Abstract
The vomeronasal system of mice is thought to be specialized in the detection of pheromones. Two multigene families have been identified that encode proteins with seven putative transmembrane domains and that are expressed selectively in subsets of neurons of the vomeronasal organ. The products of these vomeronasal receptor (Vr) genes are regarded as candidate pheromone receptors. Little is known about their genomic organization and sequence diversity, and only five sequences of mouse V1r coding regions are publicly available. Here, we have begun to characterize systematically the V1r repertoire in the mouse. We isolated 107 bacterial artificial chromosomes (BACs) containing V1r genes from a 129 mouse library. Hybridization experiments indicate that at least 107 V1r-like sequences reside on these BACs. We assembled most of the BACs into six contigs, of which one major contig and one minor contig were characterized in detail. The major contig is 630–860 kb long, encompasses a cluster of 21–48 V1r genes, and contains marker D6Mit227. Sequencing of the coding regions was facilitated by the absence of introns. We determined the sequence of the coding region of 25 possibly functional V1r genes and seven pseudogenes. The functional V1rs can be arranged into three groups; V1rs of one group are novel and substantially divergent from the other V1rs. The genomic and sequence information described here should be useful in defining the biological function of these receptors.
The olfactory system provides sensory information about the chemical composition of the external world (Farbman 1992). Olfactory chemoreception initiates in mammals at the level of sensory neurons that are located in either of two sites. Sensory neurons of the main olfactory epithelium detect volatile odorants (Buck 1996); those of the vomeronasal organ (VNO) are believed to respond mainly, but not exclusively, to pheromones (Halpern 1987; Keverne 1999; Holy et al. 2000; Leinders-Zufall et al. 2000). Pheromones are chemical signals that provide information about gender, dominance, and reproductive status between individuals of the same species (Karlson and Lüscher 1959; Keverne 1983). They elicit innate and stereotyped reproductive and social behaviors in the recipients, along with profound neuroendocrine and physiological changes.
The chemosensory receptors of the main olfactory system are called odorant receptors (ORs). They are seven-transmembrane proteins, encoded by a family that may comprise as many as 1000 genes in rat and mouse (Buck and Axel 1991; for review, see Mombaerts 1999a,b). The chemosensory receptors of the vomeronasal system are termed vomeronasal receptors (Vrs). Two families of Vr genes, also encoding putative seven-transmembrane proteins, are proposed to represent genes for pheromone receptors, but direct evidence has yet to be delivered. The first discovered family of Vr genes was estimated initially to consist of 30–100 V1r genes, expressed selectively in sensory neurons of the apical zone of the epithelium of the VNO (Dulac and Axel 1995; Saito et al. 1998). The family of V2r genes comprises between 30–140 genes that are expressed in neurons of the basal zone (Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997). There are no conserved motifs between the V1r and V2r gene families, and Vrs have no significant sequence homology with ORs. Differential expression of Vr genes in neurons of the apical and basal zones correlates with that of G protein subunits Gαi2 and Gαo, (Halpern et al. 1995; Berghard and Buck 1996), but it is not clear if these G proteins are involved in chemosensory signal transduction in these neurons.
The V1r genes were initially cloned in rats, and seven full-length open reading frames (ORFs) were reported (Dulac and Axel 1995). Subsequently mouse V1r homologs were identified by cross-hybridization to a VNO cDNA library (Saito et al. 1998), a YAC library (Belluscio et al. 1999), and BAC library (Rodriguez et al. 1999). Recently we described the first human V1r-like gene, termed V1RL1, mapping to chromosome 19 (Rodriguez et al. 1999). The genomic location of only a single mouse V1r gene, termed VRi2, has been determined; it maps to chromosome 6 (Rodriguez et al. 1999). Only five sequences of mouse V1r ORFs are publicly available (Saito et al. 1998; Rodriguez et al. 1999). Thus, little information is available about this multigene family, whose biological function is poorly understood. Here, we have made an attempt to fill this void, taking a strictly genomic approach. The absence of introns in the ORFs of V1rs (Dulac and Axel 1995) permits sequence determination from genomic clones.
RESULTS
Construction of a Major BAC Contig Containing a V1r Gene Cluster
We first screened a 129 mouse BAC library with probes corresponding to rat V1r coding regions (Dulac and Axel 1995), and isolated eight BACs that were confirmed to contain V1r genes by Southern blot hybridization. Because these BACs could not be assembled into a contig, we initiated multiple chromosome walks using nonrepetitive BAC insert end probes. We also determined the sequence of several V1r ORFs located on these eight BACs, allowing us to design degenerate PCR primers corresponding to the most conserved regions. With these primers we obtained a pool of PCR products from mouse genomic DNA (designated as pool #1; see Methods for details). The chromosome walks were complemented with a new screening of the BAC library using pool #1 probes. By combining these two approaches, we isolated 48 BACs that contain V1r genes, as evidenced by Southern blot hybridization and sequencing of several ORFs.
