Abstract
Many compounds being considered as candidates for advanced biofuels are toxic to microorganisms. This introduces an undesirable trade-off when engineering metabolic pathways for biofuel production because the engineered microbes must balance production against survival. Cellular export systems, such as efflux pumps, provide a direct mechanism for reducing biofuel toxicity. To identify novel biofuel pumps, we used bioinformatics to generate a list of all efflux pumps from sequenced bacterial genomes and prioritized a subset of targets for cloning. The resulting library of 43 pumps was heterologously expressed in Escherichia coli, where we tested it against seven representative biofuels. By using a competitive growth assay, we efficiently distinguished pumps that improved survival. For two of the fuels (n-butanol and isopentanol), none of the pumps improved tolerance. For all other fuels, we identified pumps that restored growth in the presence of biofuel. We then tested a beneficial pump directly in a production strain and demonstrated that it improved biofuel yields. Our findings introduce new tools for engineering production strains and utilize the increasingly large database of sequenced genomes.
Keywords: biofuel, efflux pump, synthetic biology, tolerance engineering
Introduction
Existing biofuels, such as ethanol and esters of linear fatty acids, can be used in only limited quantities in current gasoline, diesel and jet engines. Recently, there have been several reports of efforts to engineer microorganisms to produce advanced biofuels that can be used in larger quantities in our existing transportation infrastructure. Short to medium chain (C4–C12) alcohols such as butanol, isopentanol and geraniol are superior to ethanol as gasoline replacements (Peralta-Yahya and Keasling, 2010). Longer, branched-chain compounds (C9–C23) such as geranyl acetate and farnesyl hexanoate are better biodiesel alternatives than linear esters because branching reduces their freezing point. In addition, cyclic alkenes such as limonene and pinene serve as precursors to jet fuel (Harvey et al, 2010; Peralta-Yahya and Keasling, 2010). While microbial biosynthetic routes to most of these compounds exist, the biofuels have known antimicrobial activity (Fischer et al, 2008). For the production to be cost effective, yields must exceed native tolerance levels, necessitating the development of stress-tolerant strains (Supplementary Text). Product toxicity is a common problem in strain engineering for biotechnology applications. Work on ethanol production has shown that alleviating toxicity is necessary for maintaining and maximizing yield (Alper et al, 2006; Jarboe et al, 2007). Thus, it is crucial that we improve tolerance in parallel with the development of metabolic pathways for the production of next-generation biofuels.
Microbes have several strategies for addressing biofuel toxicity (Isken and de Bont, 1998; Ramos et al, 2002). Here we focus on efflux pumps, a class of membrane transporters that export toxins from the cell using the proton motive force (Putman et al, 2000; Nikaido and Takatsuka, 2009). Efflux pumps in Gram-negative bacteria are composed of three proteins: an inner membrane protein responsible for substrate recognition and proton exchange, a periplasmic linker and an outer membrane channel. All subunits are essential for function and the corresponding genes are commonly arranged together in an operon. Relatively few solvent-resistant efflux pumps have been previously characterized. Examples include ttgABC, ttgDEF, ttgGHI (Rojas et al, 2001) and srpABC (Kieboom et al, 1998a) from Pseudomonas putida. These pumps appear to be specific to solvents (Kieboom et al, 1998b; Isken and de Bont, 2000), while more general multidrug efflux pumps such as acrAB-tolC from E. coli (Nikaido and Takatsuka, 2009) export a broad range of substrates, including solvents. All known solvent-resistant efflux pumps in Gram-negative bacteria fall into the hydrophobe/amphiphile efflux (HAE1) family of resistance-nodulation-division pumps (Tseng et al, 1999). Sequenced bacterial genomes include many efflux pumps and present a largely unexplored resource for discovering novel pumps with potential for use in engineering fuel tolerance. Here we take a systematic approach to screen a library of primarily uncharacterized heterologous pumps for engineering biofuel-tolerant host strains. We then demonstrate that expression of a heterologous pump can increase the yield of a biofuel production strain.
