Mathematical modeling and experimental analyses reveal that TGF-β ligand depletion has an important role in converting short-term graded signaling responses to long-term switch-like responses.
Keywords: mathematical model, Smad, TGF-β, ultrasensitivity
Abstract
Mammalian cells can decode the concentration of extracellular transforming growth factor-β (TGF-β) and transduce this cue into appropriate cell fate decisions. How variable TGF-β ligand doses quantitatively control intracellular signaling dynamics and how continuous ligand doses are translated into discontinuous cellular fate decisions remain poorly understood. Using a combined experimental and mathematical modeling approach, we discovered that cells respond differently to continuous and pulsating TGF-β stimulation. The TGF-β pathway elicits a transient signaling response to a single pulse of TGF-β stimulation, whereas it is capable of integrating repeated pulses of ligand stimulation at short time interval, resulting in sustained phospho-Smad2 and transcriptional responses. Additionally, the TGF-β pathway displays different sensitivities to ligand doses at different time scales. While ligand-induced short-term Smad2 phosphorylation is graded, long-term Smad2 phosphorylation is switch-like to a small change in TGF-β levels. Correspondingly, the short-term Smad7 gene expression is graded, while long-term PAI-1 gene expression is switch-like, as is the long-term growth inhibitory response. Our results suggest that long-term switch-like signaling responses in the TGF-β pathway might be critical for cell fate determination.
Introduction
The transforming growth factor-β (TGF-β) pathway is a prominent signaling pathway that regulates diverse aspects of cellular homeostasis including proliferation, differentiation, migration, and death (Massague, 1998). Remarkably, the pleiotropic biological effects of TGF-β are mediated by a relatively simple signaling module (Clarke and Liu, 2008). The module input is extracellular TGF-β, which binds two receptor molecules, the type I and type II receptors (TβRI and TβRII, respectively), to form a hetero-oligomeric active receptor complex. The TβRII is a constitutively active kinase that transphosphorylates serines and threonines in the TβRI, causing activation of TβRI (Shi and Massague, 2003). Signaling through TβRI is required for most TGF-β responses. The primary intracellular mediators of TGF-β signaling are the Smad proteins, which are classified functionally as the receptor-regulated Smads (R-Smads, Smad2 and Smad3), the common mediator Smad (Co-Smad, Smad4), and the inhibitory Smad (I-Smad, Smad7) (Heldin et al, 1997). TGF-β treatment results in TβRI phosphorylating Smad2 and Smad3 at the two distal serines that are part of the C-terminal SSXS motif (Abdollah et al, 1997; Liu et al, 1997; Souchelnytskyi et al, 1997). Phosphorylation of Smad2 and Smad3 promotes oligomerization of Smad2 and Smad3 and their hetero-oligomerization with Smad4 (Shi and Massague, 2003), resulting in the accumulation of these complexes in the nucleus and subsequent transcriptional regulation of target genes in a cell context-dependent manner (Shi and Massague, 2003). An interesting question is how such an apparently straightforward and simple cascade can generate a wide array of biological responses depending on the cellular context.
One way to achieve diversity in signaling outcomes is by controlling signal duration. A classical example of this can be found in the MAP kinase pathway where transient ERK activation by EGF is associated with cell proliferation while persistent ERK activation by NGF leads to cell differentiation (Marshall, 1995). The differences in the duration of MAP kinase signaling between EGF and NGF likely arise from network topology or variable feedback loops triggered by the respective ligands (Santos et al, 2007). Thus, duration of ERK activation controls the switch between proliferative and anti-proliferative responses.
Duration and amplitude of signaling can be controlled through both intracellular and extracellular mechanisms. The duration of TGF-β signaling appears to be cell type specific, although the exact mechanisms underlying such a variation are still poorly understood. It has been postulated that sustained TGF-β signaling may be required for growth inhibition, while transient signaling may result in resistance to the anti-proliferative effects of TGF-β in certain tumor cells (Nicolas and Hill, 2003). Duration of signaling could be attributed to negative feedback that involves Smad7-mediated degradation of TGF-β receptors (Kavsak et al, 2000) or transcriptional induction of TMEPAI, an inhibitor of Smad2 or Smad3 phosphorylation that limits the duration of TGF-β signaling (Watanabe et al, 2010). Whether these negative feedback mechanisms cause permanent desensitization of TGF-β/Smad signaling remains unknown.
Members of the TGF-β superfamily are frequently used as morphogens in early embryo development (Green, 2002). The best-studied examples include Dpp in Drosophila and Activin in Xenopus (Gurdon and Bourillot, 2001; Lander, 2007). In the developmental context, cells can respond to a graded ligand concentration and produce discrete biological responses (e.g., transcription of certain genes, proliferation or differentiation; Green, 2002). To convert continuous morphogen stimulation into discrete responses, mechanisms must exist to provide a threshold for the cellular response. Positive feedback is one of the best-studied mechanisms to produce switch-like biological processes. A clear example of this is seen in the case of the MAPK activation during oocyte maturation, which generates a bistable mitotic trigger (Ferrell, 2008).
Limiting exposure to ligand could be another mechanism to control signaling duration and switch-like cellular responses. It is known that differential ligand depletion and trafficking can account for the different mitogenic responses elicited by EGF and TGF-α (Reddy et al, 1996). Indeed, our previous work indicates that TGF-β depletion through receptor-mediated internalization has a significant role in determining the duration of signaling in cells exposed to continuous ligand stimulation (Clarke et al, 2009). The amplitude and duration of the phospho-Smad2 signal varied proportionally to the TGF-β dose. While several mathematical models on TGF-β/Smad signaling dynamics have been published (Clarke and Liu, 2008; Kahlem and Newfeld, 2009), none of the models in the literature can adequately account for this experimentally observed feature of TGF-β signaling. Here, we focus on further characterizing how cells transduce variable TGF-β doses into shapes of phospho-Smad, transcriptional and anti-proliferative responses. A comprehensive mathematical model taking into account TGF-β ligand dynamics, receptor trafficking and Smad nucleo-cytoplasmic shuttling dynamics has been developed. By integrating modeling and experimental analyses, we investigated Smad2 activation after TGF-β stimulation at short-term and long-term time scales and show that the early Smad signaling and gene expression responses are gradually dependent on the TGF-β dose, but long-term Smad signaling is ultrasensitive or switch-like. In an ultrasensitive response, a small change of stimulus within a certain range results in a large change in response. The switch-like anti-proliferative response by TGF-β correlates with ultrasensitivity in Smad2 phosphorylation. Thus, the ligand dose is quantitatively sensed and translated into Smad2 phosphorylation and discrete cell proliferative decisions.