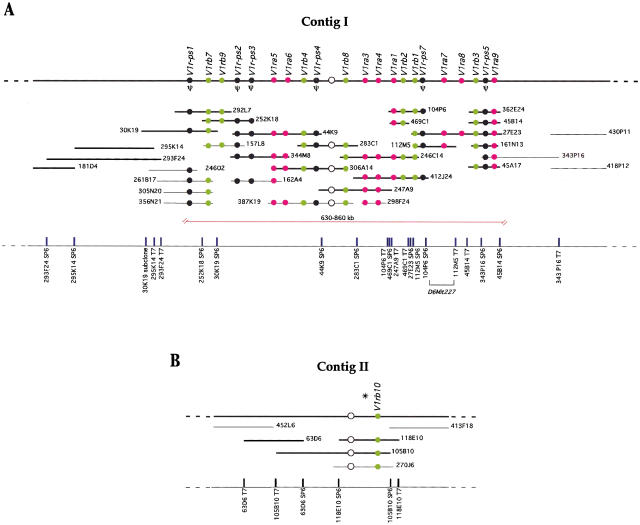
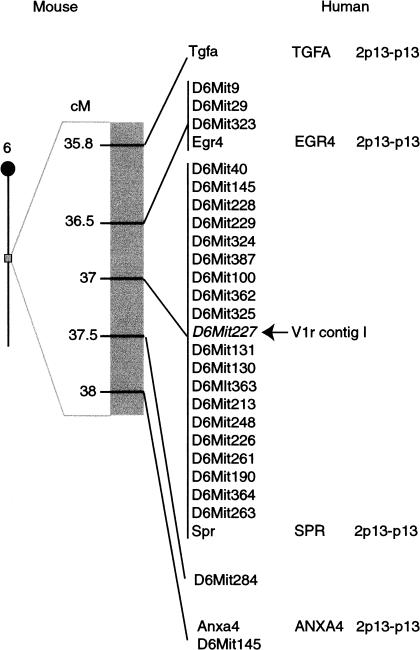
Relying largely on BAC insert end probes and V1r ORF probes, we were able to assemble most of these BACs into a single contig, with the exception of three BACs (see below). This contig, termed the major contig or contig I (Fig. 1A), was mapped with a mouse radiation hybrid panel next to marker D6Mit261 at 37cM of chromosome 6 (see also Rodriguez et al. 1999). Twenty-four MIT markers in the vicinity were tested against BACs covering the entire contig. This analysis resulted in the mapping of marker D6Mit227 (Fig. 2) to overlapping BACs 27E23 and 112M5 (Fig. 1A). By measuring the size of individual BACs in pulse-field gel electrophoresis experiments, we estimate that the BAC contig is 630–860 kb long. We walked a few hundred kb outward in both directions, but did not identify BACs that hybridize to pools of V1r ORF probes. Assuming that no major gap separates this V1r gene cluster from another one, we can tentatively assign ends to the cluster. Genes flanking the V1r cluster in the mouse have orthologous positions on human 2p13-p13 (Fig. 2).
Figure 1.
(A) Physical map of the major BAC contig I on chromosome 6. The thick black line at the top of the figure represents the chromosome. The arrangement of 22 V1r genes along the cluster is shown with circles: (solid circles)V1r coding regions that have been sequenced; (empty circle) V1r gene whose order in the cluster was determined but was not cloned or sequenced; (pink circles) members of group a; (green circles) members of group b. Pseudogenes are shown in black and indicated by the symbol Ψ. BACs, with their respective names, are represented by horizontal lines below the chromosome. BACs whose sizes were estimated by pulsed-field gel electrophoresis are indicated by thick lines and drawn to scale. Estimated sizes of BACs are: 181D4, 90 kb; 293F24, 250 kb; 295K14, 160 kb; 30K19, 170 kb; 292L7, 125 kb; 252K18, 110 kb; 44K9, 200 kb; 344M8, 130 kb; 306A14, 170 kb; 283C1, 130 kb; 247A9, 160 kb; 246C14, 160 kb; 412J24, 160 kb; 104P6, 80 kb; 469C1, 45 kb; 27E23, 200 kb; 112M5, 95 kb; 363E24, 60 kb; 45B14, 60 kb; 45A17, 60 kb; 161N13, 70 kb. BACs whose sizes were not determined are indicated by thin lines and are not drawn to scale. The red bracket below the BACs indicates the estimated extent of the V1r cluster. The positions of the BAC end sequences are indicated at the bottom as the name of the BAC clone followed by the SP6 or T7 side of the BAC vector. (B) BAC contig II on the X chromosome. The thick black line at the top of the figure represents the chromosome. V1r genes are represented by circles: (solid circle) sequenced V1rb10 gene. The green color indicates that it belongs to group b. (Open circle) V1r gene that cross-hybridizes weakly with pool #1 probe by Southern blot hybridization but was not cloned and sequenced. The asterisk above the neighboring V1r genes indicates unknown marker order. BACs whose sizes were estimated by pulsed-field gel electrophoresis are indicated by thick lines and drawn to scale. Estimated sizes of BACs are: 63D6, 130 kb; 105B10, 250 kb; 118E10, 130 kb. BACs whose sizes were not determined are indicated by thin lines and are not drawn to scale. The positions of the BAC end sequences are indicated at the bottom as the name of the BAC clone followed by the SP6 or T7 side of the BAC vector.
Figure 2.
Mapping of the major BAC contig in mouse and orthologous position in human. Linkage map at ∼37cM of mouse chromosome 6, showing the position of V1r contig I and nearby MIT markers and genes. The arrow indicates that D6Mit227 maps to the V1r contig I. Flanking genes have orthologous positions on human 2p13-p13.
Estimates of the number of V1r genes located in this cluster range from 21 to 48. The lower number represents the V1r ORFs that have been cloned and sequenced; the upper estimate represents the number of distinct bands, observed after digestion of the BACs with the restriction enzyme HindIII (see Methods) and hybridization to V1r probes at low stringency. Figure 1A shows the approximate position of 22 V1r genes within the cluster.
Localization of Two V1r Genes on the X Chromosome
The three BACs that did not fit into contig I were found to overlap and seeded contig II (Fig. 1B), containing two V1r genes as evidenced by Southern blot hybridization at low stringency (data not shown). One of these genes, termed V1rb10 (for nomenclature, see below), was cloned and its ORF sequenced, but we were unable to clone the other presumptive V1r gene. This contig was mapped between markers DXMit117 and DXMit67 with a radiation hybrid panel. Chromosome walking was performed in both directions using insert end probes from two BACs. No further BACs were found to cross-hybridize to V1r probes.
