as an “apoptotic switch” by mediating permeabilization of the mitochondrial membrane.37,38 The BCL-2 family includes both pro- and anti-apoptotic regulators of the intrinsic apoptotic pathway. Each BCL-2 family member contains at least one of four conserved domains, known as BCL-2 homology (BH) domains (BH1–4). BCL-2 family members are broadly classified into three subgroups, one anti-apoptotic and two pro-apoptotic, based on function and BH domain composition (Fig. 2a).37,39,40 Anti-apoptotic multidomain members, including BCL-2, BCL-XL, MCL1, BCL-W, A1, and BCL-B, contain three to four BH domains. Pro-apoptotic multidomain members, represented by BAX, BAK, and BOK, share substantial sequence similarities with anti-apoptotic BCL-2 proteins, as family members in both multidomain subgroups contain BH1, BH2, and BH3 domains.39,41 BH3-only proteins, the third class of BCL-2 family members, are a subset of pro-apoptotic proteins that, as indicated by their name, only share strong sequence similarity with one another and multidomain BCL-2 proteins in a single region, the BH3 domain.37–39 BH3-only family members include BID, BIM, BAD, BMF, HRK, PUMA, NOXA, and BIK.37,39
Fig. 2.
Subgroups of BCL-2 family members with representative members of each subfamily. (a) BCL-2 family members can be classified into three subgroups according to function and BH domain composition. All BCL-2 family members possess at least one of four BCL-2 homology (BH) domains, termed BH1, BH2, BH3, and BH4, and many also include a transmembrane (TM) domain. The anti-apoptotic multidomain members have three to four BH domains, with some members lacking a BH4 domain. Similar to the anti-apoptotic multidomain members, the pro-apoptotic multidomain members contain BH1, BH2, and BH3 domains. The BH3-only proteins are a subset of pro-apoptotic proteins that only bear a single BH motif, the BH3 domain. Some BH3-only proteins also include a TM domain. (b) BCL-2 proteins play a key role in mediating the delicate balance between cell survival and cell death. Disruption of this balance by cellular alterations that increase the functional activity of anti-apoptotic BCL-2 proteins relative to pro-apoptotic BCL-2 proteins can enable the evasion of apoptosis, which tips the balance to favor cell survival and thus promotes the development and progression of cancer.
The anti- and pro-apoptotic multidomain BCL-2 proteins, considered to be the core BCL-2 family members, have predicted secondary structures or experimentally determined 3D structures resembling one another, whereas BH3-only proteins, with the exception of BID, appear to lack structural similarities to the core BCL-2 family members.41 Interestingly, structural analyses of seven core BCL-2 proteins, namely BCL-XL, BCL-2, BCL-W, MCL1, BAX, BAK, as well as the BH3-only protein BID, have revealed striking similar helical bundle structures, with no discernible difference between anti- and pro-apoptotic BCL-2 family members.41 The BH1, BH2, and BH3 domains of core multidomain BCL-2 proteins form a hydrophobic groove at the surface of the protein that has been shown to bind peptides corresponding to the alpha-helical BH3 domains of BH3-only proteins and the pro-apoptotic multidomain BCL-2 protein BAK.41,42 Previous studies have firmly established that the heterodimerization of anti-apoptotic multidomain BCL-2 proteins and BH3-only proteins occurs via BH3-groove interactions,42,43 and more recent studies have indicated that the formation of homodimers of BAX or BAK also involves BH3-groove interactions.43,44 Importantly, protein–protein interactions among BCL-2 family members act to either prevent or promote apoptosis, and thus the BH3-groove mode of interaction plays a key role in the mechanisms by which BCL-2 proteins regulate apoptosis.43
While a predicted C-terminal transmembrane (TM) domain is common to most multidomain BCL-2 proteins, the subcellular localization of these proteins varies in healthy cells. For example, with regard to the well-studied pro-apoptotic multidomain BCL-2 proteins BAX and BAK, BAX is predominantly cytosolic and translocates to the mitochondria during apoptosis induction, whereas BAK is an integral membrane protein localized to the mitochondria and endoplasmic reticulum (ER).41 Differences in the subcellular localization of anti-apoptotic multidomain BCL-2 proteins also exist, as constitutively membrane-bound BCL-2, with the C-terminal tail anchored in the membrane and the remaining residues residing in the cytosol, is located at the mitochondria and ER, yet BCL-XL, BCL-W, and MCL1 are not tightly associated with membranes and thus can be located, at least in part, in the cytosol in healthy cells and translocate to the mitochondria during apoptosis.41 In the cytosolic forms of BAX, BCL-XL, BCL-W, and MCL1, the characteristic hydrophobic pocket of multidomain BCL-2 proteins that can also bind BH3 domains is occupied by the C-terminal membrane-anchoring tail.42,45 Mitochondrial translocation of these multidomain BCL-2 proteins involves disengagement of the C-terminal TM domain from the BH3 binding pocket and its insertion into the mitochondrial membrane.41,44,46
BAX and BAK activation
In response to diverse cellular stressors, BH3-only proteins fulfil their role as the most apical regulators of the intrinsic apoptotic pathway by triggering the activation of BAX-like proteins.37–39 The essential role of BAX-like proteins positioned downstream of BH3-only proteins in mitochondria-mediated apoptotic signaling has been established by studies that have shown that BH3-only proteins cannot induce apoptosis in cells deficient in both BAX and BAK.37–39 The activation of BAX and BAK involves conformational changes that enable self-association, likely through formation of homodimers followed by generation of higher order oligomers, thereby leading to homo-oligomerization and induction of MOMP.44,47,48 Despite the undisputed importance of BAX and BAK function in MOMP induction and consequent release of apoptogenic factors, the mechanism by which BAX and BAK are activated has been a matter of contention over the past several years. Previous studies have resulted in different conclusions as to whether a subset of BH3-only proteins can trigger apoptosis by directly binding to BAX/BAK and thus have led to the proposal of two different models of BAX/BAK activation, the indirect activation model and the direct activation model.37,39
The indirect activation model, also referred to as the displacement model, maintains that anti-apoptotic BCL-2 family members act primarily to bind and restrain BAX/BAK in inactive heterooligomeric complexes, while BH3-only proteins serve to neutralize anti-apoptotic BCL-2 proteins and thus can initiate apoptosis without directly interacting with BAX or BAK. In this scenario, BH3-only proteins bind to anti-apoptotic BCL-2 family members to allow the release of BAX/BAK, resulting in BAX/BAK homo-oligomerization and the consequent induction of MOMP.37,39,49 According to the indirect model, this displacement mechanism is the principal means by which BH3-only proteins stimulate apoptosis, and the robust pro-apoptotic activity of BIM and BID can be attributed to their ability to strongly bind all anti-apoptotic BCL-2 proteins, whereas the other BH3-only proteins only engage subsets.39,49
The direct activation model contends that certain BH3-only proteins, termed activators, can directly bind and activate BAX/BAK. Activator BH3-only proteins (activator BH3s) include BIM, tBID (the activated, truncated form of BID), and perhaps others such as PUMA50–54 (Fig. 1). In this model, promotion of cell survival by anti-apoptotic BCL-2 proteins involves sequestering activator BH3s in addition to binding any activated BAX/BAK protein monomers that may exist.37,54 BH3-only proteins that are not activators are categorized as sensitizers. The sensitizer BH3-only proteins are unable to directly interact with BAX/BAK, but instead bind to anti-apoptotic BCL-2 proteins (Fig. 1), freeing activator BH3s for binding to and activation of BAX/BAK.37,54
In recent years, compelling experimental evidence of BH3-only proteins directly interacting with BAX and consequent induction of BAX activation has been published.44,50,51 For example, the NMR structure of a stabilized α-helix of the BCL-2 domain (SAHB) peptide of the BH3 domain of BIM in complex with full-length BAX has been reported and binding of the BH3 ligand BIM SAHB was shown to trigger BAX activation.50 Interestingly, the BIM SAHB-BAX interaction site defined by NMR analysis involves the two helices α1 and α6 of BAX and is located on the completely opposite face of the protein from the canonical BH3-groove interaction site characteristic of anti-apoptotic BCL-2 proteins.50 Studies using Fluorescence (or Förster) Resonance Energy Transfer (FRET) analyses and liposomes to demonstrate the ordered series of events leading to tBid-induced oligomerization of BAX and subsequent membrane permeabilization have added further support for the direct activation of BAX by activator BH3s.51 These studies also attest to the importance of the lipid membrane in BAX activation; the reported sequential steps for tBID-induced BAX-mediated MOMP entail protein– membrane interactions as well as protein–protein interactions, including the direct interaction between tBid and BAX, which occur primarily on or in the lipid membrane.51 Previous evidence of the lipid membrane as a critical player in the regulation of membrane permeabilization by BCL-2 proteins formed the basis of the “embedded together” model.55 While aspects of both the indirect and direct activation models are incorporated in the “embedded together” model, the focus of the model is on the importance of the membrane in governing conformational changes of BCL-2 proteins and interactions between them that mediate MOMP.55
New insights into the molecular mechanism of BAX/BAK activation revealed by recent studies have provided the basis for a proposed model of stepwise activation of BAX and BAK by activator BH3s.44 According to this model, BAX activation is initiated by transient binding of an activator BH3 to the α1 helix of BAX, which has previously been determined by NMR analysis to be a component of a noncanonical BH3 binding site on BAX.50 In the inactive cytosolic form of BAX, the α1 helix maintains the engagement of the C-terminal α9 helix in the hydrophobic pocket, thereby preventing mitochondrial translocation of BAX. Interaction between an activator BH3 and the α1 helix of BAX results in conformational changes that involve exposure of the N-terminal α1 helix of BAX with consequent release of the C-terminal TM domain from the hydrophobic pocket and thus promotion of mitochondrial targeting of BAX.44 Subsequent to mitochondrial translocation, the activator BH3 remains associated with BAX, though rearrangement of the interaction occurs and results in the association of the activator BH3 with the BH1 domain of BAX, which drives homo-oligomerization of BAX.44 Given that BAK is an integral membrane protein with its N-terminal α1 helix constitutively exposed, activation of BAK bypasses the first activation step of mitochondrial targeting promoted by activator BH3-induced conformational changes. Nevertheless, akin to activation of mitochondrially translocated BAX, activator BH3s are critical for homo-oligomerization of BAK activation, and both BAX and BAK require BH1 and BH3 domains for homo-oligomerization.44 Previously, a major role for activator BH3s in driving the activation of BAX/BAK via direct interactions has been challenged, with arguments against direct activation including the failure to detect stable interactions between BH3-only proteins and soluble BAX in its inactive monomeric form.49 However, recent studies demonstrating the transient and dynamic nature of interactions between activator BH3s and BAX offer an explanation for the difficulty encountered in earlier studies in observing these interactions, and provide clear evidence validating direct activation of BAX/BAK by activator BH3s.44,50