Results
Cellular responses to sustained and pulsatile TGF-β stimulation
Cells respond to the absolute number of bioavailable TGF-β molecules in their environment. We developed a bioassay that enables us to count precisely the number of bioactive TGF-β molecules present in the medium (Clarke et al, 2009). Ligand molecules per cell is the input variable to which the cells respond, and ligand number per cell is the best predictor of signaling responses (Zi and Klipp, 2007a; Clarke et al, 2009). Thus, we use ligand molecules per cell as the unit of TGF-β dose for the quantitative analyses.
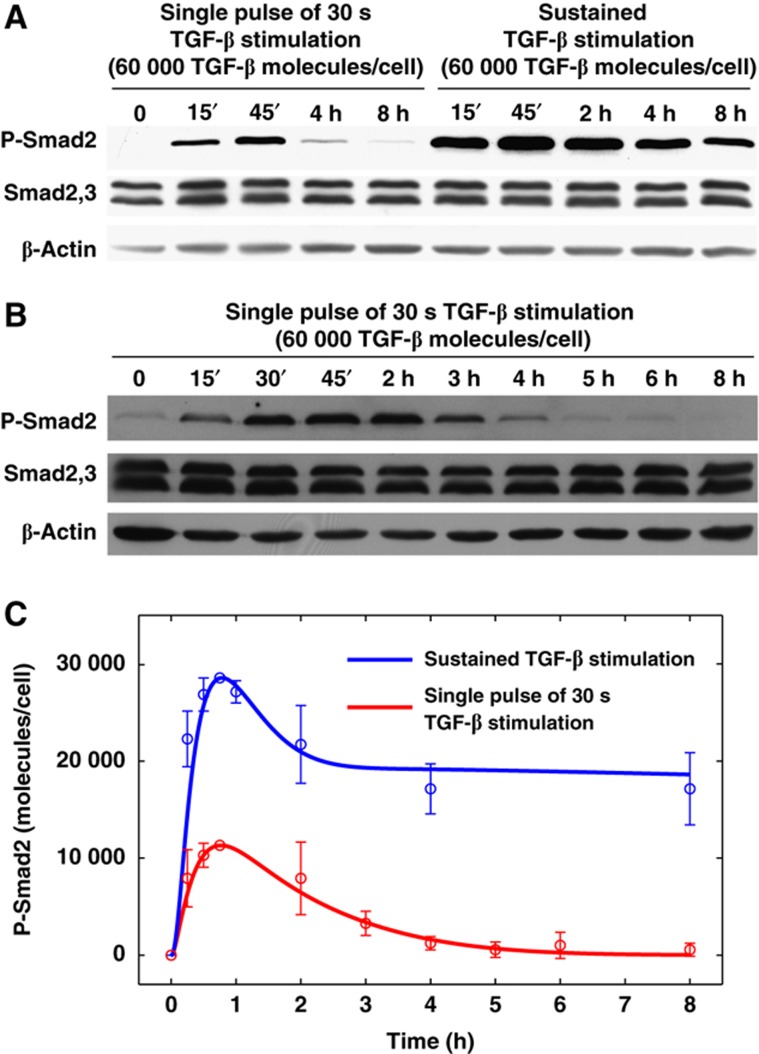
To quantitatively assess TGF-β signaling in response to short-term exposure to ligand, we treated HaCaT cells with 60 000 molecules/cell of TGF-β for 30 s followed by removal of ligand from the medium through washing and measured Smad2 phosphorylation kinetics. As shown in Figure 1, 30 s exposure is sufficient to induce Smad2 phosphorylation yet signaling is transient compared with continuous ligand stimulation, which triggers a more persistent signaling. We used our TGF-β bioassay to quantitatively determine the amount of TGF-β remaining in the medium after three washes. Our data indicate that no >500 molecules/cell are left after three washes when cells were initially treated with 60 000 molecules/cell (Supplementary Figure S1), suggesting that our washing procedure is capable of removing most, if not all, of the TGF-β in the extracellular environment. Thus, removal of ligand prevents sustained receptor activation.
Figure 1.
Experimental and modeling analysis of P-Smad2 response to very short TGF-β stimulation. (A) Western blot analysis of P-Smad2 levels with different TGF-β stimulations. For a single pulse of 30 s of TGF-β stimulation, HaCaT cells were stimulated with TGF-β for 30 s. Then, they were washed with D-PBS and incubated with normal fresh medium without TGF-β. In the sustained TGF-β stimulation, cells were stimulated with TGF-β without washout. (B) Time course western blot analysis of P-Smad2 levels in HaCaT cells with single pulse of 30 s of TGF-β stimulation. (C) Comparison of model simulation with the experimental data. Blue and red curves represent model predictions of P-Smad2 response to sustained TGF-β stimulation and to single pulses of 30 s TGF-β stimulation, respectively. Circles and error bars are shown based on the scaled mean values and standard deviations of the quantified experimental data from 2 to 6 replicates, which were scaled to compare with model simulations in terms of molecules per cell. Source data is available for this figure at www.nature.com/msb.
