Abstract
AIDS patients undergoing autologous transplantation for lymphoma were treated with gene-modified peripheral blood derived (CD34+) hematopoietic progenitor cells (HPC) expressing 3 RNA-based anti-HIV moieties (Tat/Rev shRNA, TAR decoy and CCR5 ribozyme). In vitro analysis of gene-modified HPC showed no differences in the hematopoietic potential compared with non-transduced cells. In vitro estimates of gene marking were as high as 22% but declined to ~1% over 4 weeks of culture. Ethical study design required that patients were transplanted with both gene modified and unmanipulated hematopoietic progenitor cell apheresis products (HPC-A). All 4 infused patients engrafted (ANC>500) by day 11 post-infusion and showed no unexpected infusion related toxicities. Persistent vector marking in multiple cell lineages has been observed at low levels for up to 24 months as has expression of siRNA and ribozyme. This is the first demonstration of siRNA expression in human blood cells following transplantation of autologous gene-modified CD34+ HPC. These results support the development of an RNA-based cell therapy platform for HIV.
Summary
Stem cell gene therapy for HIV results in sustained RNA expression in the blood of patients for up to 2 years following transplant.
Introduction
Highly active antiretroviral therapy (HAART) has dramatically improved survival of patients with HIV infection, but it is likely never to be curative(1). Although compliance problems has been improved with new multiple-drug formulations, virus replication continues, and the risk of development of antiviral resistance remains a concern. Importantly, medications represent up to 84% of AIDS-related healthcare costs(2), thus, development of genetic therapies could decrease the need for continuous medication and its attendant cost. An approach which would allow a single genetic manipulation to delay or prevent the progression of HIV infection to AIDS remains an important goal.
Since it was first proposed that gene transfer might “immunize” against intracellular infection(3), there has been significant investigation into the use of genetic medicine to treat HIV. Given the difficulties, risks and failures associated with human gene therapy, it has remained unclear how a single gene manipulation could have a lasting, significant impact in this disease setting. Recently however, the transplantation of allogeneic hematopoietic stem cells (HSC) with a HIV-resistant genotype based on a naturally occuring 32-bp deletion in the chemokine receptor 5 gene (Δ32CCR5), used in conjunction with myeloablative therapy for leukemia, resulted in apparent elimination of HIV in the recipient(4). This genetic “test-of-concept” supports the idea that replacing a susceptible immune system with a genetically modified, virus-resistant one would likely result in reduced viral load and perhaps prevent progression to AIDS. However, the frequency and logistics of finding matched allogeneic homozygous Δ32CCR5 donors precludes widespread use of this strategy. Alternatively, if ex-vivo genetic modification of autologous HSC rendered their progeny HIV resistant, transplantation of these cells could potentially be therapeutic.
Multiple anti-viral strategies have been proposed for AIDS gene therapy including both protein(5–11) and RNA-based approaches(12–16). Several approaches have been tested in which autologous T-cells or CD34+ cells were transduced with a retroviral vector construct encoding either a surface fusion peptide(17), a mutant Rev molecule(18, 19), or a ribozyme targeting vpr/tat(20) or tat/rev(21) and then infused into HIV individuals. All were shown to be safe and well tolerated, but, resulted in very low level of long-term genetic marking of peripheral blood cells and variable to no detectable expression of the therapeutic transcript in the peripheral blood.
Based on our experience with HSC transplantation of AIDS lymphoma patients(22), this population appears to offer a unique opportunity to evaluate RNA-based anti-HIV strategies in an ethically acceptable clinical setting including marrow ablation. Long-term expression of the RNA transgenes will be necessary for success of this procedure, and thus, gene integration will be a requirement in the progeny cells if the therapeutic effect is to be sustained. In this regard, lentiviral vectors have been promoted as an ideal gene-delivery system since they have been reported to integrate into non-dividing cells and do not preferentially locate near gene promoters(23–25). We describe here a clinical trial in which autologous hematopoietic progenitor cells (HPCs) are programmed with an expressed short hairpin RNA (shRNA) in combination with two novel forms of HIV-specific RNA-based inhibitors (a nucleolar localizing RNA decoy and hammerhead ribozyme) (26). We show for the first time long-term expression of an ectopically expressed siRNA and ribozyme in the multiple peripheral blood cells lineages of patients transplanted with gene modified progenitor cells
Results
Participants Enrolled
Seven patients were consented to the study and treated according to the schema shown in Figure 1. All patients had non-Hodgkin lymphoma (NHL), two had relapsed disease and two were induction failures, two in first partial remission and one in high risk first remission. Secondary exclusion criteria required that patients could mobilize sufficient HPCs for collection by apheresis (HPC-A) of two cell products of at least 2.5 × 106 CD34+ cells/kg each; one a therapeutic product that was otherwise unmanipulated (HPC-A-Rx) and the other an experimental research product for genetic transduction (HPC-A-Exp). Two patients were removed from the study prior to HPC collection due to either progressive NHL (UPN0302) or to inadequate HPC-A collections (UPN0303). The remaining five patients mobilized an adequate number of cells for the investigational portion of the trial (Table 1). One of these (UPN 301), however, received only HPC-A-Rx because the HPC-A-Exp failed to pass the viability release test. The 4 patients that underwent transplantation with both transduced and untransduced cell products were a median age of 42.5 years (range 25–55), had CD4 counts at study entry ranging from 18–577 cell/μl, and a viral load ranging from below the limit of detection − 25,000 copies/ml. Following transplantation, HIV RNA in plasma and CD4 counts were followed at monthly intervals (Table S1). All patients were maintained on HAART and HIV loads were undetectable (<50 gc/ml) except for patient UPN0306 who had a viral load of 270 copies/ml at the time of transplant (Day 0) and patient UPN0304 who temporarily discontinued HAART therapy between 12 and 15 months post transplant resulting in a transient viral load of 890 copies/ml at 15 months after which he resumed prior anti-retroviral dosing.