The extrinsic apoptotic pathway
Overview of the extrinsic apoptotic pathway
Stimulation of the extrinsic apoptotic pathway is initiated by the association of cell surface death receptors, a subset of the tumor necrosis factor (TNF) receptor superfamily, with their respective activating cytokine ligands (also known as death ligands), which are members of the TNF family of proteins.13,26,56 TNF family members are mostly synthesized as type-II transmembrane proteins that assemble into biologically active homotrimers.13,29 Widely studied death receptor-ligand signaling systems include TNF receptor 1 (TNFR1)-TNF (also known as TNF-alpha), FAS (APO-1, CD95)-FASL, TNF-related apoptosis inducing ligand (TRAIL) receptor 1 (TRAILR1, DR4)-TRAIL, and TRAIL receptor 2 (TRAIL-R2, DR5)-TRAIL.13,29
Signal transduction through death receptors entails receptor oligomerization involving aggregation of an intracellular motif common to the death receptor family members, known as the death domain (DD).13,57 Trimerization of death receptors was initially thought to be prompted by ligand binding, but later studies have provided evidence in favor of the formation of preassembled receptor oligomers in a ligand-independent manner.13 Ligand binding leads to the recruitment of DD-containing adaptor proteins, such as FAS-associated death domain protein (FADD) and TNFR1-associated death domain protein, to the aggregated receptor domains via DD–DD interactions.13,29 These adaptor proteins include an additional homotypic protein interaction module, the death effector domain (DED), that sequesters the inactive zymogen of initiator caspases with a DED in their prodomain, namely caspases-8 and -10, to form the Death Inducing Signaling Complex (DISC) (Fig. 1).29 DISC formation mediates the oligomerization and consequent activation of caspase-8.35 Activation of the executioner caspases, including caspase-3, caspase-6, and caspase-7, by the activated upstream initiator caspases results in the cleavage of numerous caspase substrates and the subsequent demise of the cell.56,58,59
Integration of the extrinsic and intrinsic apoptotic signaling pathways
In response to death receptor stimulation, the extent of caspase-8 activation at the DISC can be insufficient to elicit apoptosis and thus effective apoptotic signaling requires engagement of the mitochondria-mediated intrinsic pathway for full activation of executioner caspases, such as caspase-3, and consequent induction of apoptosis downstream of MOMP.29 The cleavage and consequent activation of BID by caspase-8 to generate the activated BID fragment (tBID) serves as the conduit through which the apoptotic signal is relayed from the extrinsic to the intrinsic signaling pathway.60,61 In this scenario, caspase-8 activity induced by death receptor-mediated signaling is adequate to cleave cytosolic, full-length BID to result in the production of tBID and its translocation to the mitochondria, which leads to the release of the apoptogenic factors (Fig. 1).26,29,60,61 These pro-apoptotic molecules ultimately promote activation of executioner caspase-3 and subsequent activation of caspase-8, thereby generating a positive feedback loop to amplify the apoptotic response.29,62
The requirement of the intrinsic pathway to commit cells to death in response to FAS activation defines two types of cells, type I and type II. In type I cells, active caspase-8, generated by extensive processing of procaspase-8 at the DISC, directly activates the executioner caspases, including caspase-3, to carry out FAS-initiated apoptosis, whereas in type II cells a low concentration of active caspase-8 is generated at the DISC. Accordingly, although FAS-mediated death signaling triggers activation of the mitochondrial apoptotic pathway in both cell types, only type II cells depend on mitochondria to complete the apoptotic response induced by FAS activation.13,63 This fundamental difference between cell types is evidenced by reports demonstrating that overexpression of BCL-2 or BCL-XL, anti-apoptotic BCL-2 family members which inhibit MOMP and cytochrome c release, blocks activation of caspase-8 and caspase-3 as well as FAS-mediated apoptosis in type II cells, but not type I cells.13,63 Classification of cells based on the requirement of mitochondria for induction of apoptosis in response to death receptor signaling has been extended beyond FAS to other death receptors, including TRAIL death receptors.13,64 Highlighting the important roles that BCL-2 proteins can play in mediating apoptosis, the rate of BID cleavage has been implicated as the determining factor in defining TRAIL-sensitive cells as mitochondria-dependent or independent.64 Various other BCL-2 family members, both pro-apoptotic and anti-apoptotic, have been shown to promote or inhibit TRAIL-induced apoptosis, respectively, in a cell type-specific fashion.65
Control points in the apoptotic pathways
Control point 1: apoptosis control via regulation of MOMP by BCL-2 family proteins
BH3-only proteins serve as sensitive detectors of apoptotic stimuli that transmit death signals to downstream BAX-like proteins, the principal effectors of the intrinsic apoptotic pathway, thereby promoting apoptosis.37–39 The activation of certain BH3-only family members can entail distinct transcriptional or post-translational modifications under specific conditions.39,41,54,66 For example, the pro-apoptotic activity of BAD and BIM is controlled by phosphorylation, and as mentioned above, the full activity of BID is unleashed through proteolytic cleavage.67,68 The relatively large number of BH3-only proteins, together with the unique activation mechanisms of particular BH3-only proteins, have been proposed to contribute to the ability of cells to decipher the signals from a wide range of apoptotic stimuli by enabling specialized response to different death signals.39,66 In addition, the concept that BH3-only proteins can act in a tissue- and/or signal-specific manner has been supported by genetic deletion studies.68
As with the BH3-only proteins, apoptotic signals can regulate the functional activity of multidomain BCL-2 proteins.68 The regulatory mechanisms of BCL-2 family members can serve to alter their stability or their ability to bind other family members.68 Given that interactions between BCL-2 family members govern BAX and BAK activation and consequent induction of MOMP, shifts in the binding activities of pro- and anti-apoptotic BCL-2 family members in response to many diverse stimuli play a fundamental role in determining whether a cell will live or die.39,54 The specific contribution of an individual BCL-2 family member to promote or inhibit apoptosis is mediated by its binding profile. Anti-apoptotic proteins inhibit apoptosis by sequestering pro-apoptotic BCL-2 proteins, including BAX and BAK, as well as activator BH3s. For example, an investigation of the inhibition of tBID-mediated BAX activation by BCL-XL has provided evidence that multiple mechanisms, involving both BCL-XL-tBID and BCL-XL-BAX interactions, play a part in the anti-apoptotic function of BCL-XL.46 BH3-only proteins promote apoptosis by neutralizing anti-apoptotic BCL-2 proteins and/or by binding to and thereby directly activating BAX and BAK. Characterization of the binding affinities and specificity of the interactions between BH3-only proteins and multidomain BCL-2 family members has revealed that BID, BIM, and PUMA can bind to all anti-apoptotic multidomain BCL-2 family members while other BH3-only proteins, such as NOXA and BAD, are only able to engage a certain subset of the anti-apoptotic BCL-2 proteins.66,68 The differences in the observed potency of the BH3-only proteins have been attributed to the wide range of differences in their binding affinities.69–71 Using a genomic approach to examine the pro-apoptotic function of BIM in vivo, the maximal pro-apoptotic potential of BIM, found to be significantly enhanced relative to other BH3-only proteins, seems to require its ability to bind all anti-apoptotic BCL-2 family members as well as directly interacting with BAX.71
Disruption of the intricate balance between pro- and anti-apoptotic BCL-2 proteins can render cells resistant to apoptotic stimuli and thereby promote cancer cell survival (Fig. 2b).9 As oncogenic signaling is linked to apoptotic signaling pathways through mechanisms involving critical apoptosis regulators such as BCL-2 family members, 72 alterations that increase the functional activity of anti-apoptotic BCL-2 proteins relative to pro-apoptotic BCL-2 proteins can significantly contribute to cancer development and progression by enabling cells to escape from apoptosis. Both suppressed activation of pro-apoptotic members73–76 and overexpression of anti-apoptotic members9,77 have been reported in various cancers. Genetic and epigenetic alterations targeting genes encoding pro-apoptotic BCL-2 proteins, including BAX as well as several BH3-only proteins, have been identified in several types of cancer cells. Frameshift mutations along with functional inactivation of BAX have been found in colorectal cancers with microsatellite instability,78,79 and these mutations have been suggested to be associated with disease progression.80 DNA methylation has been reported to downregulate the expression of BAD, BAK, BIK, and BAX in a multiple myeloma cell line.74 Silencing of BIM by homozygous deletions and promoter hypermethylation has been observed in mantle cell lymphoma (MCL)73,81 and Burkitt lymphoma (BL),73 respectively. The significance of frequent BIM inactivation in BL has been demonstrated by the reversal of BL chemoresistance upon restoration of BIM function.82 Similarly, the critical role of a specific BH3-only protein in governing a death response has been shown in pancreatic ductal adenocarcinoma (PDAC) cells, as reversal of epigenetically-mediated NOXA silencing sensitizes PDAC cells to apoptosis by induced by etoposide.83
Besides genetic and epigenetic changes, another mechanism of gene silencing involves microRNAs (miRNAs), small non-coding RNA molecules (20–24 nucleotides long) that base-pair to target mRNAs and generally repress protein synthesis.84 miRNAs known to be aberrantly expressed in a variety of cancers have been linked to multiple oncogenic processes, including inhibition of apoptosis,85,86 and have been found to regulate the expression of many key factors involved in the apoptotic pathways, including BCL-2 family proteins.87,88 For example, several miRNAs have been found to mediate the suppression of BIM expression in a number of cancers;87 BIM has been reported to be targeted by miRNAs encoded by the miR-17-92 cluster in neuroblastoma89 and lymphomas90,91 and the miR-106b-25 cluster, specifically miR-25, in hepatocellular carcinoma (HCC)92 and esophageal adenocarcinoma.93 Moreover, as with other mechanisms of BIM silencing, treatment with agents that upregulate BIM expression, such as inhibitors of BIM-targeting miRNA, can induce apoptosis in therapy-resistant cancer cells.89