Results and discussion
Using E. coli as our engineering host, we asked whether heterologously expressed efflux pumps could reduce toxicity by exporting biofuel from the cell. We constructed a database of all HAE1 pumps from sequenced bacterial genomes (Materials and methods). Using this set, we performed a bioinformatics screen to compare regions that are predicted to be responsible for substrate specificity to those of TtgB, a well-characterized solvent-resistant pump. This metric allowed us to rank the complete set of pumps and select a subset that represented a uniform distribution of candidates (Supplementary Figure S1, Supplementary Methods). To construct the library, efflux pump operons were amplified from the genomic DNA of the selected bacteria, cloned into a vector, and transformed into an E. coli host strain (Materials and methods). In total, our library contains 43 efflux pumps, most of which have not been previously characterized for biofuel or solvent tolerance.
Although tolerance and export of intracellularly produced biofuel is the ultimate goal, we hypothesized that testing for tolerance to exogenous biofuels would identify pumps with the potential to export biofuel from the cell. Similar strategies have been used previously to improve production. For example, mutations in Saccharomyces cerevisiae that improved ethanol tolerance led to an increase in production (Alper et al, 2006). In addition, an evolved isobutanol-tolerant strain of E. coli improved growth and production when grown under isobutanol stress (Atsumi et al, 2010). It should be noted that yields from production strains can exceed the inhibitory concentrations of exogenous biofuels. For example, isobutanol inhibits E. coli growth at 8 g/l, but strains continue to produce up to 20 g/l in stationary phase after growth stops (Atsumi et al, 2008).
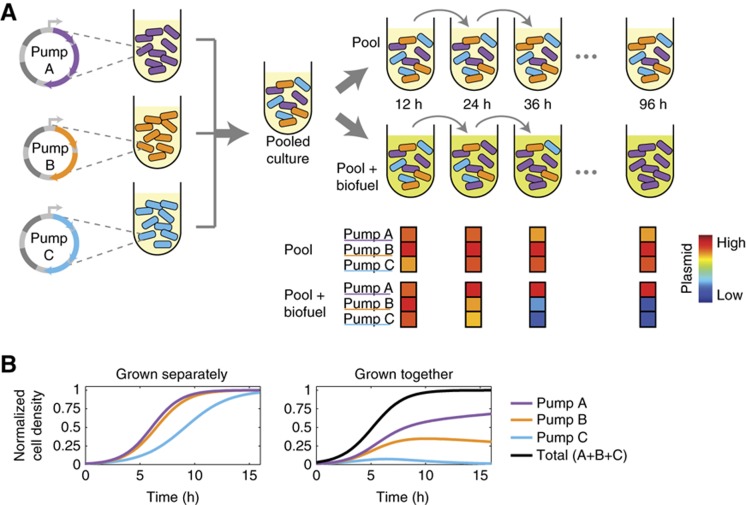
In order to efficiently screen the efflux pumps against biofuel candidates, we devised a competition-based strategy to select for pumps that improved biofuel tolerance (Figure 1A). When a survival or fitness phenotype can be used, competitive growth experiments provide an effective selection strategy (Lynch et al, 2007; Ho et al, 2009). Efflux pump expression strains were grown individually and then pooled, so that all strains were represented in equal proportion. This pooled culture was then grown both with and without biofuel and maintained through serial dilutions every 10–14 h. At each dilution time point, plasmids from the culture were isolated and a custom microarray was used to quantify the amount of each efflux pump plasmid remaining in the culture (Materials and methods). A mathematical model of competitive growth was used to guide experimental design (Figure 1B). Because strains expressing pumps that help to mitigate biofuel toxicity will have a growth advantage, these strains will dominate the co-cultures after only a small number of dilution cycles (Supplementary Figure S2).
Figure 1.
