Abstract
Compounds that modulate microtubule dynamics include highly effective anticancer drugs, leading to continuing efforts to identify new agents and improve the activity of established ones. Here, we demonstrate that [3H]-labeled halichondrin B (HB), a complex, sponge-derived natural product, is bound to and dissociated from tubulin rapidly at one binding site per αβ-heterodimer, with an apparent Kd of 0.31 μM. We found no HB-induced aggregation of tubulin by high-performance liquid chromatography, even following column equilibration with HB. Binding of [3H]HB was competitively inhibited by a newly approved clinical agent, the truncated HB analogue eribulin (apparent Ki, 0.80 μM) and noncompetitively by dolastatin 10 and vincristine (apparent Ki’s, 0.35 and 5.4 μM, respectively). Our earlier studies demonstrated that HB inhibits nucleotide exchange on β-tubulin, and this, together with the results presented here, indicated the HB site is located on β-tubulin. Using molecular dynamics simulations, we determined complementary conformations of HB and β-tubulin that delineated in atomic detail binding interactions of HB with only β-tubulin, with no involvement of the α-subunit in the binding interaction. Moreover, the HB model served as a template for an eribulin binding model that furthered our understanding of the properties of eribulin as a drug. Overall, these results established a mechanistic basis for the antimitotic activity of the halichondrin class of compounds.
INTRODUCTION
Because the sponge-derived halichondrin B (HB) (Figure 1A) modulates microtubule dynamics by a novel mechanism of action relative to other known antitubulin agents,1,2 it is an attractive anticancer therapeutic candidate. However, its clinical development was hampered by its scarcity and its lengthy total synthesis.3–5 Nonetheless, work on the halichondrin family has borne fruit with the recent approval of the truncated HB analogue eribulin6 (Figure 1B) for the treatment of metastatic breast cancer.7,8 Despite progress in understanding the mechanism of action of the halichondrin family of compounds, the binding site for these agents on tubulin remains uncertain and is the focus of our studies.
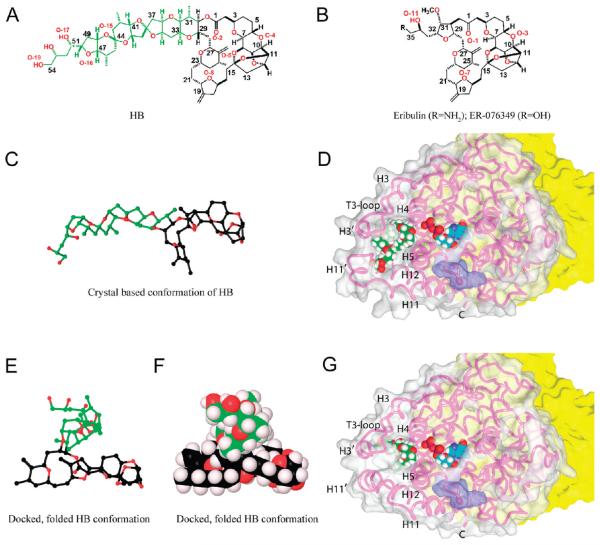
Figure 1.
Molecular structures of HB, eribulin, and ER-076349 and the binding poses of HB and eribulin on β-tubulin. (A) Structure of HB (color code in E/F). (B) Structures of eribulin and ER-076349. (C) Ball-and-stick structure of HB in a conformation based on the crystal structure of NBE (color code in E/F). (D) HB bound to β-tubulin. The surface of β-tubulin is gray (α-tubulin is yellow), except for the portion colored purple that interacts with vinblastine in the 1Z2B structure.26 The β-tubulin polypeptide chain is shown as a pink strand, with key features indicated. The E-site GDP and HB are shown with cyan and green carbons, respectively. Other atoms: hydrogen, white; oxygen, red; nitrogen, blue; and phosphorus, orange. (E and F) Folded conformation of HB, obtained from docking studies presented here, rendered in ball-and-stick and Corey–Pauling–Koltun (CPK), respectively. Carbon atoms of the side chain are green to illustrate the major conformational change HB undergoes from the derived crystal structure to the bound conformation. Macrocycle carbon atoms are black, and oxygen atoms are red. The tight packing of the HB side chain on top of the macrocycle demonstrates the hydrophobic collapse in the folded conformations, so that solvent exposure of HB is minimized. Hydrogen atoms (light lavender) are shown in the CPK diagram. (G) Eribulin bound to β-tubulin. Details as in panel D.
Inhibitors of microtubule assembly usually inhibit the binding of colchicine or vinblastine to tubulin. The latter compounds are often highly cytotoxic natural products that show noncompetitive patterns of inhibition of vinblastine binding, implying distinct binding sites on tubulin. These include the peptides and the depsipeptides dolastatin 10 (D10),9 hemiasterlin,10 cryptophycin 1,11 and vitilevuamide12 and the macrocyclic polyether compounds HB1 (Figure 1A) and spongistatin 1.13 Antimitotic peptides act as competitive inhibitors of the binding of D10 to tubulin,10,11 while, in contrast, eribulin and spongistatin 1 are noncompetitive inhibitors of D10 binding.2,13
While noncompetitive inhibition implies allosteric effects causing reduced ligand affinity, all inhibitors of vinblastine binding also inhibit GDP/GTP entry into and exit from the exchangeable nucleotide site (E-site) on β-tubulin. This led us to postulate that there is a region on tubulin near the E-site that should be designated the “vinca domain” that was involved in causing these complex inhibitory effects.9
We had [3H]HB prepared to explore HB binding to tubulin and to examine effects of other vinca domain drugs on this binding interaction. Our findings led us to extensively revise the binding model for HB and eribulin previously proposed.2 The revised models provide a common binding site for HB and eribulin on β-tubulin and explain why the pharmacophore for the antimitotic properties of the halichondrin class of compounds resides in the macrocycle.
MATERIALS AND METHODS
Materials
Bovine brain tubulin14 was freed of unbound nucleotide.15 HB was from Lissodendoryx species sponges16 and tritiated at AmBios Laboratories (0.9 Ci/mmol). Eribulin, vincristine, and maytansine were from the Drug Synthesis & Chemistry Branch, National Cancer Institute. Spongistatin 1,17 D10,18 hemiasterlin,19 auristatin PE,20 and diazonamide A21 were isolated or prepared as described previously. Cryptophycin 1, vitilevuamide, and N,N-dimethylvalylvalyl-N-methylvalylprolylproline were generous gifts, respectively, from Merck Research Laboratories, Dr. C. M. Ireland (University of Utah), and the Genzyme Corporation.
Biochemical Methods
Binding studies were performed by centrifugal gel filtration in tuberculin syringe barrels with Sephadex G-50 (superfine) swollen in 0.5 mM MgCl2–0.1 M 4-morpholineethanesulfonate (Mes) (1 M stock solution adjusted to pH 6.9 with NaOH),22 which was also the reaction buffer. All operations were at room temperature (21 °C). The hydrated bed volume for the syringe columns was 1.0 mL, and excess buffer was removed by a 4 min centrifugation at 2000 rpm in a Beckman Allegra 6KR centrifuge with a GH-3.8A horizontal rotor. Reaction volume was 0.3 mL, and, following incubation, two 0.145 mL aliquots were applied to precentrifuged syringe columns. Centrifugation was as before.
Analysis by size exclusion high-performance liquid chromatography (HPLC) was performed with an Agilent Technologies system at room temperature. Either one or two Shodex protein KW-803 columns (8 × 300 mm), together with a Shodex KWG guard column (6 × 50 mm), were used. Column outflow was analyzed for A280 and radiolabel content, if appropriate, with an IN/US β-RAM model 3 flow detector.
Molecular Modeling
The Schrodinger Maestro 9 program running on a Dell Precision 690 with Red Hat Enterprise Linux 4 was used in the modeling studies. Minimizations and dynamics simulations were performed as described previously,23 using the OPLS 2005 force field. For constrained simulations, the force constant for distance constraints was 100 kcal/mol, and the restrain function was a flat-bottomed well.