In addition to inactivation of pro-apoptotic BCL-2 family proteins, overexpression of anti-apoptotic family members is a prominent mechanism of apoptosis dysregulation in cancer.7,94 Increased expression of BCL-2 has been observed in many human malignancies, including acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), diffuse large B-cell lymphoma (DLBCL), glioblastoma, melanoma, and cancers of the prostate and lung.95–102 BCL-XL overexpression has also been reported in a number of cancers, including multiple myeloma, Kaposi’s sarcoma, HCC, colorectal adenocarcinoma, and pancreatic cancer.103–108 Elevated levels of BCL-2 and BCL-XL have frequently been associated with aggressive disease and/or chemoresistance.101,106,109–114 Furthermore, given that some cell types, namely type II cells, require activation of the mitochondria-mediated intrinsic pathway for commitment to apoptosis in response to TRAIL, the anti-apoptotic effect of overexpression of anti-apoptotic BCL-2 proteins can extend to the death-receptor mediated pathway and render these cells resistant to TRAIL-induced apoptosis.12,65,115
The widespread overexpression of BCL-2 in human cancer arises from several different types of cellular defects.116 The involvement of BCL-2 in the t(14;18) chromosomal translocation, which frequently occurs in follicular lymphomas, led to its initial identification117 and is now known to drive elevated BCL-2 expression.101 Gene amplification has also been implicated as an important mechanism for BCL-2 overexpression in DLBCL118 and small cell lung cancer (SCLC).119 Furthermore, loss or reduced expression of the tumor suppressor miRNAs miR-15a and miR-16-1 through the deletion or down-regulation of their encoding gene cluster, the miR-15a–miR-16-1 cluster, can serve as yet another mechanism promoting elevated levels of BCL-2 expression in cancer cells.120 Markedly reduced levels of miR-15a and miR-16-1 have been observed in approximately 70% of chronic lymphocytic leukemia (CLL) patient samples, as compared with their normal counterparts,121 along with inversely correlated levels of BCL-2 expression.122 Indeed, dysregulated posttranscriptional repression of BCL2 by miR-15a and miR-16-1 is currently regarded as one of the main causes of BCL-2 overexpression in CLL.120
The known tumor suppressor function of miR-15a and miR-16-1 in CLL is supported by their ability to target multiple oncogenes, including multiple negative regulators of apoptosis, as indicated by the report that these miRNAs downregulate the expression of another anti-apoptotic BCL-2 family member, MCL-1.123 Compelling evidence indicates that aberrantly low levels of miR-15a and miR-16-1 may significantly contribute to the pathogenesis of various other cancers as well.120 Moreover, within the last few years, several studies have implicated the dysregulated expression of other miRNAs that have also been suggested to play a tumor suppressor role as an important contributing factor to the frequent aberrant expression of BCL-2 and/or MCL-1 expression in cancer cells.86 For example, inactivation of miR-34 family members (miR-34a, miR-34b, and miR-34c) has been found to occur in many cancers;124 miR-34 family members have been shown to target BCL-2 in numerous types of cancer cells, including neuroblastoma,125 non-small cell lung cancer (NSCLC),126 gastric cancer,127 and pancreatic cancer cells;128 and ectopic expression of miR-34a can induce apoptosis.124 In addition, the miR-29 family members (miR-29a, miR-29b, and miR-29c), particularly miR-29b, have been proposed to serve as tumor suppressors in various cancers, as restoration of mir-29b in cancer cells exhibiting downregulation of this miRNA has been shown to induce a pro-apoptotic effect that is associated with its ability to target, and thereby repress the expression of, MCL-1 in cholangiocarcinoma129 and AML130 and both BCL-2 and MCL-1 in HCC.131
Control point 2: IAP-mediated inhibition of apoptosis
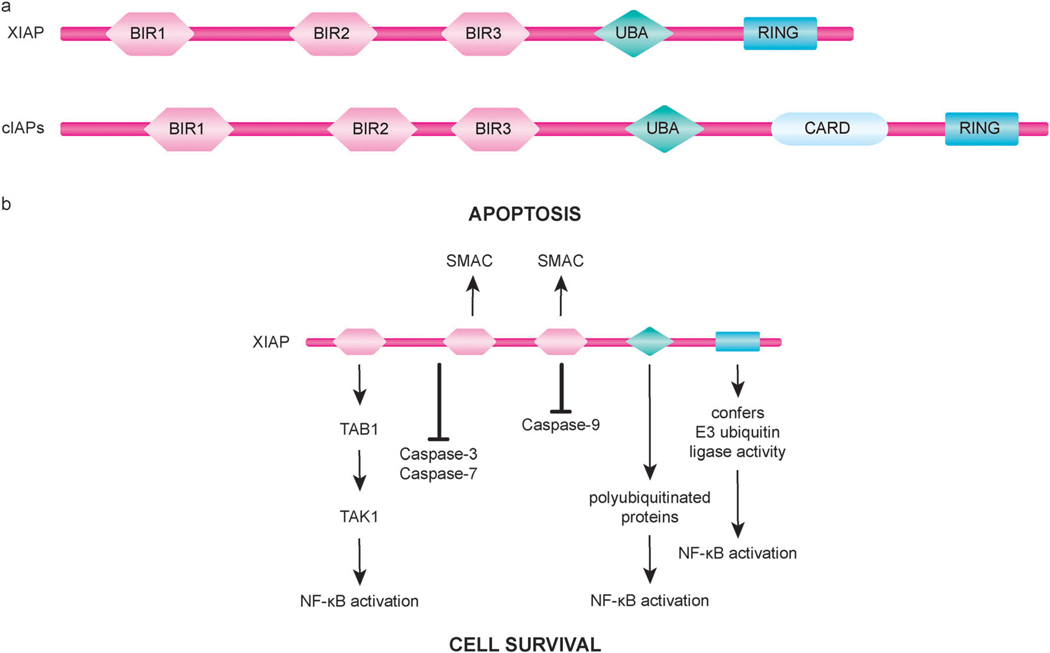
Considering that proteolysis is an irreversible process, strict control of caspase-mediated proteolytic cleavage is imperative to prevent inappropriate cell destruction.21,24 Accordingly, cells use multiple layers of regulation to modulate caspase activation.132 One such layer of caspase regulation involves inhibition of caspase activity by IAP proteins.132 All members of the IAP family of proteins (with the human IAP family including; NAIP, XIAP, cIAP1, cIAP2, ILP2, livin, survivin, and BRUCE) contain at least one baculovirus IAP repeat (BIR) domain and many have multiple copies of this defining motif of IAPs.132,133 Three tandem BIR domains (BIR1–BIR3) are present in the N-terminal region of the mammalian IAPs XIAP, cIAP1, and cIAP2.133 In addition to BIR domains, these IAPs also contain a C-terminal RING finger domain that confers E3 ubiquitin ligase activity and an ubiquitin-associated (UBA) domain for binding ubiquitinated proteins (Fig. 3a).134 Furthermore, cIAP1 and cIAP2 include a caspase recruitment domain (CARD) domain of unknown function. BIR domains of IAPs can mediate interactions with various proteins and their ability to directly bind to caspases can serve as a mechanism for IAP-mediated caspase inhibition.133
Fig. 3.
Domain organization and function of inhibitors of apoptosis (IAP) proteins. (a) XIAP, a well studied human IAP family member, and the structurally similar family members cIAP1 and cIAP2 (cIAPs) each have three tandem BIR domains followed by an ubiquitin-associated (UBA) domain and a C-terminal RING finger domain. cIAPs also possess a caspase recruitment domain (CARD) of unknown function located between the UBA and the RING domains. (b) The BIR2 domain of XIAP, along with residues in its N-terminal flanking linker region, mediates the binding and inhibition of caspase-3 and caspase-7. Inactivation of caspase-9 by XIAP involves the BIR3 domain of XIAP binding to caspase-9. In addition to blocking caspase activity, XIAP can also promote cell survival through regulation of important cellular signaling pathways, including signaling mechanisms of NF-κB activation. IAP-binding motif (IBM)-containing proteins, such as SMAC, interact with the BIR2 and BIR3 domains of XIAP to neutralize its anti-apoptotic activity.
XIAP is recognized as the most potent IAP family member with respect to its anti-apoptotic activity, which involves inhibition of active executioner caspases as well as prevention of initiator caspase-9 activation (Fig. 3b).94,133 In XIAP, the linker region between BIR1 and BIR2, along with BIR2, contributes to the direct binding and inhibition of caspase-3. As with caspase-3, the same linker region of XIAP also binds to and thus obstructs the substrate-binding groove of caspase-7.133 Although structural studies have found some contacts between the BIR2 domain of XIAP and caspase-3, but not caspase-7, the inability of the linker region alone to effectively inhibit caspases suggests that BIR2 does play a role in binding.133 A distinct mode of binding occurs between XIAP and caspase-9; the BIR3 domain of XIAP mediates interactions with caspase-9 to keep it in an inactive monomeric state.133
Notably, cIAP1 and cIAP2 are able to bind caspases but lack critical elements required for direct caspase inhibition.135,136 In fact, although direct inhibition of caspase activity was previously thought to be a significant function of several IAP proteins, later studies suggest that XIAP is the only IAP family member that serves as a direct caspase inhibitor.136 Nevertheless, IAP family members have been shown to be key regulators of signaling pathways important in determining whether cells live or die.132,133 In particular, several IAPs have been found to play a complex role in modulating the signaling pathways that activate NF-κB transcription factors.132,134,137 The upstream signaling mechanisms leading to their activation can generally be classified as classical (also known as canonical) or alternative (also known as non-canonical) NF-κB signaling pathways.137,138 Members of the NF-κB family of transcription factors are key regulators of a range of physiological processes, including innate and adaptive immune responses, inflammation, and cell survival, and their established ability to induce the expression of genes encoding anti-apoptotic proteins has been recognized as an important means of suppressing apoptosis and promoting cell survival.137 Interestingly, while XIAP, cIAP1, and cIAP2 have been reported to mediate NF-κB activation, these IAPs are also among the recognized anti-apoptotic target genes of NF-κB, thus indicating a positive feedback loop for activation of NF-κB.134,137
The ability of XIAP overexpression to induce NF-κB activation has been commonly observed in previous studies, yet the mechanistic details involved have not been fully elucidated and the physiological role of XIAP in regulating NF-κB activity is not yet well understood.134,139 Nevertheless, reports using ectopic expression of XIAP provide evidence that XIAP can induce NF-κB activation through the activation of transforming growth factor-β (TGF-β)-activated kinase (TAK1),134,139 a major player in classical NF-κB signaling;138,140 direct interaction of the BIR1 domain of XIAP with the adaptor protein TAK1-binding protein (TAB1) has been reported to play a central role in XIAP-mediated TAK1 and NF-κB activation (Fig. 3b).134,139 In at least some contexts, efficient activation of NF-κB by XIAP has been shown to require functionally intact UBA and RING finger domains.134,139 In addition to TAK1, the ubiquitin ligase activity of XIAP has also been implicated as an important regulator of other components involved in signaling mechanisms of NF-κB activation.139,141
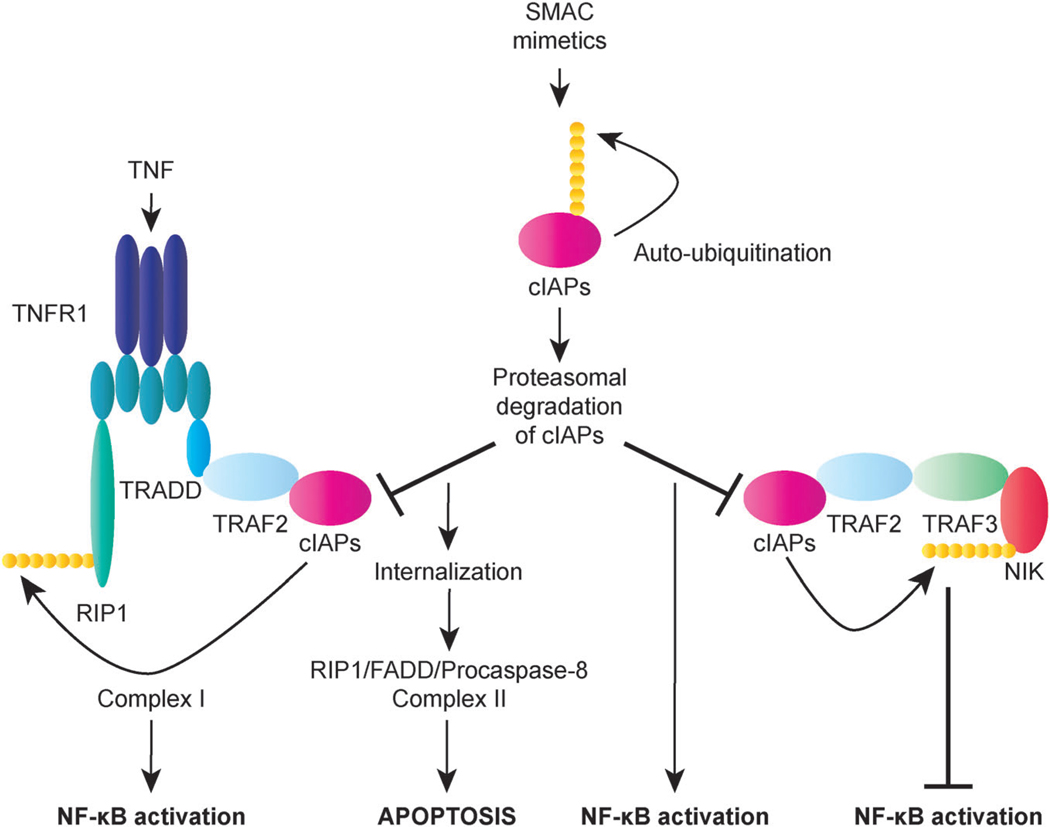
While the role of XIAP in cellular signaling pathways is less well-defined,139 a number of studies within the last few years have confirmed the physiological involvement of cIAP1 and cIAP2 in receptor-initiated signaling mechanisms.132,134 These studies have established cIAP1 and cIAP2 as critical regulators of both classical and alternative NF-κB signaling pathways that act in a functionally redundant manner.132,134 Interestingly, cIAPs are essential for efficient activation of classical NF-κB signaling in response to stimulation of the death receptor TNFR1, yet they also function as vital participants in the suppression of alternative NF-κB signaling in resting cells (Fig. 4).132,134 The ubiquitin ligase activity of cIAP1 and cIAP2, along with their BIR1-mediated binding to the adaptor protein TNF receptor-associated factor 2 (TRAF2), plays a major role in their function as both positive regulators of TNF-mediated NF-κB activation and negative regulators of alternative NF-κB signaling pathways.132–134
Fig. 4.
