Abstract
The oral cavity harbors several hundred different bacterial species that colonize both hard (teeth) and soft tissues, forming complex populations known as microbial biofilms. It is widely accepted that the phenotypic characteristics of bacteria grown in biofilms are substantially different from those grown in suspensions. Because biofilms are the natural habitat for the great majority of oral bacteria, including those contributing to oral diseases, a better understanding of the physiology of adherent populations is clearly needed to control oral microbes in health and disease. In this chapter, we use oral streptococci as examples for studying the physiology of oral biofilms.
Keywords: Biofilm, oral streptococci, Streptococcus, enzymatic assays, stress, production of polysaccharides
1. Introduction
Oral biofilms normally exist in dynamic equilibrium with host defenses and are important for preventing colonization by undesirable organisms (1, 2). However, changes in the composition and metabolic activities of biofilm communities that lead to increases in the proportion of pathogenic species can lead to oral diseases, including dental caries and periodontitis. Despite the importance of oral biofilms to health and disease, studies on the physiology and genetics of oral bacteria were primarily conducted using planktonic populations of bacteria. In recent years, the development of in vitro and in vivo biofilm methodologies to study sessile populations have demonstrated that there are many physiologic and molecular differences between planktonic and surface-bound bacteria, suggesting that the organisms can acquire a “biofilm phenotype” (2–4).
Starting with the premise that “all models are wrong, but some are useful,” a quote attributed to the British statistician George Box, there are a variety of in vitro systems to study oral streptococci biofilms. These include simple and economical models in which bacteria are cultivated in batch systems using different surfaces such as glass, plastic, or hydroxyapatite (HA), the latter being used as a surrogate of tooth enamel. These systems can give reproducible results and can be scaled up to provide sufficient biomass for physiologic and genetic studies. In addition to batch and static systems, the use of shear force in continuous flow systems is considered ideal for the analysis of the dynamics of cell attachment to surfaces and the initial stages of biofilm development. Yet another commonly used system is the so-called constant depth film fermentor in which a scraper intermittently passes over the grown biofilms in wells to achieve constant biofilm depth. Batch and continuous feed systems can also be used to generate more complex multispecies biofilms of known composition, or microcosms can be formed from starter biofilm samples from the body and subsequent cultivation of biofilms in vitro. Because of the natural heterogeneity of these more complex biofilms, the interpretation of the behavior of these populations is very challenging.
Here, we focus on batch-culture systems that our laboratories have routinely used for studying the physiology of oral biofilms, with a particular emphasis on Streptococcus mutans biofilms. One of the advantages of the models presented in this chapter is that multiple biofilms can be formed simultaneously, which provides significant benefit in establishing reproducibility of the data and reducing variance. In addition, test agents can be applied and removed from the system instantaneously allowing a tightly controlled substance exposure time. Moreover, biofilms formed on glass slides or HA are amenable to confocal and electron microscopy and can yield a quantity of bacterial biomass that is sufficient for enzymatic assays. Finally, these model systems can be easily adapted for studies with non-streptococcal species.
2. Materials
2.1. Biofilm Medium (BM)
Base medium (5) per liter
| K2HPO4 | 10 g |
| KH2PO4 | 2 g |
| (NH4)2SO4 | 1.3 g |
| NaCl | 2 g |
| MnCl2·4H2O | 0.02g |
| FeSO4·7H2O | 0.001 g |
| Casamino acids | 2 g |
Dissolve all components in deionized (Milli-Q) water. Autoclave (121°C for 20 min) and store at room temperature (seeNote 1, Note 2).
| 100× amino acid stock solution | per 100 mL |
| l-glutamate (l-glutamic acid) | 5 g |
| l-arginine·HCl | 2 g |
| l-cysteine·HCl | 2 g |
| l-tryptophan | 0.2 g |
Dissolve all components in Milli-Q water. Filter-sterilize (0.22 μm pore size) and store wrapped in aluminum foil (components are light-sensitive) at 4°C for up to 4 weeks.
