Summary
Short hairpin RNAs (shRNAs) provide powerful experimental tools by enabling stable and regulated gene silencing through programming of endogenous microRNA pathways. Since requirements for efficient shRNA biogenesis and target suppression are largely unknown, many predicted shRNAs fail to efficiently suppress their target. To overcome this barrier, we developed a “Sensor assay” that enables the biological identification of effective shRNAs at large scale. By constructing and evaluating 20,000 RNAi reporters covering every possible target site in 9 mammalian transcripts, we show that our assay reliably identifies potent shRNAs that are surprisingly rare and predominantly missed by existing algorithms. Our unbiased analyses reveal that potent shRNAs share various predicted and previously unknown features associated with specific microRNA processing steps, and suggest a new model for competitive strand selection. Together, our study establishes a powerful tool for large-scale identification of highly potent shRNAs and provides new insights into sequence requirements of effective RNAi.
Introduction
RNA interference (RNAi) provides a programmable mechanism for targeted suppression of gene expression. Through a highly conserved pathway, the RNAi machinery recognizes and processes double-stranded RNAs into small RNAs that guide the repression of complementary genes [for review see (Bartel, 2004; Hannon, 2002)]. Experimental RNAi acts by providing exogenous sources of double-stranded RNA that mimic endogenous triggers and has paved the way for rapid loss-of-function studies that range from exploring the function of single genes to large-scale genetic screens. Moreover, RNAi is being developed into new therapies that can, in principle, inhibit any gene product.
In animals, somatic RNAi is mainly programmed by microRNAs (miRNAs), small non-coding RNAs that regulate gene expression [for review see (Bartel, 2004; Filipowicz et al., 2008)]. miRNAs are produced through a coordinated processing program whereby primary miRNA transcripts (pri-miRNAs) are cleaved by the nuclear Drosha/DGCR8 complex, resulting in the formation of precursor miRNAs (pre-miRNAs). These short hairpin-like molecules are actively exported to the cytoplasm, where Dicer excises mature small RNA duplexes that are incorporated into the RNA induced silencing complex (RISC). Following strand selection, AGO2 discards the passenger (Leuschner et al., 2006; Matranga et al., 2005) and uses the guide for selection of complementary target mRNA substrates, whose expression is suppressed by accelerated mRNA degradation and/or translational inhibition.
Synthetic sources of double-stranded RNA can enter the RNAi pathway at various points. The most basic approach involves transfection of small interfering RNA (siRNA) duplexes (Elbashir et al., 2001) that resemble Dicer products. Although often potent, siRNA effects are transient and limited to transfectable cell types. An alternative approach relies on vectors that express stem-loop short hairpin RNAs (shRNAs), which resemble pre-miRNAs and enable stable and heritable gene silencing (Brummelkamp et al., 2002; Paddison et al., 2002). shRNAs can also be embedded in the context of endogenous miRNA transcripts – a configuration that creates a natural substrate for miRNA pathways (Silva et al., 2005; Zeng et al., 2002), enables stable and regulated expression from polymerase-II promoters (Dickins et al., 2005; Stegmeier et al., 2005), and reduces shRNA associated toxicity (Castanotto et al., 2007; McBride et al., 2008). Such miRNA-mimetics provide a versatile tool for long-term gene suppression in vitro and in vivo, as well as pool-based RNAi screening [see, for example (Dickins et al., 2007; Schlabach et al., 2008; Silva et al., 2008; Zender et al., 2008; Zuber et al., 2011)].
While powerful, RNAi technology has some limitations. Besides suppressing the intended target gene, synthetic RNAi triggers can evoke off-target effects by suppressing unintended transcripts due to sequence homologies of either the sense or the antisense strand. Generally, the potential for misinterpreting such false positive results can be minimized through the use of several independent RNAi triggers targeting the same transcript. In addition, high intracellular levels of synthetic small RNAs can result in toxicities related to saturation of the RNAi machinery (Grimm et al., 2006). Such effects can be reduced by the use of microRNA-based RNAi triggers (Castanotto et al., 2007; McBride et al., 2008) and, in principle, would be eliminated through the use of shRNAs that effectively repress gene expression at low concentrations.
Beyond off target effects, it remains difficult to identify potent shRNAs from among hundreds or thousands of possibilities within a given transcript. Consequently many shRNAs are ineffective, leading to false-negative results in functional studies and screens. The precise sequence requirements of efficient RNAi remain incompletely understood, hampering the establishment of rational shRNA prediction rules. Studies using siRNA datasets indicate that RISC loading and target repression are dictated by sequence features in both the mature small RNA as well as the targeted mRNA region (Ameres et al., 2007; Khvorova et al., 2003; Schwarz et al., 2003). These include a preference for thermodynamic asymmetry (Khvorova et al., 2003; Schwarz et al., 2003), low G/C content (Reynolds et al., 2004) and a strong bias for A/U at the 5'-end of the guide strand (Tomari and Zamore, 2005). Nonetheless, these features are not sufficient to accurately distinguish between potent and weak RNAi triggers.
Machine-learning based applications trained on siRNA datasets have produced algorithms that facilitate prediction of potent siRNAs (Huesken et al., 2005; Vert et al., 2006). However, such analyses have not been applied to shRNAs, which may require more stringent criteria as they rely on transcription and multistep miRNA processing for the production of small RNA duplexes. Indeed, experience indicates that siRNA algorithms are inefficient for predicting potent shRNAs, leaving their identification to laborious testing (Bassik et al., 2009; Li et al., 2007). Moreover, key RNAi applications such as pooled shRNA screening and RNAi transgenics require shRNAs that are effective even when expressed from a single genomic locus (“single copy”). Since most currently available shRNA reagents are not designed or tested to fulfill such stringent criteria, studies using shRNAs often rely on suboptimal reagents and libraries contain many ineffective shRNAs that complicate the execution and interpretation of genetic screens.
Here, we describe a high-throughput assay to evaluate shRNA potency in a massively parallel format. Our approach is based on a single-vector reporter assay that functionally monitors the interaction of shRNAs with their specific target sites, and thereby takes into account all aspects of shRNA biogenesis and target repression. This simple strategy reliably identifies rare potent shRNAs, most of which are not predicted using existing algorithms. By tracking the behavior of 20,000 shRNAs through all steps of microRNA biogenesis, we uncovered novel sequence preferences that contribute to potent and specific RNAi. Such information will advance the use of RNAi in functional studies and lays the groundwork for validated shRNA libraries.
Results
Single-vector Sensor assay for functional shRNA evaluation
Synthetic RNAi triggers can be accurately evaluated in functional assays by placing their cognate target site (“Sensor”) in the 3'UTR of a reporter gene and quantifying its RNAi-mediated repression (Du et al., 2004; Kumar et al., 2003). In previous systems, the reporter construct and RNAi trigger were delivered separately and, thus, had to be assayed in a one-by-one format. We reasoned that physically linking shRNAs and their cognate target sites in a single vector would enable multiplexed analysis of shRNA-target pairs. Therefore, we constructed a reporter vector (pSENSOR; Figure 1A) harboring an shRNA expressed under the control of a Tet-responsive element (TREtight) (Gossen and Bujard, 1992; Sipo et al., 2006) and its cognate target sequence (Sensor) in the 3'UTR of a constitutively expressed fluorescent reporter (Venus) (Nagai et al., 2002). Since the adjacent context of target sites may affect RNAi potency (Ameres et al., 2007), we designed Sensors as 50 nt fragments of the endogenous mRNA, harboring the 22 nt target in the center (Figure S1A). In reporter cells expressing the reverse Tet-transactivator (rtTA) (Gossen et al., 1995), doxycycline (Dox) induces shRNA expression, which in turn represses the Venus reporter to an extent that reflects the potency of the shRNA (Figure 1A).