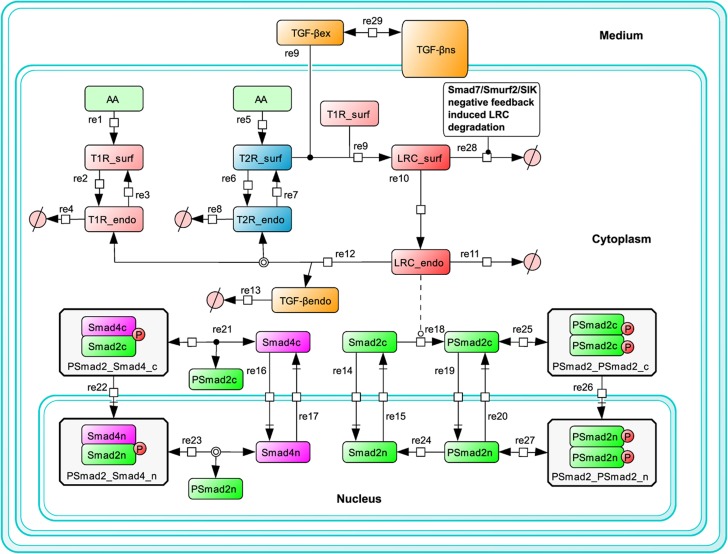
The dynamic behavior of Smad2 phosphorylation in response to alternating ligand exposure cannot be predicted by the existing mathematical models of TGF-β signaling pathway, which have not explicitly taken into account the ligand dynamics in the medium (Clarke et al, 2006; Vilar et al, 2006; Schmierer et al, 2008). Therefore, we formulated a new and more comprehensive mathematical model to investigate dynamic properties of TGF-β signaling. Based on our previous model (Zi and Klipp, 2007b), we simplified the receptor trafficking submodules and integrated the submodules of Smad2 nucleo-cytoplasmic shuttling, phosphorylation and dephosphorylation mainly based on the model of Schmierer et al (2008). Distinct from the models developed by Schmierer et al (2008) and Vilar et al, (2006), this new model has taken into account TGF-β depletion. More importantly, the new model was calibrated and refined with quantitative experimental data sets from different TGF-β stimulation profiles. The detailed model scheme is described in Figure 2.
Figure 2.
Scheme of the mathematical model. A detailed description of mathematical model is given in the Supplementary information and Supplementary Tables S1–S4.
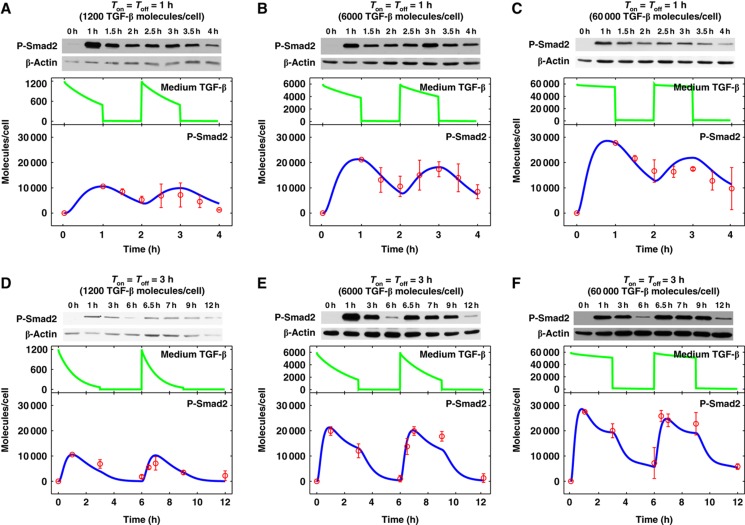
Although signaling decays upon the ligand removal, it is not clear whether there is strong adaptation or desensitization of Smad2 activation under these conditions. To address this question, we performed a sequential pulse stimulation experiment with varying doses and treatment time of TGF-β. HaCaT cells were exposed to the indicated doses of TGF-β for 1 or 3 h (Ton=1 or 3 h) then washed out followed by adding fresh medium without TGF-β for 1 or 3 h (Toff=1 or 3 h). At the end of the ligand-free incubation period, another dose of TGF-β was added and cells were incubated for the same duration as the initial treatment. Smad2 phosphorylation kinetics was determined for each treatment schedule (Figure 3). It is quite evident that cells can periodically respond to pulses of TGF-β stimulation with different doses and different periods. The 3-h on-off regiment produced more dramatic dynamic changes than the 1-h regiment. These results further support the reversibility of short-term TGF-β signaling.
Figure 3.
Dynamics of P-Smad2 response to pulses of TGF-β stimulation. For pulses of TGF-β stimulation, HaCaT cells were stimulated with TGF-β for a specific duration (TGF-β on time, Ton), then TGF-β was washed out and cells were immediately incubated with normal fresh medium for another specific duration (TGF-β off time, Toff). TGF-β was added and washed out at the indicated times. Green and blue curves represent model simulation results for medium TGF-β and P-Smad2 profiles, respectively. Experimental data from two replicates are scaled and plotted in red circles. (A–C) Pulses of TGF-β stimulation with 1 h stimulation and 1 h washout. (D–F) Pulses of TGF-β stimulation with 3 h stimulation and 3 h washout. Source data is available for this figure at www.nature.com/msb.
The availability of time courses of phospho-Smad2 data sets with different TGF-β stimulation profiles have allowed us to refine and improve the new model we have developed. The model fits the nine experimental data sets that were used for parameter estimation very well (Supplementary Figure S2). In addition, the model is validated by its ability to predict the Smad2 phosphorylation dynamics for different TGF-β stimulation profiles (Figure 3A–E).