Identification of a Novel Group of V1r Genes
When the BAC filters were screened with pool #1, two types of signals were detected: strong or very faint, almost undistinguishable from background. The strong signals correspond to the 45 BACs that form the major contig on chromosome 6 (Fig. 1A), and to the three BACs of the minor contig on the X chromosome (Fig. 1B). When the BACs that yielded very faint signals were subjected to Southern blot hybridization with pool #1, very weakly cross-hybridizing bands were detected. The correspondence of these faint signals to genuine V1r members was confirmed by shotgun subcloning of the BACs and sequencing the entire insert of the smaller subclones. However, these genes appear to be very divergent from the previously sequenced V1r genes.
Therefore, a second set of probes (designated as pool #2) was prepared. These probes correspond to the most conserved sequences within the ORFs of this novel V1r group and were generated by PCR with a set of distinct pairs of primers (see Methods for details). Screening of the BAC library with pool #2 resulted in 59 new positive BACs. Overlaps between the BACs and relative positions of V1r ORFs were assessed by Southern blot hybridization. Most of the BACs could be assembled into four minor contigs (III–VI, see Table 1). The map location of these contigs could not be determined with certainty. The number of V1r genes in contigs III, IV, V, and VI is estimated to be 21, 15, 9, and 12, respectively, or 57 in total (data not shown). These estimates reflect the number of different HindIII bands detected by Southern blot hybridization with pool #2. Strong bands were not detected with pool #1, suggesting that these BACs do not contain V1r genes of the groups that are located within the major contig (groups a and b, see below). Conversely, no signals were detected on Southern blots of BACs from contig I with pool #2, indicating that genes of the novel group are not present in this major contig.
Table 1.
Minor Contigs III-VI: BAC IDs and Gene Content
| Number | Representative BACs of the contig | Cloned Genes | Cloned Pseudogenes |
|---|---|---|---|
| Contig III | 269A9, 428F20, 357I4, 101J4, 101J3, 251G22, 137C17, 386K13, 356L5, 352B3, 418N24, 279M5, 420P18, 137P5, 137O5, 267N10, 268B11 | V1rc1, V1rc3, V1rc4, V1rc5, V1rc6, V1rc7 | V1r-ps6 |
| Contig IV | 426F15, 430H17, 425D21, 272J09, 269J06, 241A14, 404N12 | V1rc8 | — |
| Contig V | 254L24, 361J10, 381J14, 248E17, 402L06, 397O20, 264C24 | V1rc2, V1rc9 | — |
| Contig VI | 269K23, 249F9, 353K13, 424O7, 243B13, 353K13, 393L01 | — | — |
Sequence Diversity of V1r Genes
The ORFs of V1r genes are intronless (Dulac and Axel 1995). Therefore contiguous sequence information can be obtained directly from cloned genomic DNA. Clones containing V1r ORFs were obtained by shotgun subcloning of BACs followed by dot blot analysis. We determined the nucleotide sequence of the ∼1 kb ORF of 32 V1r genes: 21 genes from contig I; one gene from contig II on the X chromosome; and 10 genes from the contigs III–VI (Table 1). In contig I, six of the 21 sequences (29%) correspond clearly to pseudogenes: the ORFs are interrupted by stop codons and/or frameshifts. In contigs III–V, only one sequence corresponds to a pseudogene.
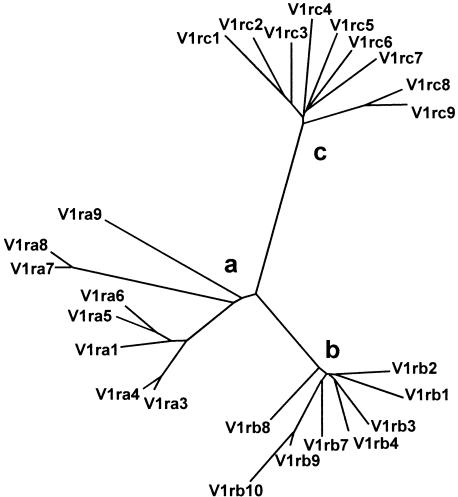
Phylogenetic analysis of the predicted amino acid sequences of the 25 V1rs studied here reveals that they fall into three distinct groups, termed groups a, b, and c (Fig. 3). In consultation with The International Committee on Standardized Genetic Nomenclature for Mice, we devised a rational nomenclature that is based on this division in three groups. The five mouse V1r genes whose sequences were available in GenBank were renamed; for instance the VRi2 gene that we reported previously (Rodriguez et al. 1999) now corresponds to V1rb2. Table 2 lists new and old names, accession numbers, and references.
Figure 3.
Phylogenetic tree of predicted amino acid sequences of mouse V1r genes. The unrooted tree shows the phylogenetic relationships between predicted V1r amino acid sequences. V1r genes can be classified in groups a, b, and c.
Table 2.