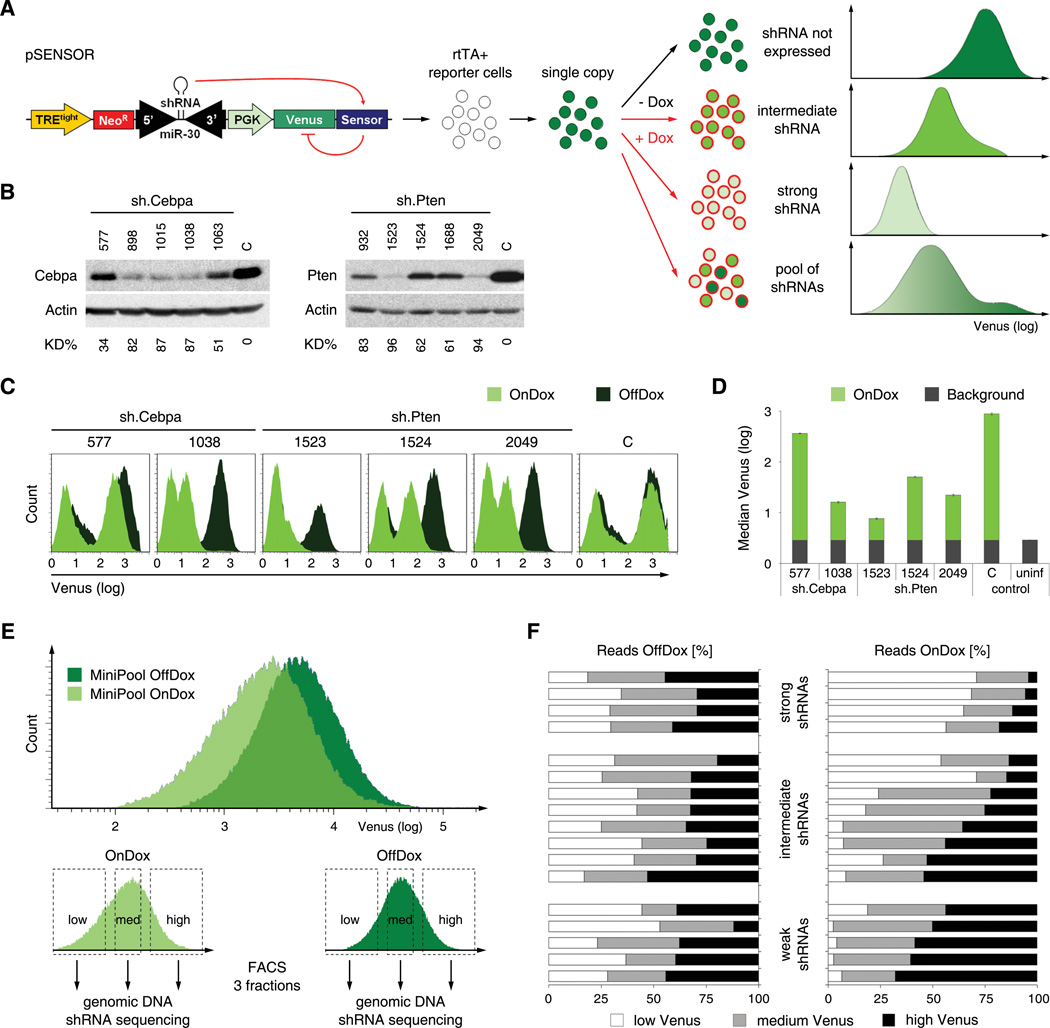
Figure 1. Sensor assay for assessment of shRNA potency.
(A) Schematic of the Sensor assay. The pSENSOR vector harbors a Tet-inducible shRNA and its cognate target sequence in the 3'UTR of a PGK-driven Venus reporter. Upon infection of cells expressing rtTA, Dox treatment induces shRNA expression. In turn, the extent of Venus knockdown directly reports shRNA potency. Flow cytometry plots depict predicted fluorescence intensity distributions for shRNAs of different potencies.
(B) Immunoblotting of Cebpa and Pten in NIH3T3s transduced with Cebpa or Pten shRNAs of different potencies. C, sh.Luci.1309 control shRNA. KD%, knockdown level relative to C and normalized to Actin.
(C) Flow cytometry analysis of ERC reporter cells transduced with pSENSOR carrying indicated shRNA-Sensor cassettes and treated with or without Dox (On-/OffDox). The leftmost peaks represent uninfected cells. C, control construct harboring an shRNA with a non-corresponding Sensor.
(D) Quantification of Venus fluorescence intensity of OnDox cells shown in C and uninfected reporter cells (uninf) used to define background levels. Error bars represent the standard deviation of triplicates.
(E) Flow cytometry plots of rtTA reporter cells transduced with a MiniPool of 17 shRNA-Sensor constructs and treated +/− Dox for 7 days. The lower panels illustrate Venus-based cell sorting of each population into three equal subpopulations.
(F) Quantification of shRNA sequence reads within the sorted populations outlined in E. For each shRNA, the distribution of reads among low/medium/high Venus fractions is plotted as a percentage of total reads of that shRNA. The shRNAs are clustered according to their pre-annotated groups (see Figure S1E for details).
To determine the dynamic range of the assay, we constructed a set of pSENSOR vectors harboring 17 pre-existing shRNAs of different potency, which were re-evaluated by western blotting and classified into groups of strong, intermediate and weak shRNAs (Figure 1B, Figure S1B–E). Following transduction into rtTA-reporter cells, we quantified changes in Venus expression after Dox treatment for all 17 shRNAs (Figure 1C, 1D and data not shown). Induction of strong shRNAs resulted in dramatic reduction of Venus fluorescence; conversely, intermediate and weak shRNAs induced only a moderate or slight reduction of Venus intensity, respectively. Overall, Venus repression reflected the efficacy of individual shRNAs in suppressing their endogenous target, indicating that the Sensor assay accurately quantifies shRNA potency.
Pooled evaluation of shRNAs
Since each shRNA and its corresponding Sensor are delivered in a single vector, our assay is adaptable to a pooled format. In such a setting, pooled shRNA-Sensor constructs must be transduced at single copy to ensure that Venus fluorescence of each cell reports the activity of a single shRNA. Upon shRNA induction, cells harboring potent shRNAs should display strong Venus repression, enabling their identification through fluorescence activated cell sorting (FACS) followed by sequencing of proviral shRNA cassettes. To evaluate this approach, we transduced a pool of 17 pre-tested pSENSOR constructs into rtTA reporter cells and sorted equal fractions of low, medium and high Venus expressing cells in the absence and presence of Dox (Figure 1E and Table S1). Next, genomic DNA was isolated from sorted cells in each fraction, and the abundance of each shRNA was determined by capillary sequencing (288 reads for each fraction). In the absence of Dox, each shRNA was equally distributed among the three fractions (Figure 1F). Following Dox addition, potent shRNAs were enriched in the low Venus fraction and underrepresented in the high Venus fraction, while weak shRNAs were shifted to the high Venus fraction and almost absent in the low Venus fraction. Thus, the Sensor assay can be used to select shRNAs based on their potency in a pooled format.
Optimization of the Sensor assay
In pilot experiments we observed that potent shRNA-Sensor constructs showed decreased viral titers, potentially reducing their representation in the population. We hypothesized that this was due to potent shRNAs targeting their Sensor on proviral transcripts, thereby inducing their degradation. To circumvent this, we transiently suppressed shRNA biogenesis in packaging cells by co-transfecting a potent DGCR8 siRNA. Indeed, this modification normalized the packaging and transduction efficiency of pSENSOR constructs (Figure S2A–C).
We also realized that effects of shRNAs on their endogenous target might alter the proliferation and/or viability of reporter cells and thereby bias the assay. For example, potent shRNAs targeting essential genes will deplete reporter cells, and thereby escape identification in a pooled setting. Since RNAi utilizes an evolutionary conserved machinery, we reasoned that an avian reporter cell line would provide an accurate system for evaluating mammalian shRNAs, where biases induced by effects on endogenous targets would be minimized due to divergence at the nucleic acid level. We therefore engineered DF-1 chicken embryonic fibroblasts (Himly et al., 1998) to express the ecotropic retroviral receptor and an improved reverse Tet-transactivator (rtTA3) (Das et al., 2004). When tested using different shRNA-Sensor constructs, “Eco-rtTA-chicken (ERC)” reporter cells accurately reported shRNAs of different potency (Figure S2D–F), indicating that shRNA processing is similar between ERC and mammalian cells (see Figure S5I for large scale confirmation). Therefore, avian ERC cells are accurate reporters for the Sensor assay and less sensitive to the biological effects of mammalian shRNAs.
Generation of a high-complexity Sensor library
To evaluate the ability of the Sensor assay to simultaneously evaluate the potency of thousands of shRNAs, we constructed and surveyed a library of ~20,000 shRNA-Sensor constructs comprising every possible shRNA for 9 mammalian transcripts (Table S2). To ensure that individual shRNAs were cloned together with their specific Sensor, we applied large-scale on-chip oligonucleotide synthesis (Cleary et al., 2004) to produce ~20,000 185-mers each harboring an shRNA and its target sequence separated by cloning sites, and used them to assemble the Sensor library in a pooled two-step procedure (Figure 2A). Serving as internal controls, all 17 previously characterized shRNAs were included at 15-fold representation to ensure their presence in the final pool. Deep sequencing of the library after cloning revealed that >99% of all designed shRNAs were present (Figure 2D).