Periodic short pulse exposure of ligand can produce a sustained TGF-β response
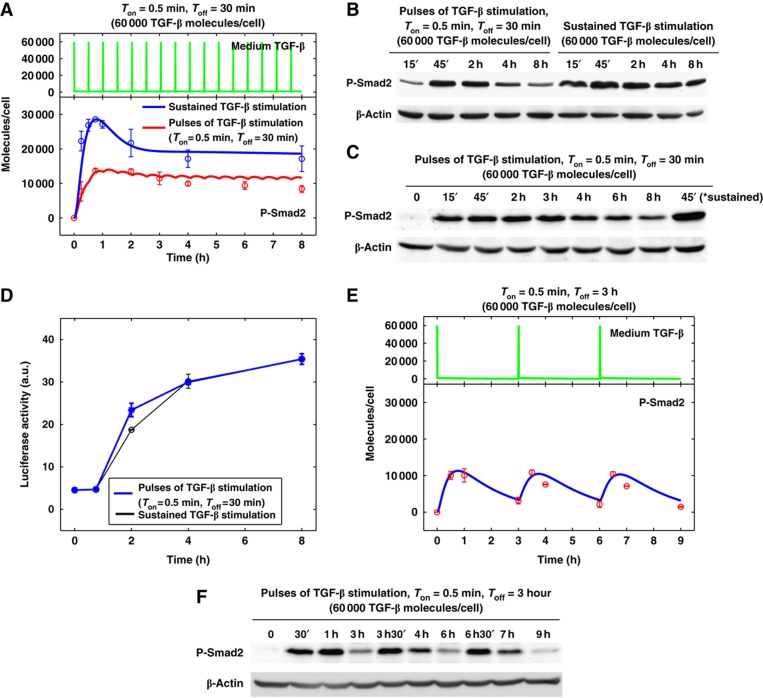
With the data-calibrated mathematical model, we can make some non-intuitive predictions about TGF-β signaling dynamics. One of the predictions is that if we subject HaCaT cells to multiple short pulses (30 s) of TGF-β stimulation, we should be able to observe a sustained TGF-β signaling output in the phosphorylation of Smad2 (Figure 4A). To test this prediction, we treated HaCaT cells with 60 000 molecules/cell of TGF-β for 30 s followed by ligand removal and a 30-min ligand-free incubation before addition of fresh ligand. This cycle of treatment was executed for 16 cycles over a period of 8 h. As a control, a continuous 60 000 molecules/cell TGF-β stimulation was set up in parallel. The model predicted TGF-β profiles in the medium during the treatment and Smad2 phosphorylation kinetics are shown in Figure 4A. The experimental results of the Smad2 phosphorylation are shown in Figure 4B and C. The total exposure time of TGF-β is only 4 min during the entire 8-h time course, yet this repeated short-term exposure to TGF-β resulted in a sustained Smad2 phosphorylation comparable to cells exposed to a full 8 h of TGF-β stimulation.
Figure 4.
Signaling response to continuous short pulses of TGF-β stimulation. (A) Model prediction for P-Smad2 response to sustained TGF-β stimulation (the blue curve) and to pulses of 30 s TGF-β stimulation at 30 min intervals (the red curve). Circles and error bars are shown based on the scaled mean values and standard deviations of the quantified experimental data from six replicates (blue circles) and two replicates (red circles), respectively. The green curve on the top shows the medium TGF-β time course profile with pulses of 30 s TGF-β stimulation at 30 min intervals. (B) Western blot analysis of P-Smad2 response to pulses of 30 s TGF-β stimulation at 30 min intervals and sustained TGF-β stimulation in HaCaT cells. (C) Time course western blot analysis of P-Smad2 response to pulses of 30 s TGF-β stimulation at 30 min intervals in HaCaT cells. The last lane on the right was loaded with the cell lysates from 45 min of sustained TGF-β stimulation. (D) In HaCaT-lux cells, time course CAGA luciferase gene reporter activity with continuous short pulses of TGF-β stimulation is similar to those with sustained TGF-β stimulation. Standard deviations are shown from two experimental replicates. (E) Model prediction for P-Smad2 response to pulses of 30 s TGF-β stimulation at 3 h intervals. Experimental data from two replicates are scaled and plotted in red circles. (F) Time course western blot analysis of P-Smad2 response to pulses of 30 s TGF-β stimulation at 3 h intervals in HaCaT cells. Source data is available for this figure at www.nature.com/msb.
HaCaT-lux cells (a variant of HaCaT cell line) stably express a TGF-β responsive luciferase reporter containing 12 copies of CAGA elements (Inman et al, 2002). To determine if cycles of short-term exposure can trigger a stable transcriptional response, we measured the luciferase activity at 0, 1, 2, 4, and 8 h time points of the TGF-β treatment in HaCaT-lux cells. As shown in Figure 4D, the transcriptional reporter activity from the periodic stimulation matched the continuous treatment, suggesting that periodic ligand treatment can resemble a sustained high-dose ligand stimulation.
We next investigated the underlying mechanism that explains how the TGF-β pathway can integrate the repeated short pulses of ligand stimulation. The model analysis suggested that the integration of repeated short pulses of TGF-β signal is achieved by receptor signal processing. When the TGF-β signal is removed, ligand–receptor complex (LRC) deactivation and disassembly take some time. There is a short memory of LRC activity after TGF-β is removed. When a new short pulse of TGF-β is coming, it will form new LRC by interacting with the receptors that are newly produced or recycled to the cell surface. The gain of new activated LRC compensates the loss of LRC activity when TGF-β is removed (Supplementary Figure S3). To test the hypothesis that LRC activity can be ‘remembered’ for some time after the removal of the TGF-β signal, we made a model analysis of the time course of Smad2 phosphorylation upon treatment with SB431542, a direct inhibitor of type I receptor kinase activity, or ligand washout after 1 h of TGF-β stimulation. The model predicts that phospho-Smad2 will last longer when TGF-β is removed than when SB431542 is applied, a direct inhibitor of type I receptor kinase activity (Supplementary Figure S4A). The performed experimental results are consistent with the model predictions (Supplementary Figure S4B).
In order to study whether the interval between short pulses of TGF-β stimulation (Toff) can affect the maintenance of the sustained P-Smad2 response, we performed model simulations for analyzing the P-Smad2 response to 30 s pulses at 3 h intervals (Ton=30 s, Toff=3 h; Figure 4E). In agreement with the model prediction, the corresponding experimental data indicate that P-Smad2 undergoes periodic changes with response to the addition of 30 s TGF-β stimulation and decreases to a low level over the 3-h TGF-β washout interval (Figure 4F). Cells cannot integrate short pulses of TGF-β stimulation when the duration between pulses is too long. This result is in agreement with our view that the cell is able to tell the difference in duration of ligand withdrawal. With a strong TGF-β stimulation, the pre-bound receptors are capable of sustaining signaling for at least half an hour and bridging signaling provided that the next stimulus is received within this period of time.