Regulation of NF-κB signaling by cIAP1 and cIAP2 (cIAPs). cIAPs play a key role in TNF-induced NF-κB activation by functioning as E3 ligases that ubiquitinate RIP1, leading to the stabilization of complex I and activation of NF-κB. In the presence of SMAC mimetics, cIAPs undergo auto-ubiquitination and proteasomal degradation. The loss of cIAPs sensitizes cells to apoptosis in response to TNF by allowing the formation of complex II, which requires deubiquitination of RIP1. Complex II contains RIP1, FADD, and procaspase-8 and serves to trigger apoptosis by promoting activation of caspase-8. Interestingly, cIAPs can also act as negative regulators of NF-κB activation by targeting NF-κB-inducing kinase (NIK) for proteasomal degradation. In this context, the loss of cIAPs induced by SMAC mimetics promotes NIK stabilization and consequent NF-κB activation.
Based on a number of recent findings and reassessment of previous data, a newly revised model of NF-κB activation by TNF involves the recruitment of the protein kinase receptor-interacting protein 1 (RIP1) as well as TRADD and TRAF2, which in turn recruit cIAP1, to the activated TNFR1 receptor to form the membrane-bound complex I (Fig. 4).142 Subsequently, cIAP ubiquitinates RIP1 as well as other components of complex I, which triggers the recruitment of additional components to complex I, thus stabilizing it and activating kinases that lead to NF-κB activation.142 While TNFR1 signaling typically serves to stimulate inflammation and promote cell survival, under certain circumstances, TNF can elicit a death response.13 Importantly, cIAP-mediated K63-linked polyubiquitination of RIP plays a central role in protecting cells from TNF-induced apoptosis.132,134 The pro-apoptotic potential of TNF can be switched on through the formation of complex II, a cytoplasmic complex that is derived from the internalization of complex I in a temporally distinct step of TNFR1 signaling; with the loss of both cIAP1 and cIAP2, complex II can recruit FADD and procaspase-8 to induce apoptosis in a manner that requires deubiquitination of RIP1 (Fig. 4).13,134,143 Accordingly, cIAPs can significantly contribute to cell survival by promoting NF-κB activation as well as preventing caspase-8-mediated apoptosis in response to TNF. Nevertheless, previous studies have also elucidated an important role for cIAPs in maintaining low levels of NF-κB-inducing kinase (NIK), a central mediator of alternative NF-κB signaling, thereby restraining NF-κB activation in resting cells (Fig. 4).134,137 Constitutive degradation of NIK occurs through its recruitment to the adaptor protein TNF receptor-associated factor 2 (TRAF3), which also binds to the complex TRAF2–cIAP1 and/or TRAF2–cIAP2, resulting in the targeting of NIK for proteasomal degradation by cIAP-mediated K48-linked polyubiquitination.134,137 Given their multifaceted function in regulating signal transduction pathways, IAPs may play a role in both promoting and antagonizing cell survival, depending on the cellular context.132
In response to apoptotic stimuli that trigger MOMP, endogenous inhibitors of IAPs, including SMAC (also known as DIABLO) and OMI (also known as HTR2A), are released from the mitochondria to counter the activity of IAPs and thereby promote apoptosis.26,29 In their mature form, these proteins contain a conserved N-terminal IAP-binding motif (IBM) that interacts with a conserved groove found in the BIR2 and BIR3 domains of IAPs (Fig. 3b).133,136 SMAC has been found to function as a homodimer, with the four residues (AVPI) that constitute the IBM of each SMAC molecule acting in a bivalent manner so that one IBM binds to BIR2 and the other to BIR3 of a single XIAP molecule to block caspase inhibition.132,144 In addition to XIAP, SMAC has been shown to bind most IAPs,132 and IAP antagonists that mimic the IBM of SMAC (SMAC mimetics) have been found to effectively kill cancer cells through a mechanisms involving auto-ubiquitination and proteasomal degradation of cIAP1 and cIAP2 (Fig. 4).133,134
Aberrant expression or activity of IAPs has been reported in numerous types of human cancer.132,144,145 Elevated IAP expression levels have frequently been observed in a variety of hematological malignancies as well as solid tumors and have been associated with advanced disease and poor prognosis.132,144,145 An examination of the NCI 60 cancer cell line panel has revealed high mRNA levels of XIAP in nearly all of the cell lines.146 Increased expression of XIAP has also been reported in several different types of tumor samples and has been linked to chemoresistance, tumor progression, and/or shorter survival in some cancers, including HCC, renal cell carcinoma, colorectal, ovarian and prostate cancer.132,145,147–149 Various mechanisms can play a role in the dysregulation of XIAP in cancer cells.145 One proposed mechanism of enhanced XIAP activity involves co-expression of survivin, another IAP family member, which can bind to SMAC to block SMAC-mediated inhibition of XIAP.145,150 Furthermore, survivin can interact with XIAP to form a complex that increases XIAP stability against ubiquitin-mediated proteasomal degradation and enhances caspase inhibition in a synergistic manner.150 Besides its function as a potent inhibitor of apoptosis, which has been associated with accelerating tumor growth,151 this IAP–IAP complex has also been reported to promote the metastasis of tumor cells via activation of NF-κB signaling.152 Notably, survivin, unlike other IAPs, is over-expressed in nearly all human cancers but is generally undetectable in most adult tissues.132,150 While the prognostic value of XIAP has been found to vary with tumor type,132,147 overexpression of survivin has consistently been implicated as a negative prognostic factor.132,145,150 As with XIAP, multiple mechanisms have been described by which survivin expression can be upregulated in cancer.150 The prominent overexpression of XIAP and survivin, along with many studies demonstrating that inhibition of these IAPs can promote apoptosis and sensitize cancer cells to various therapies, strongly suggests that their anti-apoptotic activity can significantly contribute to cancer pathogenesis.132,145
cIAP1 and cIAP2 expression is also commonly dysregulated in cancer. A chromosomal region containing the genes encoding cIAP1 (BIRC2) and cIAP2 (BIRC3), 11q22, has been found to be amplified in several cancers,134 with overexpression of cIAP1 and/or cIAP2.153–155 Additional genetic evidence in support of an oncogenic role for cIAPs includes the involvement of BIRC3 in the t(11q21 : 18q21) translocation, the most common translocation event in mucosa-associated lymphoid tissue (MALT) lymphoma.156 The resulting cIAP2–MALT1 fusion protein triggers constitutive activation of classical NF-κB signaling, leading to B cell transformation and lymphoma progression.134 Although the aberrant expression of IAPs that has been associated with tumor-promoting process typically involves their upregulation, inactivating genetic mutations of cIAPs have also been linked to oncogenesis.134,145 Specifically, biallelic deletions and correlated reduced expression levels of cIAP1 and cIAP2, resulting in NIK stabilization and activation of alternative NF-κB signaling, has been identified in multiple myeloma.137,157
Control point 3: regulation of apoptosis by c-FLIP
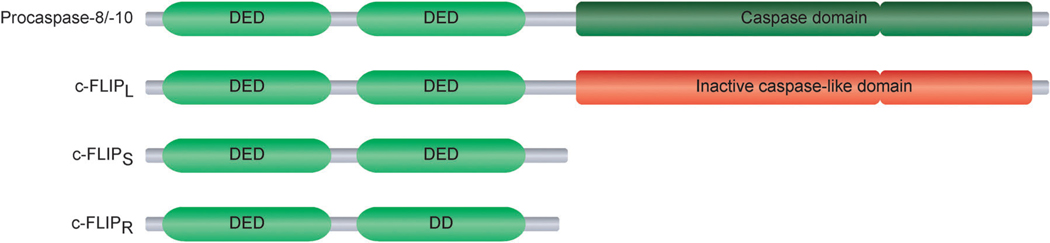
c-FLIP is a DED-containing protein that is recruited to the DISC and regulates the activation of caspase-8 in death receptor signaling pathways (Fig. 1).18 Three c-FLIP protein isoforms derived from distinct mRNA splice variants have been identified, namely c-FLIPL, c-FLIPS, and c-FLIPR.18,158 c-FLIPL, the long c-FLIP isoform, is structurally similar to procaspase-8, with two tandem DEDs at its N-terminus and a catalytically inactive caspase-like domain at its C-terminus.18,158 The short c-FLIP isoforms c-FLIPS and c-FLIPR also possess two tandem DEDs at their N-termini, but differ structurally from procaspase-8 at their C-termini, which are defined only by a short stretch of amino acids (Fig. 5).18,158
Fig. 5.
Structural features of the FLICE-inhibitory protein (c-FLIP) isoforms. Each of the three c-FLIP isoforms, c-FLIPL, c-FLIPS, and c-FLIPR, contain two tandem death effector domains (DEDs) at its N-terminus. c-FLIPL, the long c-FLIP isoform, is similar in overall structure to procaspase-8, but its caspase-like domain is functionally inactive. Other than the two DEDs, the short c-FLIP isoforms c-FLIPS and c-FLIPR only include a short C-terminus.
c-FLIP was initially characterized as a potent inhibitor of death receptor-mediated apoptosis.159 At high expression levels, all three isoforms are recruited to the DISC via DED interactions and subsequently impair death receptor-induced apoptotic signaling by blocking the activation of caspase-8.18,158,159 Nevertheless, some early studies reported conflicting evidence regarding the function of c-FLIPL in mediating apoptosis.160 Later studies have clarified the previous, seemingly contradictory, findings by clearly revealing a dual functional role for c-FLIPL as an inhibitor or activator of caspase-8 activation, depending on various factors, including its expression level relative to that of caspase-8.18,160,161 In contrast to its anti-apoptotic role at high levels of expression, c-FLIPL has been shown to act at the DISC to enhance caspase-8 activation and promote apoptosis at low and more physiologically relevant expression levels.160,161 Modulation of caspase-8 activity by c-FLIPL is further complicated by evidence that in certain contexts c-FLIPL may induce only moderate procaspase-8 activation, which results in a level of caspase-8 activity that is too low to trigger apoptosis but is sufficient for cleavage of c-FLIPL, ultimately leading to the activation of NF-κB survival signaling and other non-apoptotic signaling pathways.161,162 Still, the function of c-FLIP in blocking caspase-8 activation at the level of the DISC is recognized as an important check on caspase activity to protect against unwanted cellular destruction.21,24
Despite the identified dual functionality of c-FLIPL as a pro- or anti-apoptotic factor in normal tissues, c-FLIPL has generally been shown to act as a key negative regulator of apoptosis in human cancer cells.18,163 Increased expression of c-FLIP has been commonly found in a number of human malignancies, including melanoma, hepatocellular carcinoma,164 non-small cell lung carcinoma,165 endometrial,166 colon,167 and prostate cancer.168–170 Previous studies have clearly demonstrated that in various types of cancer cells, elevated c-FLIP expression levels serve to block caspase-8 activation at the DISC, thereby rendering cells resistant to death receptor-mediated apoptosis.18 Notably, c-FLIP overexpression has been associated with disease progression and/or poor prognosis in BL, HCC, ovarian, endometrial, colon, and prostate cancer.18,163,164,171
Besides blocking apoptotic signaling, c-FLIP has been implicated in contributing to tumor-promoting processes through its influence on key cellular survival signaling mechanisms.163 In a number of diverse cellular contexts, death receptor stimulation triggers nonapoptotic signaling cascades with important roles in cell proliferation, migration, and survival.13 Overexpression of c-FLIPL has been reported to bind proteins involved in these signaling pathways, including TRAF1, TRAF2, RIP, and RAF1, and thereby promote the activation of the downstream signaling molecules NF-κB and ERK.163 In TRAIL-resistant NSCLC cells, c-FLIP and RIP have been shown to be essential for TRAIL-induced formation of the DISC in non-raft domains of the plasma membrane and consequent activation of NF-κB and ERK cell survival signals. In contrast, knockdown of c-FLIP has been found to redistribute the DISC to lipid rafts and switch DISC signaling to TRAIL-induced apoptosis.172 Furthermore, c-FLIPL-mediated ERK activation has been shown to be responsible for the capacity of TRAIL to induce proliferation of TRAIL-resistant glioma cells.173 Recently, c-FLIPL has also been reported to modulate signaling downstream of the pro-survival kinase AKT by means of direct interaction with AKT.174 A novel mechanism of c-FLIPL-mediated TRAIL resistance in cancer cells has been proposed based on evidence indicating that c-FLIPL overexpression markedly reduces phosphorylation of a major AKT substrate, GSK3β, resulting in reduced levels of p27 and caspase-3 expression along with decreased sensitivity to TRAIL-induced apoptosis.174
Given that c-FLIP expression in cancer cells can be regulated at the transcriptional level by a variety of transcription factors, as well as at the translational and post-translational levels,18,163 numerous opportunities exist for dysregulation of this key apoptosis regulator. One noteworthy regulatory mechanism by which c-FLIP expression can be upregulated involves activation of NF-κB transcriptional activity.175,176 As NF-κB constitutive activation has been linked to the pathogenesis of many human cancers137 and inhibition of NF-κB-induced transcription of c-FLIP has been shown to sensitize cells to death receptor-mediated apoptosis,13,177 NF-κB-mediated upregulation of c-FLIP is implicated as an important factor in the evasion of cell death by cancer cells.