References of V1r Genes
| New Nomenclature of V1r Genes and Pseudogenes | New Accession Numbers | Old Nomenclature of Previously Reported V1r Genes | Accession Numbers of Previously Reported Genes | References for Previously Reported Genes |
|---|---|---|---|---|
| V1ra1 | AF291481 | mpr1 | Y12725 | Saito et al. 1998 |
| V1ra2 | mpr2 | Y12724 | Saito et al. 1998 | |
| V1ra3 | AF291482 | |||
| V1ra4 | AF291483 | |||
| V1ra5 | AF291484 | |||
| V1ra6 | AF291485 | |||
| V1ra7 | AF291486 | |||
| V1ra8 | AF291487 | |||
| V1ra9 | AF291488 | |||
| V1rb1 | AF291489 | |||
| V1rb2 | VRi2/V1r5 | AF132114/NP036041 | Rodriguez et al. 1999 | |
| V1rb3 | AF291491 | |||
| V1rb4 | AF291492 | |||
| V1rb5 | mV1R5 | Y17566 | Saito et al. 1998 | |
| V1rb6 | mV1R6 | Y17567 | Saito et al. 1998 | |
| V1rb7 | AF291493 | |||
| V1rb8 | AF291494 | |||
| V1rb9 | AF291495 | |||
| V1rb10 | AF291496 | |||
| V1rc1 | AF291497 | |||
| V1rc2 | AF291498 | |||
| V1rc3 | AF291499 | |||
| V1rc4 | AF291500 | |||
| V1rc5 | AF291501 | |||
| V1rc6 | AF291502 | |||
| V1rc7 | AF291503 | |||
| V1rc8 | AF291504 | |||
| V1rc9 | AF291505 | |||
| V1r-ps1 | AF291506 | |||
| V1r-ps2 | AF291507 | |||
| V1r-ps3 | AF291508 | |||
| V1r-ps4 | AF291509 | |||
| V1r-ps5 | AF291510 | |||
| V1r-ps6 | AF291511 |
Both a and b groups are present in contig I. The one gene we sequenced from contig II, V1rb10, belongs to group b. Although there is no apparent distribution or segregation of groups a and b within contig I (Fig. 1A), adjacent genes tend to be more closely related in sequence; this becomes more obvious when nucleotide sequences are compared, including those of the pseudogenes (data not shown). Group c genes are absent from contigs I and II. Conversely, the four minor contigs only contain group c genes. Amino acid identity within each group ranges from 65% to 94%, with the exception of genes V1ra7, V1ra8, and V1ra9, which share between 50% and 55% amino acid identity with other group a members. Long branches in the phylogenetic tree (Fig. 3) reflect this lower degree of sequence similarity. The percentage of amino acid identity between group a and b genes ranges from 40%–53%, whereas members of group c share only 22%–31% amino acid identity with any gene member of the other two groups.
Deduced amino acid sequences of the ORFs of the seven rat V1r genes (Dulac and Axel 1995) and five mouse V1r genes (Saito et al. 1998; Rodriguez et al. 1999) available in public databases were compared with the sequences produced here. These belong to groups a and b, except for VN6 (from the rat) which does not belong to any of these groups (<35% amino acid identity). Two published mouse sequences, mPR1 (Saito et al. 1998) and VRi2 (Rodriguez et al. 1999), are identical to our genes V1ra1 and V1rb2, respectively, with the only difference being that the reported mPR1 amino acid sequence has an extra amino-terminal methionine; this discrepancy may be explained by allelic variation between the strains BALB/C (Saito et al. 1998) and 129 (this paper).
Sequence Alignment
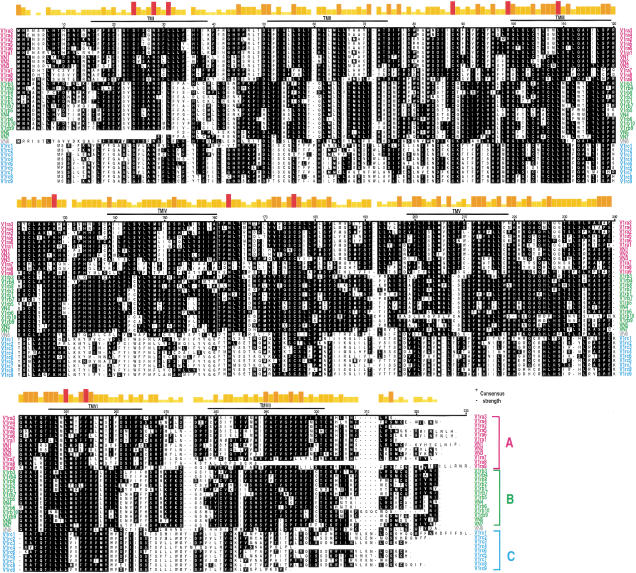
Figure 4 shows an alignment of the publicly available and the new sequences. Putative transmembrane (TM) domains were predicted using three independent methods (see Methods for details). V1ra8, V1rc4, and V1rc9 are considerably shorter at the carboxyl terminus. As TM7 does not appear to exist in V1ra8, we do not know whether this truncated V1r is biologically active. It is interesting to note that the three programs predict for V1ra7, V1ra8, V1rc3, V1rc5, and V1rc8 an extra transmembrane helix (not shown in Figure 4) between TM IV and TM V.
Figure 4.
Alignment of predicted amino acid sequences of mouse and rat V1r genes. Predicted amino acid sequences from V1r sequences were aligned with the other mouse (V1ra2, V1rb5, and V1rb6) and rat (VN1, VN2, VN3, VN4, VN5, VN6, and VN7) V1r sequences available in public databases. The alignment was performed using the Clustal V method of the MegAlign program. The V1r sequences were aligned by the first methionine common to all open reading frames. Predicted consensus positions of TM domains are indicated as TM I-VII. Consensus residues are shown as white lettering on black background. Consensus strength is shown on top. The shortest, yellow bars represents the least conserved amino acids; the tallest, red bars represent amino acids that are common to all sequences. V1r genes from groups a, b, and c are indicated in pink, green, and blue, respectively. VN6 from rat does not belong to any of the three groups and is indicated in gray. The publicly available sequences of VN5 and VN7 from rat seem to be incomplete, precluding full-length alignment.
The alignment in Figure 4 indicates that 11 amino acid residues are 100% conserved in the 35 mouse and rat V1r genes. The conserved residues are distributed among TM I, II, and VI, extracellular loops (EC) 1 and 2, and intracellular loops (IC) 2. Several features can be discerned that V1rs share with other 7TM proteins: the asparagine in TM I (N28); two cysteines in EC 1 and 2 (C88 and C176); the phenylalanine in TM VI (F254); and the highly conserved proline in TM VII (P295). There are no conserved motifs shared with other 7TM proteins, as was initially reported (Dulac and Axel 1995).