Initially, we used a conformation of HB (Figure 1C) based on the crystal structure of norhalichondrin A p-bromophenacyl ester (NBE) (Cambridge Structural Database entry DAPRUH).24 That of eribulin was derived from the HB structure. The protein template was derived from the electron crystallographic model of β-tubulin25 [Protein Data Bank (PDB) code 1JFF]. Other 3-dimensional β-tubulin structures26–29 were analyzed, and no significant differences were found versus 1JFF.
Analysis with Sitemap of these β-tubulin structures failed to identify a binding site for HB consistent with all experimental data. Therefore, the HB and the 1JFF structures underwent molecular dynamics simulations to sample their solution behavior. This revealed a site in the vinca domain susceptible to melting that could become intermittently available for HB binding. Figure 1D shows the location of this potential HB site on β-tubulin, with the HB carbons colored green and the E-site GDP shown nearby (magenta carbons). The surface of β-tubulin interacting with vinblastine in the 1Z2B structure26 is indicated in purple.
Only conformations of HB that were highly divergent from that derived from the crystal structure of NBE (Figure 1C) would bind in this β-tubulin site. A collection of “folded,” low-energy HB conformations that represented reasonable solution structures were therefore used in this study.
The proposed HB binding site is formed by large loop regions (residues 94–110 and 399–414) on two sides and by helices H4 and H5 as its base. The 94–110 loop region contains the T3-loop and the short helix H3′, while the 399–414 loop region is composed in part of the short helix H11′ (nomenclature as in ref 25). The conformational flexibility of these two loop regions is augmented by the fact that the 399–414 loop is contiguous with the terminal helix H12 of the β-tubulin structure. The core of this site is formed by the hydrophobic collapse of three key residues from different structural elements: Trp103 of the T3-loop, Leu189 of helix H5, and Tyr408 of helix H11′ (residue numbering as in ref 25).
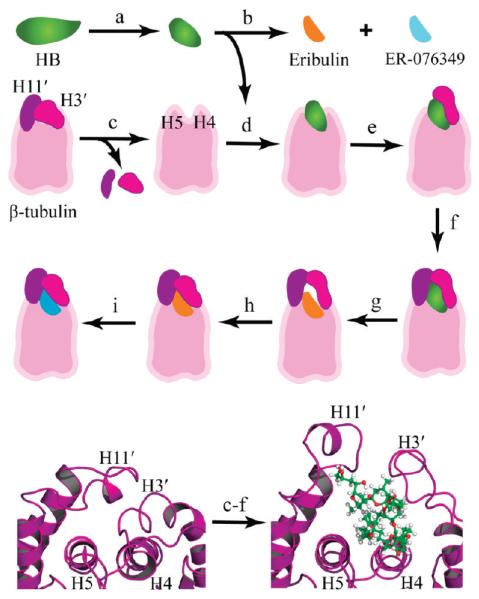
Figure 2 outlines the stepwise modeling methodology used to generate the binding models. We had developed and previously described a modeling methodology for engineering a large protein binding pocket for a complex natural product.23 Following this approach, residues 94–110 and 399–414 were removed from β-tubulin to allow HB access to the binding site. A “folded” HB molecule was then manually docked onto helix H4, which formed the base of the binding site. The macrocycle showed an excellent stereoelectronic fit to the glycine-rich helix H4, due in part to the fact that the small Gly144 and Gly148 hydrogen side chains are dovetailed into the central opening of the macrocycle. With GDP and β-tubulin fixed in Cartesian space, HB was subjected to iterative cycles of energy minimization, resulting in an optimal docking of HB onto the truncated β-tubulin.
Figure 2.
Modeling the HB binding site. Top three rows: stepwise approach used to model the binding modes. (a) The crystal-based structure of HB was subjected to molecular dynamics, and folded conformations were selected for docking studies. (b) Eribulin and ER-076349 structures were generated from the HB folded conformations. (c) H3′ and H11′ loops were removed from a β-tubulin model to unmask the HB binding site for ligand docking. (d) HB was docked and refined. (e) The H3′ loop was reattached to β-tubulin, and the protein–ligand model was refined. (f) The H11′ loop was reattached to β-tubulin, and the atom–atom interactions were optimized to yield the HB binding model. (g) Eribulin was docked based on superimposition of the common macrocycle shared with HB. (h) Constrained simulations were used to fold the H3′ and H11′ loops onto eribulin. (i) ER-076349 was docked. Bottom row: cartoon rendering showing the key secondary structures of the HB binding site and the conformational changes associated with HB binding. The β-tubulin is rendered in purple ribbon, and HB is shown in ball-and-stick with carbon, oxygen, and hydrogen atoms colored green, red, and white, respectively. Left panel: The protein fold in the 1JFF β-tubulin structure shows the packing of the H3′ and H11′ loops onto helices H4 and H5, which occludes the HB binding site. Right panel: The protein fold in the HB binding model shows the conformational switch of the H3′ and H11′ loops upon HB binding.
The next steps involved refolding first the 94–110 loop and then the 399–414 loop onto bound HB. In its conformation from the 1JFF structure, the 94–110 loop was returned to the model. However, in its position from the 1JFF structure, the 94–110 loop occupies the same Cartesian space as the docked HB molecule. Accordingly, the 94–110 loop was translated away from the β-tubulin–HB complex to remove steric bumps as indicated by a 0.89 setting in van der Waals overlap. The 94–110 loop was then reattached to β-tubulin via an elongated covalent bond from residue 94 to residue 93. With the bound HB and β-tubulin structures outside of the loop segments fixed in Cartesian space, minimization and dynamics simulations were used to generate a biochemically realistic 93–94 bond length and refine the loop conformation. In order to determine a bioactive fold of the H3′ segment, distance constraints were used to generate different folding scenarios for evalution. Since the hydrophobic effect is the main impetus for protein folding and ligand binding, interactions involving Trp103, the largest hydrophobic residue in the 94–110 segment, were found to be central to determining the bioactive fold of this loop segment. An optimal 94–110 loop conformation was obtained using distance constraints that brought the Trp103 Cγ atom to within van der Waals packing distance of the C-42 methyl group of HB during molecular dynamics. The refolding of the H3′ segment was completed by reconnecting residue 110 with residue 111 in β-tubulin via an elongated covalent bond and energy minimizing the complex.
A similar approach was used to model the folding of the 399–414 loop onto HB. The 399–414 segment from the 1JFF structure was returned to the model and found to occupy the Cartesian space of the bound HB molecule and the H3′ segment. Accordingly, the 399–414 segment was translated in space to remove steric bumps. The 399–414 loop was reattached to β-tubulin by linking residue 399 with residue 398 using an elongated covalent bond. With atoms outside of the 399–414 segment fixed in Cartesian space, minimization and dynamics simulations were employed to generate a biochemically realistic 398–399 bond length and refine the 399–414 loop conformation. Distance constraints that packed the Met418 Cγ atom onto the C-46 methyl group of HB during simulations were found to provide an optimal fold of this H11′ segment. Forming an elongated bond between residues 414 and 415 and using energy minimization to generate a biochemically realistic bond length completed the refolding of the H11′ segment.
With the H3′ and H11′ segments refolded onto bound HB, the model was subjected to iterative cycles of energy minimization without distance constraints. Structural analysis of the β-tubulin model showed that it maintained its overall secondary structure content relative to the starting 1JFF structure and that the structural quality was maintained as evident by the fact that there were no highly unfavorable φ/ψ bond angles detected by Procheck.30 The final binding pose for HB had a favorable Gscore of −12.1, and the ligand is shown undocked in Figure 1E and F as ball-and-stick and space-filling models, respectively.
To determine the energy cost for the conformational changes in the H3′ and H11′ loops upon HB binding, we calculated the binding energies of these two loops on the protein surface of an energy minimized model of the 1JFF structure in which the backbone was fixed in Cartesian space during refinement and on the protein surface of the HB model with bound HB. Calculations were performed using the Calculate Binding Energies protocol in Discovery Studio 2.5 with implicit solvent model set to distance-dependent dielectric, a nonbond list radius of 14.0 Å, electrostatics set to spherical cutoff, and force field set to CHARMM. To define the H3′ and H11′ loops as substrate for these calculations, residues 93, 111, 398, and 415 in both models were removed, and the H3′ and H11′ loop structures were identified as the input ligands. For the 1JFF model, the H3′ and H11′ loops had calculated binding energies of −120.3 and −87.0 kcal/mol, respectively. For the HB model, similar values for the H3′ and H11′ loops were −94.3 and −119.0 kcal/mol, respectively. These results show that HB binding stabilizes the H11′ loop by 32 kcal/mol but destabilizes the H3′ loop by 26 kcal/mol. With a sum for the two loops of −207.3 and −213.3 kcal/mol in the 1JFF model and the HB model, respectively, the H3′ and H11′ conformations in the HB model are overall 6 kcal/mol more stable than in the unliganded 1JFF model, an indication that this conformational switch is energetically feasible.