| 100× vitamin stock solution | per 100 mL |
| Pyridoxine HCl | 240 mg |
| Nicotinic acid | 46 mg |
| Pantothenic acid | 24 mg |
| Riboflavin | 4 mg |
| Thiamine HCl | 1 mg |
| d-Biotin | 0.12 mg |
Dissolve all components with Milli-Q water. Filter-sterilize (0.22 μm pore size) and store wrapped in aluminum foil at 4°C.
| Final BM medium composition | per liter |
| Base medium | 950 mL |
| MgSO4·7H2O (0.1 g/mL stock) | 5 mL |
| CaCl2·2H2O (0.03 g/mL stock) | 5 mL |
| 100× vitamin stock | 10 mL |
| 100× amino acid stock | 10 mL |
| 1 M glucose or 0.5 M sucrose (see Note 3) | 20 mL |
Adjust mixed solution to pH 7 and filter-sterilize (0.22 μm filter). Use immediately or store wrapped in aluminum foil at 4°C for up to 1 week.
2.2. Tryptone-Yeast Extract (TY) Medium
per liter
| Bacto tryptone | 30 g |
| Bacto yeast extract | 5 g |
Dissolve in Milli-Q water, autoclave (121°C, 20 min), and adjust pH to 7 aseptically with NaOH. Add glucose or sucrose (20% stock solution) to a final concentration of 1% after autoclaving to avoid caramelization.
2.3. Low Molecular Weight (LMW) Medium
per liter
| Bacto tryptone | 25 g |
| Bacto yeast extract | 15 g |
Filter solution through a Millipore Prep/Scale-TFF cartridge (10 kDa cut-off) using a peristaltic pump. Add KH2PO4 (25 mM final concentration) and MgSO4 (4 mM final concentration) and adjust the pH to 7. Autoclave (121°C, 20 min). Add glucose or sucrose (20% stock solution) to a final concentration of 1%.
2.4. Adsorption Buffer (AB)
50 mM KCl, 1 mM potassium phosphate (0.35 mM K2HPO4 plus 0.65 mM KH2PO4), 1 mM CaCl2, 0.1 mM MgCl2. Adjust pH to 6.5. Store at room temperature.
2.5. Clarified Saliva
Collect 50 mL of whole saliva on ice from one donor. Mix saliva with AB buffer (1:1 ratio, v/v). Add 50 μL 0.1 M phenylmethyl-sulfonyl fluoride (PMSF, store at 4°C for up to 9 months). Centrifuge the mixture (5,500g, 4°C, 10 min). Collect supernatant (clarified whole saliva) and filter through a 0.22 μm PES low protein-binding filter.
2.6. Buffers
Glycine buffer. 0.1 M glycine adjusted to desired pH with 1 N HCl or 1 N KOH.
Phosphate buffer (pH 7): Mix 60 mL of 1 M K2HPO4 with 40 mL of 1 M KH2PO4. The pH of the mixed solution should be pH 7.
2.7. 2× ATPase Assay Buffer
100 mM Tris–maleate buffer (pH 7). Prepare a 200 mM stock solution of Trizma-maleate. Adjust 50 mL of stock solution to desired pH with 0.1 M NaOH. Make up to 100 mL with Milli-Q water.
2.8. Bencini Reagent
100 mM ZnCl2 plus 15 mM ammonium molybdate.
2.9. Salt Solution
50 mM KCl plus 1 mM MgCl2.
2.10. Other Materials or Equipment
96-well, flat-bottom microtiter plate (Costar, Corning Inc. USA) – required when saliva-coated plates are used for biofilm growth (see below).
One milliliter (1 mL) disposable cuvettes for spectrophotometric readings.
Camera for photographing biofilms.
Hydroxyapatite disks with a surface area of (Clarkson Chromatography Products, Inc., South Williamsport, PA, USA) – this is required for growing biofilms on a hydroxyapatite matrix.
24-well tissue culture plate (Corning, NY, USA).
Ultrasonic bath.
14-mL (Falcon) centrifuge tubes.
50 mL conical tubes.
Sonicator (e.g., Branson Sonifier 150 or equivalent).
pH probe to measure pH changes in broth media.