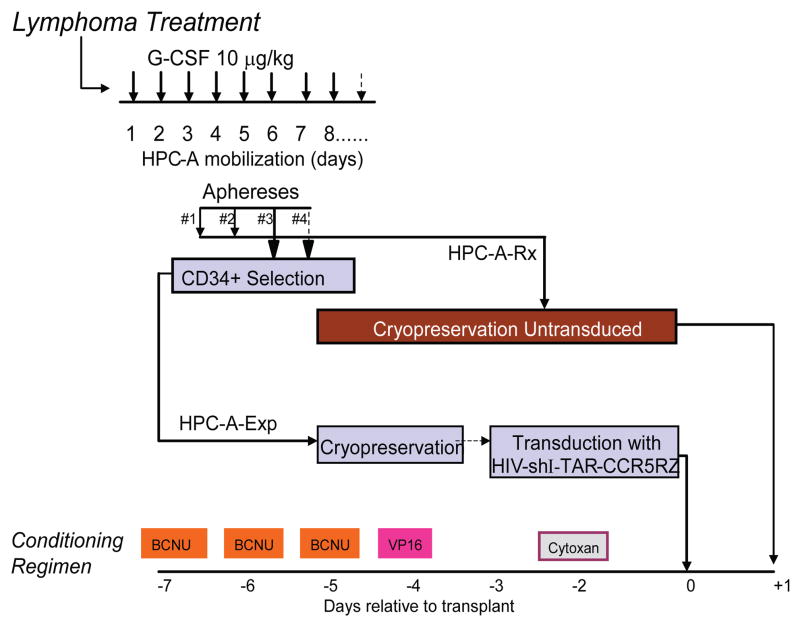
Figure 1. Clinical Trial Design.
Following Lymphoma Therapy, patients underwent hematopoietic progenitor cell mobilization with G-CSF as shown. The resulting apheresis product (HPC-A) was collected for up to 4 days; first to collect 2–5 ×105 CD34+ cells/kg for the unmanipulated transplantation product (Fraction A) and then to collect CD34+ cells for the genetic modification (Fraction B). Unmanipulated HPC-A collections were cryopreserved immediately while HPC-A for genetic modification were enriched for CD34 content using a CliniMACS™ device and then cryopreserved. The patients were treated with a myeloablative conditioning regimen (BCNU/VP16/Cytoxan as shown). Three days prior to transplantation (day -3) CD34-enriched HPC-A is thawed and transduced with the HIV-shI-TAR-CCR5Rz lentiviral vector. On day 0, the patients are infused with the gene modified HPC-A product followed by the unmanipulated HPC-A product on Day+1.
Table 1.
Patient Characteristics.
| CD34+ Cells/Kg | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UPN | Sex | Diagnosis | Age | Cell Products Infused | HIV Load Pre-Transplant (gc/ml) | CD4 Level Pre-Transplant (Cells/ml) | Days to Engraftment (ANC>500) | Follow Up (months) | HPC-A-RX Infused | HPC-A-Exp Infused | Total Infused | % Gene Marked cells infused* |
| 301 | M | Diffuse Large B Cell Lymphoma | 42 | Rx only | 9830 | 138 | 11 | NA | 2.8E+06 | NA | NA | NA |
| 304 | M | Diffuse Large Cell Type (Immunoblastic Plasmacytoid) | 55 | Exp/Rx | <400 | 366 | 11 | 24 | 3.9E+06 | 7.7E+05 | 4.7E+06 | 0.14% |
| 305 | M | Diffuse Large B-Cell (Anaplastic) | 45 | Exp/Rx | <400 | 206 | 11 | 18 | 3.4E+06 | 7.3E+05 | 4.1E+06 | 0.14% |
| 306 | M | Plasmablastic Lymphoma | 45 | Exp/Rx | 2100 | 18 | 11 | 18 | 5.6E+06 | 1.2E+06 | 6.8E+06 | 0.11% |
| 307 | M | Diffuse Large B Cell Lymphoma | 25 | Exp/Rx | <400 | 577 | 11 | 12 | 6.5E+06 | 1.6E+06 | 8.1E+06 | 0.15% |
Abbreviations: UPN – Unique patient number; M = male; HPC-A-Rx - minimally manipulated Hematopoietic Progenitor Cell Apheresis product; HPC-A-Exp - CD34-enriched, ex-vivo transduced HPC-A. gc/ml = gene copies per milliliter serum; ANC = absolute neutrophil count.
% gene marked cells = % gene marked (estimated from 4 week in vitro culture analysis) × HPC-A-Exp Infused/(HPCA-Exp Infused + HPC-A-Rx Infused) × 100.
CD34 cell selection and lentiviral transduction
CD34-selection resulted in products highly enriched (Avg. = 121-fold, range 63–205) for CD34+ cell frequency (N=5). Starting with an average of 312 × 106 CD34+ cells (range 122–618 ×106), 151 × 106 CD34+ cells (range 47–317 × 106) were recovered (48% average yield) (Figure S1). CD34-selected cells were cryopreserved in the vapor phase of liquid nitrogen until 3 days prior to infusion at which time they were thawed and transduced.
In vitro analysis of Transduced HPC-A Products
In vitro analysis was conducted to determine the effects of transduction on hematopoietic potential and extent of gene marking of cells. In methyl cellulose culture, the frequency of colony forming units for transduced and non-transduced control cells, as a measure of toxicity of the transduction process, showed no significant differences in either the frequency or types of colonies formed (Figure S2). Cells were also placed in liquid culture with a cytokine cocktail designed to promote myelo/erythropoiesis or on a murine stromal cell line previously demonstrated to support the growth of B-lymphoid cells(26, 27). Samples were taken at weekly intervals for phenotypic analysis of myeloid, erythroid, B-lymphocytes and CD34+ stem/progenitor cells. No differences in the kinetics or magnitude of lineage development were observed between the transduced and non-transduced cell cultures (Figure S3). Taken together, these results support the overall safety of the cell transduction process with respect to in vitro hematopoietic potential.