Competition assay efficiently identifies efflux pumps that provide biofuel tolerance. (A) Plasmids containing the operons for individual pumps were transformed into cells. These strains were grown independently and then pooled in equal proportion. The pooled culture was grown both with and without biofuel for 96 h, with dilution into fresh medium every 10–14 h. At selected time points, cultures were saved, plasmids isolated, and the relative levels of each plasmid were quantified. If certain plasmids provide a growth advantage, they become overrepresented in the culture. (B) Simulations of competitive exclusion. Grown separately, the growth curve of a strain is dependent only on its growth rate. When grown together, as in the competition assay, strains with higher growth rates dominate the population.
In order to experimentally validate our predictions, we first asked if the composition of the competing cultures changed over time. When the pooled culture was grown without any biofuel, all pumps were represented equally, indicating that no strain had a particular advantage (Figure 2A). This remained true over the course of the 96-h experiment, showing that under these induction conditions, any burden of pump expression was roughly equivalent for all strains. In contrast, when the pooled culture was grown in the presence of an inhibitory biofuel such as geranyl acetate, some efflux pumps conferred a distinct advantage (Figure 2B). Although all strains started out with equal representation, after 38 h the population composition changed, with cells containing the advantageous pumps becoming an increasingly large proportion of the population. The efflux pumps that enhanced tolerance to geranyl acetate originated from a variety of hosts and include both known and previously uncharacterized pumps.
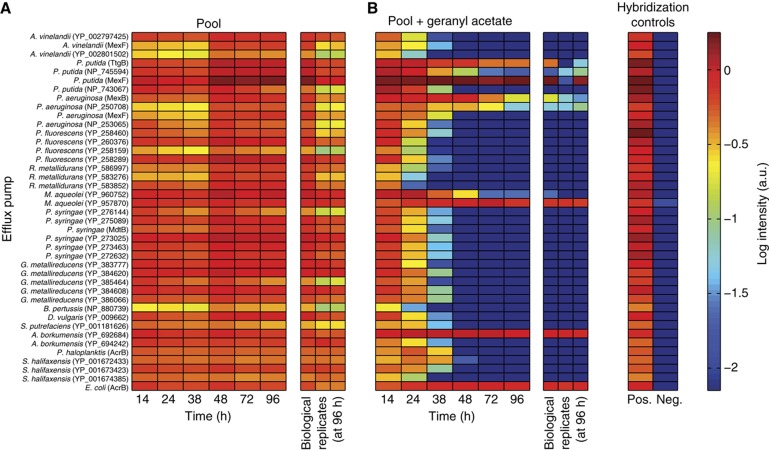
Figure 2.
When grown with biofuel, strains with beneficial pumps dominate the culture. (A) The pooled culture shows performance of all pumps to be similar in the absence of an inhibitor. Each row represents the quantity of a different pump-containing plasmid; columns show time points in the competition assay. For each pump, the host and GenBank accession number (or protein name, if annotated) of the inner membrane protein are given (Supplementary Table S1). (B) When the pooled culture was grown with 2% geranyl acetate, some pumps improved survival. Positive and negative hybridization controls label all pump plasmids and a pump-free plasmid, respectively (Materials and methods). All data are shown in log scale with arbitrary units (a.u.).
Next, we asked whether competing the cultures in the presence of different biofuels would identify unique sets of resistant efflux pumps for each biofuel. With respect to the compounds tested, our results fall broadly into two classes: (i) biofuels that are toxic, but where pumps do not reduce toxicity and (ii) biofuels where the pumps do reduce toxicity. Both n-butanol and isopentanol fall into the first class of fuels (Figure 3A and B). This could be because none of the pumps in the library export these compounds or, alternatively, the rate of export may not be sufficient to counteract intracellular accumulation (Lim and Nikaido, 2010). Pumps improved tolerance for the second class of fuels. We saw a distinct separation between the competition winners and those pumps that were not beneficial (Figure 3C). The competition survivors and their relative abundances in the culture were biofuel-specific.