It should be noted that the presence of multiple methionines at the amino terminus of several V1rs complicates precise assignment of the start site for translation. Five mouse V1r ORFs (V1ra1, V1ra2, V1rb9, V1rb10, and V1rc7) have an additional methionine immediately upstream of the one shown in the sequence alignment of Figure 4. V1rb10 contains two additional methionines: one immediately upstream, and another one 18 residues upstream to the one presented in Figure 4. Thus, V1rb10 could potentially have a considerably longer amino terminus. Two of the reported rat V1rs (VN4 and VN2) exhibit an amino terminus pattern similar to V1rb10. It has been shown for some genes that alternative ATG codons can be used as the start site of translation giving rise to multiple proteins with different amino-terminal lengths (Ossipow et al. 1993). This may also be the case for some V1r genes.
Expression of Group c V1r Genes in the VNO
In situ hybridizations were performed with several members of the three groups. As expected, all genes were expressed selectively in neurons located within the apical zone of the vomeronasal epithelium, which correlates with the expression of the Gαi2 subunit. This is also the case for members of the novel group c (Fig. 5), indicating that these are truly V1r genes by the criterion of zone-specific expression.
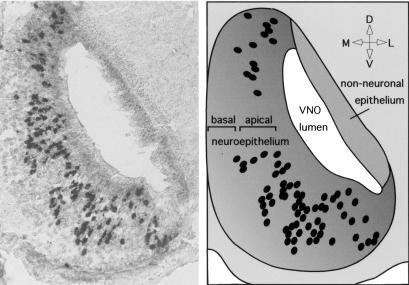
Figure 5.
In situ hybridization with a V1r gene of group c. (Left) Coronal section through the vomeronasal organ of an adult male mouse annealed with a digoxigenin-labeled antisense RNA-probe for a conserved region of gene V1rc7. Each dark spot represents the cell body of a vomeronasal sensory neuron expressing a cross-hybridizing V1r gene. The probe recognizes multiple bands in Southern blot hybridization of genomic DNA. (Right) Drawing representing the image on the left. Cell bodies of cells expressing V1rc7-like genes are located in the apical zone of the vomeronasal organ. Orientation: L, lateral; M, medial; D, dorsal; V, ventral.
DISCUSSION
Genomic Organization of the V1r Gene Repertoire
The V1r gene family was discovered in rat by differential screening of cDNA libraries, constructed with the aid of PCR from single vomeronasal sensory neurons that express the Gαi2 subunit (Dulac and Axel 1995). Extending our initial characterization of the mouse V1r repertoire (Rodriguez et al. 1999), we have isolated by cross-hybridization and chromosome walking a total of 107 BACs that contain V1r genes; 68 of these BACs are described in this paper. The BACs were arranged into six contigs. The major contig, located on chromosome 6, contains between 21 and 48 V1r genes, exclusively of groups a and b. A contig on chromosome X contains two V1r genes, one of which has been sequenced and shown to be a member of group b. Genes belonging to group c reside within four minor contigs; these detailed characterizations were not completed. Gene density in these clusters appears to be high: In the major contig, 21–48 genes reside within a region of 630–860 kb, resulting in distances of 13–41 kb between the coding regions. We sequenced the intergenic region between the poly(A) signal of the V1rb2 and the putative translation start site of the downstream V1r (data not shown), and found that the distance between is 2.5 kb, indicating that some V1r genes are very close to each other.
Complexity of the V1r Gene Repertoire
Considering the number of hybridizing HindIII fragments to be a reliable indicator of the number of V1r genes residing on a given BAC, the complexity of the mouse V1r repertoire exceeds 107 genes. Similar estimates with genomic DNA instead of BAC DNA are not possible, due to the large number of hybridizing bands in the genome. Our minimum estimate of 107 is intermediate between the estimates of 35–100 in rat (Dulac and Axel 1995) and 150–200 in mouse (Saito et al. 1998). Because group c genes cross-hybridize only very weakly with genes of groups a and b and have thus far remained unnoticed, it is conceivable that the V1r repertoire comprises additional groups or families. The number 107 is thus clearly a minimum estimate. We have provided the first evidence of the presence of pseudogenes in the V1r gene family: 7/32 sequences, or 22%. The presence of pseudogenes has not been reported in previous studies (Dulac and Axel 1995; Saito et al. 1998), perhaps because these studies were carried out with cDNA instead of genomic DNA. It remains to be seen whether or not these pseudogenes are expressed.
The complexity of the V2r family may be of similar magnitude (100–140 genes), and about two thirds are reported to be pseudogenes (Bargmann 1997; Herrada and Dulac 1997; Matsunami and Buck 1997). The mouse genome may thus contain 83 functional V1rs (i.e., 78% of 107 genes) and 33–47 functional V2rs (i.e., 33% of 100–140 genes), or 116–130 functional Vrs in total. It is not obvious and it was not anticipated that pheromone detection requires so many receptors. Perhaps some of these receptors have other chemosensory functions.
Diversity of the V1r Gene Repertoire
Phylogenetic analysis of the sequences provided here indicates that V1rs fall into three groups (Fig. 3), prompting us to devise for the first time a rational nomenclature for mouse V1r genes. One scheme for classifying multigene families (Nebert et al. 1991; Lancet and Ben-Arie 1993) defines 40% amino acid identity as a threshold for forming a “family”; according to this definition, group a and b genes are members of the same family, whereas group c would constitute a novel family of V1r genes. The V1r repertoire would thus have to be regarded as a “superfamily” consisting of at least two families: a + b and c. Further division of these families into “subfamilies”, defined with a threshold of 60% amino acid identity (Nebert et al. 1991; Lancet and Ben-Arie 1993), is complicated by the borderline sequence identity of genes V1ra7, V1ra8, and V1ra9, although they clearly belong to group a (Fig. 4). It is clear that these definitions are in flux and may have to be revisited when the V1r repertoire is further characterized.