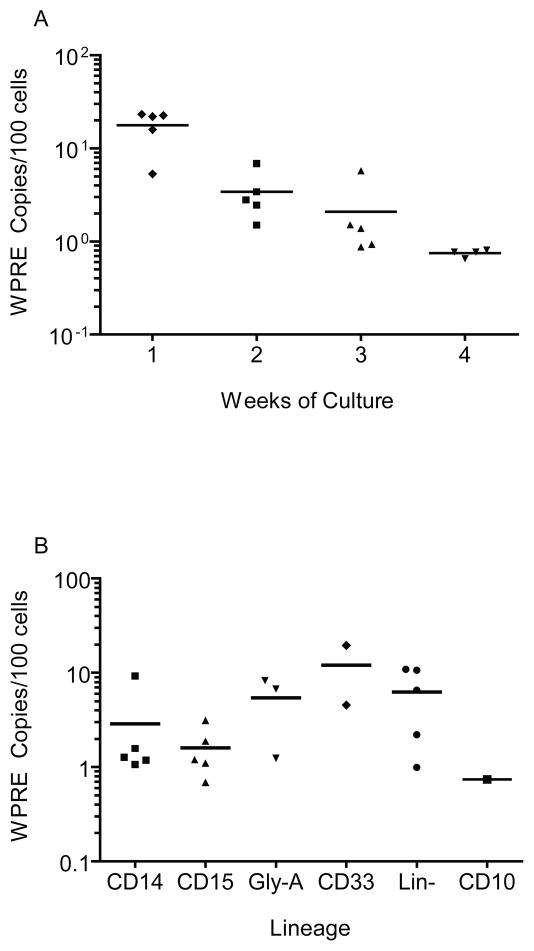
Cells from these cultures were also analyzed weekly for the presence of the rHIV7-shITAR- CCR5RZ transgene. Quantitative PCR analysis was performed using primers specific for the WPRE-marker of the transgene and for apolipoprotein B (ApoB), a single copy housekeeping gene to normalize for cell number. By calculating the ratio of copies of WPRE:ApoB, we observed gene marking levels as high as 23% (Avg. = 18% range 5–23%, N=5) after one week of culture (Figure 2a). However, the percentage of gene-marked cells declined rapidly over 4 weeks of culture to approximately 1% (average = 0.75, range 0.65–0.81, N=4). Lineage-specific cells were isolated by flow cytometric cell sorting at 2 weeks of culture (erythroid/myeloid cells) or 4 weeks (B-cells) and evaluated for gene marking. While the average percentage of marked cells varied by lineage, gene marking of all subsets was observed at levels similar to that in of the bulk population (0.7–19.5%) (Figure 2b). Since most of the cells used to initiate culture were negative for mature myelo-erythroid lineage markers (CD14, CD15, CD33, Gly-A) and mature cells do not contribute significantly to 4 week cultures, we believe these results indicate that minimally, multi-lineage HPC were transduced with the viral vector and differentiated into erythroid, monocytic, granulocytic and lymphoid lineages, each carrying the transgene.
Figure 2. Genetic Marking of patient samples during in vitro culture.
CD34-enriched HPC-A products were cultured in vitro following transduction for four weeks in liquid culture or on a murine stromal cell layer line in the presence of GM-CSF, IL3, IL6, Flt3L, erythropoietin, thrombopoietin. Cells were harvested weekly and DNA was extracted for qPCR analysis of the transgene. The average percentage of cells transduced with the HIV-shI-TAR-CCR5Rz genetic construct is defined as described in Methods. Copies of WPRE derived from a standard curve of known quantities of pHIV-shI- TAR-CCR5Rz plasmid spiked into the background of non transduced peripheral blood mononuclear cells. Apo B gene copies are determined from qPCR analysis of a standard curve of DNA from 10–1,000,000 peripheral blood mononuclear cells. (A) Average gene marking (bar) and individual values of gene marking (symbols) in bulk populations of cells from each patient over the 4 weeks of bulk liquid culture is shown. (B) For each patient, cells from in vitro culture were also sorted at 2 weeks (CD14, CD15, CD33, Gly-A and Lin-) or 4 weeks (CD10) for lineage specific marking analysis as described above (b).
We also analyzed cells derived from limiting dilution assays to verify the gene marking estimates from bulk PCR analysis and to determine the number of integrated copies of the lentiviral vector per cell. Using cultures of cells plated at 50, 10 or 5 cells/well and maintained for four to five weeks, we evaluated ≥500 growth positive wells from 4 patients and found only 6 wells positive for transgene from three of the patients (2/169 from UPN0305, 1/125 from UPN0307 and 3/125 from UPN0307). Of interest, positive wells had 1, 2 or 3 copies per cell of the integrated transgene (Figure S4). While this low number of transgene positive wells is consistent with the average frequency of transduced cells estimated from bulk culture, it suggests that the actual percentage of transduced cells in bulk culture is lower than estimated when assuming a single copy per cell.
Northern analysis was used to evaluate the expression of the CCR5 ribozyme, TAR decoy and siRNA sequences in the HPC-A-Exp cell products following in vitro differentiation and expansion. As expected, RNA was present early (day 13) for all 3 transgene species, but was not detectable by Northern blot analyses in cells from four-week cultures where the frequency of gene marking was only 1% (Figure 3a). Nevertheless, using the RT-PCR assays described in methods, we were able to detect between 105 and 106 copies of siRNA/8ng of total cellular RNA in samples from three time points of in vitro culture of transduced cells from UPN0306 (Figure 3b).
Figure 3. RNA Expression in Pre-Infusion Transduced Cell Product.
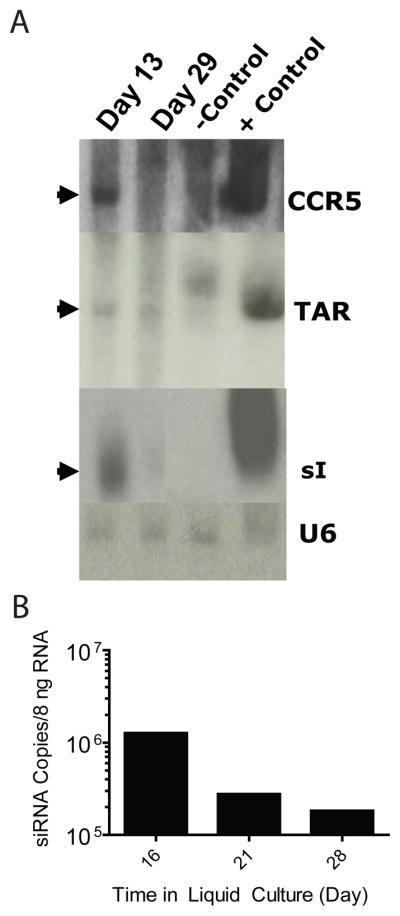
(A) Northern blot analysis of RNA expression in Patient 305 autologous CD34+ cells transduced with rHIV7-shI-TAR-CCR5RZ. Total RNA was extracted after transduction at day 13 and day 29. CEM cells were transduced with rHIV7-shI-TAR-CCR5RZ as a positive control. Non-transduced CD4+ cells were used as a negative control. 4 μg of total RNA was electrophoresed in an 8% polyacrylamide gel with 8 M urea, blotted onto a nylon membrane,and hybridized with a 32P-labeled probe for the siRNA, U16TAR decoy and anti-CCR5 ribozyme. (B) Quantitative PCR analysis of in vitro cultures for siRNA expression. Estimated siRNA copies per 8 ng of total cellular RNA is shown over time. Numbers of copies are estimated from a standard curve using known amounts of chemically synthesized tat/rev siRNA.