Eribulin was modeled into the HB site by superimposing it onto the HB macrocycle. Since the amino group of eribulin has a calculated pKa of 8.8, it exists predominantly as the ammonium cation under physiological conditions. Accordingly, the protonated, cationic form of eribulin was used in modeling its binding mode and docking trajectory. The absence of the 300 Da side chain in eribulin meant that segments of the β-tubulin structure had to be remodeled in order to generate a structurally reasonable binding pocket for eribulin.
Because the H11′ loop region (residues 399–414) contains large hydrophobic residues in Trp407, Tyr408, and Met413, the H11′ segment was folded onto bound eribulin first, and this was followed by refolding the H3′ (residues 94–110) segment.
Constrained dynamics and minimization simulations were used to generate different folding scenarios for the H11′ segment. During simulations, eribulin and residues outside of the H3′ and H11′ segments were fixed in Cartesian space. An optimal H11′ fold was obtained by using distance constraints to pull the aromatic side chain of Trp407 to within van der Waals packing of the C-5 and C-18 atoms of eribulin and, concomitantly, pull the aliphatic side chain of Met413 to within packing of the C-22 and C-28 atoms of eribulin. In this fashion, the side chains of Trp407 and Met413 are wedged against the central opening of the eribulin macrocycle.
The H3′ segment was subsequently modeled via evaluation of different folding scenarios. The optimal folding of the H3′ segment was driven by hydrogen-bonding constraints on opposite sides of the binding pocket. At the plus end of β-tubulin, the hydrogen-bonding distance constraint was between the side chain carbonyl of Asn101 and the side chain amine of Lys402. At the other end of the binding site, the hydrogen-bonding distance constraint was between the imidazole nitrogen of His107 and the carboxylate oxygen of Asp414. These two hydrogen-bond constraints brought the H3′ and H11′ segments together to close opposite ends of the binding site.
The complex was refined using iterative cycles of energy minimization. The overall conformation of the docked eribulin-β-tubulin complex is shown in Figure 1G, and the final eribulin pose had a favorable Gscore of −9.1. Unconstrained molecular dynamics of both the HB and eribulin complexes with tubulin for 100 ps at 300 K showed that both ligands remained in their binding pockets.
The binding model of ER-076349 was generated from the eribulin model by exchanging the C-35 amine of eribulin with a hydroxyl group and subsequently minimizing the structure with GDP and β-tubulin fixed in Cartesian space. The resulting pose for ER-076349 was scored using Glide XP, yielding a Gscore of −8.1.
The binding events of eribulin and ER-076349 were studied using constrained molecular dynamics and hydropathic analysis as previously described.31 Constrained molecular dynamics was used to undock the ligands from their binding poses in β-tubulin, and the resulting trajectories were reversed in order to simulate the binding event. In brief, a methane molecule was positioned 40 Å from the aperture of the bound ligands to serve as a constraining target. While the methane molecule and tubulin peptide backbone were fixed in Cartesian space, distance constraints between the methane carbon and the C-10 atom of eribulin or ER-076349 were decreased in 0.1 Å increments during molecular dynamics until the final distance of 20 Å was reached. At each 0.1 Å increment, molecular dynamics was performed using 600 fs simulations with a time step of 0.2 fs at 300 K. The undocking trajectory was saved as individual frames, and the 1000 collected frames were reversed to produce a docking trajectory of either eribulin or ER-076349 onto β-tubulin.
Hydropathic analysis of the docking trajectories was accomplished using the HINT program,32 which was adapted as a customized module in the Pipeline Pilot platform. The individual conformations of eribulin and ER-076349 were extracted from the 1000 frames of the docking trajectories and analyzed using the HINT program, which calculated six descriptive interaction terms for each frame: (1) hydrogen bonding, (2) acid–base, (3) hydrophobic, (4) acid–acid, (5) base–base, and (6) hydrophobic-polar.
RESULTS
Biochemical Studies Binding Mechanism
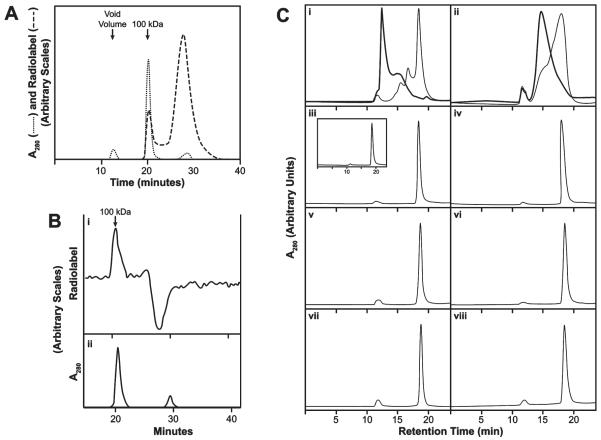
We prepared [3H]HB to study its binding mechanism and interrelationships between different binding regions of the vinca domain. Centrifugal gel filtration was used to assess the reaction, but binding and dissociation were too rapid for rate measurements. Scatchard analysis indicated one HB binding site per αβ-dimer, but the derived Ka values varied with experimental conditions, implying failure to achieve equilibrium. This failure was demonstrated by variability in stoichiometry dependent on column volume, with 10 μM HB and tubulin. With the syringe columns (bed volume, 1 mL), binding stoichiometry was 0.48 ± 0.03; with one HPLC column (16.5 mL), 0.16 ± 0.01; with two HPLC columns in series, 0.01.
The one-column HPLC experiments showed that [3H]HB eluted with 100 kDa tubulin at several tubulin and [3H]HB concentrations, as shown in Figure 3A with both components at 5 μM. Hummel–Dreyer33 HPLC was successfully used to obtain binding constants for vinca alkaloids,34 and we utilized this equilibrium method to determine a Kd value for HB. In the Figure 3B experiment, 50 μg of tubulin was injected into a column equilibrated with 1.0 μM[3H]HB. A classic Hummel–Dreyer pattern was obtained: radiolabel rose above the equilibrium value as 100 kDa protein emerged from the column, returned to the equilibrium value, followed by an offsetting trough. In experiments with varying tubulin and HB concentrations, apparent Kd values of 0.28–0.40 μM were obtained (average, 0.31 ± 0.07 μM).
Figure 3.
HB binds to 100 kDa αβ-heterodimer. (A) Nonequilibrium binding of [3H]HB to tubulin. Dashed line, radiolabel; dotted line, A280. A 0.1 mL sample containing 5 μM each of tubulin and [3H]HB was injected into a single Shodex KW-803 column with a guard column. Running buffer: 0.1 M Mes –0.5 mM MgCl2 at 0.5 mL/min. First absorbance peak (12 min) is at the void volume (denatured tubulin aggregates). Second peak (20 min) emerges at 100 kDa.10 Third peak (29 min) contains unbound small molecule(s). (B) HPLC Hummel–Dreyer evaluation of the binding of [3H]HB to tubulin. (i) Radiolabel. (ii) A280. Chromatography as in panel A. Equilibration with 1.0 μM [3H]HB. Injected sample: 50 μg tubulin (0.1 mL). (C) Differences in nonequilibrium versus equilibrium HPLC gel filtration occur with vinblastine or D10 but not HB, spongistatin 1, or maytansine. Two Shodex KW-803 columns with a guard column were used in series. Flow rate: 1.0 mL/min. Injection sample: 0.1 mL, containing 10 μM tubulin. “Unequilibrated columns” means the column system had been equilibrated with 0.1 M Mes–0.5 mM MgCl2; “equilibrated columns” means the columns had been equilibrated with the buffer containing the indicated drug. Drug concentration, except with D10 (panel ii), was 20 μM in the injected sample and when the column system was equilibrated with drug (darker curve in i; panels iv, vi, and viii). The D10 concentration was 3 μM in the injected samples and equilibrated columns (panel ii, darker curve). (i) Vinblastine. Unequilibrated columns, lighter curve. Equilibrated columns, darker curve. (ii) D10. Unequilibrated columns, lighter curve. Equilibrated columns, darker curve. (iii) HB, unequilibrated columns. Inset: protein elution profile following injection of tubulin only into unequilibrated columns. (iv) HB, equilibrated columns. (v) Spongistatin 1, unequilibrated columns. (vi) Spongistatin 1, equilibrated columns. (vii) Maytansine, unequilibrated columns. (viii) Maytansine, equilibrated columns.