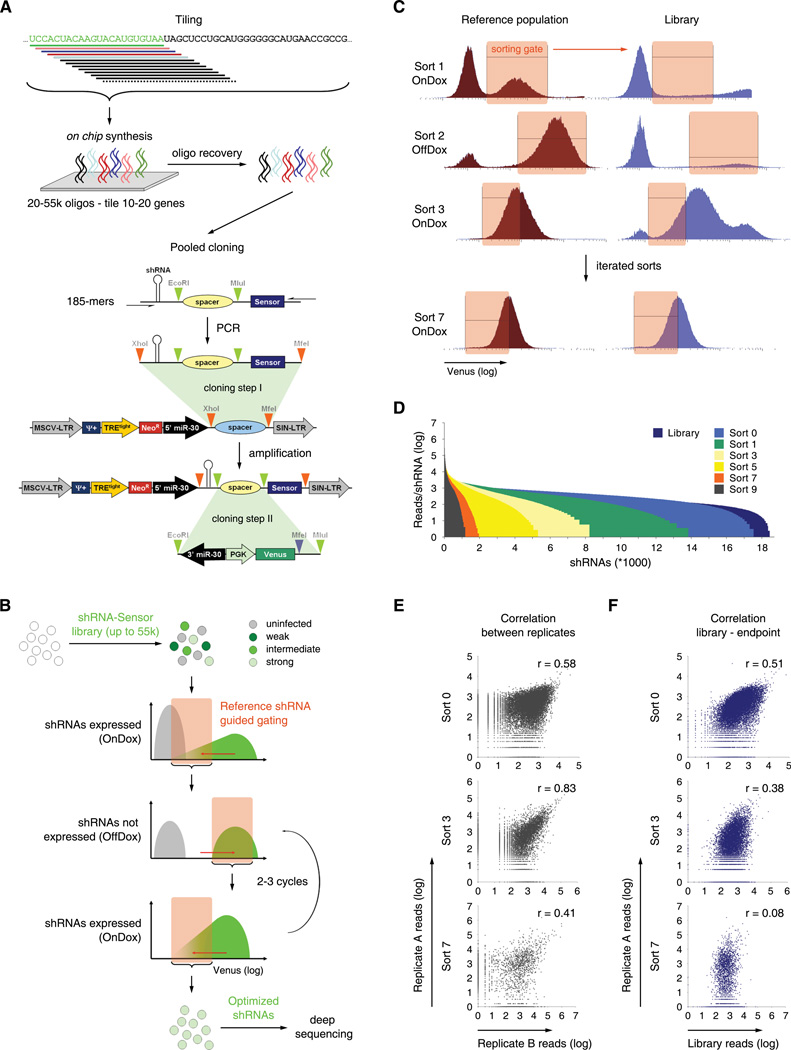
Figure 2. Sensor Ping-Pong strategy for deconvolution of complex shRNA-Sensor libraries.
(A) Schematic of design and cloning of shRNA-Sensor libraries. A library of 20,000 constructs tiling every possible target site in nine mammalian transcripts was designed. 185-mer oligonucleotides containing shRNAs and cognate Sensors were synthesized and cloned into a 5’miR30 recipient vector. In a second step, the 3'miR30-PGK-Venus reporter cassette was cloned between shRNAs and their cognate Sensor to reconstitute complete pSENSOR vectors.
(B) Schematic of the Sensor Ping-Pong sorting strategy. Reporter cells infected with an shRNA-Sensor library at single copy are cultured sequentially in presence or absence of Dox. According to reference populations, sorting gates are drawn to include only cells harboring potent shRNAs (see Figure S2G and S2H for details). Through iterative rounds of shRNA induction and FACS-based selection, the initial library is reduced to a pool of functional shRNA-Sensor constructs that can be identified by deep sequencing.
(C) Representative flow cytometry histograms of Top5 reference and library populations at sorting step 1, 2, 3 and 7. ERC reporter cells were infected with the Library, Top5 or Bottom5 pools, grown repeatedly for 6–7 days On- then OffDox, and sorted according to the indicated gates. Data is presented as Venus intensity histograms; actual sorts were done using Venus/FSC dot plots (see Figure S2H for details).
(D) Histogram of library complexity over sort cycles. Shown are normalized read numbers in one replicate for each shRNA represented within the pool after the indicated sorts.
(E) Correlation of reads per shRNA between two replicates after the indicated sorts. r, Pearson correlation coefficient.
(F) Correlation of reads per shRNA between the initial library pool and the pools after the indicated sorts. r, Pearson correlation coefficient.
Multiplexed evaluation of shRNA potency using Sensor Ping-Pong sorting
To evaluate shRNA potency in this complex library, we initially applied fractionated sorting paralleling our analysis of small pools (Figure 1E). However, at an increased complexity level this strategy failed to distinguish strong and weak control shRNAs (data not shown). Reasoning that iterative rounds of selection could be used to strongly enrich potent shRNAs and eliminate background noise, we developed a FACS strategy (Sensor Ping-Pong, Figure 2B) that involves sequential cycles of shRNA induction and withdrawal, each followed by sorting for reporter cells displaying Venus levels similar to potent shRNA-Sensor controls. In this approach, OnDox sorts for “Venus-low” reporter cells exclude cells harboring dysfunctional shRNAs (thus maintaining high Venus levels); conversely, OffDox steps for “Venus-high” reporters eliminate cells with constitutively defective reporters, e.g. due to positional effects of the retroviral integration. In each sort, FACS gating was guided by parallel analysis of two small reference pools containing five strong (Top5) and five weak (Bottom5) control shRNAs. By four cycles of enrichment (Sort 7) the OnDox FACS profile of the library became more uniform and resembled that of the Top5 reference population (Figure 2C, Figure S2G and S2H).
To monitor the representation of individual shRNAs throughout the procedure, genomic DNA was extracted after every sort and shRNA guide strands were amplified and quantified by deep sequencing. While more than 98% of all cloned constructs were initially represented in infected ERC reporter cells, each sort led to a reduction of library complexity such that less than 2,000 shRNAs remained after 7 sorts (Figure 2D). Importantly, the shRNA composition of independent duplicates correlated throughout the procedure (Figure 2E), while their correlation to the initial population was progressively lost (Figure 2F). Therefore, the decrease in pool complexity that occurred throughout the procedure results from a non-random enrichment of specific shRNAs.
Next, we quantified the abundance of our 17 internal control shRNAs throughout the experiment. After the second cycle (Sort 3), strong shRNA controls already showed significant enrichment and weak shRNAs were depleted (Figure 3A). By the fourth cycle (Sort 7), all strong shRNAs were robustly enriched, while all weak and most intermediate shRNAs were virtually eliminated (Figure 3A and 3B). The initial overrepresentation of some weak control shRNAs in the library did not prevent their eventual depletion (Figure 3B), suggesting the assay can tolerate imbalances in the initial pool composition. Optimal enrichment was obtained by 4 cycles, after which we did not observe any additional changes in the overall representation of our control shRNAs (data not shown). We also explored the use of barcoded shRNA-Sensor libraries in conjunction with micro-array based monitoring of shRNA representation and found that this approach can stratify controls of known potency (Figure S3A and S3B). Collectively, the behavior of our 17 control shRNAs indicates that the Sensor Ping-Pong assay strongly enriches for potent shRNAs while robustly depleting non-functional and weak shRNAs.