Dissolved oxygen meter (e.g., VWR Model 4000).
Acid-washed 0.1 mm diameter glass beads (Biospec, Bartlesville, OK).
Beadbeater (e.g., Biospec, Bartlesville, OK) for homogenizing cells.
Vacuum concentrator (e.g., SpeedVac).
A desiccator containing phosphorus pentoxide (P2O5) or Drierite®. (under vacuum) for drying samples.
2.11. Other Chemicals or Media
0.1% crystal violet: Dissolve 0.1 g crystal violet in 100 mL deionized water.
33% acetic acid: To 33 mL glacial acetic acid, add deionized water to make up to 100 mL.
0.89% NaCl (filter-sterilized or autoclaved)
Brain heart infusion (BHI) medium (per liter): 37.5 g broth powder. Autoclave for 20 min at 121°C to sterilize. If solid medium is required, add agar to 15 g/L.
Toluene–acetone (1:9 ratio).
20% tricarboxylic acid (TCA).
100 mM adenosine triphosphate (ATP), pH 7.0.
2 mM NADH for NADH assays.
Bovine liver catalase (50 mg/mL stock) for NADH assays.
0.2% iodine in 2% potassium iodide solution – required for measuring intracellular polysaccharides.
3. Methods
3.1. Biofilm Growth
3.1.1. Quantitative Growth of Biofilms on Microtiter Plates
This method is particularly useful to assess the ability of different strains to form biofilms (seeNote 4).
Prepare overnight cultures, in triplicates, in 5 mL of TY plus 1% glucose [or Brain Heart Infusion (BHI)] broth and incubate at 37°C in a 5% CO2 atmosphere.
Transfer 100 μL of the overnight culture to a tube containing 5 mL of pre-warmed fresh TY (or BHI) medium and incubate at 37°C in a 5% CO2 atmosphere to an optical density at 600 nm (OD600) of 0.5. Chill on ice and keep cells on ice until the last culture reached the desired OD600.
Prepare fresh BM containing the desired amount of carbohydrate and pre-warm media at 37°C for 1 h.
Optional. Coat the wells of a 96-well, flat-bottom microtiter plate (Costar 3595, Corning Inc. USA) with 50 μL of clarified saliva for 1 h at 37°C. Remove unbound saliva by blotting the plate on a clean absorbent paper.
Dilute cultures 1:100 in fresh BM containing the desired carbohydrate source. Dispense 200 μL of each diluted culture into six wells. Wells containing uninoculated growth medium should serve as negative controls.
Incubate plate for 24 h at 37°C in a 5% CO2 atmosphere without agitation.
Optional. To measure total growth yield of each strain, remove both planktonic and biofilm cells by scraping the bottom of one well using a pipette tip. Transfer to a 1 mL cuvette. Blank with one of the cuvettes containing uninoculated medium.
Blot the plate on a paper towel to remove culture media. To remove loosely bound cells, carefully immerse the microtiter plate in a large dish with distilled water. Blot the plate on a paper towel. Repeat this step twice.
Add 50 μL of 0.1% crystal violet to the test wells, including the negative control wells. Incubate for 15 min.
Repeat step 8.
Air dry plate and photograph.
Add 200 μL of 33% acetic acid solution to the wells. Leave for 10 min. Keep the plate covered. Transfer the acetic acid solution to a 1 mL cuvette (seeNote 5).
Bring the volume of all cuvettes to 1 mL using Milli-Q water (including controls). Mix well.
For biofilm growth, read at a wavelength of 575 nm (OD575). Blank with a cuvette containing the solution from a crystal violet-stained uninoculated well.
3.1.2. Growth of Biofilms in Gram-Staining Boxes
This method is excellent for experiments that normally require a large bacterial biomass, particularly enzymatic assays.
Prepare overnight cultures in 10 mL of TY broth and incubate overnight at 37°C in a 5% CO2 atmosphere.