Therapeutic strategies modulating the induction of apoptosis
Over the past decade, significant advances have been made in discovery and validation of several novel cancer therapeutics. However, inevitably resistance to therapy leads to treatment failures. As highlighted in the preceding section, regulators of apoptosis are major factors contributing to resistance to chemotherapy. Considering the established ability of cellular alterations that lead to the evasion of apoptosis to contribute to the development and progression of cancer as well as resistance to various anticancer therapies,6,7,94 these advances have provided new drug targets for promising apoptosis-inducing therapeutic strategies that can be used most likely in combination with other apoptosis-inducing therapeutic agents. Several review articles have called attention to the great potential of apoptosis-targeted therapies,8,10,178 some of which focus on BCL-2 family proteins,9,179,180 IAPs,144,181,182 or c-FLIP18,183 and the approaches that have been used to modify their activity to reactivate apoptosis and thus eradicate cancer cells. We refer readers to these reviews for specific details regarding the development of novel therapeutic agents directed against these targets, which have been shown to demonstrate enhanced apoptotic killing and sensitize resistant cancer cells to antineoplastic agents. Nevertheless, we outline below a few of the most promising therapeutic strategies for the targeted induction of apoptosis.
The recognition of the association between overexpression of BCL-2 and BCL-XL and aggressive disease and/or chemoresistance has marked these proteins as attractive therapeutic targets and spurred efforts to develop pharmacological interventions aimed at downregulating anti-apoptotic BCL-2 family members.9,179 Strategies to inhibit anti-apoptotic BCL-2 proteins include reducing protein expression by targeting the corresponding mRNA with an antisense oligonucleotide compound as well as blocking anti-apoptotic activity by targeting at the protein level.8,9,179 Small-molecule inhibitors that mimic the BH3 domain of BH3-only proteins and thus act to directly bind and neutralize anti-apoptotic BCL-2 proteins can inhibit multiple BCL-2 family members and seem to be especially promising anti-cancer therapeutics, both alone and in combination with other therapies, based on preclinical and early clinical study results.179
As with anti-apoptotic BCL-2 proteins, IAPs have also been strongly pursued as therapeutic targets for apoptosis-inducing anti-cancer strategies184 due to evidence of their overexpression in many cancers and their association with poor prognosis and chemoresistance in some of these cancers.132,144,145 Various IAP-targeted therapies have been developed, including antisense oligonucleotides against XIAP and survivin8,144 and small-molecule inhibitors that bind to the BIR2 or BIR3 domain of XIAP, cIAP1, and cIAP2.184 Most small-molecule IAP antagonists are SMAC mimetics, several of which are in advanced preclinical or early clinical development.132,184 Bivalent SMAC mimetics, which bind to both the BIR2 and BIR3 domains within a single molecule of XIAP as well as the BIR3 domains of two cIAP molecules to induce their dimerization and subsequently trigger their proteasome-mediated degradation, have been shown to be significantly more potent than their monovalent counterparts.132,184 Despite evidence indicating that SMAC mimetics show great promise for cancer therapy,132,184 their potential value as anti-cancer therapeutics has been called into question as a result of their established ability to induce loss of cIAP1 and cIAP2, resulting in NIK stabilization and consequent NF-κB activation (Fig. 4).132,137
Downregulation of c-FLIP appears to be a highly promising therapeutic strategy to lower the apoptotic threshold of cancer cells, as evidenced by several reports demonstrating that c-FLIPL overexpression can protect cancer cells from both TRAIL- and chemotherapy-induced apoptosis, while small interfering RNA (siRNA)-mediated downregulation of c-FLIP is able to sensitize cancer cells to FASL, TRAIL, and chemotherapeutic agents.18,163,183 FLIP-targeted siRNAs have also been shown to induce spontaneous apoptosis in a variety of cancer cell types, underscoring their potential as therapeutic agents.183 Nevertheless, obstacles against the effective and safe systemic delivery of siRNA-based therapies pose a significant challenge to their widespread use in clinical settings.185 Despite this, intense research efforts have been focused on the development of RNA interference (RNAi) therapeutics, leading to important advances in the field.186,187 In May 2008, the first Phase I clinical trial for the treatment of solid tumors involving systemic delivery of siRNA via targeted nanoparticles was initiated (ClinicalTrials.gov identifier: NCT00689065).187 Recently, data from this trial demonstrated that systemically delivered siRNA can reduce mRNA and protein levels of a specific gene through an RNAi mechanism in humans.188 The exciting breakthrough offers great hope for the future application of siRNA as a gene-specific therapeutic for cancer treatment. Given that therapies aimed at inhibiting the expression of anti-apoptotic proteins are expected to improve the effectiveness of cancer treatment, in addition to c-FLIP, other anti-apoptotic proteins are promising targets for the development of RNAi therapeutics.183 Indeed, as with c-FLIP, RNAi-mediated downregulation of BCL-2, BCL-XL, XIAP, and survivin has also been shown to sensitize cancers cells to a range of cancer therapies, including chemotherapeutic drugs and TRAIL.8,183,189
Challenges with validating the therapeutic strategies that modulate apoptosis
The low rate of FDA approval for cancer therapeutics in the past several years necessitates more clinically relevant models for preclinical studies for validation and development of novel targeted therapeutics, including Bcl-2, IAP and FLIP inhibitors. The major concerns with the current preclinical studies are that the cell-based assays and animal models that are being used do not closely resemble the human disease.190–192 The majority of cell-based studies used for analysis of apoptotic pathways and their modulators are carried out in two-dimensional models, which differ significantly from tumors in vivo. It is now well established that tumors behave like organs, consisting of tumor, stroma, inflammatory and endothelial cells with complex interactions.193 Moreover, it is now apparent that 3D models developed for a variety of cancers, including breast and melanoma, more closely resembles the in vivo setting.194,195 Indeed, studies using 3D models (i.e., breast cancer) indicate that additional levels of regulation of the apoptotic machinery exist.196–200 For instance, various components of the extracellular matrix, including integrins, also modulate the response of cells to apoptotic stimuli.192,200,201 Therefore, in therapeutic consideration of TRAIL, TRAIL-receptor agonist antibodies, modulators of the Bcl-2 family of proteins, inhibitors or IAPs and FLIPs, we should bear in mind the complexity of tumor microenvironment and investigate the validity and efficacy of these therapeutics in 3D models that more closely resemble the physiological conditions.192,196
Another concern in the validation of apoptosis-inducing agents as therapeutics is the use of in vivo animal models. Currently, preclinical in vivo studies for efficacy and mechanism of action of therapeutic targets are carried out mostly in xenograft and sometimes in orthotopic mouse tumor models. However, these models do not adequately represent human cancer, as the interaction between tumor and its microenvironment and the multi-step tumor progression is not recapitulated.191 Recent evaluation of genetically engineered mouse models (GEMMS) as platforms for testing cancer therapeutics has provided solid evidence to the value of these models in preclinical studies.190 It is now apparent that the GEMMS can more accurately predict the efficacy and mechanism of action of anti-cancer therapeutics. Despite the high upfront cost of GEMMS for preclinical validation, these models can potentially significantly reduce the enormous cost of unsuccessful clinical trials and will allow selection of therapeutics that will have greater potential of success in the clinic. Therefore, the utility of these models needs to be considered in evaluation of anti-cancer agents that modulate apoptosis.
Conclusion
Evasion of apoptosis has been established as a hallmark of cancer and the impaired apoptotic signaling characteristic of cancer cells is frequently linked to tumor development and progression as well as resistance to treatment. Intense research efforts to elucidate the mechanisms by which cancer cells are protected from apoptosis have led to the identification of several alterations that result in the aberrant activity of key apoptosis regulators, including BCL-2 family proteins, IAPs, and c-FLIP. These insights into the dysregulation of apoptosis in cancer have fuelled the pursuit of various targeted therapeutic strategies for reactivating apoptosis, which hold great promise for translation into anti-cancer therapeutics that will significantly benefit cancer patients. Nevertheless, the heterogeneity among tumors, even of the same type, necessitates a continued effort to further investigate mechanisms of apoptosis dysregulation in distinct cancer cell types. We are confident that future advances in our understanding of the underlying cellular defects that are responsible for cancer cell resistance to apoptosis and use of more stringent preclinical models will lead to more effective cancer treatments.
Acknowledgements
We thank Jane Hayward, media specialist, Beth Israel Deaconess Medical Center, for assistance with figure preparation and Susan Glueck for editing this manuscript. J.P. is supported by a T32, HL007893. O.B. received a fellowship from the Lady TATA Memorial Trust, London, U.K. This work was also supported by the National Institutes of Health grants CA105306, CA131664, HL080192 awarded to R.K. R.K. is the recipient of a Melanoma Research Alliance Award and Harry Lloyd Trust Award.
Footnotes
Published as part of an Integrative Biology themed issue in honour of Mina J. Bissell: Guest Editor Mary Helen Barcellos-Hoff.