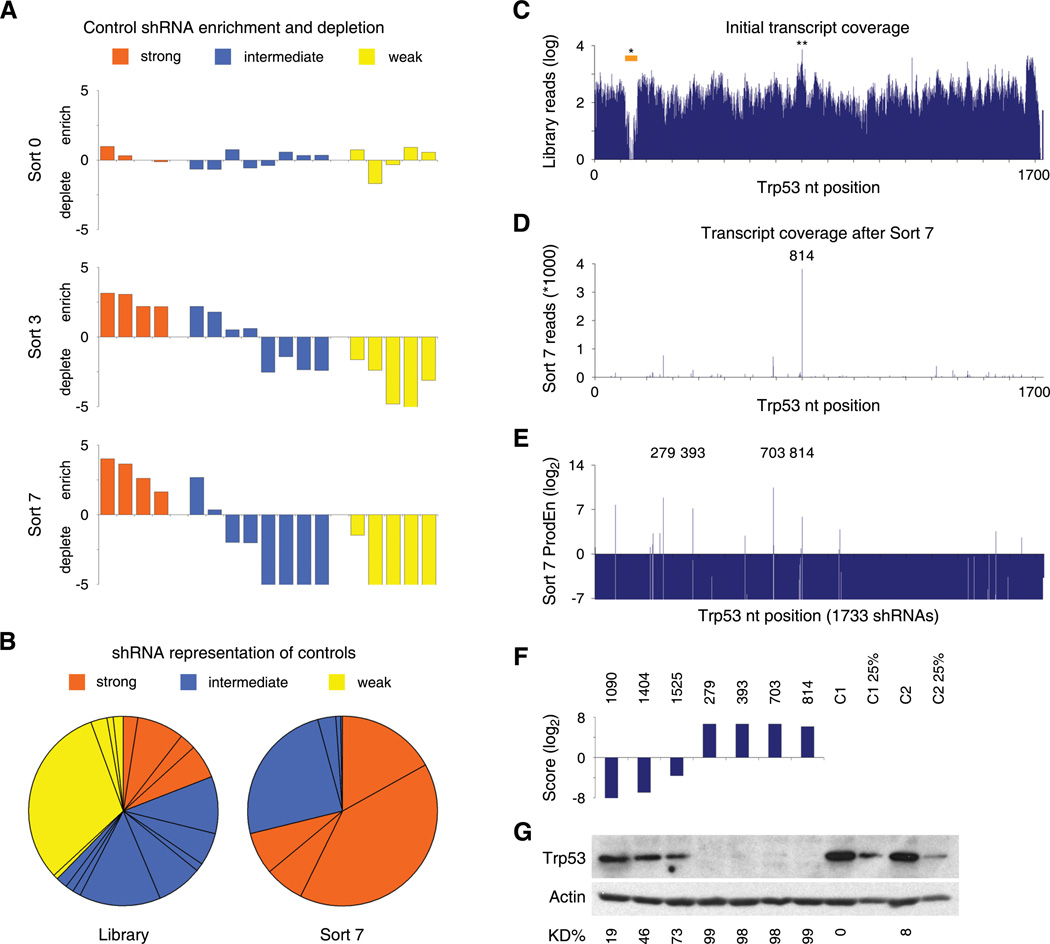
Figure 3. Assay performance of control shRNAs and shRNA-Sensor constructs tiling Trp53.
(A) Enrichment or depletion of 17 control shRNA-Sensor constructs in transduced reporter cells before sorting (top), after Sort 3 (middle) and Sort 7 (bottom). Values denote the log2 ratio of reads at the indicated stage compared to reads in the initial shRNA-Sensor plasmid library.
(B) Representation of control shRNA-Sensor constructs in the initial plasmid library and after seven sorts. Pie wedges represent mean values of technical (Library) or biological (Sort 7) duplicates.
(C) Trp53 transcript coverage in the initial library. Shown are absolute reads (mean of technical duplicates) for 1733 Trp53 shRNA-Sensor constructs in the plasmid pool. *, XhoI restriction site affecting the cloning of 45 shRNAs. **, most abundant shRNA, sh.p53.816.
(D) Read numbers (mean of biological duplicates) for Trp53 shRNA-Sensor constructs after 4 Ping-Pong cycles (Sort 7). 814, most abundant shRNA, sh.p53.814.
(E) Product enrichment scores (ProdEn, representing the product of the relative enrichment in each replicate at Sort 7 compared to the initial library) of all 1733 Trp53 shRNA-Sensor constructs. Numbers (279, 393, 703, 814) pinpoint highly enriched shRNAs.
(F) Integrated Score for selected shRNAs analyzed by western blotting.
(G) Western blot analysis of Trp53 levels in adriamycin-treated NIH3T3s stably expressing the shRNAs indicated above from a single genomic integration. C1 and C2, control shRNAs (sh.Bcl2.906, sh.Bcl2.1132). C1 25% and C2 25%, 1:4 diluted control samples. KD%, knockdown level relative to C1 and normalized to Actin.
Validation of Sensor-identified shRNAs
Sequence analysis indicated that the Sensor assay can identify potent shRNAs from complex libraries de novo. For example, all 1733 possible Trp53 shRNAs were represented at the beginning of the assay, with the exception of 11 shRNAs containing a restriction site used for cloning and 5 shRNAs in the poly(A) tail (Figure 3C). Conversely, after 4 cycles most shRNAs were completely absent from the pool (Figure 3D), while only a few were enriched. Strikingly, the most prominent hit based on total reads was sh.p53.814 (a.k.a. sh.p53.1224) – an shRNA that was previously identified empirically and shown to be extremely potent (Dickins et al., 2005).
To rank and select shRNAs for further validation, we developed two complementary scoring systems. The quantitative product enrichment (ProdEn), defined as the product of enrichment ratios in independent replicates, takes both the initial representation and consistency between replicates into account (Figure 3E). A second semi-quantitative score uses a logistic function to integrate the initial representation of each shRNA, the consistency between replicates, and the trend for shRNA enrichment or depletion throughout all sorts (Figure 3F and Table S3). Based on these readouts, we examined the potency of four top scoring and three non-scoring Trp53 shRNAs by immunoblotting (Figure 3G). All three newly-identified Trp53 shRNAs showed similar potency to sh.p53.814, suppressing Trp53 expression to virtually undetectable levels, while the non-scoring shRNAs had no effect. These results validate the Sensor assay's ability to identify potent shRNAs and suggest that these RNAi triggers are very rare and equally distributed over a given transcript.
These observations were confirmed by Sensor results from other tiled transcripts. While the initial transcript coverage was nearly complete (98.1% overall), only a small number of shRNAs were enriched for each transcript after four Sensor Ping-Pong cycles (2.4% of all shRNAs had a Score >10; Figure 4A, 4E, 5A, 5E and Figure S4C). The vast majority of scoring shRNAs examined (85–90%) showed strong knockdown of their target protein when expressed at single copy (Figure 4C, 4G, 5C, 5G, Figure S4E and Table S4). Importantly, non-scoring shRNAs that were ineffective at single copy often showed substantial knockdown when transduced under conditions that lead to multiple proviral integrations (Figure 4D and data not shown). Hence, the Sensor assay accurately distinguishes between shRNAs that work at single versus high copy – the latter of which are useless in pool-based shRNA screens or other applications where only single integrations are achievable or desirable.
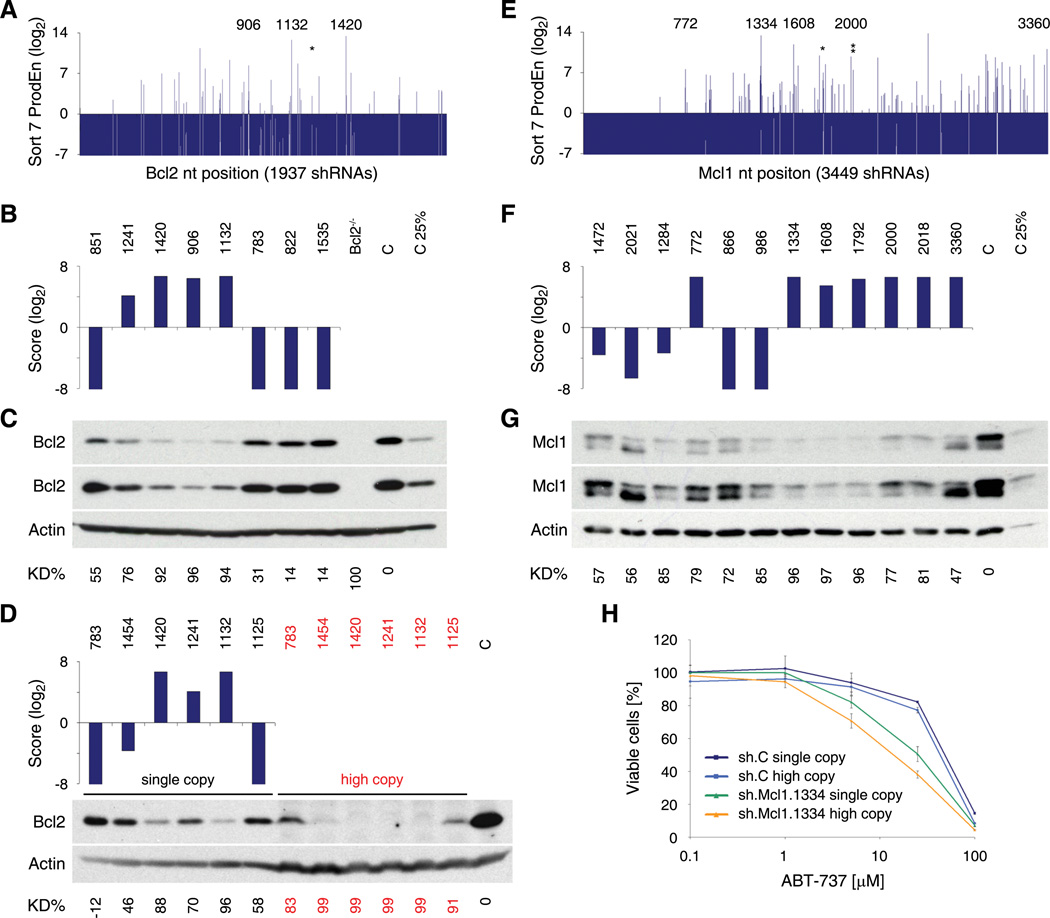
Figure 4. Analysis of Sensor-identified shRNAs targeting Bcl2 and Mcl1.