These experiments, with relatively low tubulin and HB concentrations, showed no HB-induced tubulin aggregation. Because eribulin was reported to induce tubulin aggregation,35 we performed an equilibrium study with higher concentrations of protein and HB. Two columns in series were used because better resolution of included oligomer peaks occurs.10 Our findings are summarized in Figure 3C. The tubulin profile without drug is shown in Figure 3C-iii (inset). The injection volume was always 0.1 mL containing 10 μM tubulin and 20 μM drug, except for D10 studies, where only 3 μM drug was used. This was because the tight binding of D10 to tubulin36 shifts most of the protein to the void volume with only 5 μM D10.
Vinblastine and D10 were examined as controls known to cause tubulin aggregation. In Figure 3C-i, studies with vinblastine are shown. The lighter line shows the effect of mixing vinblastine and tubulin in the reaction mixture and injecting the sample with no vinblastine in the column bed. The darker line shows the same experiment, except that the columns had been pre-equilibrated with 20 μM vinblastine. Figure 3C-ii presents similar studies with 3 μM D10. Equilibration with either compound caused the protein profile to shift to larger aggregates.
With HB, there was no difference whether the tubulin–drug mixture was injected into the column system equilibrated with buffer (Figure 3C-iii, principal tracing) or into the columns equilibrated with HB (Figure 3C-iv). HB did not induce detectable tubulin aggregation.
We also evaluated spongistatin 1 and maytansine, which also do not promote tubulin aggregation. This is confirmed here following injection of a mixture of tubulin and spongistatin 1 (Figure 3C-v) or tubulin and maytansine (Figure 3C-vii) into the column system equilibrated with buffer. No change in the protein profile occurred when the tubulin-spongistatin 1 or tubulin-maytansine mixture was injected into columns equilibrated with spongistatin 1 or maytansine (Figure 3C-vi and -viii, respectively).
Biochemical Studies: Inhibition of [3H]HB Binding
HB is a noncompetitive inhibitor of [3H]vinblastine binding to tubulin, and eribulin is a noncompetitive inhibitor of [3H]vinblastine and [3H]D10 binding.1,2 [3H]HB allowed us to perform the reverse experiments and evaluate other vinca domain compounds for potential inhibitory effects on HB binding. We examined HB analogues (eribulin and isohalichondrin B), peptides/depsipeptides (D10, auristatin PE, hemiasterlin, cryptophycin 1, vitilevuamide, diazonamide A, dolastatin 15, and N,N-dimethylvalylvalyl-N-methylvalylprolylproline), vinca alkaloids (vinblastine and vincristine), competitive inhibitors of vinca alkaloid binding (maytansine and rhizoxin),9 and spongistatin 1. Strong inhibition of [3H]HB binding occurred with eribulin, D10, auristatin PE, and hemiasterlin; moderate inhibition with isohalichondrin B, vinblastine, and vincristine; weak inhibition with spongistatin 1; and negligible inhibition with the other compounds.
Eribulin, vincristine, and D10 (Figure 4A–C, respectively) underwent detailed studies. Experimental data were examined in the Hanes format,37 with competitive inhibition generating a family of parallel lines and noncompetitive inhibition a family of lines intercepting on the negative abscissa, and by the Dixon method37 to obtain apparent Ki values. A competitive Hanes pattern was obtained with eribulin (Figure 4A) (apparent Ki value, 0.80 ± 0.3 μM), while vincristine (Figure 4B) and D10 (Figure 4C) yielded noncompetitive Hanes patterns (apparent Ki values, 5.4 ± 2 μM for vincristine, 0.35 ± 0.2 μM for D10).
Figure 4.
Hanes plots of inhibition of the binding of [3H]HB to tubulin by (A) eribulin, (B) vincristine, or (C) D10. Each 0.3 mL reaction mixture contained 2.5 μM tubulin and the indicated concentrations of [3H]HB and inhibitor. Reaction mixtures were incubated 30 min. (A) Reaction mixtures contained eribulin:○, none; Δ, 1 μM;▽, 2 μM; □, 3 μM; ◇, 4 μM. (B) Reaction mixtures contained vincristine:○, none; Δ, 3 μM;▽, 9 μM; □, 12 μM. (C) Reaction mixtures contained D10: o, none; Δ, 2 μM; ▽, 3 μM; □, 4 μM; ◇, 6 μM.
Molecular Modeling: the HB-β-Tubulin Complex
Previously,2 taking into account a description of eribulin-induced tubulin aggregates,35 we docked eribulin and a 30 Å long conformation of HB, based on the crystal structure of NBE (Figure 1C),24 between two different αβ-heterodimers in the 1Z2B structure.26 Here, since we demonstrated binding of HB only to the 100 kDa αβ-heterodimer, with no aggregate formation, we concluded that our previous binding model was no longer consistent with experimental data and constructed a model with the HB binding site within β-tubulin only. This involved generating a folded conformation of HB and determining conformational changes within β-tubulin to produce a site that could accommodate both HB and eribulin. The β-tubulin structure underwent conformational changes in two large loop regions, which includes the short helices H3′ and H11′, to unmask the HB site. Analysis of other reported three-dimensional tubulin structures26–29 revealed that the HB site was similarly masked in each structure. In the folded structure of HB (Figure 1E and F), about 8 by 15 Å, the polyether tail folds over and flattens the macrocycle and minimizes HB exposure to the aqueous environment (Figure 5A).
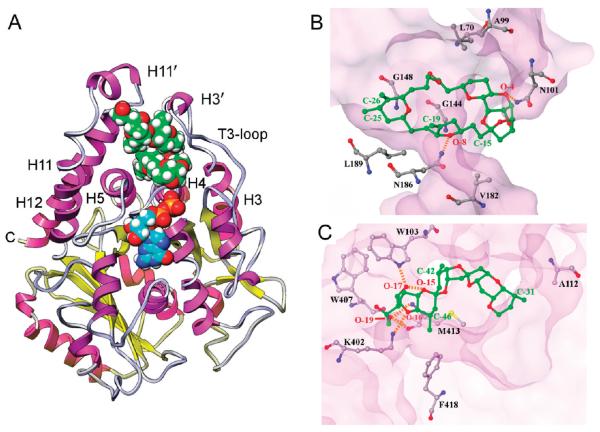
Figure 5.
(A) HB binding site in β-tubulin. β-tubulin, rendered in ribbon, with helices in magenta, sheets in yellow, and loops in gray. E-site GDP and HB are drawn in CPK. Nitrogen, oxygen, phosphate, and hydrogen atoms are blue, red, orange, and white, respectively. The carbon atoms of HB are green; those of GDP are cyan. The HB site is at the junction of a helix-rich tertiary structure adjacent to the E-site. (B and C) Crucial HB interactions at the binding site involving the macrocycle and side chain, respectively. β-tubulin is rendered as a transparent surface, with important amino acid residues labeled and rendered in ball-and-stick, as is the (B) HB macrocycle and the (C) HB side chain. Oxygen atoms are red, nitrogen blue, and sulfur yellow. Residue carbons of β-tubulin are gray and HB carbons green. HB atom positions are labeled as in Figure 1A. Hydrogen bonds are indicated by orange dashed lines.