Treatment, Gene Marking and Expression in Patients Post-Transplant
Four patients received both the HPC-A-Exp and HPC-A-Rx at the doses listed (Table 1). All patients engrafted at 11 days post-transplant (engraftment defined as ANC>500 for three consecutive days). Excluding cytopenias, expected serious transplant-related adverse events (AE) in the first 30-day period post-transplant included grade 3 hypotension (2), grade 3 hypoxia (2), grade 3 fever (1) or central line infection (1). At the six month restaging, UPN0307 was found to have a syndrome consistent with Chlamydia and Pneumocystis jirovecii pulmonary infections which were treated with appropriate antibiotics without sequelae. The only unexpected AE was an asymptomatic (grade 2) occurrence of a solitary lytic scapular bone lesion histologically determined to be Langerhans cell histiocytosis which subsequently resolved without interevention. No vector sequences were detectible by PCR from a biopsy of this lesion.
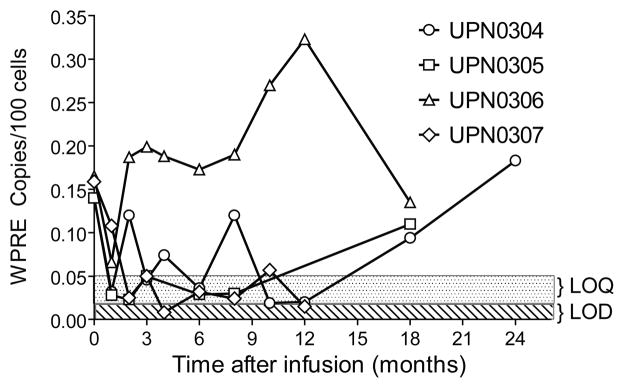
Median length of follow up is 18 months (range 6–24). All patients remain in remission from their lymphoma. Gene-marking levels of between 0.02% and 0.32% (200–3200 copies per 106 cells) of peripheral blood mononuclear cells have been observed for multiple patients and time points (Figure 4). Patients UPN0304 and UPN0306 had variable but quantifiable levels of gene marking (0.04%–0.12%) between 2 and 12 months post infusion, while UPN0305 and UPN0307 showed detectable (≥0.02%) but not quantifiable (≤ 0.05%) levels of marking over the same time frame. At 18 and 24 months however, UNP0304 and UPN0305 showed significant increases in the level of gene marking from non-quantifiable to 0.18% and 0.11% respectively while UPN0306 dropped to less than half of the 12 month marking level. Thus, our estimates of the frequency of gene marked, 4 week culture-initiating cells infused (0.11% –0.15%) correspond well with the levels of long term (>12 months) in vivo gene marking seen in UPN0304, UPN0305 and UPN0306 (0.09%–0.18%).
Figure 4. Gene Marking in the peripheral blood following HSC transplantation.
DNA was isolated directly from peripheral blood samples obtained pre-apheresis and at 1–4, 6, 8, 10, 12, 18 and 24 months post infusion. Real-time PCR was performed to detect the presence of the Woodchuck post-transcriptional regulatory element (WPRE) sequence using primers specific for a 174 nt long fragment contained within the pHIV7-shITAR-CCR5RZ DNA construct and against a standard curve derived by titration of DNA isolated from a clone that contains a single-copy integration of the lentivirus into DNA isolated from PBMC. A parallel set of qPCR amplifications of all test and reference samples using primers specific for the human p21 promoter was used to normalize input DNA for the WPRE amplifications. Sample values were considered quantifiable if they were at or above the limit of quantification (LOQ), defined as the lowest dilution of the H9c1 DNA standard curve for which at least 2 of the 3 replicates provide a measurable Ct value with a single expected melting curve. They were considered detectable if they were below the LOQ but above the limit of detection (LOD) defined as the lowest sample concentration where at least 1 of the 3 replicates generated a detectable Ct value with a single expected melting curve. LOQ and LOD values were determined for each amplification reaction and typically were in the range of 0.05% (500 cells/million) and 0.01% (100 cells/million), respectively. Open symbols on the Y axis (0 timepoint) indicate level of gene marking estimated for each patient as shown in Table 1.
Expression of the anti-tat/rev siRNA and CCR5 ribozyme in the peripheral blood was also followed for up to 24 months post infusion. Despite the low level of gene marking, we were able to detect vector expressed RNAs in multiple patient’s peripheral blood and bone marrow for up to 24 months post infusion in UPN0304 (Table 2). For example, UPN0304 whose gene marking varied between 0.05 and 0.1 percent had detectable levels of siRNA and CCR5 ribozyme at 3 and 6 months but siRNA alone at 1, 2 and 12 months and CCR5 ribozyme only at 4, 8 and 10 months. UPN0305, on the other hand, showed expression of siRNA and CCR5 ribozyme at 1 month and only siRNA sporadically (6 and 8 months) thereafter. However, in patient UPN0306 higher levels of gene marking were observed and we were able to quantify siRNA expression at between 103–105 copies/8 ng total cellular RNA (Figure S5a). Expression of the CCR5 ribozyme was also evident in peripheral blood samples from patients UPN0306 and UPN0307 for up to 10 months (Figure S5b).
Table 2.
Analysis of RNA expression in peripheral blood and bone marrow of patients following transplantation. Peripheral blood and or bone marrow was harvested between 1 and 24 months (1M-24M) post transplant and separated into peripheral blood mononuclear cells (PBMC), peripheral blood granulocytic cells (PBGC) bone marrow mononuclear cells (BMMC) or Bone marrow granulocytes (BMGC) by density separation (Ficoll). RNA was isolated from each fraction and analyzed for the expression of siRNA and CCR5 Ribozyme RNA as described in methods.
| 1M | 2M | 3M | 4M | 6M | 8M | 10M | 12M | 18M | 24M | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UPN | Cells | siRNA | CCR5RZ | siRNA | CCR5RZ | siRNA | CCR5RZ | siRNA | CCR5RZ | siRNA | CCR5RZ | siRNA | CCR5RZ | siRNA | CCR5RZ | siRNA | CCR5RZ | siRNA | CCR5RZ | siRNA | CCR5RZ |
| 304 | PBMC | + | − | + | − | + | + | − | + | + | + | − | + | − | + | + | − | − | + | + | + |
| PBGC | + | + | + | + | + | + | − | + | − | + | − | − | − | − | |||||||
| BMMC | + | − | + | + | |||||||||||||||||
| BMGC | − | + | − | − | |||||||||||||||||
| 305 | PBMC | + | + | − | − | − | − | − | − | + | − | + | − | − | − | − | − | ||||
| PBGC | + | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| BMMC | + | − | − | − | − | − | |||||||||||||||
| BMGC | − | − | − | − | − | − | |||||||||||||||
| 306 | PBMC | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||
| PBGC | + | + | − | − | + | + | + | + | + | + | + | + | |||||||||
| BMMC | + | + | |||||||||||||||||||
| BMGC | + | + | |||||||||||||||||||
| 307 | PBMC | + | + | + | + | − | + | + | + | + | + | − | + | ||||||||
| PBGC | + | + | + | + | + | − | − | − | − | − | − | − | |||||||||
| BMMC | + | + | |||||||||||||||||||
| BMGC | − | − | |||||||||||||||||||
Samples with detectable levels of RNA are marked + while those with no detectable RNA are marked −. Blanks indicate no sample scheduled or available for analysis.