References
- 1.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat. Rev. Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 2.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 5.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability-an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 6.Fulda S. Evasion of apoptosis as a cellular stress response in cancer. Int. J. Cell. Biol. 2010;2010:370835. doi: 10.1155/2010/370835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plati J, Bucur O, Khosravi-Far R. Dysregulation of apoptotic signaling in cancer: Molecular mechanisms and therapeutic opportunities. J. Cell. Biochem. 2008;104,:1124–1149. doi: 10.1002/jcb.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gimenez-Bonafe P, Tortosa A, Perez-Tomas R. Overcoming drug resistance by enhancing apoptosis of tumor cells. Current Cancer Drug Targets. 2009;9:320–340. doi: 10.2174/156800909788166600. [DOI] [PubMed] [Google Scholar]
- 9.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin. Cancer Res. 2009;15:1126–1132. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson TR, Johnston PG, Longley DB. Anti-apoptotic mechanisms of drug resistance in cancer. Current Cancer Drug Targets. 2009;9:307–319. doi: 10.2174/156800909788166547. [DOI] [PubMed] [Google Scholar]
- 11.Kruyt FA. TRAIL and cancer therapy. Cancer Lett. 2008;263:14–25. doi: 10.1016/j.canlet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Mahalingam D, Szegezdi E, Keane M, Jong S, Samali A. TRAIL receptor signalling and modulation: Are we on the right TRAIL? Cancer Treat. Rev. 2009;35:280–288. doi: 10.1016/j.ctrv.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Guicciardi ME, Gores GJ. Life and death by death receptors. FASEB J. 2009;23:1625–1637. doi: 10.1096/fj.08-111005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brumatti G, Salmanidis M, Ekert PG. Crossing paths: interactions between the cell death machinery and growth factor survival signals. Cell. Mol. Life Sci. 2010;67:1619–1630. doi: 10.1007/s00018-010-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duronio V. The life of a cell: apoptosis regulation by the PI3K/PKB pathway. Biochem. J. 2008;415:333–344. doi: 10.1042/BJ20081056. [DOI] [PubMed] [Google Scholar]
- 16.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bitomsky N, Hofmann TG. Apoptosis and autophagy: Regulation of apoptosis by DNA damage signalling-roles of p53, p73 and HIPK2. FEBS J. 2009;276:6074–6083. doi: 10.1111/j.1742-4658.2009.07331.x. [DOI] [PubMed] [Google Scholar]
- 18.Bagnoli M, Canevari S, Mezzanzanica D. Cellular FLICE-inhibitory protein (c-FLIP) signalling: a key regulator of receptor-mediated apoptosis in physiologic context and in cancer. Int. J. Biochem. Cell Biol. 2010;42:210–213. doi: 10.1016/j.biocel.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Smith AJ, Karpova Y, D’Agostino R, Jr, Willingham M, Kulik G. Expression of the Bcl-2 protein BAD promotes prostate cancer growth. PLoS One. 2009;4:e6224. doi: 10.1371/journal.pone.0006224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 21.Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 25.Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- 26.Khosravi-Far R, Esposti MD. Death receptor signals to mitochondria. Cancer Biol. Ther. 2004;3:1051–1057. doi: 10.4161/cbt.3.11.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bucur O, Nat R, Cretoiu D, Popescu LM. Phagocytosis of apoptotic cells by microglia in vitro. J. Cell. Mol. Med. 2001;5:438–441. doi: 10.1111/j.1582-4934.2001.tb00181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon GM, Dix MM, Cravatt BF. Comparative assessment of large-scale proteomic studies of apoptotic proteolysis. ACS Chem. Biol. 2009;4:401–408. doi: 10.1021/cb900082q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol. Ther. 2005;4:139–163. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- 30.Bouchier-Hayes L, Lartigue L, Newmeyer DD. Mitochondria: pharmacological manipulation of cell death. J. Clin. Invest. 2005;115:2640–2647. doi: 10.1172/JCI26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroemer G. Introduction: mitochondrial control of apoptosis. Biochimie. 2002;84:103–104. doi: 10.1016/s0300-9084(02)01382-2. [DOI] [PubMed] [Google Scholar]
- 32.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 33.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 34.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 35.Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol. Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 36.Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, Green DR, Martin SJ. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giam M, Huang DC, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene. 2008;27 Suppl 1:S128–S136. doi: 10.1038/onc.2009.50. [DOI] [PubMed] [Google Scholar]
- 38.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danial NN. BCL-2 family proteins: critical checkpoints of apoptotic cell death. Clin. Cancer Res. 2007;13:7254–7263. doi: 10.1158/1078-0432.CCR-07-1598. [DOI] [PubMed] [Google Scholar]
- 40.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 41.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 42.Hinds MG, Day CL. Regulation of apoptosis: uncovering the binding determinants. Curr. Opin. Struct. Biol. 2005;15:690–699. doi: 10.1016/j.sbi.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Dewson G, Kluck RM. Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis. J. Cell Sci. 2009;122:2801–2808. doi: 10.1242/jcs.038166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guillemin Y, Lopez J, Gimenez D, Fuertes G, Valero JG, Blum L, Gonzalo P, Salgado J, Girard-Egrot H, Aouacheria H. Active fragments from pro- and antiapoptotic BCL-2 proteins have distinct membrane behavior reflecting their functional divergence. PLoS One. 5:e9066. doi: 10.1371/journal.pone.0009066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Billen LP, Kokoski CL, Lovell JF, Leber B, Andrews DW. Bcl-XL inhibits membrane permeabilization by competing with Bax. PLoS Biol. 2008;6:e147. doi: 10.1371/journal.pbio.0060147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bleicken S, Classen M, Padmavathi PV, Ishikawa T, Zeth K, Steinhoff HJ, Bordignon E. Molecular details of Bax activation, oligomerization, and membrane insertion. J. Biol. Chem. 2010;285:6636–6647. doi: 10.1074/jbc.M109.081539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dewson G, Kratina T, Sim HW, Puthalakath H, Adams JM, Colman PM, Kluck RM. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol. Cell. 2008;30:369–380. doi: 10.1016/j.molcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluck RM, Adams JM, Huang DC. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 50.Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, Walensky LD. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Gallenne T, Gautier F, Oliver L, Hervouet E, Noel B, Hickman JA, Geneste O, Cartron PF, Vallette FM, Manon S, Juin P. Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J. Cell Biol. 2009;185:279–290. doi: 10.1083/jcb.200809153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letai A. Puma strikes Bax. J. Cell Biol. 2009;185:189–191. doi: 10.1083/jcb.200903134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leber B, Lin J, Andrews DW. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fulda S. Caspase-8 in cancer biology and therapy. Cancer Lett. 2009;281:128–133. doi: 10.1016/j.canlet.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 57.Boldin MP, Mett IL, Varfolomeev EE, Chumakov I, Shemer-Avni Y, Camonis JH, Wallach D. Self-association of the “death domains” of the p55 tumor necrosis factor (TNF) receptor and Fas/APO1 prompts signaling for TNF and Fas/APO1 effects. J. Biol. Chem. 1995;270:387–391. doi: 10.1074/jbc.270.1.387. [DOI] [PubMed] [Google Scholar]
- 58.Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene. 2003;22:8543–8567. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- 59.Mazumder S, Plesca D, Almasan A. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods Mol. Biol. 2008;414:13–21. doi: 10.1007/978-1-59745-339-4_2. [DOI] [PubMed] [Google Scholar]
- 60.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 61.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 62.Engels IH, Stepczynska A, Stroh C, Lauber K, Berg C, Schwenzer R, Wajant H, Janicke RU, Porter AG, Belka C, Gregor M, Schulze-Osthoff K, Wesselborg S. Caspase-8/FLICE functions as an executioner caspase in anticancer drug-induced apoptosis. Oncogene. 2000;19:4563–4573. doi: 10.1038/sj.onc.1203824. [DOI] [PubMed] [Google Scholar]
- 63.Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, Debatin KM, Krammer PH, Peter ME. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ozoren N, El-Deiry WS. Defining characteristics of Types I and II apoptotic cells in response to TRAIL. Neoplasia. 2002;4:551–557. doi: 10.1038/sj.neo.7900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 66.Ghiotto F, Fais F, Bruno S. BH3-only proteins: the death-puppeteer’s wires. Cytometry A. 2010;77:11–21. doi: 10.1002/cyto.a.20819. [DOI] [PubMed] [Google Scholar]
- 67.Billen LP, Shamas-Din A, Andrews DW. Bid: a Bax-like BH3 protein. Oncogene. 2008;27 Suppl 1:S93–S104. doi: 10.1038/onc.2009.47. [DOI] [PubMed] [Google Scholar]
- 68.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol. Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 70.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Merino D, Giam M, Hughes PD, Siggs OM, Heger K, O’Reilly LA, Adams JM, Strasser A, Lee EF, Fairlie WD, Bouillet P. The role of BH3-only protein Bim extends beyond inhibiting Bcl-2-like prosurvival proteins. J. Cell Biol. 2009;186:355–362. doi: 10.1083/jcb.200905153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 73.Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, Beltran E, Agirre X, Marugan I, Marín M, Rosenwald A, Sugimoto KJ, Wheat LM, Karran EL, García JF, Sanchez L, Prosper F, Staudt LM, Pinkel D, Dyer MJ, Martinez-Climent JA. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–280. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- 74.Pompeia C, Hodge DR, Plass C, Wu YZ, Marquez VE, Kelley JA, Farrar WL. Microarray analysis of epigenetic silencing of gene expression in the KAS-6/1 multiple myeloma cell line. Cancer Res. 2004;64:3465–3473. doi: 10.1158/0008-5472.CAN-03-3970. [DOI] [PubMed] [Google Scholar]
- 75.Sturm I, Stephan C, Gillissen B, Siebert R, Janz M, Radetzki S, Jung K, Loening S, Dorken B, Daniel PT. Loss of the tissue-specific proapoptotic BH3-only protein Nbk/Bik is a unifying feature of renal cell carcinoma. Cell Death Differ. 2006;13:619–627. doi: 10.1038/sj.cdd.4401782. [DOI] [PubMed] [Google Scholar]
- 76.Zantl N, Weirich G, Zall H, Seiffert BM, Fischer SF, Kirschnek S, Hartmann C, Fritsch RM, Gillissen B, Daniel PT, Hacker G. Frequent loss of expression of the pro-apoptotic protein Bim in renal cell carcinoma: evidence for contribution to apoptosis resistance. Oncogene. 2007;26:7038–7048. doi: 10.1038/sj.onc.1210510. [DOI] [PubMed] [Google Scholar]
- 77.Buggins AG, Pepper CJ. The role of Bcl-2 family proteins in chronic lymphocytic leukaemia. Leuk. Res. 2010;34:837–842. doi: 10.1016/j.leukres.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 78.Miquel C, Borrini F, Grandjouan S, Auperin A, Viguier J, Velasco V, Duvillard P, Praz F, Sabourin JC. Role of bax mutations in apoptosis in colorectal cancers with microsatellite instability. Am. J. Clin. Pathol. 2005;123:562–570. doi: 10.1309/JQ2X-3RV3-L8F9-TGYW. [DOI] [PubMed] [Google Scholar]