Conserved Residues
Eleven of ∼296 amino acid residues are 100% conserved among V1rs, including the human V1RL1 (Rodriguez et al. 2000). Of these, five (N28, C88, C176, F254, P295) correspond to residues that are generally conserved generally in 7TM proteins. The two conserved cysteines are located in EC 1 and 2, where they are thought to participate in a disulphide bridge (Probst et al. 1992; Baldwin 1994); site-directed mutagenesis studies have invoked the presence of the disulphide bonds in the stabilization of the active form of the receptor (Davidson et al. 1994).
The remaining seven residues that are 100% conserved among V1rs (G24, L31, R99, L109, K128, N163, and L250) seem to be unique to this superfamily of 7TM proteins, and may provide a signature for classification of novel 7TM proteins to the V1r superfamily. One of these residues, N163 in EC 2, forms part of a putative N-glycosylation site (N [^P][ST][^P]) that is present in all V1rs, including the human V1RL1 (Ro driguez et al. 2000). (Two other genes, V1rb2 and V1ra5, have a second putative N-glycosylation site, in EC 1 and 2, respectively.) One conserved residue is located in an intracellular loop (K128 in IC 2); this loop, together with IC 3 and the carboxyl terminus, has been implicated in G-protein interaction (Baldwin 1994). Finally, five V1r-specific conserved residues (G24, L31, R99, L109, and L250) are within TM domains or very close to the membrane in the extracellular side. 7TM proteins are thought to interact with their ligands in an internal pocket formed by the TM domains (for reviews, see Schwartz 1994; Baldwin 1994); conserved residues in this region may be involved in contributing to the structure of the pheromone-binding pocket.
Group c V1r Genes and V1rb10
V1rs from group c appear to have a shorter amino-terminal domain than those from the other two groups. Group c V1rs are the most divergent when compared as a group to a or b. There are three stretches of conserved amino acid motifs within group c that differ from the other two groups: in TM IV, EC 2, and EC 3. These regions have been implicated in ligand-binding (Baldwin 1994), suggesting that the different groups of receptors may be involved in the recognition of different classes of pheromones. V1rs from group c have single amino acid insertions or deletions that are unique to this group when compared to the consensus of aligned sequences.
Insertions and deletions are also observed in V1rb10, the gene located on the X chromosome. V1rb10 is further unique in that TM V contains a very distinguishable stretch of amino acids: MASLSHSVSLPCA. The distinct localization on the X chromosome together with the existence of unique features in its amino acid sequence make V1rb10 an attractive gene to study its biological function and gene regulation. It will be interesting to determine whether V1rb10 exhibits monoallelic expression, as has been shown for V1rb2 (Rodriguez et al. 1999), and if so, if this pattern correlates with X chromosome inactivation.
Perspectives
The information reported here should prove useful in defining the biological function of these receptors. Progress has been slow, and to date their role in pheromone perception has not been proven. Our partial characterization of the V1r repertoire will enable the application of chromosome engineering techniques that permit the deletion of large regions of the mouse genome by genetic manipulation of embryonic stem cells (Ramirez-Solis et al. 1995). Also, selected genes can be overexpressed in transgenic mice and may cause a behavioral phenotype. Furthermore, these sequences could be used to define motifs that will be useful to search databases, for instance to identify additional functional human V1rs (Rodriguez et al. 2000). Specific PCR primers or specific probes can then be designed to search for genes that are expressed only in males or females, or at a particular developmental stage. Finally, the cloned genes can be inserted into functional expression systems to identify natural or synthetic ligands for V1rs.
METHODS
Genomic Libraries and Screening
Filters from a mouse ES-129 BAC genomic library (Release I and II) were purchased from Genome Systems. The screening was performed as follows: filters were prewet in hybridization buffer (5× SSPE, 5× Denhardts, 0.1% SDS and 10 μg/mL salmon sperm DNA) and hybridized overnight at either 55°C (low stringency) or 65°C (high stringency). Filters were then washed twice for 20 min in 2× SSC/0.1% SDS at the same temperature. Positive clones were usually identified after overnight film exposure.
Establishment of BAC Contigs
The method used to establish the different contigs entails a combination of chromosome walking and screening with V1r ORF probes. The chromosome walking strategy was based on BAC end probes. Because sequencing directly from BAC DNA works poorly in our hands, we developed a method to isolate BAC end clones. It consists of subcloning small restriction fragments of BAC DNA in a vector (pBluescript) as follows: BACs are digested with NotI together with BglII, SpeI, ClaI or XhoI. At both ends of the insert in the BAC vector backbone are restriction sites for NotI, which cuts mouse genomic DNA very infrequently. Digestion with NotI together with any of the other restriction enzymes will generate, among others, fragments containing the ends of the BAC insert. Plasmids that have incorporated an end insert are screened by colony PCR using specific primers for the polylinker of the BAC vector and for pBluescript. The positive end subclones are then sequenced to generate probes for chromosome walking.
Hybridization with V1r ORF probes was carried out with two types of probes termed pool #1 and pool #2. Pool #1 consists of group a and b V1r sequences, and was obtained by PCR amplification of genomic DNA, using 39 cycles of 95°C for 1 min, 45°C for 3 min, and 72°C for 3 min, with 6 sec extension per cycle. Degenerate PCR primers correspond to the conserved regions of the cloned V1r genes of type a and type b between transmembrane domains 4–7. Primer nucleotide sequences are:
sense 5′ ATMGCIACICCIAAYTTRAC 3′,
reverse 5′ ACRAAIGGRCTIACIGTIGCRTA 3′.