Peripheral blood was isolated from patient UPN0306 at 18 months and separated into mononuclear and granulocytic fractions by density centrifugation. The mononuclear cells were further fractionated into CD3+ (T-cells), CD14+ (monocytes), CD19+ (B-cells) and cells that are negative for all three markers (Lin−) by fluorescence activated cell sorting. All subsets showed gene marking at levels consistent with the whole blood marking analysis (Figure S6). These data support the in vitro findings that mutli-lineage progenitor cells (in this case capable of sustaining hematopoiesis in vivo for 18 months) were in fact transduced.
Discussion
We have previously demonstrated that our standard HSC transplant procedure for high risk or relapsed AIDS related lymphoma (ARL) resulted in very low mortality and long term survival(22). We have also suggested a role for RNA based gene therapy as a potential therapeutic approach to controlling HIV infection(28, 29). Based on these results, we designed and conducted a clinical trial to assess the safety and feasibility of HPC based lentiviral gene therapy for HIV in the context of treatment for ARL. Secondary objectives included monitoring gene marking and RNA expression in the peripheral blood and marrow of treated patients and disease outcome. We describe here the results of a five patient study that establishes compelling evidence for the feasibility of the approach.
Unlike the lentivirus-based stem cell transplantation in adrenoleukodystrophy patients (30) in which only transduction processed HPC-A cells were infused after complete myeloablation, our study included an unmanipulated HPC-A graft. This was done to comply with regulatory requirements related to the then unknown toxicity and engraftment potential of the transduced cells. While we were successful in mobilizing enough peripheral blood stem cells in this patient population to prepare both unmanipulated and gene modified products for infusion in four of five patients, the infusion of the un-manipulated HPC-A product one day after the gene modified product contributed to a reduction in the frequency of gene modified progenitor cells in vivo. An additional possible explanation for the low level of marking of the more immature cells is the lack of entry into cell cycle during transduction. Although there is evidence that lentiviral vectors do not need cells to enter cycle to be transfected(31), other reports indicate that cytokines that induce cell proliferation enhance transduction of engrafting cells (32–34). Process development studies to optimize CD34+ cell transduction with minimal loss of hematopoietic potential are currently under investigation in our laboratories.
Importantly, there was no short term toxicity associated with the infusion of the genetically modified HPC-A product, and observed toxicities were procedure-related events consistent with standard autologous HCT. It has been noted, however, that siRNA can be toxic to cells in vitro(35), and therefore, a major concern for this study was whether expression of siRNA would be observed in the mature cell compartment of treated individuals. We demonstrated with both in vitro and in vivo studies that there was no overt hematopoietic toxicity associated with the lentiviral transduction process. Specifically, the persistent levels of shRNA expression observed in UPN0304 and UPN306 up to 24 and 18 months respectively strongly supports a conclusion that constitutive expression of the tat/rev siRNA is not toxic to peripheral blood cells. In addition, given the genetic marking of T cells, B cells, and granulocytes, there is no evidence for lineage specific toxicity, consistent with in vitro analysis and pre-clinical studies with this vector(13, 36).
Interestingly, patients UPN0306 and UPN0304 showed increased levels of gene marking immediately following a viremia (UPN0306 at day 0, UPN0304 at 15 months). Although we can not formally attribute the increase in gene modified cell frequency to viremia, the selective expansion of disease resistant cells in the face of viral selective pressure has been observed in a previous HIV gene therapy trial(19). While it is not possible at this time to predict what percentage of gene-marked cells would be required for clinical benefit in AIDS, a long term goal of this approach is the selection of genetically modified cells. If HIV could be a selection factor, then treatment interruption of HAART could become a component of gene transfer approaches. Alternatively, the inclusion of a selectable genetic marker in the anti-viral construct may allow for prospective chemotherapeutic selection of disease resistant cells. Support for this strategy comes from studies in large animal models that have demonstrated substantial increases in the percentage of gene marked cells in peripheral blood following transplant of gene modified HSC and drug selection (37–40).
In conclusion, we have developed methods for the isolation, genetic modification and infusion of CD34+ cells that support clinical investigation of stem cell gene therapy strategies for HIV. The sustained expression of siRNA and ribozyme for up to 24 months post-infusion marks an initial milestone in development of a genetic therapy for HIV infection using stem cells. Development of improved transduction processes and revising transplant procedures to preferentially infuse only transduced cells are likely to lead to higher levels of engrafted genetically modified cells. This would provide a setting for delivery and/or selection of therapeutic levels of HIV-resistant cells.
Methods
Vector Production
The lentiviral vector used in this study was manufactured according to cGMP requirements in the Center for Biomedicine and Genetics at City of Hope and was fully released tested according to FDA guidelines prior to enrolling patients. A self-inactivating lentiviral vector was designed to encode 3 RNAs consisting of an siRNA targeting a common exon shared by HIV tat/rev, a nucleolar localizing TAR decoy, and an anti-CCR5-specific hammerhead ribozyme as previously described(29). The packaging system used 4 separate plasmids developed at City of Hope as previously described(41).