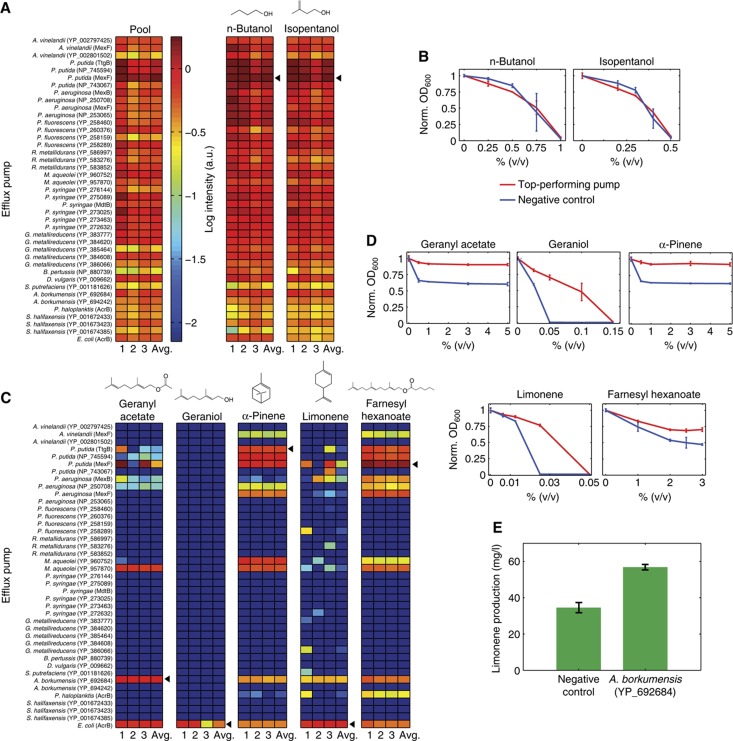
Figure 3.
Efflux pumps can improve tolerance and production yields for selected biofuels. (A) Plasmid levels for each of the efflux pumps in the library after 72 h of competition. The pool culture was grown without an inhibitor. For all others, biofuel was added at an inhibitory level (Materials and methods). Data from three biological replicates and their average are shown. (B) Inhibition experiments with the top-performing pump (shown with an arrow in (A)) showed that for n-butanol and isopentanol the pump provides no growth advantage over the negative control. Taken together, (A) and (B) demonstrate that none of the efflux pumps we tested improved tolerance to n-butanol or isopentanol. (C) Competition results for biofuels where efflux pumps provided a growth advantage. (D) Top-performing pumps (shown with an arrow in (C)) showed improvements over the negative control strain. Measurements shown in (B) and (D) were taken in triplicate and averaged; error bars show standard deviation. Note that all efflux pumps that survived the α-pinene competition showed good performance when grown individually with α-pinene (Supplementary Figures S3, S4). (E) Limonene yield in a production strain expressing an efflux pump compared with an identical strain without the pump. Error bars show standard deviation between biological replicates.
To control for the possibility that pumps were mutated over the course of the competition assay and to further characterize their individual performance, we retransformed sequenced pump plasmids into the E. coli host strain and retested for tolerance improvements after 14 h of growth. For each biofuel, we tested strains expressing the top-performing pumps individually and compared their survival relative to a pump-free control. All competition winners outperformed the control strain (Figure 3D). Furthermore, comprehensive tests with one fuel verified that all competition survivors had increased tolerance relative to the control (Supplementary Figure S3) and outperformed non-winning strains (Supplementary Figure S4). The pumps that survived the competition are ideal candidates for future biofuel engineering efforts.
Next, we asked whether pumps that improved tolerance also enhanced biofuel production. Two pumps consistently survived the limonene competition: the native E. coli pump AcrAB and a previously uncharacterized pump from Alcanivorax borkumensis. We focused on the A. borkumensis pump and tested it in a limonene production strain. Notably, strains expressing the pump produced significantly more limonene than those with no pump (Figure 3E). These results provide an important proof-of-principle demonstration that efflux pumps that increase tolerance to exogenous biofuel can improve the yield of a production host. It should be noted that current limonene production levels are not yet toxic to cells. Irrespective of the toxicity, an effective export pump may serve to relieve end product inhibition of metabolic pathway enzymes and result in an improvement in production. As yields improve, export pumps may play an increasingly important role.