As described in detail in the Materials and Methods Section and outlined in Figure 2, complementary structural changes were made to the HB and β-tubulin structures to generate a HB binding model. The HB site is demarcated on one side by helix H3′ and the T3-loop (residues 94–110), on a second side by helices H11 and H11′ (residues 399–414), and on a third side, as a base closer to the interior of the protein, by helices H4 and H5 (Figure 5A). To model the HB site, initially residues 94–110 and 399–414 were removed from the 1JFF structure to expose the binding site, and the HB macrocycle was packed onto helix H4 (Figure 5B). In a stepwise process, the two removed loop regions were reattached and refolded onto the HB-containing β-tubulin structure, using constrained modeling simulations to generate multiple scenarios for evaluation and selection of bioactive folds based on stereoelectronic complementarity.
At the end, the HB model shows that the macrocycle portion of the compound forms two hydrogen bonds to tubulin. The first is between the Asn101 side chain amide and O-4, and the second is between the Asn186 side chain amide and O-8 (Figure 5B).
In addition to these hydrogen bonds, stabilization is provided by favorable hydrophobic contacts on opposite sides of the macrocycle (Figure 5B). One side of the HB macrocycle is packed against the hydrophobic side chains of Val182 and Leu189 in helix H5, and the other side against the side chains of Leu70 from the T2-loop and Ala99 from the T3-loop.
The side chain of HB also forms hydrophobic contacts with β-tubulin (Figure 5C). The C-31 methyl group is packed against the Ala112 side chain from helix H3, the C-42 methyl group against the Trp103 indole ring of the T3-loop, and the C-46 methyl group against the Met413 aliphatic side chain and the Phe418 aromatic ring. The side chain binding is further stabilized by two hydrogen bonds involving the terminal O-19 hydroxyl group of HB and the amide NH and the carbonyl moiety of Met413. The O-17 hydroxyl group and the tetrahydrofuran O-16 oxygen atom are hydrogen bonded to the Trp103 indole -NH and the Lys402 ε-amino group, respectively. The fold of the side chain of HB is stabilized by an intramolecular hydrogen bond between the O-17 hydroxyl and the tetrahydropyran O-15 oxygen atom.
The HB site location is consistent with its ability to inhibit vinblastine binding and nucleotide exchange. The closest heavy atom-to-atom distance to bound vinblastine (Figure 1D) is about 16 Å, consistent with the mutually noncompetitive effects of HB and vinca alkaloids. The bound HB is much closer to the E-site (Figures 1D and 5A), with the closest heavy atom-to-atom distance to GDP being 5 Å. Helix H4 is an important component of both the HB and E-sites. The longer side of HB rests on top of helix H4 in a parallel fashion and interacts largely with its N-terminal end. The N-terminal of helix H4 also interacts with the β-phosphate of GDP. Because the N-terminal of helix H4 has glycine residues at positions 142–144, 146, 148, and 150, this minimizes unfavorable side chain interactions with the terminal nucleotide phosphate group and allows the protein structure to flex to accommodate GDP or GTP. The binding of HB takes advantage of this topology in helix H4. The HB macrocycle bears few substituents from C-2 to C-23, providing a relatively flat binding surface. Helix H4 is also glycine rich in the direction of HB binding, and the small central opening in the HB macrocycle is docked over Gly144 and Gly148 (Figure 5B).
Molecular Modeling: the Eribulin-β-Tubulin Complex
Because of its clinical use, competitive inhibition of [3H]HB binding (Figure 4A), and three-fold stronger interaction with tubulin as compared with HB,2 we modeled eribulin into the HB site, using the HB model as an initial template, with the identical macrocycles superimposed. Since eribulin lacks virtually the entire side chain of HB, this left a large gap between eribulin and β-tubulin. This was closed via hydrophobic collapse of the H3′ and H11′ moieties, using constrained simulations (Figure 6A). While eribulin and residues outside of the H3′ and H11′ segments were fixed in Cartesian space, constrained dynamics and minimization simulations generated different folding scenarios for the H11′ motif onto eribulin. An optimal H11′ fold was obtained by using distance constraints to pack the aromatic side chain of Trp407 with the eribulin C-5 and C-18 atoms and the aliphatic side chain of Met413 against the eribulin C-18, C-21, and C-22 atoms (Figure 6B). Optimal H3′ packing was achieved by distance constraints that led to formation of hydrogen bonds between the side chain amide carbonyl of Asn101 and the ε-amino group of Lys402 and between an imidazole nitrogen of His107 and the carboxylate oxygen of Asp414 (Figure 6C). The overall changes between the eribulin and HB models can be visualized by comparing Figures 1G and 6A (eribulin) with Figures 1D and 5A (HB).
Figure 6.
(A) Eribulin binding site in β-tubulin. β-tubulin structure and the CPK structures of eribulin and GDP are shown as described in the Figure 5 legend. (B and C) Crucial eribulin interactions at the binding site. Panel C also shows new intra-β-tubulin hydrogen bonds (orange dashed lines) introduced by refolding the protein over the macrocycle.
Eribulin binding is stabilized by five hydrogen bonds to β-tubulin (Figure 6C). Two hydrogen bonds are similar to ones formed by HB and occur in the macrocycle. The first is between the side chain amide moiety of Asn101 and O-3, and the second is between the side chain amide of Asn186 and O-7. The three remaining hydrogen bonds are unique to the eribulin model. The first is between the O-11 hydroxyl at C-34 and the Thr109 carbonyl oxygen atom. The other two hydrogen bonds are formed by the amino group (N-1) at C-35 with the His107 and Thr109 carbonyl oxygen atoms.
Hydrophobic contacts also are critical in the eribulin model (Figure 6B). The C-13-to-C-21 segment of eribulin is packed against helix H5, particularly the hydrophobic side chains of Val182 and Leu189. On the opposite side of the macrocycle, the C-2-to-C-6 segment of eribulin is packed against the side chains of Leu70 from the T2-loop and Ala99 from the T3-loop. Moreover, the specificity of eribulin binding is defined by the interactions of its substituent olefin and methyl groups with β-tubulin. The C-26 olefin is packed against the side chain of Leu152, the C-25 methyl against the side chain methyl of Thr151 and the isobutyl side chain of Leu189, the C-19 olefin against the side chains of Leu189, Met413, and Phe418, and the C-31 methoxy against the Ala99 methyl moiety.
A conformational switch of two large hydrophobic residues of β-tubulin is a major difference between the HB and eribulin models. In the HB model, the aromatic side chain of Trp407 is positioned at the solvent interface and is stacked face-to-face with the Trp103 side chain, which is itself packed against the HB side chain (Figure 5C). In contrast, the roles of Trp103 and Trp407 are reversed in the eribulin model (Figure 6B), with Trp103 positioned at the solvent interface and Trp407 packed against the eribulin macrocycle.
Modeling the Trajectories of Drug Binding
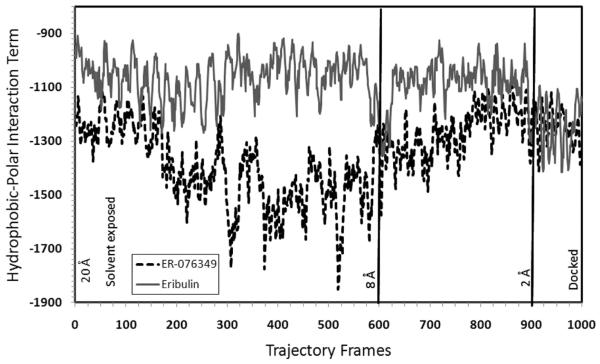
The docking trajectories of eribulin and ER-076349 (Figure 1B), a close eribulin congener in which the C-35 amine is replaced by a hydroxyl group, were modeled. Constrained molecular dynamics was used to undock eribulin and ER-076349 from their respective binding poses in β-tubulin, and the resulting trajectories were reversed in order to simulate the different binding events. From each docking trajectory, 1000 frames were produced and analyzed. Figure 7 shows intramolecular hydrophobic-polar interaction terms for 1000 conformations of eribulin and ER-076349 in their docking trajectories. As we had previously shown,31 the intramolecular hydrophobic-polar interaction term, which characterizes unfavorable atom-to-atom interactions, is an important determinant in the biological activity of a molecule. From frames 1–600, when eribulin and ER-076349 were 20–8 Å away from their docked poses and were solvent exposed, the overall magnitude of this negative interaction was less for eribulin than for ER-076349. This suggests that eribulin exhibited a more favorable solution behavior than ER-076349. From frames 600–900, when the two compounds were 8–2 Å away from their docked poses and were near or at the protein–solvent front, the hydrophobic-polar term for ER-076349 became progressively less unfavorable, while still not eclipsing that for eribulin over a wide range of time frames. From frames 900–1000, when the two compounds were entering their respective binding sites, the hydrophobic-polar terms of the two compounds converged to within a band range of −1400 to −1100. In the latter phase of the binding event, for a significant number of frames or time periods, ER-076349 seemed to be experiencing more favorable intramolecular binding interactions than eribulin. These docking simulations revealed more favorable dynamic properties for eribulin in solution and at the protein–ligand interface than for ER-076349, and, conversely, slightly more favorable dynamic properties were evident for ER-076349 in the binding pocket than was observed for eribulin. The convergent hydrophobic-polar terms of eribulin and ER-076349 when the two compounds are in their binding pockets are consistent with their similar in vitro activity against tubulin.