Pool #2 consists of group c V1r sequences, and was obtained by PCR amplification (29 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 90 sec) of DNA from BAC clones 272J9, 248E17, 356L5, and 418N24 as template. The primer corresponds to the conserved regions between transmembrane domains 4 and 7. Their nucleotide sequences are:
sense 5′ TCTTCTATGTTGGTGGTTTTA 3′,
reverse 5′ CAATTACTCCTTTGGTACAAATCA 3′.
To assemble the various contigs and estimate the number of V1r genes, BACs were digested with restriction enzymes and subjected to Southern blot hybridization. Because the BAC library was constructed by partial HindIII digestion and cloning of size-selected restriction fragments into the HindIII restriction site of the vector backbone, the BACs were digested preferentially with HindIII to determine fragment sizes accurately. A combination of different probes (ends of BACs, V1r coding sequences, and B1 repeats) were used to assess overlaps.
Mouse/Hamster Radiation Hybrid Mapping
A T31 Mouse/Hamster radiation hybrid panel (male) was purchased from Research Genetics. This panel was constructed by fusing irradiated mouse embryo primary cells (129aa) with hamster cells (McCarthy et al. 1997). The 100 cell lines from the panel were typed by PCR with three independent pairs of primers that yielded diagnostic fragments in the mouse, of different sizes than in the hamster. The primers used were the following:
“30K19 subclone”:
sense 5′ CACTTGAAGGCAAGGTACC 3′,
reverse 5′ GGGTGTTTCAGTTAGATG 3′
PCR product size: mouse, 196 bp; hamster, 300 bp.
“44K9 subclone”:
sense 5′ CTGACACTTGACACGTGCTT 3′
reverse 5′ TTCCAACTGCACATGGGATCA 3′
PCR product size: mouse, 146 bp; hamster, none.
“104P6 T7”:
sense 5′ GTCGCTTTTTCTGGGCTTAG 3′
reverse 5′ AGAGGATTCTGGTCCATGG 3′
PCR product size: mouse, 191 bp; hamster, none.
All reactions were done at least in duplicate.
The PCR products were loaded on a 4% agarose gel (Agarose for the Separation of GeneAmp PCR products, PEXPRESS). Gels were scored for the presence or absence of positive mouse PCR products at the expected size.
Data from the T31 Mouse radiation hybrid panel were submitted to the Jackson Laboratory where they were mapped against the T31 database.
Cloning and Sequencing of ORFs of V1r Genes
BAC fragments digested with the restriction enzyme HindIII, BamHI or EcoRI, were shotgun-subcloned in pBluescript vector (Stratagene), which was digested with the appropriate enzyme and dephosphorylated with calf intestine alkaline phosphatase (New England BioLabs). Single colonies of the BAC subclones were isolated from agar plates containing Luria broth (LB)/ampicillin, and grown for 4 h at 37°C in 96-well plates containing 50 μL of the same media. The grown colonies (40 μL from each of the 96 wells) were spotted onto nylon membranes by capillary action using denaturing buffer (0.5 M NaOH, 1 M NaCl) and neutralizing buffer (0.5 M TrisBase at pH 7.4, 3 M NaCl). The dotted membranes were then washed with 1× SSC and irradiated at 254-nm UV light for 60 sec. Prior to hybridization, the dotted membranes were prehybridized with QuickHyb (Stratagene) for 20 min at 65°C.
BAC subclones containing V1r sequences were identified by hybridizing the dotted membrane for 2 h at 65°C to a probe corresponding to pools #1 or #2 (described above) depending on the contig to which the BAC of interest corresponds. Positive subclones were sequenced using a ABI310 Prism automated DNA sequencer. The primers used for sequencing the positive clones were derived in parallel with our sequencing efforts from conserved regions of the sequenced mouse V1rs. Typically, eight primers were sufficient to cover both strands of the entire coding region of each gene and to generate overlapping regions.
In Situ Hybridization
Tissue was obtained from adult mice and frozen in Tissue Tec (Miles, Inc.). Sections (10 μm) were attached to silanized slides and prepared for in situ hybridization as described in Schaeren-Wiemers and Gerfin-Moser (1993). Digoxigenin-labeled antisense and control sense RNA probes were synthesized that correspond partially or completely to V1r ORFs. The probe used for the in situ hybridization shown in Figure 5 comprised 420 nucleotides (positions 446–866 from start codon) of V1rc7. This probe recognizes multiple bands in Southern blot hybridization of genomic DNA. In situ hybridization was performed at 55°C overnight, and washed twice in 0.1× SSC at 60°C. Signals were developed using alkaline phosphatase-conjugated antibodies to digoxigenin and standard chromogenic substrates (Boehringer Mannheim). Controls for the hybridizations were performed using a sense probe for each antisense probe tested; no signal was detected for any of the sense RNA probes.
Phylogenetic Analyses, Sequence Alignment, and Transmembrane Domains Predictions
The unrooted tree diagram was plotted with the DRAWTREE program of the PHYLIP package (Felsenstein 1993). The predicted amino acid sequences from the V1r genes were aligned using the Clustal V method of the MegAlign program (DNASTAR). The multiple alignment algorithm was described in Higgins and Sharp (1989). The TM domains were predicted by three independent methods: the THMM1.0 server from the Center for Biological Sequence Analysis of The Technical University of Denmark; the PHDhtm, PHDtopology programs of the Predict Protein server from Columbia University (Rost et al. 1995, 1996); and the SOSUI system (ver. 1.0/10, March, 1996) of the Tokyo University of Agriculture and Technology (Hirokawa et al. 1998).