Beneficial efflux pumps can come from a variety of unrelated bacteria. Our results identified marine microbes such as A. borkumensis, Marinobacter aqueolei and Pseudoalteromonas haloplanktis to be valuable sources of biofuel-tolerant efflux pumps. Pumps from these organisms are similar (40–69% sequence homology) to several known solvent-tolerant pumps, but it is not clear whether their substrate specificity is narrow (similar to TtgB) or broad (similar to AcrB) (Supplementary Table S2). Interestingly, several of the biofuel-tolerant pumps have only the inner membrane and periplasmic proteins present in their operons (Materials and methods), meaning that they must successfully recruit a native E. coli outer membrane protein for export. As pumps can work in concert to export substrates (Lee et al, 2000; Segura et al, 2003), multi-pump constructs may produce further increases in tolerance. Additional methods for improvement, such as codon optimization or directed evolution, can be used to fine-tune these pumps for a specific target. This opportunity for optimization is underscored by the fact that the native E. coli pump AcrB appeared as a competition winner for many of the biofuels assayed. It is possible that increases in expression by sequence optimization may improve non-native pumps. Furthermore, for biofuel production from renewable materials, toxic by-products of lignocellulosic biomass pretreatment comprise another factor that affects biofuel yields (Jarboe et al, 2007; Pienkos and Zhang, 2009). Pumps may also provide a potential mechanism for excluding these inhibitors.
As metabolic engineering efforts continue to increase biofuel production titers, it will be crucial to develop strategies for increasing tolerance. This is especially important for production of bulk commodities such as biofuels, where relatively high titers of an inhibitory compound are required, as was found to be the case in industrial production of 1,4-butanediol and 1,3-propanediol in E. coli (Burk, 2010; Zeng and Biebl, 2010). Efflux pumps show great promise as biofuel transporters and are a valuable tool for engineering production strains. Our strategy of bioprospecting for heterologous tolerance mechanisms also provides a widely applicable method for the rapidly advancing field of biofuel research.
Materials and methods
Efflux pump selection
Pumps were sorted based on their homology to the solvent-resistance pump TtgB from P. putida (Rojas et al, 2001) (Supplementary Figure S1). The search space included all efflux pump genes annotated as members of the HAE1 family (TIGR00915) from all sequenced bacterial genomes, as listed in http://microbesonline.org/. To avoid biases due to homology in the transmembrane regions, the homology search was limited to the amino acids in the two large periplasmic loops, which are the regions primarily responsible for substrate recognition (Elkins and Nikaido, 2002; Eda et al, 2003). Periplasmic loops were identified using the TMHMM software; the homology searches were conducted using BLAST. Custom scripts were written to automate the periplasmic loop identification and subsequent homology searches.
From the list of efflux pumps, representatives were selected that span the range of homology, including proteins with both low and high similarity to TtgB (Supplementary Figure S1). For each pump construct, we included all pump genes encoded in their native operons. In some cases, only genes for the inner membrane and periplasmic linker proteins were present on the operon (e.g., NP_745594), and in a few cases there were additional copies of pump components (e.g., YP_274832). A complete list of the efflux pump genes used in this experiment is provided in Supplementary Table S1.
Strain and plasmid construction
E. coli DH1 ΔacrAB was used as the base strain for all experiments. Efflux pump operons were cloned into a vector containing the medium-copy p15A origin of replication, kanamycin resistance marker, lacI repressor and IPTG-inducible lacUV5 promoter (Supplementary Figure S5, Supplementary Methods).
The limonene production pathway was constructed on two plasmids: (i) pTSL-0040, which converts acetyl-CoA to isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP) and (ii) pTSL-0076, which converts IPP and DMAPP to limonene. pTSL-0040 contains seven genes, including the mevalonate pathway genes from Staphylococcus aureus and S. cerevisiae, and enzymes from E. coli. pTSL-0076 contains two genes that are variants of the geranyl diphosphate synthase from Abies grandis and the limonene synthase from Mentha spicata. Further details on strain construction and pathway genes are available in Supplementary Methods.