Figure 7.
Calculated intramolecular hydrophobic-polar interaction terms for the 1000 collected conformations of eribulin and ER-076349 during their simulated binding to β-tubulin. In frame 1, each ligand starts at a position that is 20 Å away from its docked pose in the binding site on β-tubulin, and in frame 1000, each ligand assumes its docked pose in the binding pocket at a position described as 0 Å. Based on the HINT program,32 the hydrophobic-polar term is characterized by a negative value. Higher negative values indicate a larger magnitude of unfavorable atom-to-atom interactions.
Moreover, the difference in dynamic properties of eribulin and ER-076349 outside of their binding pockets may explain their varying in vivo activities. ER-076349 was shown to be more potent than eribulin in human cancer cell lines but, in contrast, was less efficacious than eribulin in tumors in mice. Okouneva et al.38 had proposed that this was explained by their observations that ER-076349 entered cells more rapidly than did eribulin but that ER-076349 also was very rapidly removed from the cell by washout.
The propensity of ER-076349 for rapid washout may be explained by our docking trajectory simulations. From our modeling study, eribulin appears to be more stable than ER-076349 near or on the protein surface. This suggests that eribulin exists at a higher concentration on or near the tubulin protein surface than ER-076349 and would consequently remain longer in both cells and tumors. Conversely, the simulations indicate that ER-076349 exists at a higher concentration in the unbound form than eribulin, leading to more rapid cellular efflux.
DISCUSSION
With the approval of eribulin for the treatment of metastatic breast cancer, gaining a greater understanding of the interactions of the halichondrin class of compounds with tubulin becomes more important. Such knowledge may ultimately prove useful in reducing toxicity or overcoming tubulin-based resistance. In the work presented here, we determined an apparent Kd of 0.31 μM for HB by Hummel–Dreyer HPLC. We unequivocally demonstrated that HB bound to the αβ-tubulin heterodimer and, in 0.1 M Mes–0.5 mM MgCl2, did not cause formation of larger tubulin assemblies.
We surveyed compounds known or suspected to bind in the vinca domain for inhibitory effects on [3H]HB binding to tubulin. Adequate inhibition to pursue kinetic inhibitory studies occurred with D10 ≈ auristatin PE > eribulin ≈ hemiasterlin > vincristine ≈ isohalichondrin B > vinblastine (order of relative activity). Detailed studies were performed with eribulin, D10, and vincristine. Eribulin yielded a competitive pattern (apparent Ki, 0.80 μM), while D10 and vincristine yielded noncompetitive patterns (apparent Ki’s, 0.35 and 5.4 μM, respectively), consistent with previous studies showing HB and/or eribulin were noncompetitive inhibitors of [3H]vinblastine and [3H]D10 binding to tubulin.1,2
HB, spongistatin 1, and maytansine had no ability to cause formation of multiunit aggregates of tubulin under equilibrium conditions, unlike vinblastine and D10. It is tempting to ascribe the noncompetitive inhibitory patterns obtained with HB and spongistatin 1 versus vincristine/vinblastine and D10 to interference with aggregate formation. However, this is clearly not the explanation. Nonaggregators, such as maytansine and rhizoxin, can display competitive inhibitory patterns against vinca alkaloid binding.9 Similarly, aggregators, such as D10, auristatin PE, phomopsin A, hemiasterlin, and cryptophycin 1, display noncompetitive inhibitory patterns against the vinca alkaloids.9–11,39 With the latter compounds, this occurs despite apparent partial overlap in the binding sites for peptide antimitotics and vinca alkaloids, as shown by Cormier et al.27 in studies in which phomopsin A or auristatin PE interacted with crystals containing two αβ-heterodimers.
In our HPLC studies with [3H]HB and with columns equilibrated with nonradiolabeled HB, we found binding of the ligand only to 100 kDa αβ-tubulin. We therefore developed a binding model for HB that was localized solely to β-tubulin. Initial modeling failed to indicate a potential binding site, so we extensively analyzed the conformations of both HB and β-tubulin. Binding HB to tubulin in a chemically feasible manner required that the HB side chain be folded onto the macrocycle. With β-tubulin, development of a binding pocket required transient movements of two polypeptide regions (loops containing residues 94–110 and 399–414). Figure 2 shows the overall approach in modeling the HB binding site and generating the reported binding models.
The resulting binding model provides a high-quality match between HB and β-tubulin. The HB macrocycle docks onto helix H4, with Gly144 and Gly148 fitted snugly into the cavity of the macrocycle. More importantly, hydrophobic residue side chains of β-tubulin are wrapped around each methylene and methyl substituent on HB, providing substantial stability to the ligand–protein complex, as do six hydrogen bonds between protein and ligand.
Among these six hydrogen bonds in the HB model, five tie the H3′ and H11′ loops to HB (Figure 5). In the 1JFF β-tubulin structure, the H3′ and H11′ loops are linked to the base of the HB binding site by only a single hydrogen bond between the hydroxyl of Tyr408 in H11′ and the side chain of Asn186 in helix H5. The greater stability of the H3′ and H11′ loops in the HB binding model was confirmed by calculations which showed that the sum of the H3′ and H11′ energies in the HB model is 6 kcal/mol more stable than in the unliganded 1JFF model. This energy difference confirms that a conformational switch in the H3′ and H11′ loops upon HB binding is energetically feasible.
Moreover, the folded conformation of HB affords the molecule a markedly different binding mode than the one we previously proposed.2 In the earlier model, HB was docked as an extended conformation and bound to a protein–protein site that was composed of residues from α- and β-subunits from different heterodimers, while, in the model described here, HB assumes a folded conformation and binds to a protein pocket composed entirely of β-tubulin residues. In the earlier model, the HB macrocycle is stabilized by hydrophobic contacts to α-subunit residues Phe244, Ala247, Leu248, and Tyr357 and to β-subunit residues Gly81, Pro82, Thr223, and Gly225. In contrast, in the model presented here, the HB macrocycle is mainly stabilized by its hydrophobic packing with its own folded polyether chain and by its dovetail packing with the glycine-rich helix H4.
The HB binding model led to an analogous model for eribulin binding to tubulin. The chief difference between the two models is that the smaller structure of eribulin required a partial hydrophobic collapse of the H3′ and H11′ loops to avoid leaving a “gap” in the protein–ligand structure (Figure 2).
Our current models provide a clear explanation for why the macrocycle portion contains the pharmacophore for activity of the halichondrin chemotype, since, with both HB and eribulin, the molecular footprint for their interactions with rigid structural elements of tubulin is virtually the same, despite their significant molecular size and weight differences. Additionally, the two models may establish a structural basis for the three-fold greater activity of eribulin with tubulin as compared with HB.2 The eribulin model is characterized by five hydrogen bonds between ligand and β-tubulin, compared with six such bonds in the HB model. However, the significantly lower molecular weight of eribulin indicates a greater contribution of each hydrogen bond to ligand binding efficiency as compared with those of HB. More importantly, the entropic cost associated with eribulin binding to β-tubulin is less than that for HB due to the larger conformational landscapes that the HB and β-tubulin structures must sample in order to reach their bioactive conformations in solution.