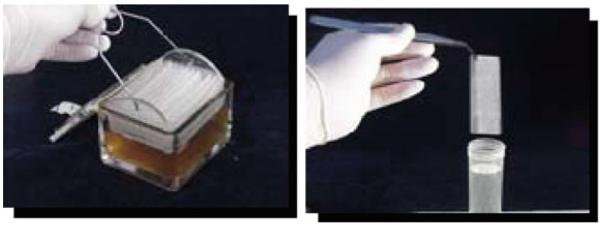
Autoclave a Gram-staining box (see Fig. 7.1) containing a slide holder basket and the desired number of microscope glass slides.
Fill the sterile Gram-staining box to completely cover the slides with TY broth containing 1% (w/v) of the desired (tested) carbohydrate.
Add 10 mL of the overnight culture and incubate for 48 h at 37°C in a 5% CO2 atmosphere.
Aseptically transfer the slide holder to another sterile Gram-staining box filled with medium (see Fig. 7.1). From now on, repeat this step once a day for 4–5 days (seeNote 6).
Fig. 7.1.
Typical Streptococcus mutans mature biofilm grown on glass slides placed in a Gram-staining box using TY medium supplemented with 1% sucrose.
3.1.3. Growth and Processing of Biofilms on Saliva-Coated Hydroxyapatite (HA) Disks
This method uses hydroxyapatite disks coated with saliva (mimicking the presence of salivary pellicle), placed in a vertical position.
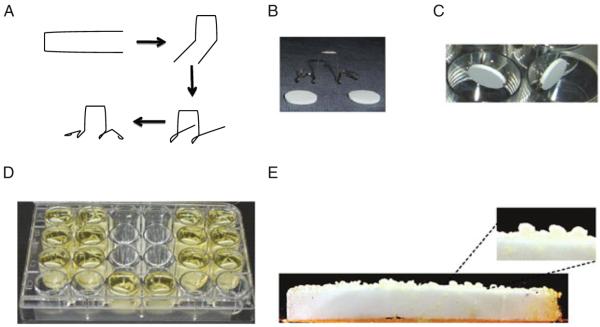
Select HA disks and place it in the custom-made holder (see Fig. 7.2).
Place the disks vertically (attached in the holder) into a 24-well culture plate. Pipette clarified saliva into wells until disks are submerged (approximately 2.8 mL saliva per well).
Incubate disks in saliva at 37°C for 1 h on an orbital shaker. Remove disks, shake off excess saliva, and dip-wash once in sterile AB buffer. The saliva-coated HA (sHA) disks are now ready for use.
Prepare overnight culture in 5 mL of LMW medium containing 1% glucose (LMWG). Dilute overnight culture 1:20 in LMWG and grow the cells at 37°C in 5% CO2 to an OD600 of 0.5.
Dilute culture 1:250 in LMW containing 1% sucrose (LMWS: 3 mL of medium per disk), which will provide approximately 5×106 colony-forming units (CFU) per mL. Transfer 2.8 mL of the inoculated medium into a 24-well tissue culture plate. Place a freshly prepared sHA disk into each well. Check the disk position (should be vertical) and the disks should be completely submerged. Incubate the plate for 24 h at 37°C in 5% CO2.
After 24 h incubation, transfer each of the sHA disks to a new plate containing fresh LMWS. Repeat this procedure daily for 4–6 days. Biofilms are considered mature after day 4.
Biofilm processing: Dip-wash the mature biofilms in 0.89% NaCl solution three times. Carefully release each disk from the holder and drop it into a sterile glass tube containing 1 mL of sterile 0.89% NaCl. Pipette an additional 1 mL of 0.89% NaCl to each tube. Place the tubes in an ultrasonic bath for 10 min.
Remove each disk from its tube using a spatula and check if all biomass has been removed.
Transfer suspension of glass tubes to 14-mL sterile centrifuge tubes. Wash each glass tube with 1 mL of sterile 0.89% NaCl using a pipettor and transfer the content to the centrifuge tube. Repeat this procedure two more times. Final volume of cell suspension should be approximately 5 mL.