For production assays, the A. borkumensis (YP_692684) efflux pump genes were subcloned from the p15A vectors used in the library into a vector containing the low-copy origin SC101**, kanamycin resistance marker, araC, and the arabinose-inducible promoter PBAD. Pump genes were amplified by PCR using the forward primer 5′-gaattcaaaagatcttttaagaaggagatatacatatg-3′ and the reverse primer 5′-tatttgatgcctggagatccttactcgagtttggatcc-3′, and the vector was amplified using the forward primer 5′- ggatccaaactcgagtaaggatctccaggcatcaaata-3′ and the reverse primer 5′-catatgtatatctccttcttaaaagatcttttgaattc-3′. Constructs were confirmed by DNA sequencing.
Preparing pooled cultures
Cells containing the efflux pump plasmids were individually adapted to M9 minimal medium (per liter: 200 ml 5 × M9 salts, 2 ml 1 M MgSO4, 50 ml 20% glucose, 20 ml 5% Casamino acids, 100 μl 0.5% Thiamine, 100 μl 1 M CaCl2), as described in the Supplementary Methods. At the conclusion of the adaptation period, the optical densities (absorbance at 600 nm) of the cultures were normalized and cultures were combined in equal proportion. This pooled culture was used to prepare single-use glycerol stocks.
Mathematical modeling
The competitive Lotka–Volterra equation (Strogatz, 1994) was used to model growth of the pooled culture:
 |
where Ni is the cell density of strain i, di is the growth rate of strain i, C is the number of unique strains, and K is the carrying capacity of the entire culture. A is the interaction matrix, where we assume
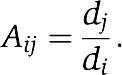 |
Simulation parameters for Figure 1B are dA=0.75, dB=0.7, dC=0.5 and K=1; parameters for Supplementary Figure S2 are described in the figure caption. Individual (non-competitive) simulations set the initial conditions to zero for all but one strain; competitive growth simulations set the initial conditions to be equal for all strains. All simulations were done in Matlab (MathWorks Inc.).
Competitive growth assays
Pooled culture glycerol stocks were used to inoculate M9 minimal medium supplemented with 30 μg/ml kanamycin and 10 μM IPTG. A low induction level was selected to avoid potential toxicity due to pump overexpression (Dunlop et al, 2010). The culture was then divided into tubes to a final volume of 5 ml and biofuel was added directly to the tubes. All conditions were prepared and assayed in triplicate. Cultures were grown at 37°C with orbital shaking at 200 r.p.m. for all experiments.
In all cases, biofuel was added at a level that inhibited growth (v/v: 0.5% n-butanol; 0.25% isopentanol; 2% geranyl acetate; 0.05% geraniol; 2% α-pinene; 0.025% limonene; 2.5% farnesyl hexanoate), see also Supplementary Methods. Owing to the different solubility and toxicity profiles of the different compounds, no single criterion was applied to all fuels. In general, we aimed to select an inhibitory concentration that reduced growth by 25%, while maximizing the difference between the survival of the pool and negative control. Figures 3B and D show toxicity profiles for cells with the best efflux pump and the negative control plasmid for each of the compounds tested.
Every 10–14 h, the cultures were diluted 1:100 into 5 ml of fresh M9 minimal medium supplemented with kanamycin, IPTG and biofuel (where applicable). For the time-course assay, cultures were pelleted by centrifugation at the completion of a dilution cycle and plasmids were isolated. For endpoint competition assays, plasmids were isolated from the culture after 72 h of serial dilution (i.e., six dilution cycles).
Microarray design and methods
Custom microarrays (NimbleGen, Roche, USA) were used to measure the relative quantities of the efflux pump genes present in the plasmid DNA isolated from the competition cultures. Arrays (12 × 135K) that contained probes for all annotated HAE1 genes were designed, allowing for 12 experiments per slide.