(A) Product enrichment scores (ProdEn) of 1937 shRNA-Sensor constructs tiling the common region of both murine Bcl2 transcripts. *, sh.Bcl2.1241. Numbers highlight enriched shRNAs that were analyzed by immunoblotting.
(B) Integrated Score for selected Bcl2 shRNAs.
(C) Western blot analysis of Bcl2 levels in NIH3T3s expressing the shRNAs indicated above from a single genomic integration. Bcl2−/− MEFs served as control. C, sh.Luci.1309. Two different exposures are shown. KD%, knockdown level relative to C and normalized to Actin.
(D) Integrated Score for selected Bcl2 shRNAs and western blot analysis of Bcl2 levels in NIH3T3s expressing the indicated shRNAs from a single (single copy) or multiple (high copy, red) genomic integrations. C, uninfected cells.
(E) Product enrichment scores (ProdEn) of 3449 shRNA-Sensor constructs covering the mouse Mcl1 transcript. *, sh.Mcl1.1792. **, sh.Mcl1.2018. Numbers highlight enriched shRNAs that were analyzed by immunoblotting.
(F) Integrated Score for selected Mcl1 shRNAs.
(G) Western blot analysis of Mcl1 expression in NIH3T3s expressing the shRNAs indicated above from a single genomic integration. C, sh.Luci.1309. Two different exposures are shown.
(H) Synthetic lethal assay using the BH3-mimetic ABT-737 in combination with a potent Sensor-identified Mcl1 shRNA (sh.Mcl1.1334) or a control shRNA (C, sh.Luci.1309). NIH3T3s expressing the indicated shRNA from a single or multiple genomic integrations were treated with ABT-737 for 48 h and subsequently analyzed for viable cell numbers using flow cytometry (FSC/SSC and propidium iodide staining). DMSO (1%) treated cells were used for normalization. Error bars represent the standard deviation of duplicate experiments.
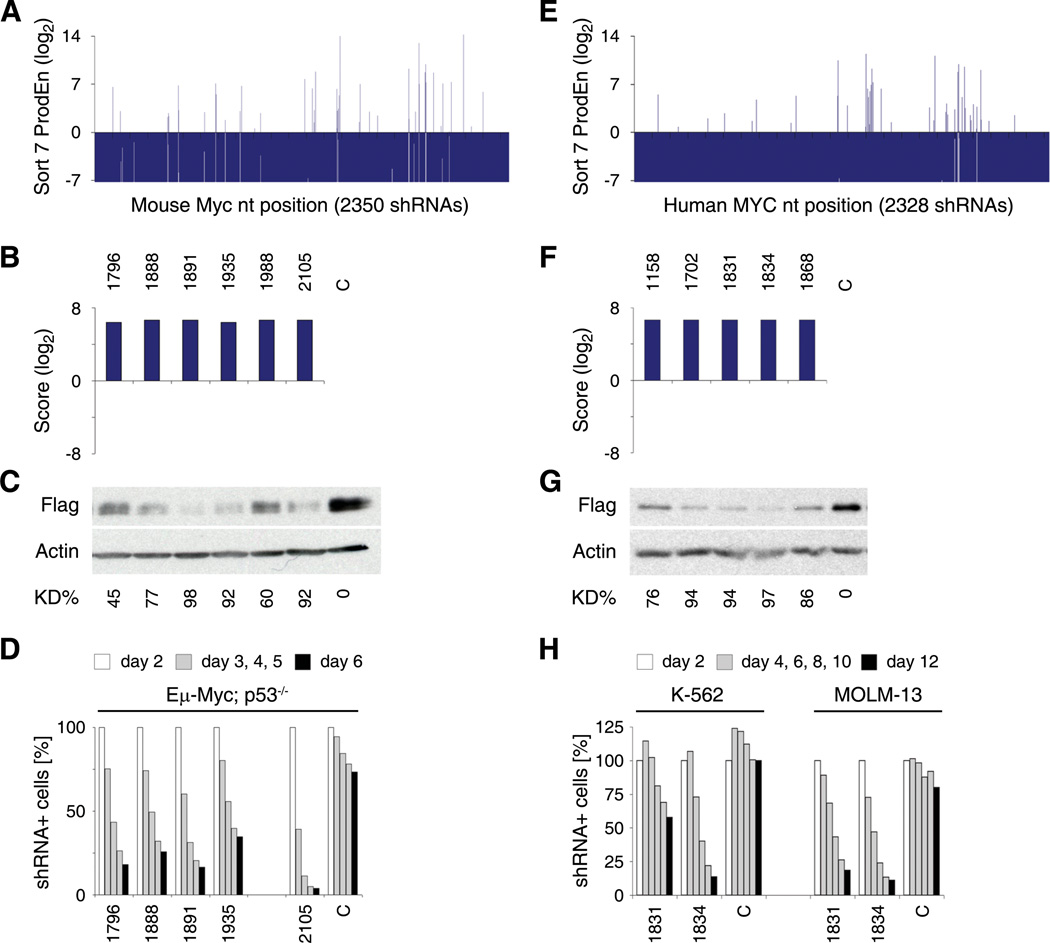
Figure 5. Analysis of Sensor-identified shRNAs targeting mouse and human MYC.
(A) Product enrichment scores (ProdEn) of 2350 shRNA-Sensor constructs tiling the mouse Myc transcript.
(B) Integrated Score for selected scoring Myc shRNAs.
(C) Western blot analysis of immortalized Rosa26-rtTA-M2 (RRT) MEFs expressing Flag-tagged murine Myc and shRNAs indicated above at single copy. Overexpression of Myc lacking the 3’UTR rescues knockdown by sh.Myc.1988 and 2105 (Figure S4B). C, sh.Luci.1309. KD%, knockdown level relative to C and normalized to Actin.
(D) Competitive proliferation assay of Eμ-Myc; p53−/− lymphoma cells expressing the indicated shRNAs. The relative percentage of shRNA expressing cells at indicated days following retroviral transduction is shown. C, sh.Luci.1309.
(E) Product enrichment scores (ProdEn) of 2328 shRNA-Sensor constructs tiling the human MYC transcript.
(F) Integrated Score for selected scoring MYC shRNAs.
(G) Flag-tag western blot analysis in RRT MEFs expressing Flag-tagged human MYC and shRNAs indicated above at single copy. C, sh.Luci.1309.
(H) Competitive proliferation assay of K-562 and MOLM-13 human leukemia cell lines expressing the indicated shRNAs. The relative percentage of shRNA expressing cells at the indicated days following shRNA induction is shown. C, sh.Luci.1309.
To functionally validate selected shRNAs, we developed a series of simple biological readouts for several of the genes. Generally, shRNAs that showed potent knockdown by immunoblotting displayed the most pronounced biological effects. For Mcl1, an anti-apoptotic protein, we transduced NIH3T3s at single or multiple copies with sh.Mcl1.1334 or a control shRNA and treated them with various concentrations of ABT-737 (Oltersdorf et al., 2005), an inhibitor of Bcl-2, Bcl-XL and Bcl-w that is known to synergize with Mcl-1 inactivation to promote cell death (van Delft et al., 2006). As predicted, knockdown of Mcl1 sensitized NIH3T3s to ABT-737 in a dose-dependent manner (Figure 4H).
For Rpa3 and Myc, proteins involved in DNA replication and cell proliferation, respectively, we examined shRNA potency using competitive proliferation assays. All five tested top-scoring shRNAs targeting mouse Myc rapidly depleted B-cell lymphoma cells isolated from diseased Eμ-Myc; p53−/− transgenic mice (Figure 5A–D). Similarly, the most potent human MYC shRNAs displayed deleterious effects in two human leukemia cell lines (Figure 5E–H). Such potent shRNAs can be readily applied in Tet-regulated expression systems, where Dox titration can be used to generate hypomorphic states (Figure S4A). All strongly scoring Rpa3 shRNAs tested impaired proliferation of fibroblasts, while several randomly selected non-scoring shRNAs were neutral (Figure S4C–F). A few functional Rpa3 shRNAs that were previously identified empirically (Zuber et al., 2011) were not identified using the Sensor assay, suggesting it does not identify every potent shRNA. However, all other previously characterized functional and non-functional shRNAs reported correctly (data not shown).