Place tubes on ice. Immerse the tip of the probe of a sonicator at least 1 in. deep into the solution (without touching the bottom or the side walls). Sonicate the samples with three 30-s pulses at an output of 7 W.
The biofilm homogenate can be used for several assays, including determination of biomass (dry weight), protein content, and extracellular and intracellular polysaccharide composition (seeSection 3.7 below).
Fig. 7.2.
Growth of biofilm on saliva-coated hydroxyapatite (sHA) disks. A Schematic showing how to make the disk holder using orthodontic wire, B hydroxyapatite disks and custom-made holder, C sHA disks placed in a vertical position inside the wells of a microtiter plate, D growth of biofilms using LMWS medium, e typical 5-day-old biofilm formed on sHA disk.
3.2. Acid-Mediated Killing
Since most microbes in nature grow in biofilms, the use of biofilms for assessing cidal action is generally more appropriate than the use of suspensions. The example presented here is for acid-mediated killing. This assay can be performed with intact biofilms grown on the surface of microtiter plates for 24–48 h, glass slides, or HA disks. Below we describe a standard protocol using biofilms grown on microtiter plates.
Grow biofilms for 24 h using BM medium containing the desired sugar source as described in Section 3.1.1 (seeNote 7).
Blot the plate on absorbent paper, e.g., paper towel, to remove culture media.
Trying not to disturb the biofilm, slowly add 200 μL of 0.1 M glycine buffer adjusted to pH 2.7 to each well (see Note 8). The control biofilm group should receive 200 μL of 0.1 M glycine buffer pH 7 and processed in the first and last time points.
At each desired time point, remove biofilm by scraping the bottom with a pipette tip and transfer cell suspension into a microfuge tube and disperse cells either by vortexing at maximum speed or by sonication for 30 s.
Prepare serial dilutions in sterile 1% Difco peptone broth of dispersed cells and plate dilutions in triplicate in Brain Heart Infusion plates.
Incubate plates for 24–48 h at 37°C in a 5% CO2 atmosphere before colonies are counted. Cell viability at each time point is expressed as the log percentage of viable cells [colony-forming units (CFU) per mL] relative to the control group at time zero. The killing course should follow the relationship N = N0 ekt, where N0 is the initial count (in CFU/mL)at time zero, N is the count at time t, t is the sampling time, e is the base for natural logarithms, and k is the killing rate constant. An alternative equation is N = N0 2.3 log10kt.
3.3. pH Drop (Glycolytic Profile)
Grow biofilms on glass slides in Gram-staining boxes for 5–6 days as described in Section 3.1.2.
Remove slides from the box with a sterile forceps and dip three times into a 50 mL sterile conical tube containing 45 mL of 50 mM KCl/1 mM MgCl2 solution.
Transfer the slide to a 50 mL conical tube containing 49 mL of 50 mM KCl/1 mM MgCl2 solution and a small magnetic stir bar (the volume of salt solution can be reduced to prevent overflow). Place the tube in a beaker on a stir plate.
Insert a pH probe into the KCl/MgCl2 solution and titrate the solution pH to 7.2 using 0.1 N KOH (it may take several min to stabilize the pH at 7.2). Reduce agitation if biofilms start to detach from the glass slides.
Add 900 μL of 1 M glucose. The final concentration of glucose in the tube should be 55.6 mM. Record the pH drop every 30 s for the first 5 min and then, every 5 min for the next 85 min.
3.4. F-ATPase Assay
Scrape biofilm cells from the glass slides or HA disks into a 50 mL conical tube containing 5 mL of 1× ATPase buffer and sonicate the cell suspension as described in step 10 of Section 3.1.3.
Centrifuge at 4,000g for 12 min at 4°C. Carefully discard the supernatant. Resuspend pellet in 5 mL of 1× ATPase buffer and repeat centrifugation.
Resuspend the pelleted cells in 2 mL of 1× ATPase buffer.
Chill the cells on ice and add 100 μL of toluene–acetone (1:9).
Vortex for 2 min, then chill on ice for 2 min. Repeat.
Freeze the cells in dry ice ethanol bath and thaw at 37°C. Repeat.