The HB and eribulin binding models extend our understanding of the biological activity of the halichondrin class of compounds. We have begun to explore whether other active analogs of HB are readily accommodated in the HB binding pocket described here. Thus far, we have found that homohalichondrin B1 and halistatin 140 fit well into the binding pocket of the HB model. The C-10 hydroxyl group of halistatin 1 interacts with solvent in our model, consistent with the minimal effect on activity of adding a hydroxyl group at this position. Homohalichondrin B differs from HB in the terminal portion of the polyether side chain, and these modifications were accommodated within the model. With the eribulin model, we found that the active eribulin congener ER-076349 has a similar predicted binding affinity to that of eribulin, which is consistent with their similar in vitro activity.41 ER-076349 has a slightly lower calculated binding affinity due to the exchange of a stronger hydrogen-bond donor in the C-35 amine of eribulin with a weaker donor in the C-35 hydroxyl moiety of ER-076349. In addition, we used the binding models of eribulin and ER-076349 as templates for modeling their docking trajectories. Despite being close analogues, eribulin and ER-076349 exhibited distinct dynamic properties at three stages of binding: in solution, near the protein surface, and in the binding pocket. Whereas the most complete cocrystal structure or binding model may not provide such insights, our dynamic simulations revealed that the solution behavior of these two compounds correlates with differences in their in vivo activity.
The binding pocket of HB places the ligand within 5 Å of the E-site, consistent with its inhibition of nucleotide exchange. The binding pocket is about 16 Å from the vinblastine site, explaining the noncompetitive inhibition obtained with both HB1 and eribulin2 of vinblastine binding and with vincristine of HB binding. Similarly, the close proximity of the binding sites of auristatin PE and phomopsin A to the vinblastine site27 is consistent with the noncompetitive inhibition of D10 binding by eribulin2 and of HB binding by D10 (Figure 4C).
We note two recent studies with eribulin. Alday and Correia42 studied the interaction of eribulin and ER-076349 with tubulin by analytical ultracentrifugation. They found that eribulin inhibited not only Mg2+-induced tubulin oligomer formation but also disrupted the stabilization by stathmin of paired αβ-tubulin heterodimers. With ER-076349, Mg2+-induced oligomer formation was enhanced, but formation of the tubulin-stathmin complex was still inhibited. These differences could be related to the tighter binding of eribulin revealed in our trajectory study (Figure 7). Smith et al.43 examined the interaction of [3H]eribulin with microtubules and with tubulin and reported a single site for eribulin on tubulin. Their overall stoichiometry was much lower than we observed with lower concentrations of [3H]HB. They only reached higher stoichiometries with [3H]eribulin ≥30 μM. Consequently, they reported an overall apparent Kd of 46 μM, with a subset of tubulin molecules binding eribulin more tightly, with an apparent Kd of 0.4 μM, similar to our Hummel–Dreyer Kd for HB of 0.31 μM and our apparent Ki value for eribulin of 0.80 μM.
ACKNOWLEDGMENT
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This research was supported in part by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute.
REFERENCES
- (1).Bai R, Paull KD, Herald CL, Malspeis L, Pettit GR, Hamel E. Halichondrin B and homohalichondrin B, marine natural products binding in the vinca domain of tubulin: discovery of tubulin-based mechanism of action by analysis of differential cytotoxicity data. J. Biol. Chem. 1991;266:15882–15889. [PubMed] [Google Scholar]
- (2).Dabydeen DA, Burnett JC, Bai R, Verdier-Pinard P, Hickford SJH, Pettit GR, Blunt JW, Munro MHG, Gussio R, Hamel E. Comparison of the activities of the truncated halichondrin B analog NSC 707389 (E7389) with those of the parent compound and a proposed binding site on tubulin. Mol. Pharmacol. 2006;70:1866–1875. doi: 10.1124/mol.106.026641. [DOI] [PubMed] [Google Scholar]
- (3).Aicher TD, Buszek KR, Fang FG, Forsyth CJ, Jung SH, Kishi Y, Matelich MC, Scola PM, Spero DM, Yoon SK. Total synthesis of halichondrin B and norhalichondrin B. J. Am. Chem. Soc. 1992;114:3162–3164. [Google Scholar]
- (4).Choi HW, Demeke D, Kang FA, Kishi Y, Nakajima K, Nowak P, Wan ZK, Xie CY. Synthetic studies on the marine natural product halichondrins. Pure Appl. Chem. 2003;75:1–17. [Google Scholar]
- (5).Zheng W, Seletsky BM, Palme MH, Lydon PJ, Singer LA, Chase CE, Lemelin CA, Shen YC, Davis H, Tremblay L, Towle MJ, Salvato KA, Wels BF, Aalfs KK, Kishi Y, Littlefield BA, Yu MJ. Macrocyclic ketone analogues of halichondrin B. Bioorg. Med. Chem. Lett. 2004;14:5551–5554. doi: 10.1016/j.bmcl.2004.08.069. [DOI] [PubMed] [Google Scholar]
- (6).Jackson KL, Henderson JA, Phillips AJ. The halichondrins and E7389. Chem. Rev. 2009;109:3044–3079. doi: 10.1021/cr900016w. [DOI] [PubMed] [Google Scholar]
- (7).Arnold SM, Moon J, Williamson SK, Atkins JN, Ou S-HI, LeBlanc M, Urba SG. Phase II evaluation of eribulin mesylate (E7389, NSC 707389) in patients with metastatic or recurrent squamous cell carcinoma of the head and neck: Southwest Oncology Group trial S0618. Invest. New Drugs. 2009:1–8. doi: 10.1007/s10637-009-9348-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Vahdat LT, Pruitt B, Fabian CJ, Rivera RR, Smith DA, Tan-Chiu E, Wright J, Tan AR, DaCosta NA, Chuang E, Smith J, O’Shaughnessy J, Shuster DE, Meneses NL, Chandrawansa K, Fang F, Cole PE, Ashworth S, Blum JL. Phase II study of eribulin mesylate, a halichondrin B analog, in patients with metastatic breast cancer previously treated with an anthracycline and a taxane. J. Clin. Oncol. 2009;27:2954–2961. doi: 10.1200/JCO.2008.17.7618. [DOI] [PubMed] [Google Scholar]
- (9).Bai R, Pettit GR, Hamel E. Binding of dolastatin 10 to tubulin at a distinct site for peptide antimitotic agents near the exchangeable nucleotide and vinca alkaloid sites. J. Biol. Chem. 1990;265:17141–17149. [PubMed] [Google Scholar]
- (10).Bai R, Durso NA, Sackett DL, Hamel E. Interactions of the sponge-derived antimitotic tripeptide hemiasterlin with tubulin: comparison with dolastatin 10 and cryptophycin 1. Biochemistry. 1999;38:14302–14310. doi: 10.1021/bi991323e. [DOI] [PubMed] [Google Scholar]
- (11).Bai R, Schwartz RE, Kepler JA, Pettit GR, Hamel E. Characterization of the interaction of cryptophycin 1 with tubulin: binding in the vinca domain, competitive inhibition of dolastatin 10 binding, and an unusual aggregation reaction. Cancer Res. 1996;56:4398–4406. [PubMed] [Google Scholar]
- (12).Edler MC, Fernandez AM, Lassota P, Ireland CM, Barrows LR. Inhibition of tubulin polymerization by vitilevuamide, a bicyclic marine peptide, at a site distinct from colchicine, the vinca alkaloids and dolastatin 10. Biochem. Pharmacol. 2002;63:707–715. doi: 10.1016/s0006-2952(01)00898-x. [DOI] [PubMed] [Google Scholar]
- (13).Bai R, Taylor GF, Cichacz ZA, Herald CL, Kepler JA, Pettit GR, Hamel E. The spongistatins, potently cytotoxic inhibitors of tubulin polymerization, bind in a distinct region of the vinca domain. Biochemistry. 1995;34:9714–9719. doi: 10.1021/bi00030a009. [DOI] [PubMed] [Google Scholar]