- 79.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 80.Yashiro M, Hirakawa K, Boland CR. Mutations in TGFbeta-RII and BAX mediate tumor progression in the later stages of colorectal cancer with microsatellite instability. BMC Cancer. 2010;10:303. doi: 10.1186/1471-2407-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, Morishima Y, Nakamura S, Seto M. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 82.Richter-Larrea JA, Robles EF, Fresquet V, Beltran E, Rullan AJ, Agirre X, Calasanz MJ, Panizo C, Richter JA, Hernandez JM, Roman-Gomez J, Prosper F, Martinez-Climent JA. Reversion of epigenetically-mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood. 2010;116:2531–2542. doi: 10.1182/blood-2010-02-268003. [DOI] [PubMed] [Google Scholar]
- 83.Fritsche P, Seidler B, Schüler S, Schnieke A, Göttlicher M, Schmid RM, Saur D, Schneider G. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut. 2009;58:1399–1409. doi: 10.1136/gut.2009.180711. [DOI] [PubMed] [Google Scholar]
- 84.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 85.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garofalo M, Croce CM. microRNAs: Master Regulators as Potential Therapeutics in Cancer. Annu. Rev. Pharmacol. Toxicol. 2010;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 87.Garofalo M, Condorelli G, Croce CM, Condorelli G. MicroRNAs as regulators of death receptors signaling. Cell Death Differ. 2010;17:300–308. doi: 10.1038/cdd.2009.105. [DOI] [PubMed] [Google Scholar]
- 88.Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J. Cell Physiol. 2010;223:289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- 89.Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, Peschle C, Fruci D. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002236. e2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 91.van Haaften G, Agami R. Tumorigenicity of the miR-17-92 cluster distilled. Genes Dev. 2010;24:1–4. doi: 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 93.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, Selaru FM, Hamilton JP, Yang J, Abraham JM, Mori Y, Meltzer SJ. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fulda S. Tumor resistance to apoptosis. Int. J. Cancer. 2009;124:511–515. doi: 10.1002/ijc.24064. [DOI] [PubMed] [Google Scholar]
- 95.Colombel M, Symmans F, Gil S, O’Toole KM, Chopin D, Benson M, Olsson CA, Korsmeyer S, Buttyan R. Detection of the apoptosis-suppressing oncoprotein bc1-2 in hormone-refractory human prostate cancers. Am. J. Pathol. 1993;143:390–400. [PMC free article] [PubMed] [Google Scholar]
- 96.Kitagawa Y, Wong F, Lo P, Elliott M, Verburgt LM, Hogg JC, Daya M. Overexpression of Bcl-2 and mutations in p53 and K-ras in resected human non-small cell lung cancers. Am. J. Respir. Cell Mol. Biol. 1996;15:45–54. doi: 10.1165/ajrcmb.15.1.8679221. [DOI] [PubMed] [Google Scholar]
- 97.Ramsay JA, From L, Kahn HJ. bcl-2 protein expression in melanocytic neoplasms of the skin. Mod. Pathol. 1995;8:150–154. [PubMed] [Google Scholar]
- 98.Venditti A, Del Poeta G, Maurillo L, Buccisano F, Del Principe MI, Mazzone C, Tamburini A, Cox C, Panetta P, Neri B, Ottaviani L, Amadori S. Combined analysis of bcl-2 and MDR1 proteins in 256 cases of acute myeloid leukemia. Haematologica. 2004;89:934–939. [PubMed] [Google Scholar]
- 99.Abramson JS, Shipp MA. Advances in the biology and therapy of diffuse large B-cell lymphoma: moving toward a molecularly targeted approach. Blood. 2005;106:1164–1174. doi: 10.1182/blood-2005-02-0687. [DOI] [PubMed] [Google Scholar]
- 100.Deininger MH, Weller M, Streffer J, Meyermann R. Antiapoptotic Bcl-2 family protein expression increases with progression of oligodendroglioma. Cancer. 1999;86:1832–1839. doi: 10.1002/(sici)1097-0142(19991101)86:9<1832::aid-cncr27>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 101.Reed JC. Bcl-2-family proteins and hematologic malignancies: history and future prospects. Blood. 2008;111:3322–3330. doi: 10.1182/blood-2007-09-078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Campana D, Coustan-Smith E, Manabe A, Buschle M, Raimondi SC, Behm FG, Ashmun R, Arico M, Biondi A, Pui CH. Prolonged survival of B-lineage acute lymphoblastic leukemia cells is accompanied by overexpression of bcl-2 protein. Blood. 1993;81:1025–1031. [PubMed] [Google Scholar]
- 103.Foreman KE, Wrone-Smith T, Boise LH, Thompson CB, Polverini PJ, Simonian PL, Nunez G, Nickoloff BJ. Kaposi’s sarcoma tumor cells preferentially express Bcl-xL. Am. J. Pathol. 1996;149:795–803. [PMC free article] [PubMed] [Google Scholar]
- 104.Krajewska M, Moss SF, Krajewski S, Song K, Holt PR, Reed JC. Elevated expression of Bcl-X and reduced Bak in primary colorectal adenocarcinomas. Cancer Res. 1996;56:2422–2427. [PubMed] [Google Scholar]
- 105.Tu Y, Renner S, Xu F, Fleishman A, Taylor J, Weisz J, Vescio R, Rettig M, Berenson J, Krajewski S, Reed JC, Lichtenstein A. BCL-X expression in multiple myeloma: possible indicator of chemoresistance. Cancer Res. 1998;58:256–262. [PubMed] [Google Scholar]
- 106.Bai J, Sui J, Demirjian A, Vollmer CM, Jr, Marasco W, Callery MP. Predominant Bcl-XL knockdown disables antiapoptotic mechanisms: tumor necrosis factor-related apoptosis-inducing ligand-based triple chemotherapy overcomes chemoresistance in pancreatic cancer cells in vitro. Cancer Res. 2005;65:2344–2352. doi: 10.1158/0008-5472.CAN-04-3502. [DOI] [PubMed] [Google Scholar]
- 107.Watanabe J, Kushihata F, Honda K, Mominoki K, Matsuda S, Kobayashi N. Bcl-xL overexpression in human hepatocellular carcinoma. Int. J. Oncol. 2002;21:515–519. [PubMed] [Google Scholar]
- 108.Sharma J, Srinivasan R, Majumdar S, Mir S, Radotra BD, Wig JD. Bcl-XL protein levels determine apoptotic index in pancreatic carcinoma. Pancreas. 2005;30:337–342. doi: 10.1097/01.mpa.0000160282.64451.f1. [DOI] [PubMed] [Google Scholar]
- 109.Castilla C, Congregado B, Chinchon D, Torrubia FJ, Japon MA, Saez C. Bcl-xL is overexpressed in hormone-resistant prostate cancer and promotes survival of LNCaP cells via interaction with proapoptotic Bak. Endocrinology. 2006;147:4960–4967. doi: 10.1210/en.2006-0502. [DOI] [PubMed] [Google Scholar]
- 110.Huang Z, Lei X, Zhong M, Zhu B, Tang S, Liao D. Bcl-2 small interfering RNA sensitizes cisplatin-resistant human lung adenocarcinoma A549/DDP cell to cisplatin and diallyl disulfide. Acta Biochim. Biophys. Sin. 2007;39:835–843. doi: 10.1111/j.1745-7270.2007.00356.x. [DOI] [PubMed] [Google Scholar]
- 111.Lei X, Huang Z, Zhong M, Zhu B, Tang S, Liao D. Bcl-XL small interfering RNA sensitizes cisplatin-resistant human lung adenocarcinoma cells. Acta Biochim. Biophys. Sin. 2007;39:344–350. doi: 10.1111/j.1745-7270.2007.00286.x. [DOI] [PubMed] [Google Scholar]
- 112.Meng Y, Tang W, Dai Y, Wu X, Liu M, Ji Q, Ji M, Pienta K, Lawrence T, Xu L. Natural BH3 mimetic (−)-gossypol chemosensitizes human prostate cancer via Bcl-xL inhibition accompanied by increase of Puma and Noxa. Mol. Cancer Ther. 2008;7:2192–2202. doi: 10.1158/1535-7163.MCT-08-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wacheck V, Losert D, Gunsberg P, Vornlocher HP, Hadwiger P, Geick A, Pehamberger H, Muller M, Jansen B. Small interfering RNA targeting bcl-2 sensitizes malignant melanoma. Oligonucleotides. 2003;13:393–400. doi: 10.1089/154545703322617078. [DOI] [PubMed] [Google Scholar]
- 114.Mason KD, Vandenberg CJ, Scott CL, Wei AH, Cory S, Huang DC, Roberts AW. In vivo efficacy of the Bcl-2 antagonist ABT-737 against aggressive Myc-driven lymphomas. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17961–17966. doi: 10.1073/pnas.0809957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hetschko H, Voss V, Horn S, Seifert V, Prehn JH, Kogel D. Pharmacological inhibition of Bcl-2 family members reactivates TRAIL-induced apoptosis in malignant glioma. J. Neurooncol. 2007;86:265–272. doi: 10.1007/s11060-007-9472-6. [DOI] [PubMed] [Google Scholar]
- 116.Yip KW, Reed JC. Bcl-2 family proteins and cancer. Oncogene. 2008;27:6398–6406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 117.Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- 118.Monni O, Joensuu H, Franssila K, Klefstrom J, Alitalo K, Knuutila S. BCL2 overexpression associated with chromosomal amplification in diffuse large B-cell lymphoma. Blood. 1997;90:1168–1174. [PubMed] [Google Scholar]
- 119.Olejniczak ET, Van Sant C, Anderson MG, Wang G, Tahir SK, Sauter G, Lesniewski R, Semizarov D. Integrative genomic analysis of small-cell lung carcinoma reveals correlates of sensitivity to bcl-2 antagonists and uncovers novel chromosomal gains. Mol. Cancer Res. 2007;5:331–339. doi: 10.1158/1541-7786.MCR-06-0367. [DOI] [PubMed] [Google Scholar]
- 120.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 121.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, Alder H, Volinia S, Rassenti L, Liu X, Liu CG, Kipps TJ, Negrini M, Croce CM. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 125.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 127.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, Fearon ER, Lawrence TS, Xu L. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006816. e6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, Andreeff M, Croce CM. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 132.LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 133.Mace PD, Shirley S, Day CL. Assembling the building blocks: structure and function of inhibitor of apoptosis proteins. Cell Death Differ. 2010;17:46–53. doi: 10.1038/cdd.2009.45. [DOI] [PubMed] [Google Scholar]
- 134.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat. Rev. Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 135.Eckelman BP, Salvesen GS. The human anti-apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J. Biol. Chem. 2006;281:3254–3260. doi: 10.1074/jbc.M510863200. [DOI] [PubMed] [Google Scholar]
- 136.Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discovery. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 139.Galban S, Duckett CS. XIAP as a ubiquitin ligase in cellular signaling. Cell Death Differ. 2010;17:54–60. doi: 10.1038/cdd.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, Yamada G, Akira S, Matsumoto K, Ghosh S. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Winsauer G, Resch U, Hofer-Warbinek R, Schichl YM, de Martin R. XIAP regulates bi-phasic NF-kappaB induction involving physical interaction and ubiquitination of MEKK2. Cell. Signalling. 2008;20:2107–2112. doi: 10.1016/j.cellsig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 142.Bianchi K, Meier P. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol. Cell. 2009;36:736–742. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 143.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 144.Fulda S. Inhibitor of apoptosis proteins in hematological malignancies. Leukemia. 2009;23:467–476. doi: 10.1038/leu.2008.329. [DOI] [PubMed] [Google Scholar]
- 145.Fulda S. Targeting inhibitor of apoptosis proteins (IAPs) for cancer therapy. Anticancer Agents Med. Chem. 2008;8:533–539. doi: 10.2174/187152008784533107. [DOI] [PubMed] [Google Scholar]
- 146.Fong WG, Liston P, Rajcan-Separovic E, St Jean M, Craig C, Korneluk RG. Expression and genetic analysis of XIAP-associated factor 1 (XAF1) in cancer cell lines. Genomics. 2000;70:113–122. doi: 10.1006/geno.2000.6364. [DOI] [PubMed] [Google Scholar]
- 147.Kashkar H. X-linked Inhibitor of Apoptosis: A Chemoresistance Factor or a Hollow Promise. Clin. Cancer Res. 2010;16:4496–4502. doi: 10.1158/1078-0432.CCR-10-1664. [DOI] [PubMed] [Google Scholar]
- 148.Xiang G, Wen X, Wang H, Chen K, Liu H. Expression of X-linked inhibitor of apoptosis protein in human colorectal cancer and its correlation with prognosis. J. Surg. Oncol. 2009;100:708–712. doi: 10.1002/jso.21408. [DOI] [PubMed] [Google Scholar]