Into a glass tube, pipette 0.75 mL of 2× ATPase buffer, 0.475 mL of Milli-Q water, 0.2 mL of permeabilized cells. Mix well and incubate in the water bath at 37°C for 2 min.
Then, add 75 μL of pre-warmed 100 mM ATP (pH 7) solution.
Incubate at 37°C for 15 min.
Stop the reaction by adding 0.5 mL 20% tricarboxylic acid (TCA). Centrifuge for 14,000g for 10 min. Collect the supernatant.
For the blank tube (negative control), do the same procedure, but add TCA at time zero to inactivate the enzyme first.
-
To quantify the inorganic phosphate released by the ATPase, prepare the following mix in a 2 mL plastic cuvette (light path = 1 cm):
- 0.15 mL H2O
- 0.15 mL supernatant containing ATP
- 0.9 mL of Bencini reagent
Mix well and read the absorbance at 350 nm (A350). Calculate the ΔA350 (ΔA350 = A350 of experimental – A350 of control).
Phosphate concentration can be calculated with use of a standard curve of inorganic phosphate prepared in the same way as described above using the concentration range from 0.5 to 6 μg of standard phosphate in the reaction mixture.
To calculate the results, One unit of ATPase activity is defined as the amount of the enzyme that can release 1 μmol phosphate per min. Relative rates can be expressed per unit of dry biomass or per mg of protein.
3.5. Respiration
Grow biofilms on glass slides in Gram-staining boxes for 5–6 days as described in Section 3.1.2 (seeNote 9).
Remove slides from the box with a sterile forceps and wash biofilms rapidly by immersion in salt solution (50 mM KCl/1 mM MgCl2).
Transfer the slide to a 50 mL conical tube containing approximately 35 mL of air-saturated 50 mM potassium phosphate buffer pH 7 containing 0.5% (wt/vol) glucose.
Start measuring the oxygen uptake at room temperature by using a dissolved oxygen meter (e.g., VWR Model 4000).
Record the readings every min for 15 min or until the reading approaches zero.
After readings are completed, collect 10 μL of cell suspension to assess protein concentration by using standard protein concentration assays.
Calculate the respiration rate by estimating the value differences between each interval (rate = nmol O2/min/mg of protein).
3.6. NADH Oxidase Activity
Grow biofilms on glass slides for 5–6 days as described in Section 3.1.2.
Remove slides from the medium with a sterile forceps and scrape biofilms with a sterile spatula into a 50-mL conical tube containing 10 mL of 20 mM Tris–HCl buffer (pH 7) containing salt solution.
Sonicate as described in Section 3.1.3.
Centrifuge at 4,000g for 12 min at 4°C and carefully discard supernatant.
Resuspend cell pellet in 10 mL of salt solution and repeat steps 3 and 4 once.
Resuspend pellet in 600 μL salt solution.
Transfer pellet to a 1.5 mL screw cap tube containing approximately 600 μL of acid-washed 0.1 mm diameter glass beads.
Homogenize the cells by using a Beadbeater for six cycles of 30 s with incubation on ice for 2 min between cycles.
Centrifuge at 14,000g for 10 min at 4°C.
Collect the clear supernatant. This is your crude extract and it should be kept on ice to prevent proteolysis.
In a 2 mL plastic cuvette (1 cm light path), prepare the following mix: 100 μL of 1 M potassium phosphate buffer (pH 7), 6 μL of 50 mM EDTA, 10 μL of 50 mg/mL bovine liver catalase, and 10–100 μL of crude extract. Bring the volume to 920 μL Milli-Q water.
Blank the spectrophotometer at 340 nm with the cuvette containing the reagents listed above. Start the reaction by adding 80 μL of 2 mM NADH.
Record the decrease in absorbance at 340 nm (OD340) every 15 s for 3 min, during which time the reaction is linear.
Use 10 μL of the crude extract to obtain protein concentration using standard methods.
One unit of NADH oxidase is defined as the amount of enzyme that catalyzed the oxidation of 1 μmol NADH/min. NADH activity is measured as (units/mg protein).