Ultrasensitivity in the TGF-β signaling responses
The system response to variable doses of TGF-β has yet to be investigated in depth in mammalian cells. Therefore, we implemented model simulations to determine whether Smad2 phosphorylation is graded or switch-like with respect to TGF-β doses. The model predicts that the short-term Smad2 phosphorylation (after 45 min of TGF-β stimulation) is a graded response, while long-term Smad2 activation (after 24 h of TGF-β stimulation) is a switch-like response (Figure 5A and B). To test the model predictions, Smad2 phosphorylation levels in HaCaT cells treated with variable TGF-β doses both in the short-term (45 min) and long-term (24 h) were measured. As shown in Figure 5A–D, both short- and long-term Smad2 phosphorylation can be saturated but doses of TGF-β that cause maximum response are quite different. Additionally, the shapes of response curves were different. The short-term Smad2 activation was a graded (Michaelis–Menten-like) response with a very low apparent Hill coefficient of about 0.8 (Figure 5A and C) while the long-term Smad2 activation (P-Smad2 at 24 h) yielded a switch-like response with an apparent Hill coefficient of about 4.5 (Figure 5B and D). Thus, the Smad2 response is initially graded and sharpens over time to become ultrasensitive.
Figure 5.
Short-term and long-term signaling responses to different doses of TGF-β stimulation. (A) Mathematical modeling predicts that short-term P-Smad2 response (near maximum at 45 min) is a graded response to different doses of sustained TGF-β stimulation. Experimental data from three replicates are scaled and plotted in red circles and they are fitted to Hill equation curve. The Hill coefficient (nHill) is 0.8 with 95% confidence interval of 0.7–0.9. (B) Mathematical modeling predicts that long-term P-Smad2 response (quasi steady state, at 24 h) is an ultrasensitive switch-like response to different doses of TGF-β stimulation. Experimental data from four replicates are scaled and plotted in red circles and they are fitted with a Hill function. The Hill coefficient (nHill) is 4.5 with 95% confidence interval of 4.0–5.1. (C) Western blot of P-Smad2 response at 45 min to different doses of TGF-β stimulation. (D) Western blot of P-Smad2 response at 24 h to different doses of TGF-β stimulation. The arrows in panels (A–D) indicate the dose of 60 000 TGF-β molecules/cell. (E) Quantitative PCR assay for Smad7 gene expression at 45 min with different doses of sustained TGF-β stimulation. The data are expressed as fold change by normalizing to the Smad7 gene expression level before TGF-β stimulation. The averages and standard deviations from two biological replicates are plotted and fitted with the Hill equation curve. The Hill coefficient (nHill) is 1.3 with 95% confidence interval of 1.1–1.4. (F) Western blot of PAI-1 protein expression at 24 h to different doses of TGF-β stimulation. Experimental data from two replicates are plotted in red circles and they are fitted with a Hill function. The Hill coefficient (nHill) is 5.3 with 95% confidence interval of 4.9–5.6. (G) Western blot of p21 protein expression at 24 h to different doses of TGF-β stimulation. Experimental data from two replicates are plotted in red circles and they are fitted using a Hill function. The Hill coefficient (nHill) is 2.0 with 95% confidence interval of 1.9–2.1. (H) BrdU assay of HaCaT cell proliferation with different doses of sustained TGF-β stimulation. HaCaT cells were seeded for 24 h and then were treated with TGF-β for 24 h. BrdU was added at 18 h after TGF-β stimulation. The data from three experimental replicates are plotted and fitted with a Hill function. The Hill coefficient (nHill) is 4.3 with 95% confidence interval of 4.1–4.5. Source data is available for this figure at www.nature.com/msb.
We next asked whether TGF-β-inducible gene expression responses are graded or switch-like in the short and long term. For the short-term transcriptional response, we measured mRNA levels of Smad7, an early response gene known to be rapidly induced by TGF-β 45 min after exposure to different doses of sustained TGF-β stimulation by quantitative PCR (QPCR). The experimental data show that Smad7 induction exhibits a graded response with corresponding Hill coefficients of about 1.3 (Figure 5E), which is consistent with the graded P-Smad2 response at 45 min (Figure 5A and C). Unlike Smad7, induction of PAI-1 expression by TGF-β is considerably delayed and can be measured at 24 h post TGF-β treatment. As shown in Figure 5F, PAI-1 induction in response to variable doses of TGF-β for 24 h is highly ultrasensitive with an apparent Hill coefficient of ∼5.3. Finally, we also measured TGF-β dose–response induction of p21 whose levels rise to steady state at around 8 h after TGF-β treatment in HaCaT cells (Datto et al, 1995). Compared with Smad7 and PAI-1, p21 induction is only modest ultrasensitive (nHill≈2) (Figure 5G). These results suggest short-term gene induction by TGF-β appears to be graded while long-term targets are more switch-like.
Since Smad2 is an essential signal transducer of the TGF-β signal, the dose–response pattern of Smad phosphorylation prompted us to ask whether the long-term cell decision to growth arrest in response to TGF-β is also switch-like. A Bromodeoxyuridine (BrdU) incorporation assay was used to determine the growth inhibitory response of HaCaT cells to variable doses of TGF-β. As shown in Figure 5H and Supplementary Figure S5, the level of BrdU incorporation is also ultrasensitive with an apparent Hill coefficient of about 4.3. Therefore, the long-term TGF-β growth inhibitory response also shows a switch-like behavior.
TGF-β depletion affects long-term Smad phosphorylation
To analyze which signaling step is responsible for the ultrasensitivity of long-term Smad2 activation, we implemented perturbation experiments of the model by monitoring the Smad2 phosphorylation level at 24 h to varying doses of TGF-β with respect to the changes of the parameter values that are involved in receptor activation, Smad2 activation and the negative feedback on receptor turnover. As shown in Supplementary Figure S6, perturbation on the activation of LRC (ka_LRC) has a significant effect on the sharpness of the long-term phospho-Smad2 dose–response curve. In contrast, the switch-like response of long-term Smad2 phosphorylation is robust and not affected by the perturbations of other signaling steps involved in ligand-induced receptor degradation (klid), Smad2 phosphorylation (kpho_Smad2) and dephosphorylation (kdepho_Smad2), the oligomerization of Smad2, its hetero-oligomerization with Smad4 (kon_Smads, koff_Smads) and the nuclear import rates of Smad complexes (kimp_Smads). Changing these parameter values leads to a shift in the dose–response curve and to a change in the saturated response amplitude (Supplementary Figure S6).