HPC collection
Hematopoietic progenitor cell-apheresis products (HPC-A) were collected after standard salvage chemotherapy or after cyclophosphamide (2g/m2) plus granulocyte colony stimulating factor (G-CSF 10 μg/kg). A minimum of 2.5 × 106 CD34+ cells/kg were collected and cryopreserved without further manipulation (HPC-A-Rx). An additional one to two apheresis collections were performed to collect cells for the genetic modification (HPC-A-Exp). CD34+ cells were enriched from the HPC-A-Exp collection over a CliniMACS™ device according to manufacturer’s instructions and cryopreserved using a control rate freezer. CD34+ cells were thawed on day −2 before HCT and pre-stimulated for 16–20 hrs in X-vivo15 medium (Lonza, Walkersville, MD) containing 2mM L-Glutamine, 100ng/mL of Stem Cell Factor (SCF), 100ng/mL of Flt-3 ligand (Flt-3L), and 10ng/mL of Thrombopoietin (TPO) (CellGenix, Antioch, IL) at density of 2×106 cells/ml. Pre-stimulated cells were transduced with lentiviral vector (rHIV7-shI-TAR-CCR5RZ) at an MOI of 5 for 16–24 hours in 75 cm2 tissue culture flasks flasks coated with 25μg/cm2 of fibronectin (Retronectin™, Takara Bio Inc. Shiga, Japan). A sample of CD34+ cells was incubated as described but in the absence of lentiviral vector to serve as a transduction control. Transduced cells were pooled, washed with X-vivo15 medium and resuspended in final formulation buffer (CliniMACs PBS/EDTA buffer with 0.5% HSA) at density of 1×106 cells/mL for infusion as described below. A sample was taken from each product and tested for total cell count, viability, sterility and endotoxin.
Transplant eligibility criteria
Patients with HIV and Non Hodgkins Lymphoma (NHL) in first CR with high or high intermediate IPI scores were eligible for transplant(42). All patients had to be on combination antiretroviral therapy (ART) that did not include zidovudine, maintain an HIV viral load < 50,000 gc/ml, and be willing to suspend antiretroviral therapy during the period of HPC mobilization and collection. The City of Hope’s Institutional Review Board and Institutional Biosafety Committee approved the protocol, and informed written consent was obtained from each patient in the presence of a patient advocate.
Transplantation Procedure
Approximately one week after completion of aphereses to collect HPC, patients were admitted to the transplant unit and received chemotherapy exactly as previously described(22). On the day of HCT (day 0), the transduced product (HPC-A-Exp) underwent release testing for endotoxin, sterility and viability and was then infused. On day +1, the unmanipulated product (HPC-A-Rx) was thawed at the bedside and infused as per guidelines approved by the Foundation for the Accreditation of Cellular Therapy (FACT). Safety was assessed for immediate effects of the treatment using the hematologic Common Terminology Criteria for Adverse Events (CTCAE) version 3.0 grading system at 24 and 48 hours and 14 days after infusion and at all subsequent visits from 1–18 months.
In Vitro Cell Culture
Methylcellulose Cultures of CD34+ cells. Samples from each patient were assayed for hematopoietic potential using standard methylcellulose-based colony forming unit assays and 4 week liquid and stromal cell cultures. The murine stromal cell line used in these studies to support the growth of B-lymphoid lineages from patient CD34+ cells has been previously described(27). Limiting dilution assay. Limiting dilution analysis (LDA) of cells from each patient was used to isolate the progeny of primitive, clonogenic progenitor cells for gene marking and copy number analysis.
Cell Phenotype and Analysis
Aliquots of cells from bulk liquid and stromal culture were taken weekly for phenotypic and qPCR analysis. Antibodies to lineage specific cell surface antigens were used to follow hematopoiesis during culture and isolate subpopulations by fluorescent activated cell sorting. Phenotypic data was collected on an FC500 flow cytometer (Beckman Coulter, Fullerton, CA) and analyzed with FCS express V3 software (De Novo Software, Ontario, Canada). Samples labeled as described were also sorted based on surface marker expression to >98% purity using a MoFlo cell sorter (Beckman Coulter Corporation, Brea, California) for subsequent analysis of DNA marking and gene expression.
DNA Analysis of in vitro-derived cells
Samples from in vitro culture were analyzed weekly for the presence of integrated viral vector. Quantitative PCR (qPCR) was performed to detect the number of copies of integrated vector per cell. In each case (except for limiting dilution analysis) genomic DNA from approximately 20,000 cells was used in each reaction. For colonies derived from LDA, DNA from the entire colony was used. The number of copies of WPRE detected was normalized to cell number using qPCR for a housekeeping gene (apolipoprotein B). Results are reported as average percent gene marked cells or WPRE copies/100 cells. Results from qPCR of individual colonies from LDA is reported as absolute copy number per cell.
RNA Analysis
RNA analysis was performed on gene modified products from in vitro culture and peripheral blood and bone marrow cells from patients at specific time points following transduction and infusion. Northern analysis was used to detect expression of all three RNA moieties from in vitro cultures but was not sensitive enough to detect RNA in patient peripheral blood samples. More sensitive detection of expression of the CCR5RZ was accomplished by a real-time RT-PCR while expression of the tat/rev coding siRNA was analyzed by RT–PCR using the TaqMan® MicroRNA Reverse Transcription kit. RT PCR products were analyzed by gel electrophoresis in a 1% agarose gel, blotting onto nitrocellulose and hybridizing with radio-labeled sequence specific probes.
Measurement of in vivo gene marking
The presence of shI-TAR-CCR5RZ-marked cells in peripheral blood and was also assessed by qPCR analysis of WPRE sequences using a fixed amount (50 ng) of genomic DNA from peripheral blood. The average % WPRE+ DNA in samples was determined using a standard curve generated from DNA isolated from a clone with a single copy integration of the WPRE-containing lentivirus and used in a standard curve to titrate DNA isolated from patient specimens. A parallel set of qPCR amplifications was performed on all test and reference samples using primers specific for the p21 promoter as an internal control.
Supplementary Material
Acknowledgments
We thanks Nancy Gonzalez and Amira Ahmed (Div. of Hematology) for Quality Systems Support, Lan-Feng Cao (CPDM Laboratory) for QC release testing, Guadalupe Duarte and Michelle Wardlow (Div. of Hematology) for patient care. Cecilia Arbayo for assistance in patient consenting. Suenell Broyer and Rodica Stan (Center for Applied Technology Development) for assistance with the IND application and the other regulatory applications and Chy-Anh Tran and Lucy Brown for assistance with cell sorting. The authors are grateful to Alexandra Levine for her advice and encouragement and to the staff at Benitec Ltd, especially Ken Reed and Sue MacLeman, for continuing support in the preclinical and clinical phases of this project.
Funding: The authors acknowledge support from NIH grants AI42552 and HL07470 (JJR), NIH P50 CA107399 (Lymphoma SPORE), P30 CA33572-26 (CCSG) (SJF), NIH AI61839 (JAZ), NCRR S10RR025083-01 (DD), and GCRC grant M01 RR00043 (JAZ). A grant from Benitec Ltd supported vector manufacture, process development, and a portion of the clinical trial.