- (14).Hamel E, Lin CM. Separation of active tubulin and microtubule-associated proteins by ultracentrifugation and isolation of a component causing the formation of microtubule bundles. Biochemistry. 1984;23:4173–4184. doi: 10.1021/bi00313a026. [DOI] [PubMed] [Google Scholar]
- (15).Grover S, Hamel E. The magnesium-GTP interaction in microtubule assembly. Eur. J. Biochem. 1994;222:163–172. doi: 10.1111/j.1432-1033.1994.tb18854.x. [DOI] [PubMed] [Google Scholar]
- (16).Litaudon M, Hickford SJH, Lill RE, Lake RJ, Blunt JW, Munro MHG. Antitumor polyether macrolides: new and hemisynthetic halichondrins from the New Zealand deep water sponge Lissodendoryx. J. Org. Chem. 1997;62:1868–1871. [Google Scholar]
- (17).Smith AB, III, Sfouggatais C, Risatti CA, Sperry JB, Zhu W, Doughty VA, Tomioka T, Gotchev DB, Bennett CS, Sakamoto S, Atasoylu O, Shirakami S, Bauer D, Takeuchi M, Koyanagi J, Sakamoto Y. Spongipyran synthetic studies. Evolution of a scalable total synthesis of (+)-spongistatin 1. Tetrahedron. 2009;65:6489–6509. doi: 10.1016/j.tet.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Pettit GR, Singh SB, Hogan F, Lloyd-Williams P, Herald DL, Burkett DD, Clewlow PJ. The absolute configuration and synthesis of natural (−)-dolastatin 10. J. Am. Chem. Soc. 1989;111:5463–5465. [Google Scholar]
- (19).Gamble WR, Durso NA, Fuller RW, Westergaard CK, Johnson TR, Sackett DL, Hamel E, Cardellina JH, II, Boyd MR. Cytotoxic and tubulin-interactive hemiasterlins from Auletta sp. and Siphonochalina spp. sponges. Bioorg. Med. Chem. 1999;7:1611–1615. doi: 10.1016/s0968-0896(99)00089-9. [DOI] [PubMed] [Google Scholar]
- (20).Pettit GR, Srirangam JK, Barkoczy J, Williams MD, Boyd MR, Hamel E, Pettit RK, Hogan F, Bai R, Chapuis J-C, McAllister SC, Schmidt JM. Dolastatin 10 SAR probes. Anti-Cancer Drug Des. 1998;13:243–277. [PubMed] [Google Scholar]
- (21).Cruz-Monserrate Z, Vervoort HC, Bai R, Newman DJ, Howell SB, Los G, Mullaney JT, Williams MD, Pettit GR, Fenical W, Hamel E. Diazonamide A and a synthetic structural analog: disruptive effects on mitosis and cellular microtubules and analysis of their interactions with tubulin. Mol. Pharmacol. 2003;63:1273–1280. doi: 10.1124/mol.63.6.1273. [DOI] [PubMed] [Google Scholar]
- (22).Hamel E, Lin CM. Guanosine 5′-O-(3-thiotriphosphate), a potent nucleotide inhibitor of microtubule assembly. J. Biol. Chem. 1984;259:11060–11069. [PubMed] [Google Scholar]
- (23).Nguyen TL, Xu X, Gussio R, Ghosh AK, Hamel E. The assembly-inducing laulimalide/peloruside A binding site on tubulin: molecular modeling and biochemical studies with [3H]peloruside A. J. Chem. Inf. Model. 2010;50:2019–2028. doi: 10.1021/ci1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Uemura D, Takahashi K, Yamamoto T, Katayama C, Tanaka J, Okumura Y, Hirata Y. Norhalichondrin A – an antitumor polyether macrolide from a marine sponge. J. Am. Chem. Soc. 1985;107:4796–4798. [Google Scholar]
- (25).Löwe J, Downing KH, Nogales E. Refined structure of αβ-tubulin at 3.5 Å resolution. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- (26).Gigant B, Wang C, Ravelli RBG, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M. Structural basis for the regulation of tubulin by vinblastine. Nature(London) 2005;435:519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- (27).Cormier A, Marchand M, Ravelli RBG, Knossow M, Gigant B. Structural insight into the inhibition of tubulin by vinca domain peptides. EMBO Rep. 2008;9:1101–1106. doi: 10.1038/embor.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ravelli RBG, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature(London) 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- (29).Dorléans A, Gigant B, Ravelli RBG, Mailliet P, Mikol V, Knossow M. Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13775–13779. doi: 10.1073/pnas.0904223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- (31).Nguyen TL, McGrath C, Hermone AR, Burnett JC, Zaharevitz DW, Day BW, Wipf P, Hamel E, Gussio R. A common pharmacophore for a diverse set of colchicine site inhibitors using a structure-based approach. J. Med. Chem. 2005;48:6107–6116. doi: 10.1021/jm050502t. [DOI] [PubMed] [Google Scholar]
- (32).Meng EC, Kuntz ID, Abraham DJ, Kellogg GE. Evaluating docked complexes with the HINT exponential function and empirical atomic hydrophobicities. J. Comput.-Aided Mol. Des. 1994;8:299–306. doi: 10.1007/BF00126747. [DOI] [PubMed] [Google Scholar]
- (33).Hummel JP, Dreyer WJ. Measurement of protein-binding phenomena by gel filtration. Biochim. Biophys. Acta. 1962;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- (34).Singer WD, Himes RH. Cellular uptake and tubulin binding properties of four vinca alkaloids. Biochem. Pharmacol. 1992;43:545–551. doi: 10.1016/0006-2952(92)90577-6. [DOI] [PubMed] [Google Scholar]
- (35).Jordan MA, Kamath K, Manna T, Okouneva T, Miller HP, Davis C, Littlefield BA, Wilson L. The primary antimitotic mechanism of action of the synthetic halichondrin E7389 is suppression of microtubule growth. Mol. Cancer Ther. 2005;4:1086–1095. doi: 10.1158/1535-7163.MCT-04-0345. [DOI] [PubMed] [Google Scholar]
- (36).Bai R, Taylor GF, Schmidt JM, Williams MD, Kepler JA, Pettit GR, Hamel E. Interaction of dolastatin 10 with tubulin: induction of aggregation and binding and dissociation reactions. Mol. Pharmacol. 1995;47:965–976. [PubMed] [Google Scholar]
- (37).Dixon M, Webb EC, Thorne CJR, Tipton KF. Enzymes. 3rd ed Academic Press; New York: 1979. pp. 332–354. [Google Scholar]
- (38).Okouneva T, Azarenko O, Wilson L, Littlefield BA, Jordan MA. Inhibition of centromere dynamics by eribulin (E7389) during mitotic metaphase. Mol. Cancer Ther. 2008;7:2003–2011. doi: 10.1158/1535-7163.MCT-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Natsume T, Watanabe J, Tamoki S, Fujio N, Miyasaka K, Kobayashi M. Characterization of the interaction of TZT-1027, a potent antitumor agent, with tubulin. Jpn. J. Cancer Res. 2000;91:737–747. doi: 10.1111/j.1349-7006.2000.tb01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Pettit GR, Tan R, Gao F, Williams MD, Doubek DL, Boyd MR, Schmidt JM, Chapuis J-C, Hamel E, Bai R, Hooper JNA, Tackett LP. Isolation and structure of halistatin 1 from the eastern Indian Ocean marine sponge Phakellia carteri. J. Org. Chem. 1993;58:2538–2543. [Google Scholar]
- (41).Towle MJ, Salvato KA, Budrow J, Wels BF, Kuznetsov G, Aalfs KK, Welsh S, Zheng W, Seletsk BM, Palme MH, Habgood GJ, Singer LA, Dipietro LV, Wang Y, Chen JJ, Quincy DA, Davis A, Yoshimatsu K, Kishi Y, Yu MJ, Littlefield BA. In vitro and in vivo anticancer activities of synthetic macrocyclic ketone analogues of halichondrin B. Cancer Res. 2001;61:1013–1021. [PubMed] [Google Scholar]
- (42).Alday PH, Correia JJ. Macromolecular interaction of halichondrin B analogues eribulin (E7389) and ER-076349 with tubulin by analytical ultracentrifugation. Biochemistry. 2009;48:7927–7938. doi: 10.1021/bi900776u. [DOI] [PubMed] [Google Scholar]
- (43).Smith JA, Wilson L, Azarenko O, Zhu X, Lewis BM, Littlefield BA, Jordan MA. Eribulin binds at microtubule ends to a single site on tubulin to suppress dynamic instability. Biochemistry. 2010;49:1331–1337. doi: 10.1021/bi901810u. [DOI] [PMC free article] [PubMed] [Google Scholar]