Since the ligand in the medium is depleted mainly through the ligand–receptor interaction (ka_LRC), we speculated that the switch-like response of the long-term Smad2 phosphorylation might arise from the ligand depletion. To test this hypothesis, we analyzed our model with an assumption that TGF-β in the medium remains constant, similar to previous models (Vilar et al, 2006; Schmierer et al, 2008). The model simulations show that ligand depletion is important for the switch-like behavior of the long-term P-Smad2 response to varying doses of TGF-β stimulation (Supplementary Figure S7).
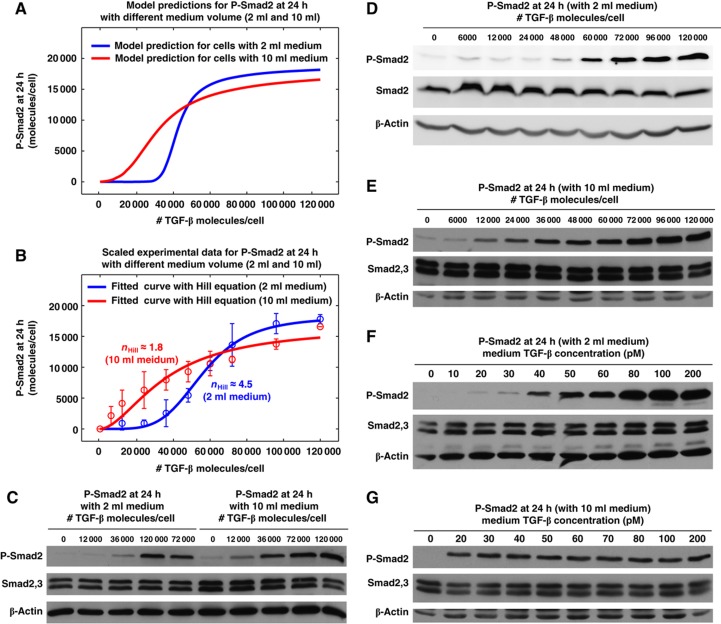
It is experimentally difficult to completely block ligand depletion in a cell culture system. However, further model analyses predict ligand depletion speed should have impact the shape of the dose–response curve. Ligand depletion speed is affected by LRC formation, which is proportional to the concentration of ligand and cell surface receptors. The speed of ligand depletion can be adjusted by varying the volume of the media while keeping the ratio of TGF-β molecule per cell constant. Our model simulation predicts that slowing down ligand depletion by increasing medium volume should decrease the ultrasensitivity of long-term P-Smad2 dose response with 24 h treatment (Figure 6A). To validate this model prediction, we stimulated the cells with the same doses of TGF-β (molecules per cell) in 10 ml medium volume compared with the 2 ml total medium volume used in all previous experiments. The experimental data shown in Figure 6B–E validate the model prediction that the alteration of TGF-β depletion accomplished by increasing the average medium volume per cell affects long-term Smad2 phosphorylation. Furthermore, we measured P-Smad2 responses to the same concentrations of TGF-β in cells growing in 10 ml medium compared with cells growing in 2 ml medium. The P-Smad2 level is very low for 20 pM TGF-β with 2 ml medium (Figure 6E), while the P-Smad2 starts to be saturated for 20 pM TGF-β with 10 ml medium (Figure 6F). This confirms that cells respond to the TGF-β doses in terms of molecules per cell and not in terms of the absolute concentration in medium.
Figure 6.
Perturbation of ligand depletion affects the ultrasensitivity of long-term P-Smad2 responses to different doses of TGF-β stimulation. (A) Model predictions for P-Smad2 responses at 24 h to different doses of sustained TGF-β stimulation with 2 and 10 ml medium. (B) Experimental data for P-Smad2 responses at 24 h to different doses of sustained TGF-β stimulation with 2 and 10 ml medium. The data were quantified from 4 to 6 replicates and were normalized to P-Smad2 signal with TGF-β dose of 120 000 molecules/cell. They are scaled and plotted in red circles in order to compare with the model predictions in (A). The Hill coefficient for P-Smad2 dose response with 10 ml medium is reduced from 4.5 (with 2 ml medium) to 1.8 with 95% confidence interval of 1.6–1.9. (C) Western blot analysis of P-Smad2 responses at 24 h to different doses of sustained TGF-β stimulation in HaCaT cells with 2 and 10 ml medium. Cell lysates were prepared from the same batch of cells that were stimulated with different volume of medium, but with the same doses of TGF-β in terms of molecules per cell. (D) Western blot analysis of P-Smad2 responses at 24 h to different doses of sustained TGF-β stimulation in HaCaT cells and used medium volume of 2 ml. (E) Western blot analysis of P-Smad2 responses at 24 h to different doses of sustained TGF-β stimulation in HaCaT cells and used medium volume of 10 ml. Cells were stimulated with similar doses of TGF-β as those in (D) in terms of molecules per cell. (F) Western blot analysis of P-Smad2 responses at 24 h to different doses of sustained TGF-β stimulation in HaCaT cells and used medium volume of 2 ml. The blots are the same as those shown in Figure 5D, but TGF-β doses were converted from TGF-β molecules per cell to TGF-β concentration in the medium. (G) Western blot analysis of P-Smad2 responses at 24 h to different doses of sustained TGF-β stimulation in HaCaT cells and used medium volume of 10 ml. Cells were stimulated with similar doses of TGF-β as those in (F) in terms of TGF-β concentration in the medium. Source data is available for this figure at www.nature.com/msb.
Discussion
Here, we have shown that the dose and time course of TGF-β stimulation have profound effects on Smad signaling dynamics. The rate of ligand depletion controls the duration of Smad2 phosphorylation. Cells can respond to a short pulse of TGF-β stimulation, and periodic short ligand exposures are sufficient to generate long-term signaling responses. Short-term TGF-β stimulation causes only transient pathway activation and can be terminated by ligand depletion. TGF-β-induced Smad2 phosphorylation is graded in the short-term but ultrasensitive (switch-like) in the long-term (Figure 7). Additionally, cell growth arrest in response to TGF-β shows switch-like rather than graded behavior. Our modeling and experimental analyses suggest that ligand depletion is likely to be involved in sharpening a graded response into a switch-like response (Figure 6).
Figure 7.