Plasmid DNA was labeled and hybridized following the protocol for NimbleGen Comparative Genomic Hybridization Microarrays with minor modifications (Supplementary Methods). Data were normalized using the standard settings for robust multi-array analysis in the NimbleScan software. Probe intensity values were averaged using a custom Matlab script (MathWorks Inc.). Data for all efflux pumps are presented with the exception of two that we were unable to measure conclusively (Supplementary Methods).
For the positive hybridization control, all library plasmids were prepared individually and combined in vitro in equal proportions. For the negative hybridization control, a plasmid with the same vector backbone as those in the library, but no efflux pump, was isolated from DH1 ΔacrAB. Both hybridization controls were labeled and hybridized to the array using the methods described above.
Single-strain biofuel toxicity assays
In all cases, plasmids were transformed into E. coli DH1 ΔacrAB and strains with a pump plasmid were compared with a negative control containing the same plasmid without a pump. Individual strains were adapted to M9 minimal medium as described in the Supplementary Methods. Adapted glycerol stocks were used to inoculate M9 minimal medium supplemented with 30 μg/ml kanamycin and 10 μM IPTG. The biofuel being tested was added directly to the cultures and they were grown overnight at 37°C with orbital shaking. For endpoint measurements, the optical density was measured after 14 h of growth in triplicate 5 ml cultures. For growth curve measurements, cultures were grown in triplicate in 24-well plates with 800 μl per well, and optical density was measured every 10 min with a plate reader (BioTek Synergy 4, USA). All data were normalized to the final cell density of the negative control strain with 0% α-pinene.
Production assays
Freshly transformed strains containing the limonene production pathway on two plasmids and the efflux pump on a third plasmid were adapted to EZ-rich medium with 1% glucose (Teknova). This strain was compared with a negative control, which included the limonene production pathway and the third plasmid without the efflux pump. Overnight cultures were diluted 1:100 in fresh EZ-rich/1% glucose medium, induced with 500 μM IPTG, overlayed with 20% dodecane, and incubated at 30°C for 72 h. At the end of the production period, dodecane layers were sampled for limonene content by gas chromatography/mass spectrometry on a DB-5 column using the internal standard caryophyllene to normalize between samples. Cultures were prepared in triplicate; in one case (negative control) one of the three cultures did not grow and was excluded from the analysis. Basal expression from the PBAD promoter was found to be sufficient to relieve biofuel toxicity (Supplementary Figure S6), thus no arabinose was added to the cultures.
Supplementary Material
Supplementary Figures S1–6, Supplementary Tables S1–2
Acknowledgments
We thank Mario Ouellet for advice on the custom microarray design; Rossana Chan for technical help with the limonene production strain; and Nathan Hillson, Pamela Peralta-Yahya and Joshua Gilmore for helpful discussions. We also thank Antoine Danchin and Jiandong Huang for providing the P. haloplanktis TAC125 strain. John Bates helped to generate the list of HAE1 efflux pump sequences. Hiroshi Nikaido provided thoughtful comments on the manuscript. This work was conducted at the US Department of Energy's Joint BioEnergy Institute, supported through contract DE-AC02-05CH11231.
Author contributions: MJD, ZYD and AM designed the research. MJD and MH developed the high-throughput cloning strategy; MJD and HCC built the library. MJD performed the bioinformatics and mathematical modeling. MJD and HLS conducted the competition and microarray experiments. TSL constructed the limonene production system. ZYD performed the limonene production/pump assays. MJD and AM wrote the manuscript with substantial input from ZYD, MZH and JDK.
Footnotes
JDK has a financial interest in Amyris and LS9.