Acknowledgments
We thank Terry Gaasterland for producing the phylogenetic tree. We thank Ken Dewar, Paul Feinstein, Terry Gaasterland, and Tzachi Pilpel for reviewing the manuscript, and Belinda Chang for discussion. Postdoctoral fellowship support to A.R. was from the National Institutes of Health, and to I.R. from the European Molecular Biology Organization, the Human Frontier Science Program Organization, and the Swiss National Science Foundation. Grant support to P.M. was from the March of Dimes Birth Defects Foundation. P.M. was an Alfred P. Sloan, Basil O'Connor, Guggenheim, Irma T. Hirschl, Klingenstein, McKnight, Rita Allen, and Searle Scholar or Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL peter@mail.rockefeller.edu; FAX (212) 327-7310.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.140600.
REFERENCES
- Baldwin J. Structure and function of receptors coupled to G proteins. Curr Opin Cell Biol. 1994;6:180–190. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. Olfactory receptors, vomeronasal receptors, and the organization of olfactory information. Cell. 1997;90:585–587. doi: 10.1016/s0092-8674(00)80518-8. [DOI] [PubMed] [Google Scholar]
- Belluscio L, Koentges G, Axel R, Dulac C. A map of pheromone receptor activation in the mammalian brain. Cell. 1999;97:209–220. doi: 10.1016/s0092-8674(00)80731-x. [DOI] [PubMed] [Google Scholar]
- Berghard A, Buck LB. Sensory transduction in vomeronasal neurons: Evidence for Gαo, Gαi2, and adenlylyl cylcase II as major components of a pheromone signaling cascade. J Neurosci. 1996;16:909–918. doi: 10.1523/JNEUROSCI.16-03-00909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck LB. Information coding in the vertebrate olfactory system. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Davidson JS, Flanagan CA, Becker II, Illing N, Sealfon SC, Millar RP. Molecular function of the gonadotropin-releasing hormone receptor: Insights from side-directed mutagenesis. Mol Cell Endocrinol. 1994;100:9–14. doi: 10.1016/0303-7207(94)90271-2. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Cell biology of olfaction. NY: Cambridge University Press; 1992. [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Seattle, WA: Department of Genetics, University of Washington; 1993. [Google Scholar]
- Halpern M. The organization and function of the vomeronasal system. Annu Rev Neurosci. 1987;10:325–362. doi: 10.1146/annurev.ne.10.030187.001545. [DOI] [PubMed] [Google Scholar]
- Halpern M, Shapiro LS, Jia C. Differential localization of G proteins in the opossum vomeronasal system. Brain Res. 1995;677:157–161. doi: 10.1016/0006-8993(95)00159-n. [DOI] [PubMed] [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Holy TE, Dulac C, Meister M. Responses of vomeronasal neurons to natural stimuli. Science. 2000;289:1569–1572. doi: 10.1126/science.289.5484.1569. [DOI] [PubMed] [Google Scholar]
- Karlson P, Lüscher M. “Pheromones”: A new term for a class of biologically active substances. Nature. 1959;183:55–56. doi: 10.1038/183055a0. [DOI] [PubMed] [Google Scholar]
- Keverne EB. Pheromonal influences on the endocrine regulation of reproduction. Trends Neurosci. 1983;6:381–384. [Google Scholar]
- ————— The vomeronasal organ. Science. 1999;286:716–720. doi: 10.1126/science.286.5440.716. [DOI] [PubMed] [Google Scholar]
- Lancet D, Ben-Arie N. Olfactory receptors. Curr Biol. 1993;3:668–674. doi: 10.1016/0960-9822(93)90064-u. [DOI] [PubMed] [Google Scholar]
- Leinders-Zufall T, Lane AP, Puche AC, Ma W, Novotny MV, Shipley MT, Zufall F. Ultrasensitive pheromone detection by mammalian vomeronasal neurons. Nature. 2000;405:792–796. doi: 10.1038/35015572. [DOI] [PubMed] [Google Scholar]
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- McCarthy LC, Terrett J, Davis ME, Knights CJ, Smith AL, Critcher R, Schmitt K, Hudson J, Spurr NK, Goodfellow PN. A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res. 1997;7:1153–1161. doi: 10.1101/gr.7.12.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Molecular biology of odorant receptors in vertebrates. Annu Rev Neurosci. 1999a;22:487–509. doi: 10.1146/annurev.neuro.22.1.487. [DOI] [PubMed] [Google Scholar]
- ————— Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999b;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Nelson DR, Coon MJ, Estabrook RW, Feyereisen R, Fujii-Kuriyama Y, Gonzalez FJ, Guengerich FP, Gunsalus IC, Johnson EF, et al. The P450 superfamily: Update on new sequences, gene mapping, and recommended nomenclature. DNA. Cell Biol. 1991;10:1–14. doi: 10.1089/dna.1991.10.1. [DOI] [PubMed] [Google Scholar]
- Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst WC, Snyder LA, Schuster DI, Brosius J, Sealfon SC. Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Greer CA, Mok MY, Mombaerts P. A putative pheromone receptor gene expressed in human olfactory mucosa. Nat Genet. 2000;26:18–19. doi: 10.1038/79124. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Feinstein P, Mombaerts P. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell. 1999;97:199–208. doi: 10.1016/s0092-8674(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Science. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Science. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba NJP, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Saito H, Mimmack ML, Keverne EB, Kishimoto J, Emson PC. Isolation of mouse vomeronasal receptor genes and their co-localization with specific G-protein messenger RNAs. Mol Brain Res. 1998;60:215–227. doi: 10.1016/s0169-328x(98)00183-1. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: In situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Schwartz T. Locating ligand-binding sites in 7TM receptors by protein engineering. Curr Opin Biotechnol. 1994;5:434–444. doi: 10.1016/0958-1669(94)90054-x. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. Discrimination of molecular signals by the olfactory receptor neuron. Neuron. 1994;13:771–790. doi: 10.1016/0896-6273(94)90245-3. [DOI] [PubMed] [Google Scholar]