Footnotes
Author Contributions: DD and AK contributed equally to this study. DD developed and supervised cell manufacturing operations and in vitro correlative studies, interpreted data and wrote the manuscript. AK was responsible for trial design, patient recruiting, enrollment and treatment on trial and wrote the manuscript. LL, AR and PY performed manufacturing of cell products and performed in vitro correlative assays. SL performed sample processing and distribution. HL performed RNA analysis on in vitro and in vivo samples. SS performed manufacturing operations. JKY conceived of and developed vector production system, LC and DH manufactured lentiviral vectors, JJR conceived and designed the triple gene vector, supervised the development of the siRNA and CCR5 ribozyme assays from clinical samples and wrote the manuscript. ML constructed the clinical vector and assisted in optimizing vector production strategies. MK supervised the development and qualification of the in vivo gene marking assays and product release testing, SM performed the in vivo marking assays, SFL supervised in vivo gene marking assays, SJF conceptualized and designed clinical studies. JAZ coordinated the clinical, laboratory, and regulatory processes, interpreted data and wrote the manuscript.
References
- 1.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science New York, NY. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 2.Chen RY, Accortt NA, Westfall AO, Mugavero MJ, Raper JL, Cloud GA, Stone BK, Carter J, Call S, Pisu M, Allison J, Saag MS. Distribution of health care expenditures for HIV-infected patients. Clin Infect Dis. 2006;42:1003–1010. doi: 10.1086/500453. [DOI] [PubMed] [Google Scholar]
- 3.Baltimore D. Gene therapy. Intracellular immunization. Nature. 1988;335:395–396. doi: 10.1038/335395a0. [DOI] [PubMed] [Google Scholar]
- 4.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 5.Bevec D, Dobrovnik M, Hauber J, Bohnlein E. Inhibition of human immunodeficiency virus type 1 replication in human T cells by retroviral-mediated gene transfer of a dominant-negative Rev transactivator. Proc Natl Acad Sci U S A. 1992;89:9870–9874. doi: 10.1073/pnas.89.20.9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonyhadi ML, Moss K, Voytovich A, Auten J, Kalfoglou C, Plavec I, Forestell S, Su L, Bohnlein E, Kaneshima H. RevM10-expressing T cells derived in vivo from transduced human hematopoietic stem-progenitor cells inhibit human immunodeficiency virus replication. Journal of virology. 1997;71:4707–4716. doi: 10.1128/jvi.71.6.4707-4716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammer D, Wild J, Ludwig C, Asbach B, Notka F, Wagner R. Fusion of Epstein-Barr virus nuclear antigen-1-derived glycine-alanine repeat to trans-dominant HIV-1 Gag increases inhibitory activities and survival of transduced cells in vivo. Human gene therapy. 2008;19:622–634. doi: 10.1089/hum.2007.095. [DOI] [PubMed] [Google Scholar]
- 8.Perez EE, Riley JL, Carroll RG, von Laer D, June CH. Suppression of HIV-1 infection in primary CD4 T cells transduced with a self-inactivating lentiviral vector encoding a membrane expressed gp41- derived fusion inhibitor. Clin Immunol. 2005;115:26–32. doi: 10.1016/j.clim.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Plavec I, Voytovich A, Moss K, Webster D, Hanley MB, Escaich S, Ho KE, Bohnlein E, DiGiusto DL. Sustained retroviral gene marking and expression in lymphoid and myeloid cells derived from transduced hematopoietic progenitor cells. Gene therapy. 1996;3:717–724. [PubMed] [Google Scholar]
- 10.Vallanti G, Lupo R, Federico M, Mavilio F, Bovolenta C. T Lymphocytes transduced with a lentiviral vector expressing F12-Vif are protected from HIV-1 infection in an APOBEC3G-independent manner. Mol Ther. 2005;12:697–706. doi: 10.1016/j.ymthe.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 11.van Griensven J, Zhan X, Van Maele B, Pluymers W, Michiels M, De Clercq E, Cherepanov P, Debyser Z. Expression of HIV-1 integrase in CEM cells inhibits HIV-1 replication. J Gene Med. 2004;6:268–277. doi: 10.1002/jgm.520. [DOI] [PubMed] [Google Scholar]
- 12.Aagaard LA, Zhang J, von Eije KJ, Li H, Saetrom P, Amarzguioui M, Rossi JJ. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene therapy. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson J, Li MJ, Palmer B, Remling L, Li S, Yam P, Yee JK, Rossi J, Zaia J, Akkina R. Safety and Efficacy of a Lentiviral Vector Containing Three Anti-HIV Genes-CCR5 Ribozyme, Tat-rev siRNA, and TAR Decoy-in SCID-hu Mouse-Derived T Cells. Mol Ther. 2007;15:1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- 14.Asparuhova MB, Barde I, Trono D, Schranz K, Schumperli D. Development and characterization of a triple combination gene therapy vector inhibiting HIV-1 multiplication. J Gene Med. 2008;10:1059–1070. doi: 10.1002/jgm.1238. [DOI] [PubMed] [Google Scholar]
- 15.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto T, Miyoshi H, Yamamoto N, Yamamoto N, Inoue J, Tsunetsugu-Yokota Y. Lentivirus vectors expressing short hairpin RNAs against the U3-overlapping region of HIV nef inhibit HIV replication and infectivity in primary macrophages. Blood. 2006;108:3305–3312. doi: 10.1182/blood-2006-04-014829. [DOI] [PubMed] [Google Scholar]
- 17.van Lunzen J, Glaunsinger T, Stahmer I, von Baehr V, Baum C, Schilz A, Kuehlcke K, Naundorf S, Martinius H, Hermann F, Giroglou T, Newrzela S, Muller I, Brauer F, Brandenburg G, Alexandrov A, von Laer D. Transfer of autologous gene-modified T cells in HIV-infected patients with advanced immunodeficiency and drug-resistant virus. Mol Ther. 2007;15:1024–1033. doi: 10.1038/mt.sj.6300124. [DOI] [PubMed] [Google Scholar]
- 18.Kang EM, de Witte M, Malech H, Morgan RA, Phang S, Carter C, Leitman SF, Childs R, Barrett AJ, Little R, Tisdale JF. Nonmyeloablative conditioning followed by transplantation of genetically modified HLA-matched peripheral blood progenitor cells for hematologic malignancies in patients with acquired immunodeficiency syndrome. Blood. 2002;99:698–701. doi: 10.1182/blood.v99.2.698. [DOI] [PubMed] [Google Scholar]