Contour landscape of the time-dependent P-Smad2 response profile to different doses of TGF-β stimulation in HaCaT cells. Model simulations indicate that when the TGF-β dose is less than a certain threshold (vertical white solid line), the stimulated cells have almost no signaling response. Within a certain dose range (between vertical white and black dashed line) of TGF-β stimulation, cells have a transient signaling response. Once the TGF-β dose is larger than a threshold (black solid line), cells rapidly switch from a transient short-term signaling response to a sustained long-term signaling response with the increase of TGF-β dose. More importantly, cells have different interpretations of signaling responses to different doses of TGF-β at different time scales. The short-term signaling response establishes a graded response to different doses of TGF-β. In contrast, the long-term signaling response (e.g., P-Smad2 at 24 h) is ultrasensitive or switch-like response. The long-term ultrasensitive signaling response is critical for cellular fate decisions, for example, cell growth arrest.
The TGF-β superfamily of ligands regulates many cellular processes. Most, if not all, of cell fate decisions regulated by TGF-β-related molecules are likely to be switch-like and irreversible (e.g., cell differentiation and apoptosis). A major question in TGF-β biology is how cells convert a continuous ligand concentration into discontinuous cellular fate decisions. Ultrasensitivity appears to be a ubiquitous phenomenon in biology yet the underlying mechanisms that are responsible for generating switch-like responses vary from pathway to pathway. Some of the most common and well-characterized ‘all-or-none’ responses are found in the mitotic trigger and the MAP kinase signaling cascade during Xenopus oocyte maturation (Ferrell, 2008). Positive and double negative feedback loops are critical for the irreversible switch of these processes (Ferrell, 2008). In the TGF-β signaling cascade, there is little evidence indicating the existence of strong positive feedback loops. Consequently, changes in ligand concentration result in faithful changes on the amount of nuclear Smad2 (Shimizu and Gurdon, 1999; Schmierer et al, 2008). In agreement with the notion that there is no signal amplification in the TGF-β signaling cascade, short-term Smad2 phosphorylation in response to changes in TGF-β concentration in the culture medium is graded. However, our modeling and experimental analyses unexpectedly revealed that the quasi steady-state phospho-Smad2 levels for short-term stimulation is quite different from that for long-term stimulation. The long-term ligand sensing is translated into a binary on/off switch, where a well-defined threshold dictates whether or not a full cellular response will be executed.
Several mathematical models have been published for TGF-β/Smad signaling dynamics (for a review, see Clarke and Liu, 2008). However, none of these published models can account for the pulsating stimulation and switch-like behavior of this system. Therefore, we developed a new mathematical model which includes ligand depletion, reversible binding at cell surface (Clarke et al, 2009) and receptor degradation kinetics (Di Guglielmo et al, 2003; Vilar et al, 2006) in addition to Smad nuclear translocation dynamics (Clarke et al, 2006; Zi and Klipp, 2007b; Schmierer et al, 2008) (Figure 2; Supplementary information). The new model produces simulations that closely approximate various aspects of the TGF-β/Smad signaling dynamics (Figures 1, 2, 3; Supplementary Figure S2). For simplicity, we plotted the experimental data and the model predictions from two representative published models in comparison with the new model (Supplementary Figure S8). For a short-term time scale or a high dose of TGF-β, TGF-β in the medium is not significantly depleted. Therefore, previous models that do not include TGF-β depletion are still able to accurately predict the pathway behavior under these conditions. However, previous models fail to predict the time course signal responses for long-term time scale or low doses of TGF-β (Supplementary Figure S8B and C). For long-term signaling response prediction, Vilar et al (2006) model predicts a graded response due to the fact that ligand depletion is not included in this model (Supplementary Figure S8E). Schmierer et al (2008) model generates a ligand-independent P-Smad2 response at 24 h because neither negative feedback nor ligand depletion is taken into account in this model (Supplementary Figure S8F). These models' simulation results are not consistent with the experimental data that shows switch-like long-term P-Smad2 response to varying doses of TGF-β stimulation.
Converting a graded short-term cellular response into a switch-like long-term one during TGF-β signaling may be a key feature associated with the TGF-β superfamily of signaling pathways. Multiple classes of mechanisms have been proposed as possible means to produce a switch-like response (Goldbeter and Koshland, 1982; Koshland et al, 1982; Ferrell and Xiong, 2001). Our modeling and experimental analyses suggest that ligand depletion is an important mechanism for terminating transient signaling and generating a long-term switch-like response. As shown in Supplementary Figure S9, the model analysis indicates that the half-life of TGF-β in the medium is also dose dependent. The low doses of TGF-β stimulations have shorter half-lives and are depleted over time, resulting in very low levels of long-term cellular responses. On the other hand, when the TGF-β dose is above a certain threshold, the half-life of TGF-β in the medium is significantly increased and the TGF-β remaining in the medium after long time periods leads to a saturated response. The filtering of low doses of TGF-β by depletion and the saturation of Smad2 phosphorylation for high doses of TGF-β make cells generate long-term ultrasensitive responses for cell fate decision-making.
Other mechanisms may also be involved in this process. Stoichiometric inhibitors are known to be responsible for generating ultrasensitivity in signaling cascades (Thron, 1994). Smad7 and Ski/SnoN could be excellent candidates as both are low abundance stoichiometric inhibitors of Smad2/3 phosphorylation and are transcriptionally induced upon TGF-β signaling (Nakao et al, 1997; Liu et al, 2001; Luo, 2004). Another possible mechanism is zero-order ultrasensitivity (Goldbeter et al, 1981). A Smad phosphatase, such as PPM1A, may operate near saturation. As more and more phospho-Smad2 accumulates during signaling, saturation of PPM1A can make Smad2 phosphorylation increasingly more effective. All of these mechanisms may not be mutually exclusive. Collectively, these features could account for the way that TGF-β ligand is quantitatively sensed and interpreted. The short-term graded response to TGF-β stimulation may enable the cells to sense a broader range of ligand concentrations and mount a short-term signaling response. The ensuing signaling allows the system to gate out low-dose stimuli and make switch-like decisions only for higher doses of TGF-β stimulation (Figure 7). Our new mathematical model should be applicable to investigate other major physiological responses to TGF-β, such as apoptosis, migration and epithelial-mesenchymal transition. Finally, it will be interesting to determine whether this feature of the pathway design is conserved among all TGF-β family signaling pathways.