References
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314: 1565–1568 [DOI] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451: 86–89 [DOI] [PubMed] [Google Scholar]
- Atsumi S, Wu T, Machado IM, Huang WC, Chen PY, Pellegrini M, Liao JC (2010) Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol 6: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk MJ (2010) Sustainable production of industrial chemicals from sugars. Int Sugar J 112: 30–35 [Google Scholar]
- Dunlop MJ, Keasling JD, Mukhopadhyay A (2010) A model for improving microbial biofuel production using a synthetic feedback loop. Sys & Syn Bio 4: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda S, Maseda H, Nakae T (2003) An elegant means of self-protection in Gram-negative bacteria by recognizing and extruding xenobiotics from the periplasmic space. J Biol Chem 278: 2085–2088 [DOI] [PubMed] [Google Scholar]
- Elkins CA, Nikaido H (2002) Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J Bacteriol 184: 6490–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CR, Klein-Marcuschamer D, Stephanopoulos G (2008) Selection and optimization of microbial hosts for biofuels production. Metab Eng 10: 295–304 [DOI] [PubMed] [Google Scholar]
- Harvey BG, Wright ME, Quintana RL (2010) High-density renewable fuels based on the selective dimerization of pinenes. Energy & Fuels 24: 267–273 [Google Scholar]
- Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JL, Porter J, Gray CA, Andersen RJ, Giaever G, Nislow C, Andrews B, Botstein D, Graham TR, Yoshida M, Boone C (2009) A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol 27: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken S, de Bont JAM (1998) Bacteria tolerant to organic solvents. Extremophiles 2: 229–238 [DOI] [PubMed] [Google Scholar]
- Isken S, de Bont JAM (2000) The solvent efflux system of Pseudomonas putida S12 is not involved in antibiotic resistance. Appl Microbiol Biotechnol 54: 711–714 [DOI] [PubMed] [Google Scholar]
- Jarboe LR, Grabar TB, Yomano LP, Shanmugan KT, Ingram LO (2007) Development of ethanologenic bacteria. Adv Biochem Eng Biot 108: 237–261 [DOI] [PubMed] [Google Scholar]
- Kieboom J, Dennis JJ, de Bont JA, Zylstra GJ (1998a) Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem 273: 85–91 [DOI] [PubMed] [Google Scholar]
- Kieboom J, Dennis JJ, Zylstra GJ, de Bont JA (1998b) Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J Bacteriol 180: 6769–6772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Mao WM, Warren MS, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O (2000) Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol 182: 3142–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SP, Nikaido H (2010) Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob Agents Ch 54: 1800–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MD, Warnecke T, Gill RT (2007) SCALEs: multiscale analysis of library enrichment. Nat Methods 4: 87–93 [DOI] [PubMed] [Google Scholar]
- Nikaido H, Takatsuka Y (2009) Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya PP, Keasling JD (2010) Advanced biofuel production in microbes. Biotechnol J 5: 147–162 [DOI] [PubMed] [Google Scholar]
- Pienkos P, Zhang M (2009) Role of pretreatment and conditioning processes on toxicity of lignocellulosic biomass hydrolysates. Cellulose 16: 743–762 [Google Scholar]
- Putman M, van Veen HW, Konings WN (2000) Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev 64: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A (2002) Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol 56: 743–768 [DOI] [PubMed] [Google Scholar]
- Rojas A, Duque E, Mosqueda G, Golden G, Hurtado A, Ramos JL, Segura A (2001) Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J Bacteriol 183: 3967–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura A, Rojas A, Hurtado A, Huertas MJ, Ramos JL (2003) Comparative genomic analysis of solvent extrusion pumps in Pseudomonas strains exhibiting different degrees of solvent tolerance. Extremophiles 7: 371–376 [DOI] [PubMed] [Google Scholar]
- Strogatz SH (1994) Nonlinear Dynamics and Chaos. Cambridge, MA: Westview Press [Google Scholar]
- Tseng T-T, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH Jr (1999) The RND Permease: Superfamily: an Acient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol 1: 107–125 [PubMed] [Google Scholar]
- Zeng A, Biebl H (2010) Bulk chemicals from biotechnology: the case of 1,3-propanediol production and the new trends. Adv Biochem Eng Biotechnol 74: 239–258 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–6, Supplementary Tables S1–2