3.7. Determination of Extracellular Polysaccharides (EPS)
Use a biofilm homogenate obtained as described in Section 3.1.3 (total volume of 5 mL). Remove 100 μL for total bacterial counting.
Centrifuge at 5,500g for 10 min at 4°C and carefully pour the supernatant into a 50 mL conical tube (save this tube to pour additional supernatant as follow).
Wash the pellet by pipetting 2.6 mL of Milli-Q water and vortex each tube until the pellet is completely dissolved.
Centrifuge at 5,500g for 10 min at 4°C. Pour supernatant in the conical tube from step 2. Repeat the wash with 2.5 mL water, as described above once more. Total volume of supernatant should be 10 mL. Save supernatant for soluble polysaccharide analysis (see below).
Resuspend the pellet in 2.55 mL water. Remove 50-μL aliquot (for determination of total protein) and dilute 1:5 in water. Pipette 50 μL of the diluted sample in micro-glass vial (in triplicates). Spin samples in a vacuum concentrator (SpeedVac) for approximately 1 h. Place tubes with the dried samples in a desiccator containing P2O5 (phosphorus pentoxide) or Drierite® under vacuum. Total protein in each sample is determined by acid digestion followed by the nin-hydrin assay (6).
3.7.1. Determination of Soluble EPS
To determine soluble EPS, transfer 3 mL from the supernatant obtained in step 4 to a 15-mL conical tube. Add 2.5 volumes of ice-cold 95% ethanol (7.5 mL), mix well, and place in freezer (−20°C) overnight to precipitate the water-soluble polysaccharides. The amount of supernatant should be enough to perform in triplicates.
Remove the samples from freezer, mix well by inverting at least five times, and centrifuge all the tubes at 9,500g for 20 min at 4°C. Discard the supernatant in a clean glass beaker (make sure the supernatant is clear).
Wash pellets three times with ice-cold 75% ethanol and resuspend pellet in 7 mL ice-cold 75% ethanol. Use a disposable inoculating loop to scrape down the water-soluble EPS adhered to the tube wall.
Vortex mixture well and centrifuge (9,500g for 20 min at 4°C). Discard supernatant. Repeat this procedure two more times.
After washing, blot-dry the pellet.
Resuspend pellet in 1 mL water by scraping down the sides. Vortex to bring pellet back into solution.
Determine total glucose and fructose using colorimetric assays (7, 8).
3.7.2. Determination of Insoluble EPS
Take 1-mL aliquot of the resuspended pellet from step 5 of Section 3.7 and centrifuge immediately (16,000g for 10 min at 4°C). Carefully remove supernatant of the tubes with a tip connected to a vacuum pump. Spin the samples in a SpeedVac vacuum concentrator for 2 h. Place tubes with the dried pellets in a desiccator containing P2O5 or Drierite® under vacuum until ready for analysis.
Weigh each sample pellet and transfer to a 1.5-mL microfuge tube. According to the weight recorded, add 300 μL of 1 N NaOH per mg dry weight to each tube. Vortex gently to disrupt cell pellet. Incubate at 37°C for 2 h.
During the first extraction, after 30 min, remove tubes from the incubator and vortex each tube until the pellet is completely dissolved. Place tube back in the incubator and complete the 2 h incubation time.
After 2 h, centrifuge the samples at 14,000g for 10 min. Transfer supernatant to a 1.5-mL tube. Do not discard pellet.
Again, add same volume of 1 N NaOH and vortex to disrupt pellet. Repeat all steps as described above.
Resuspend the pellet in NaOH, vortex, and centrifuge. Combine supernatant from each extraction in the same tube.
Neutralize pH of the extract by slowly adding 1 N HCl stepwise and checking the pH constantly until you reach pH 7 ± 0.5. After neutralizing the sample, add 3 volumes of ice-cold 95% ethanol, mix well, and precipitate the EPS in the freezer overnight (at least 18 h incubation).
After precipitation, remove tubes from freezer, mix well, and centrifuge samples for 20 min (9,500g, 4°C). Discard supernatant.