- 149.Augello C, Caruso L, Maggioni M, Donadon M, Montorsi M, Santambrogio R, Torzilli G, Vaira V, Pellegrini C, Roncalli M, Coggi G, Bosari S. Inhibitors of apoptosis proteins (IAPs) expression and their prognostic significance in hepatocellular carcinoma. BMC Cancer. 2009;9:125. doi: 10.1186/1471-2407-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Altieri DC. Survivin and IAP proteins in cell-death mechanisms. Biochem. J. 2010;430:199–205. doi: 10.1042/BJ20100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dohi T, Xia F, Altieri DC. Compartmentalized phosphorylation of IAP by protein kinase A regulates cytoprotection. Mol. Cell. 2007;27:17–28. doi: 10.1016/j.molcel.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. IAP regulation of metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A, Wu YZ, Rush LJ, Ross P, Molina JR, Otterson GA, Plass C. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum. Mol. Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- 154.Imoto I, Yang ZQ, Pimkhaokham A, Tsuda H, Shimada Y, Imamura M, Ohki M, Inazawa J. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- 155.Ma O, Cai WW, Zender L, Dayaram T, Shen J, Herron AJ, Lowe SW, Man TK, Lau CC, Donehower LA. MMP13, Birc2 (cIAP1), and Birc3 (cIAP2), amplified on chromosome 9, collaborate with p53 deficiency in mouse osteosarcoma progression. Cancer Res. 2009;69:2559–2567. doi: 10.1158/0008-5472.CAN-08-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Isaacson PG, Du MQ. Gastrointestinal lymphoma: where morphology meets molecular biology. J. Pathol. 2005;205:255–274. doi: 10.1002/path.1703. [DOI] [PubMed] [Google Scholar]
- 157.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, Braggio E, Henry T, Zhu YX, Fogle H, Price-Troska T, Ahmann G, Mancini C, Brents LA, Kumar S, Greipp P, Dispenzieri A, Bryant B, Mulligan G, Bruhn L, Barrett M, Valdez R, Trent J, Stewart AK, Carpten J, Bergsagel PL. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN. c-FLIPR, a new regulator of death receptor-induced apoptosis. J. Biol. Chem. 2005;280:14507–14513. doi: 10.1074/jbc.M414425200. [DOI] [PubMed] [Google Scholar]
- 159.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C, Rimoldi D, French LE, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 160.Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang X. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yu JW, Jeffrey PD, Shi Y. Mechanism of procaspase-8 activation by c-FLIPL. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8169–8174. doi: 10.1073/pnas.0812453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yu JW, Shi Y. FLIP and the death effector domain family. Oncogene. 2008;27:6216–6227. doi: 10.1038/onc.2008.299. [DOI] [PubMed] [Google Scholar]
- 163.Safa AR, Day TW, Wu CH. Cellular FLICE-like inhibitory protein (C-FLIP): a novel target for cancer therapy. Current Cancer Drug Targets. 2008;8:37–46. doi: 10.2174/156800908783497087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Du X, Bao G, He X, Zhao H, Yu F, Qiao Q, Lu J, Ma Q. Expression and biological significance of c-FLIP in human hepatocellular carcinomas. Journal of Experimental & Clinical Cancer Research. 2009;28:24. doi: 10.1186/1756-9966-28-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wilson TR, Redmond KM, McLaughlin KM, Crawford N, Gately K, O’Byrne K, Le-Clorrenec C, Holohan C, Fennell DA, Johnston PG, Longley DB. Procaspase 8 overexpression in non-small-cell lung cancer promotes apoptosis induced by FLIP silencing. Cell Death Differ. 2009;16:1352–1361. doi: 10.1038/cdd.2009.76. [DOI] [PubMed] [Google Scholar]
- 166.Chen HX, Liu YJ, Zhou XD, Luo RY. Expression of cellular FLICE/caspase-8 inhibitory protein is associated with malignant potential in endometrial carcinoma. Int. J. Gynecol. Cancer. 2005;15:663–670. doi: 10.1111/j.1525-1438.2005.00122.x. [DOI] [PubMed] [Google Scholar]
- 167.Korkolopoulou P, Saetta AA, Levidou G, Gigelou F, Lazaris A, Thymara I, Scliri M, Bousboukea K, Michalopoulos NV, Apostolikas N, Konstantinidou A, Tzivras M, Patsouris E. c-FLIP expression in colorectal carcinomas: association with Fas/FasL expression and prognostic implications. Histopathology. 2007;51:150–156. doi: 10.1111/j.1365-2559.2007.02723.x. [DOI] [PubMed] [Google Scholar]
- 168.Gao S, Wang H, Lee P, Melamed J, Li CX, Zhang F, Wu H, Zhou L, Wang Z. Androgen receptor and prostate apoptosis response factor-4 target the c-FLIP gene to determine survival and apoptosis in the prostate gland. J. Mol. Endocrinol. 2006;36:463–483. doi: 10.1677/jme.1.01991. [DOI] [PubMed] [Google Scholar]
- 169.Zhang X, Jin TG, Yang H, DeWolf WC, Khosravi-Far R, Olumi AF. Persistent c-FLIP(L) expression is necessary and sufficient to maintain resistance to tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in prostate cancer. Cancer Res. 2004;64:7086–7091. doi: 10.1158/0008-5472.CAN-04-1498. [DOI] [PubMed] [Google Scholar]
- 170.Zhang X, Zhang L, Yang H, Huang X, Otu H, Libermann TA, DeWolf WC, Khosravi-Far R, Olumi AF. c-Fos as a proapoptotic agent in TRAIL-induced apoptosis in prostate cancer cells. Cancer Res. 2007;67:9425–9434. doi: 10.1158/0008-5472.CAN-07-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Korkolopoulou P, Saetta AA, Levidou G, Gigelou F, Lazaris A, Thymara I, Scliri M, Bousboukea K, Michalopoulos NV, Apostolikas N, Konstantinidou A, Tzivras M, Patsouris E. c-FLIP expression in colorectal carcinomas: association with Fas/FasL expression and prognostic implications. Histopathology. 2007;51:150–156. doi: 10.1111/j.1365-2559.2007.02723.x. [DOI] [PubMed] [Google Scholar]
- 172.Song JH, Tse MC, Bellail A, Phuphanich S, Khuri F, Kneteman NM, Hao C. Lipid rafts and nonrafts mediate tumor necrosis factor related apoptosis-inducing ligand induced apoptotic and nonapoptotic signals in non small cell lung carcinoma cells. Cancer Res. 2007;67:6946–6955. doi: 10.1158/0008-5472.CAN-06-3896. [DOI] [PubMed] [Google Scholar]
- 173.Vilimanovich U, Bumbasirevic V. TRAIL induces proliferation of human glioma cells by c-FLIPL-mediated activation of ERK1/2. Cell. Mol. Life Sci. 2008;65:814–826. doi: 10.1007/s00018-008-7513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Quintavalle C, Incoronato M, Puca L, Acunzo M, Zanca C, Romano G, Garofalo M, Iaboni M, Croce CM, Condorelli G. c-FLIP(L) enhances anti-apoptotic Akt functions by modulation of Gsk3beta activity. Cell Death Differ. 2010;17:1908–1916. doi: 10.1038/cdd.2010.65. [DOI] [PubMed] [Google Scholar]
- 175.Okamoto K, Fujisawa J, Reth M, Yonehara S. Human T-cell leukemia virus type-I oncoprotein Tax inhibits Fas-mediated apoptosis by inducing cellular FLIP through activation of NF-kappaB. Genes Cells. 2006;11:177–191. doi: 10.1111/j.1365-2443.2006.00927.x. [DOI] [PubMed] [Google Scholar]
- 176.Kreuz S, Siegmund D, Scheurich P, Wajant H. NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling. Mol. Cell. Biol. 2001;21:3964–3973. doi: 10.1128/MCB.21.12.3964-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jani TS, DeVecchio J, Mazumdar T, Agyeman A, Houghton JA. Inhibition of NF-kappaB signaling by quinacrine is cytotoxic to human colon carcinoma cell lines and is synergistic in combination with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or oxaliplatin. J. Biol. Chem. 2010;285:19162–19172. doi: 10.1074/jbc.M109.091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cillessen SA, Meijer CJ, Notoya M, Ossenkoppele GJ, Oudejans JJ. Molecular targeted therapies for diffuse large B-cell lymphoma based on apoptosis profiles. J. Pathol. 2010;220:509–520. doi: 10.1002/path.2670. [DOI] [PubMed] [Google Scholar]
- 179.Patel MP, Masood A, Patel PS, Chanan-Khan AA. Targeting the Bcl-2. Curr. Opin. Oncol. 2009;21:516–523. doi: 10.1097/CCO.0b013e328331a7a4. [DOI] [PubMed] [Google Scholar]
- 180.Leibowitz B, Yu J. Mitochondrial signaling in cell death via the Bcl-2 family. Cancer Biol. Ther. 2010;9:417–422. doi: 10.4161/cbt.9.6.11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Flygare JA, Fairbrother WJ. Small-molecule pan-IAP antagonists: a patent review. Expert Opin. Ther. Pat. 20:251–267. doi: 10.1517/13543770903567077. [DOI] [PubMed] [Google Scholar]
- 182.Mannhold R, Fulda S, Carosati E. IAP antagonists: promising candidates for cancer therapy. Drug Discovery Today. 15:210–219. doi: 10.1016/j.drudis.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 183.Day TW, Safa AR. RNA interference in cancer: targeting the anti-apoptotic protein c-FLIP for drug discovery. Mini-Rev. Med. Chem. 2009;9:741–748. doi: 10.2174/138955709788452748. [DOI] [PubMed] [Google Scholar]
- 184.Mannhold R, Fulda S, Carosati E. IAP antagonists: promising candidates for cancer therapy. Drug Discovery Today. 2010;15:210–219. doi: 10.1016/j.drudis.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 185.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Baker M. RNA interference: Homing in on delivery. Nature. 2010;464:1225–1228. doi: 10.1038/4641225a. [DOI] [PubMed] [Google Scholar]
- 187.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol. Pharmaceutics. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 188.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ryan BM, O’Donovan N, Duffy MJ. Survivin: a new target for anti-cancer therapy. Cancer Treat. Rev. 2009;35:553–562. doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 190.Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, Thompson JD, Cheng JH, Bou Reslan H, Ho CC, Cao TC, Lee CV, Nannini MA, Fuh G, Carano RA, Koeppen H, Yu RX, Forrest WF, Plowman GD, Johnson L. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat. Biotechnol. 28:585–593. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 191.Singh M, Johnson L. Using genetically engineered mouse models of cancer to aid drug development: an industry perspective. Clin. Cancer Res. 2006;12:5312–5328. doi: 10.1158/1078-0432.CCR-06-0437. [DOI] [PubMed] [Google Scholar]
- 192.Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of disease: epithelial-mesenchymal transition–does cellular plasticity fuel neoplastic progression? Nat. Clin. Pract. Oncol. 2008;5:280–290. doi: 10.1038/ncponc1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Radisky D, Hagios C, Bissell MJ. Tumors are unique organs defined by abnormal signaling and context. Semin. Cancer Biol. 2001;11:87–95. doi: 10.1006/scbi.2000.0360. [DOI] [PubMed] [Google Scholar]
- 194.Mertsching H, Weimer M, Kersen S, Brunner H. Human skin equivalent as an alternative to animal testing. GMS Krankenhhyg Interdiszip. 2008;3 Doc11. [PMC free article] [PubMed] [Google Scholar]
- 195.Schmeichel KL, Weaver VM, Bissell MJ. Structural cues from the tissue microenvironment are essential determinants of the human mammary epithelial cell phenotype. Journal of Mammary Gland Biology and Neoplasia. 1998;3:201–213. doi: 10.1023/a:1018751124382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bissell MJ. Modelling molecular mechanisms of breast cancer and invasion: lessons from the normal gland. Biochem. Soc. Trans. 2007;35:18–22. doi: 10.1042/BST0350018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Roskelley CD, Srebrow A, Bissell MJ. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr. Opin. Cell Biol. 1995;7:736–747. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Weaver VM, Bissell MJ. Functional culture models to study mechanisms governing apoptosis in normal and malignant mammary epithelial cells. Journal of Mammary Gland Biology and Neoplasia. 1999;4:193–201. doi: 10.1023/a:1018781325716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Weaver VM, Fischer AH, Peterson OW, Bissell MJ. The importance of the microenvironment in breast cancer progression: recapitulation of mammary tumorigenesis using a unique human mammary epithelial cell model and a three-dimensional culture assay. Biochem. Cell Biol. 1996;74:833–851. doi: 10.1139/o96-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Werb Z, Sympson CJ, Alexander CM, Thomasset N, Lund LR, MacAuley A, Ashkenas J, Bissell MJ. Extracellular matrix remodeling and the regulation of epithelial-stromal interactions during differentiation and involution. Kidney Int. Suppl. 1996;54:S68–S74. [PMC free article] [PubMed] [Google Scholar]
- 201.Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, Bissell MJ. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]