Comparison to existing design algorithms
To compare our results to existing siRNA-based design tools, we obtained the top 50 predictions for all nine transcripts using three different algorithms (Huesken et al., 2005; Sachidanandam, 2004; Vert et al., 2006) and compared them to the 50 highest scoring Sensor-derived shRNAs for each gene. Strikingly, >70% of our scoring shRNAs were not identified in the top 50 predictions of any algorithm (Figure S5A). While such false negatives, in principle, may have little practical significance, the majority of algorithm-predicted shRNAs did not score in the Sensor assay (Figure S5B), closely resembling their low validation rate in empirical testing (J.Z. and S.W.L., unpublished data). Together, these results demonstrate that siRNA algorithms are poor at predicting potent shRNAs [see also (Bassik et al., 2009; Li et al., 2007)] and underscore the value of the Sensor approach.
Global analysis of shRNA processing
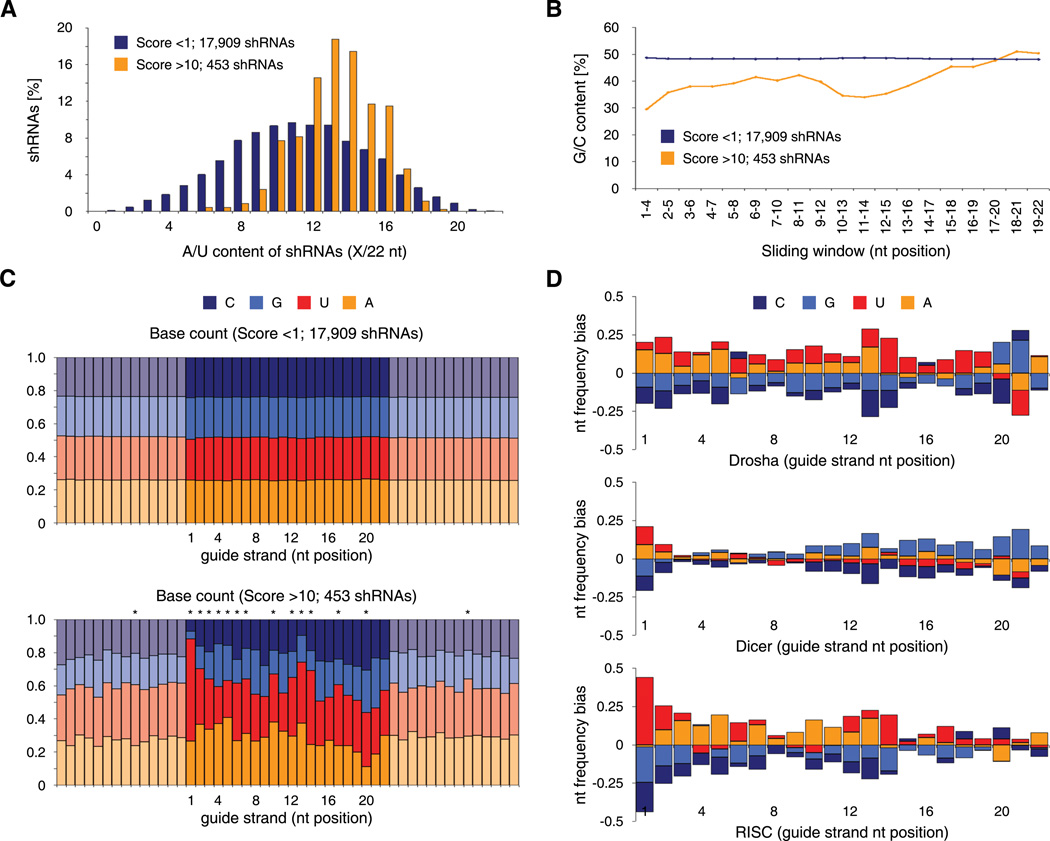
We noticed that potent shRNAs identified through our unbiased functional assay share common sequence features. Top-scoring shRNAs (Score >10, 453 in total) are predominantly A/U-rich (Figure 6A) and exhibit a strong thermodynamic asymmetry (Figure 6B) - two features that have been previously observed in studies of effective siRNAs (Khvorova et al., 2003; Reynolds et al., 2004; Schwarz et al., 2003). In contrast to non-scoring shRNAs and flanking mRNA regions, the nucleotide composition of potent guide strands shows many significant positional biases (p <0.01, Pearson’s χ2 test with Šidák correction) that progressively emerge throughout the assay (Figure 6C and S5C). Overwhelmingly, 88% of all top-scoring shRNAs carry U or A in guide position 1. Other A/U rich positions include 2, 10, 13, and 14, while position 20 and 21 are the only ones with a slight G/C bias. Position 20 also shows a remarkable depletion of A. Notably, most of these features have not been observed in siRNA-based studies.
Figure 6. Sequence features of Sensor-identified shRNAs and step-specific RNAi requirements.
(A) Overall A/U content of non-scoring (Score <1) and scoring (Score >10) shRNAs, showing enrichment of relatively A/U-rich shRNAs.
(B) Local G/C content (4 nt sliding window) of non-scoring (Score <1) and scoring (Score >10) shRNAs, indicating thermodynamic asymmetry of scoring shRNAs.
(C) Nucleotide frequency in non-scoring (Score <1, top) and scoring (Score >10, bottom) shRNA-Sensor constructs. Shown are 22 nt shRNA guide strands (dark colors, reverse complement to 22 nt target site in endogenous transcript) and adjacent mRNA regions flanking the target site (pastel colors, reverse complement to mRNA). *, p <0.01 (Pearson’s χ2 test with Šidák correction).
(D) Nucleotide bias of shRNAs that were significantly enriched (>5 fold) at the respective step and sufficiently represented (>100 reads) at the previous state. Drosha indicates sequences enriched from pri- to pre-miRNA (733 shRNAs). Dicer indicates sequences enriched from pre- to mature miRNA (931 shRNAs). RISC indicates sequences enriched from mature miRNA to shRNA representation at the genomic level after Sort 7 (root mean square value of all 4 replicates; 216 shRNAs). Data is shown for ERC cells; comparable patterns were observed in HEK293T cells.
To systematically analyze the interplay between nucleotide composition, shRNA processing, and biologic activity, we transduced the entire Sensor library into human HEK293T and chicken ERC cells, generated and quantified small RNA libraries designed to represent shRNA intermediates after major biogenesis steps (pri-, pre-, and mature miRNAs), and correlated their abundance with our functional Sensor data. At the pri-miRNA level >97% of all 18,720 shRNAs were identified and their abundances strongly correlated with those in the input library (r = 0.83 and 0.89 for ERC and HEK293T cells, respectively), indicating the absence of sequence biases in transduction and transcription. In both cell types, Drosha/DGCR8 cleavage occurred in >70% at the predicted site for most shRNAs (Figure S1A, S5D and S5E). Miscleaved pre-miRNAs were associated with G/C richness and a particular bias for C at guide position 20 (Figure S5F, p <0.01), suggesting that structural signals in pri-miRNAs guide processing to a specific site.
To examine at which pathway stage dysfunctional shRNAs are eliminated, we calculated the dropout rate for each processing step. Our data reveal that a substantial fraction of shRNAs fail processing at each level (Figure S5G), while the representation of individual precursors remained highly correlated between ERC and HEK293T cells throughout miRNA biogenesis (Figure S5I). Together, this indicates that each processing step has restrictive and specific requirements. Notably, shRNAs that score in the Sensor assay are enriched at each processing step (Figure S5H), illustrating that efficient shRNA processing is a key determinant of potency.