Resuspend pellet in 7 mL of ice-cold 75% ethanol. Use a disposable inoculating loop to scrape down the polysaccharide adhered to the tube wall to ensure resuspension. Vortex, centrifuge for 20 min (9,500g, 4°C), and discard supernatant. Repeat this procedure two more times.
After washing, blot-dry the pellet.
Resuspend pellet by scraping down the sides in 1 N NaOH (in the same total volume of the original extraction).
Determine total glucose and fructose using colorimetric assays as described elsewhere (7, 8).
3.7.3. Determination of Intracellular Polysaccharides (IPS)
Take 1-mL aliquot of a biofilm previously dispersed by sonication as described above (see step 5 of Section 3.7) and immediately centrifuge at 13,000g for 10 min at 4°C.
Carefully remove supernatant of the tubes with a tip connected to a vacuum pump. Spin samples in a SpeedVac vacuum concentrator for 2 h.
Place tubes with the dried pellets in a desiccator containing P2O5 or Drierite® under vacuum.
Weigh each pellet as described before and transfer to a clean glass culture tube (18 × 150 mm, previously rinsed with Milli-Q water).
According to the weight recorded for each pellet, add 1 mL water per 1 mg dry weight to each tube. According to the volume added, add 300 μL 5.3 M KOH per mL cell suspension. Make sure pellets are completely disrupted.
Tightly cover the tubes, individually, with aluminum foil and boil in water for 1.5 h.
After boiling, let tubes cool down. Add 5.3 M HCl to each tube in the same volume as the KOH to neutralize suspension.
-
The IPS concentration will be determined by a colorimetric assay (9) as follows:
In a 2-mL tube prepare the following mix:
800 μL sample or standard (0–100 μg glycogen) (see Note 10).
500 μL 1 M phosphate buffer pH 7. Vortex to mix.
300 μL 0.2% iodine in 2% potassium iodide solution.
Vortex each tube well and read absorbance at 520 nm (A520).
4. Notes
Unless stated otherwise, all media and chemical reagents are from Difco or Sigma, respectively.
Some precipitation in base medium can be observed because of the presence of FeSO4 and MnCl2.
Lower or higher concentrations of sugars can be used so that growth can be catabolite limited or pH limited.
This protocol can be adapted for physiologic assays that require a larger biomass using tissue culture plates.
For sucrose grown S. mutans biofilms, an additional acetic acid extraction might be required.
Biofilms are considered mature after 5- to 6-days growth. Depending on the protocol requirements, disrupted or intact biofilms can be used in the physiologic assays.
To minimize experimental variations, three independent wells containing intact biofilms should be used for each time point.
Changes as low as 0.1 pH unit result in great variations in the killing kinetics of different species of oral streptococci. A pilot experiment to determine the best pH and sampling times is recommended.
Dispersed biofilms can also be used.
The glycogen solution should always be prepared fresh.
Acknowledgments
We thank Dr. Pedro Rosalen for kindly providing images used in Fig. 7.1.
References
- 1.Wade W. Unculturable bacteria in oral biofilms. In: Newman HN, Wilson M, editors. Dental plaque revisited. Oral biofilms in health and disease. Bioline; Cardiff: 1999. pp. 313–322. [Google Scholar]
- 2.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Ann. Rev. Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 3.Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 2005;7:95–107. [PubMed] [Google Scholar]
- 4.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 5.Loo CY, Corliss DA, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore S, Stein WH. A modified ninhydrin reagent for the photometric determination of amino acids and related compounds. J. Biol. Chem. 1954;211:907–913. [PubMed] [Google Scholar]
- 7.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 8.Kulka RG. Colorimetric estimation of ketopentoses and ketohexoses. Biochem. J. 1956;63:542–548. doi: 10.1042/bj0630542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiPersio JR, Mattingly SJ, Higgins ML, Shockman GD. Measurement of intracellular iodophilic polysaccharide in two cariogenic strains of Streptococcus mutans by cytochemical and chemical methods. Infect. Immun. 1974;10:597–604. doi: 10.1128/iai.10.3.597-604.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]