- 19.Podsakoff GM, Engel BC, Carbonaro DA, Choi C, Smogorzewska EM, Bauer G, Selander D, Csik S, Wilson K, Betts MR, Koup RA, Nabel GJ, Bishop K, King S, Schmidt M, von Kalle C, Church JA, Kohn DB. Selective survival of peripheral blood lymphocytes in children with HIV-1 following delivery of an anti-HIV gene to bone marrow CD34(+) cells. Mol Ther. 2005;12:77–86. doi: 10.1016/j.ymthe.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 20.Amado RG, Mitsuyasu RT, Rosenblatt JD, Ngok FK, Bakker A, Cole S, Chorn N, Lin LS, Bristol G, Boyd MP, MacPherson JL, Fanning GC, Todd AV, Ely JA, Zack JA, Symonds GP. Antihuman immunodeficiency virus hematopoietic progenitor cell-delivered ribozyme in a phase I study: myeloid and lymphoid reconstitution in human immunodeficiency virus type-1-infected patients. Human gene therapy. 2004;15:251–262. doi: 10.1089/104303404322886101. [DOI] [PubMed] [Google Scholar]
- 21.Mitsuyasu RT, Merigan TC, Carr A, Zack JA, Winters MA, Workman C, Bloch M, Lalezari J, Becker S, Thornton L, Akil B, Khanlou H, Finlayson R, McFarlane R, Smith DE, Garsia R, Ma D, Law M, Murray JM, von Kalle C, Ely JA, Patino SM, Knop AE, Wong P, Todd AV, Haughton M, Fuery C, Macpherson JL, Symonds GP, Evans LA, Pond SM, Cooper DA. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnan A, Molina A, Zaia J, Smith D, Vasquez D, Kogut N, Falk PM, Rosenthal J, Alvarnas J, Forman SJ. Durable remissions with autologous stem cell transplantation for high-risk HIV-associated lymphomas. Blood. 2005;105:874–878. doi: 10.1182/blood-2004-04-1532. [DOI] [PubMed] [Google Scholar]
- 23.Ciuffi A, Mitchell RS, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Connolly JB. Lentiviruses in gene therapy clinical research. Gene therapy. 2002;9:1730–1734. doi: 10.1038/sj.gt.3301893. [DOI] [PubMed] [Google Scholar]
- 25.Wang GP, Levine BL, Binder GK, Berry CC, Malani N, McGarrity G, Tebas P, June CH, Bushman FD. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther. 2009;17:844–850. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szilvassy SJ, Weller KP, Lin W, Sharma AK, Ho AS, Tsukamoto A, Hoffman R, Leiby KR, Gearing DP. Leukemia inhibitory factor upregulates cytokine expression by a murine stromal cell line enabling the maintenance of highly enriched competitive repopulating stem cells. Blood. 1996;87:4618–4628. [PubMed] [Google Scholar]
- 27.DiGiusto D, Chen S, Combs J, Webb S, Namikawa R, Tsukamoto A, Chen BP, Galy AH. Human fetal bone marrow early progenitors for T, B, and myeloid cells are found exclusively in the population expressing high levels of CD34. Blood. 1994;84:421–432. [PubMed] [Google Scholar]
- 28.Li MJ, Bauer G, Michienzi A, Yee JK, Lee NS, Kim J, Li S, Castanotto D, Zaia J, Rossi JJ. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol Ther. 2003;8:196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- 29.Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 30.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, Vidaud M, Abel U, Dal-Cortivo L, Caccavelli L, Mahlaoui N, Kiermer V, Mittelstaedt D, Bellesme C, Lahlou N, Lefrere F, Blanche S, Audit M, Payen E, Leboulch P, l’Homme B, Bougneres P, Von Kalle C, Fischer A, Cavazzana-Calvo M, Aubourg P. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science (New York, NY. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 31.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science (New York, NY. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 32.Sutton RE, Reitsma MJ, Uchida N, Brown PO. Transduction of human progenitor hematopoietic stem cells by human immunodeficiency virus type 1-based vectors is cell cycle dependent. Journal of virology. 1999;73:3649–3660. doi: 10.1128/jvi.73.5.3649-3660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhoeyen E, Negre D, Cosset FL. Production of lentiviruses displaying ‘‘early-acting’’ cytokines for selective gene transfer into hematopoietic stem cells. Methods in molecular biology (Clifton, NJ. 2008;434:99–112. doi: 10.1007/978-1-60327-248-3_7. [DOI] [PubMed] [Google Scholar]
- 34.Zielske SP, Gerson SL. Cytokines, including stem cell factor alone, enhance lentiviral transduction in nondividing human LTCIC and NOD/SCID repopulating cells. Mol Ther. 2003;7:325–333. doi: 10.1016/s1525-0016(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 35.An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D, Chen IS. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerjea A, Li MJ, Bauer G, Remling L, Lee NS, Rossi J, Akkina R. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol Ther. 2003;8:62–71. doi: 10.1016/s1525-0016(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 37.Neff T, Beard BC, Kiem HP. Survival of the fittest: in vivo selection and stem cell gene therapy. Blood. 2006;107:1751–1760. doi: 10.1182/blood-2005-06-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persons DA, Allay JA, Bonifacino A, Lu T, Agricola B, Metzger ME, Donahue RE, Dunbar CE, Sorrentino BP. Transient in vivo selection of transduced peripheral blood cells using antifolate drug selection in rhesus macaques that received transplants with hematopoietic stem cells expressing dihydrofolate reductase vectors. Blood. 2004;103:796–803. doi: 10.1182/blood-2003-05-1572. [DOI] [PubMed] [Google Scholar]
- 39.Hanazono Y, Nagashima T, Takatoku M, Shibata H, Ageyama N, Asano T, Ueda Y, Dunbar CE, Kume A, Terao K, Hasegawa M, Ozawa K. In vivo selective expansion of gene-modified hematopoietic cells in a nonhuman primate model. Gene therapy. 2002;9:1055–1064. doi: 10.1038/sj.gt.3301781. [DOI] [PubMed] [Google Scholar]
- 40.Maier P, Spier I, Laufs S, Veldwijk MR, Fruehauf S, Wenz F, Zeller WJ. Chemoprotection of human hematopoietic stem cells by simultaneous lentiviral overexpression of multidrug resistance 1 and O(6)-methylguanine- DNA methyltransferase(P140K) Gene therapy. 17:389–399. doi: 10.1038/gt.2009.133. [DOI] [PubMed] [Google Scholar]
- 41.Yam PY, Li S, Wu J, Hu J, Zaia JA, Yee JK. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol Ther. 2002;5:479–484. doi: 10.1006/mthe.2002.0558. [DOI] [PubMed] [Google Scholar]
- 42.A predictive model for aggressive non-Hodgkin’s lymphoma. The International Non-Hodgkin’s Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.