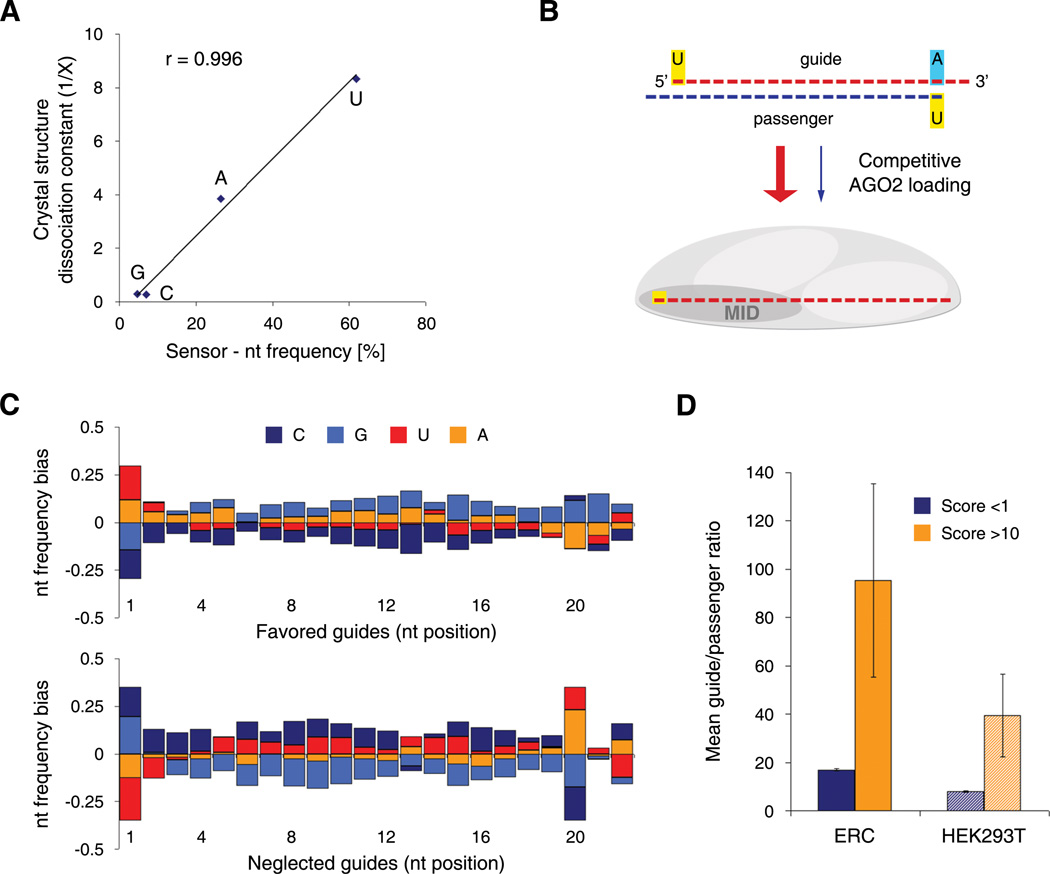
To explore specific features associated with effective processing, we analyzed the nucleotide composition of shRNAs that were enriched at each step (Figure 6D). Efficient Drosha/DGCR8 cleavage was strongly associated with a prevalence of A/U at position 13/14 and G at position 20 and 21 (p <0.01 for all). The transition from pre- to mature miRNAs, which represents Dicer/TRBP cleavage and likely AGO2 loading, shows biases for A/U in position 1 (p <0.01), while the remaining guide is characterized by a flat profile with a slightly G-rich 3’-side (nt 10–22). To monitor features associated with the terminal pathway steps (AGO2 loading, target recognition and cleavage) we analyzed shRNAs that showed an increase in their relative abundance from the mature miRNA stage to the endpoint of the Sensor assay (Sort 7). Only at this level, the structural pattern of enriched shRNAs exhibited a strong thermodynamic asymmetry (Figure 6D). Importantly, guide position 1 presented an extreme bias for U (p <0.01) and a near absence of G and C. These biases show a remarkable correlation to recently reported nucleotide binding affinities of the MID domain of human AGO2 (Frank et al., 2010) (Figure 7A), suggesting that a strong interaction between AGO2 and the 5'-end of the guide strand is a decisive prerequisite for potent RNAi.
Figure 7. Potent shRNAs show a strong strand bias dictated by guide position 1 and 20.
(A) Graphical analysis of the nucleotide bias at position 1 of the guide strand, demonstrating specific binding preferences for each nucleotide. The graph shows the correlation between dissociation constants obtained from data on crystal structures of the MID domain of human AGO2 (Frank et al., 2010) and Sensor-derived nucleotide frequencies. A linear regression indicates the trend of the dataset. r, Pearson correlation coefficient.
(B) Model for AGO2-mediated competitive guide selection. Specific binding of the 5’ nucleotide to the MID domain of vertebrate AGO2 strongly influences strand selection, thereby defining the RISC-loaded guide strand (see Figure S6C for details).
(C) Nucleotide frequency bias of favored (guide/passenger >50; 1546 shRNAs) and neglected (guide/passenger <0.02; 439 shRNAs) guide strands in ERC cells transduced with the Sensor library. Comparable results were obtained with HEK293T cells.
(D) Mean guide versus passenger ratios for scoring (Score >10) and non-scoring (Score <1) shRNAs in ERC and HEK293T cells transduced with the Sensor library. Error bars represent the standard error of the mean.
In nucleotide profiles associated with mature miRNA production and function we also noted an unusual rareness of A at position 20 (Figure 6D, p <0.05). In line with the above, shRNAs harboring A in guide position 20 will yield passenger strands carrying U at their 5'-end such that the passengers may outcompete target specific guide strands in RISC-loading due to their affinity for AGO2 binding (Figure 7B). Indeed, shRNAs showing strong guide selection are biased for U in position 1 (p <0.01) and against A in position 20 (p <0.01), while the key features of shRNAs with passenger strand preference are an absence of A/U in position 1 (p <0.01) and a strong bias for A/U in position 20 (p <0.01, Figure 7C and Figure S6A). Notably, guide:passenger ratios for individual shRNAs were highly correlated between HEK293T and ERC cells (Figure S6B), indicating that preferences in strand selection are due to a conserved and specific process. Overall, potent shRNAs identified in our assay show extreme guide selection biases (39- and 95-fold in HEK293T and ERC cells, respectively, Figure 7D), illustrating that a strong preference for utilizing target specific guide strands is a hallmark of effective RNAi.
Discussion
Here, we describe an unbiased, accurate, and scalable strategy for identifying highly potent shRNAs targeting any gene. Our approach measures the potency of shRNAs by monitoring their interaction with a surrogate target cloned into the 3’UTR of a fluorescent reporter, and thus integrates most aspects of shRNA biogenesis, target recognition and repression. Combining on-chip synthesis of long oligonucleotides with a two-step cloning procedure, we generated a library of ~20,000 shRNA-Sensor constructs representing almost every target site (>99%) in nine mammalian transcripts. Using genetically distant avian reporter cells, we simultaneously evaluated the potency of every shRNA within this library via iterative cycles of FACS-based enrichment and deep-sequencing based quantification, and thereby established a straightforward protocol for identifying potent shRNAs in a multiplexed format.
Our Sensor strategy accurately predicts the activity of shRNAs towards their endogenous targets and reliably identifies shRNAs that are effective when expressed from a single genomic integration - a criterion largely neglected in current shRNA libraries and prediction tools. As such, the assay vastly outperforms existing siRNA-based algorithms, which miss >70% of Sensor-derived shRNAs and generally necessitate the testing of many predictions to identify even a single potent shRNA. For example, despite previously testing ~15 top siRNA predictions from state of the art algorithms, we found no and only one potent shRNA targeting murine Mcl1 and Bcl2, respectively (data not shown). In contrast, the Sensor approach readily identified multiple highly effective shRNAs for both genes (Figure 4C and 4G).
Roughly 10–15% of scoring shRNAs did not efficiently suppress their endogenous target. These false positives could arise from technical problems linked to our multi-step protocol or off-target effects of the tested shRNA on the Venus transcript. Additionally, a subset of target sites could be occluded by long-range RNA interactions or protein binding events that are not reproduced on the abbreviated target site in our system. Although we presently have no estimate of false negative rates, the Sensor assay generally allowed us to easily find two or more potent RNAi triggers for every gene tested.
By surveying ~20,000 shRNAs produced in the absence of any design bias, our study describes the first systematic analysis of shRNA efficiency and provides the largest dataset of functionally annotated RNAi triggers currently available. Our data reveal that potent single-copy shRNAs are surprisingly rare, with frequencies ranging between 0.5% (Trp53) and 4.4% (Pcna) across the surveyed transcripts (2.4% on average). Except for sparing G/C-rich regions, potent shRNAs appear to be evenly distributed throughout transcripts, indicating that there is no preferential targeting of 3’UTRs.
To systematically explore the importance of efficient shRNA biogenesis for RNAi potency, we overlaid our functional data with a deep sequencing-based analysis of small RNA species at different stages of miRNA maturation. Surprisingly, a substantial fraction of shRNAs failed to be processed at each step, while potent shRNAs were consistently well represented (Figure S5G and S5H). Highly processed shRNAs shared distinct sequence features that are attributable to specific steps in miRNA biogenesis and, mostly, have not been noted previously. For example, efficient pre-shRNA production is associated with A/U in position 13/14 and G in position 20/21, while C in position 20 impairs the accuracy of Drosha/DGCR8 cleavage. Together, our findings illustrate that the multi-step process of miRNA biogenesis introduces additional structural constraints, providing an explanation for why siRNA-based algorithms often fail to predict functional shRNAs.
Other determinants of shRNA potency emerge at the end of the RNAi pathway. Strikingly, nucleotide frequencies of potent shRNAs at guide position 1 precisely mirror nucleotide binding affinities of AGO2 (Frank et al., 2010) and resemble Argonaute loading preferences for 5’-U containing strands in other organisms (Buhler et al., 2008). Together with biases at position 20, this suggests that the interaction between AGO2 and the 5’-end of both strands plays a decisive role in competitive strand selection (Figure S6C). In turn, most potent shRNAs are characterized by a strong preference for selecting the intended guide, suggesting that accurate strand selection is a key feature of effective RNAi.
Preferentially loaded strands also showed a subtle general bias for G but lacked thermodynamic asymmetry (Figure 7C), which previously has been implicated in RISC assembly (Schwarz et al., 2003). Since well selected guide strands that potently suppress their target show thermodynamic asymmetry (Figure 6D and Figure S6D), this feature may become relevant only after strand selection, e.g. by facilitating target release after cleavage and enhancing RISC turnover (Haley and Zamore, 2004; Leuschner et al., 2006). Together, our data suggest that RISC-loading is based on competitive binding of the 5’-nucleotides of both strands to AGO2, while thermodynamic asymmetry enhances the efficiency of later steps in the RNAi process.
Although the Sensor assay was designed to improve RNAi potency, its implementation may also impact RNAi specificity. First, our assay helps to control for sequence-specific off-target effects by enabling the identification of multiple potent shRNAs against any gene. Second, it will reduce passenger-mediated off-target effects by selecting potent shRNAs with a bias for incorporating the intended guide strand into RISC. Third, the identification of parameters guiding Drosha/DGCR8 processing will help to minimize off-target effects mediated by aberrant guide strands. Finally, by providing shRNAs with single-copy activity, our assay should further reduce off-target toxicities owing to saturation of the RNAi machinery. Indeed, we see that miR30-based shRNAs expressed from a single-copy promoter do not interfere with the processing of endogenous miRNAs (Premsrirut et al., submitted).
We believe that the Sensor assay provides a powerful and efficient method for identifying potent shRNAs. By taking an unbiased approach, our pilot study not only validated the Sensor assay but, unexpectedly, revealed novel insights into sequence requirements of miRNA biogenesis, strand selection and efficient target knockdown. Indeed, features deduced from our analyses provide the first shRNA-specific criteria framework for rational shRNA design (Table S5). Although these simple rules do not fully recapitulate the accuracy of the assay, they can be used to filter shRNAs prior to their Sensor-based evaluation and thereby dramatically increase the number of genes that can be surveyed in one Sensor experiment. As such, our approach lays out a practical workflow for the rapid generation of functionally validated shRNA libraries as well as the identification of potent RNAi triggers for biological studies and, eventually, RNAi therapeutics.
Experimental procedures
Vectors and library construction
The pSENSOR reporter vector, containing TREtight-NeoR-miR30-PGK-Venus-Sensor, was assembled in the pQCXIX retroviral backbone (Clontech). We designed ~20,000 185-mer oligonucleotides (each containing a 101 nt miR30-shRNA fragment, an EcoRI/MluI cloning site, the cognate 50 nt Sensor cassette and an 18 nt primer binding site), which were synthesized alongside controls on a 55k oligonucleotide array (Agilent Technologies). The shRNA-Sensor library was constructed in a two-step procedure, involving cloning of PCR-amplified shRNA-Sensor fragments into a 5'miR30-pSENSOR recipient vector, and inserting the 3'miR30-PGK-Venus cassette between shRNA and Sensor cassette. shRNAs were named according to the position of the 3’-nucleotide of the guide strand on the tiled transcript.
Reporter cell lines
RRT MEFs were generated by immortalizing Rosa26-rtTA-M2 MEFs through transduction of lentiviral SV40 large T antigen and subsequent passaging. ERC reporter cells were derived from a single-cell clone of DF-1 chicken embryonic fibroblasts (Himly et al., 1998) transduced with MSCV-rtTA3-PGK-Puro and MSCV-EcoReceptor-PGK-Hygro retroviruses, and grown in DMEM supplemented with 10% FBS, 1 mM sodium pyruvate, 100 U/ml penicillin and 100 µg/ml streptomycin. Tet-regulatable shRNAs were induced using Dox concentrations of 1.0–2.0 µg/ml in RRT MEFs and 0.5 µg/ml in ERC cells.
Sensor Ping-Pong assay
FACS procedures were carried out on a FACSAria II (BD Biosciences). ERC reporter cells were infected with pSENSOR libraries at singly copy and sorted in iterative cycles, either after treatment with Dox and G418 (500 µg/ml) for 6–7 days (OnDox) or after Dox and G418 withdrawal for 6–7 days (OffDox). The gating was guided by reference cells transduced with small pools of potent (Top5) and weak (Bottom5) control shRNA-Sensor constructs. In all sorts a representation of 1000-fold the pool complexity was maintained. Deep sequencing template libraries were generated by PCR amplification of shRNA guide strands from genomic DNA of at least 10 million cells, using primers that tag standard Illumina adaptors to the product, and sequenced using a primer reading reverse into the guide strand. Only sequences completely matching the Sensor library were retained.
Small RNA libraries
Libraries were generated as previously described (Malone et al., 2009). In brief, total RNA from HEK293T or ERC cells transduced with the pSENSOR library was extracted with TRIzol (Invitrogen) and two phenol:chloroform:IAA (Ambion) purification steps. 40 µg of total RNA was run on a 12% denaturing polyacrylamide gel and 18–26 nt mature small RNAs or 50–70 nt pre-miRNAs were selected for cloning; pri-miRNA libraries were obtained by direct amplification from total RNA using miR30-specific primers. Following Illumina sequencing, only sequences completely matching the Sensor library were retained for further analysis.
Supplementary Material
Acknowledgements
We thank A. Rappaport, C. Miething, D. Burgess, M. Spector, A. Vaseva, A. Chicas, L. Dow, M. Saborowski, S. Nuñez and H. Varmus for providing reagents. We gratefully acknowledge K. Marran, B. Ma, S. He, S. Muller, P. Moody and T. Spencer for excellent technical assistance. We also thank E. Hodges, M. Rooks and W.R. McCombie and his team for help with deep sequencing as well as Y. Erlich, A. Gordon, D. Lewis, M. Hammell and V. Thapar for bioinformatics support. This work was supported by program project grants from the National Cancer Institute and generous gifts from the Don Monti Memorial Research Foundation. J.Z. is the Andrew Seligson Memorial Fellow. S.W.L., G.J.H. and S.J.E. are Howard Hughes Medical Institute investigators.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bassik MC, Lebbink RJ, Churchman LS, Ingolia NT, Patena W, LeProust EM, Schuldiner M, Weissman JS, McManus MT. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods. 2009;6:443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MA, Kilian K, Wang Y, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- Das AT, Zhou X, Vink M, Klaver B, Verhoef K, Marzio G, Berkhout B. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J Biol Chem. 2004;279:18776–18782. doi: 10.1074/jbc.M313895200. [DOI] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature Genetics. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Thonberg H, Zhang HY, Wahlestedt C, Liang Z. Validating siRNA using a reporter made from synthetic DNA oligonucleotides. Biochem Biophys Res Commun. 2004;325:243–249. doi: 10.1016/j.bbrc.2004.09.222. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:502–506. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. PNAS. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional Activation by Tetracyclines in Mammalian Cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Himly M, Foster DN, Bottoli I, Iacovoni JS, Vogt PK. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nature Biotechnology. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kumar R, Conklin DS, Mittal V. High-Throughput Selection of Effective RNAi Probes for Gene Silencing. Genome Res. 2003;13:2333–2340. doi: 10.1101/gr.1575003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnology. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R. RNAi: design and analysis. Curr Protoc Bioinformatics. 2004;Chapter 12(Unit 12 13) doi: 10.1002/0471250953.bi1203s6. [DOI] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genome. Nature Genetics. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipo I, Picó AH, Wang X, Eberle J, Petersen I, Weger S, Poller W, Fechner H. An improved Tet-On regulatable FasL-adenovirus vector system for lung cancer therapy. J Mol Med. 2006;84:215–225. doi: 10.1007/s00109-005-0009-1. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. PNAS. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.