Materials and methods
Cell culture and cell lysis
Human keratinocytes (HaCaT) were cultured in Dulbecco's Modified Eagle Medium (GIBCO 12800-082) with 10% fetal bovine serum (SAFC Biosciences, 12303C), 2 mM L-glutamine (GIBCO 35050), 100 units/ml penicillin (ICN Biomedicals, 100548) and 100 μg/ml streptomycin (Sigma, S-6501) at 37°C and 5% CO2. Cells were seeded to a density of 850 000 cells/962 mm2 24 h before treatment with TGF-β (R&D Systems, 240-B-002). Cells were lysed in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% v/v Triton X-100, 0.1% w/v SDS, 1% w/v sodium deoxycholate, 1 mM EDTA, 1 mM PMSF, 25 mM β-glycerophosphate, and 1 mM sodium orthovanadate) and protein concentrations were determined using a BCA assay kit (Thermo Scientific #23225) according to the manufacturer's instructions.
Western blotting and quantification
SDS–PAGE gels were transferred onto nitrocellulose membrane. Phospho-SMAD2 primary antibody (Cell Signaling #3108S) was used at a dilution of 1:500 in 3% BSA-TBS-t (0.05% Tween-20) for 3 h at room temperature. SMAD1/2/3 primary antibody (Santa Cruz sc-7960, Cell Signaling #3122) was used at a dilution of 1:250 in 3% BSA-TBS-t overnight at 4°C. β-Actin primary antibody (Abcam #ab-8226100, Cell Signaling #4967) was used at a dilution of 1:1000 in 3% BSA-TBS-T. Images of western blots were quantified with ImageJ (Abramoff et al, 2004). The P-Smad2 signals were normalized with the loading control β-actin signal or total Smad2,3 signal. The basal P-Smad2 signals for control cells without TGF-β stimulation were subtracted and relative changes of P-Smad2 were used for mathematical modeling. For the western blot analysis of PAI-1 and p21 proteins, the following primary antibodies were used: p21 antibody (Santa Cruz, sc-397), PAI-1 (Santa Cruz sc-8979), and Ezrin antibody (Sigma E8897).
Luciferase assay
Promega Dual Luciferase Kit (#E1910) was used to assay cellular luciferase levels of HaCaT expressing CAGA luciferase reporter.
Cell proliferation (BrdU) assay
HaCaT cells were seeded at density of 28 000 cells/0.32 cm2 24 h before treatment with TGF-β. BrdU (50 μM final concentration) was added 8 h before harvest. Cells were fixed and imaged using a standard BrdU immunostaining protocol provided by the manufacturer (Santa Cruz Biotechnology).
RNA isolation and QPCR analysis
HaCaT cells were treated as indicated in the figures before extraction of RNA using the Arrow instrument (NorDiag ASA) according to the manufacturer's protocol. The mRNA from the samples was converted to cDNA (0.25 μg RNA per sample) using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's protocol and the DNA was amplified in QPCR (Bio-Rad C1000 thermal cycler). The total volume of the QPCRs was 25 μl: 12.5 μl IQ SybrGreen mix, (Bio-Rad), 2 μl cDNA, 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM), and 10 μl dH2O. Cycling parameters were (1) 95.0°C for 2 min, (2) 95.0°C for 10 s, (3) 60.0°C for 30 s, (4) 39 repeats of steps 2 and 3, (5) 95.0°C for 15 s, and (6) melting curve procedure: 65.0–95.0°C, with an increment of 0.5°C per 5 s. The results were analyzed using the Bio-Rad C1000 software. The primer sets used were human GAPDH, forward, 5′-GGAGTCAACGGATTTGGTCGTA-3′, reverse, 5′-GGCAACAATATCCACTTTACCA-3′; human SMAD7, forward, 5′-ACCCGATGGATTTTCTCAAACC-3′, reverse, 5′-GCCAGATAATTCGTTCCCCCT-3′.
Mathematical modeling
Parameter estimation, parameter uncertainty, and identifiability analyses were performed with the parallel parameter estimation tool SBML-PET-MPI (Zi and Klipp, 2006; Zi, 2011). Model simulations were implemented with SBML-SAT (Zi et al, 2008) in Matlab. Details of the mathematical modeling, including model development, assumptions, parameter estimation, parameter statistical analysis, and the system of ordinary differential equations, are described in the Supplementary information. The SBML model is available in the supplementary online SBML Model files. The SBML model is also deposited in BioModels database (Li et al, 2010), which can be retrieved using the following link: http://www.ebi.ac.uk/biomodels-main/MODEL1104050000
Hill coefficient calculation
The apparent Hill coefficients for the model simulation results (n) were calculated by the following equation (Legewie et al, 2005):
 |
Here, S10 and S90 equal to the ligand (TGF-β) levels required to achieve 10 and 90% of the maximum activation, respectively.
The Hill coefficients for the experimental dose–response data (nHill) were estimated by fitting the experimental data with the Hill equations:
 |
or
 |
Supplementary Material
Supplementary Information, Supplementary Tables S1-4, Supplementary Figures S1-9
Acknowledgments
We would like to thank Caroline Hill for providing us some published raw experimental data sets and Pascal Kahlem for facilitating this project. We thank Caroline Hill, Sirio Dupont, and Stefano Piccolo for the HaCaT-12xCAGA reporter cell line. We wish to thank James Ferrell, Tobias Meyer, and members of their laboratories as well as Xuedong Liu's laboratories for discussion. We thank David Clarke and Edward Kee for critical reading of the manuscript. DAC was supported by a predoctoral training grant from NIGMS (T32GM08759). This work was supported by grants from the National Institutes of Health R01GM083172 and R01CA107098 to XL, the FP6 Network of Excellence ENFIN funding (LSHG-CT-2005-518254, EK, AM, and XL), by the Excellence Initiative of the German Federal and State Governments (EXC 294, ZZ), and in part by an EMBO short-term fellowship (ASTF 237-2009, ZZ).
Author contributions: ZZ and XL designed the experiments; ZF, MD, AM, DD, and ZZ conducted the experiments; ZZ performed model analyses; ZZ, ZF, DC, MD, AM, EK, and XL analyzed the data; ZZ and XL wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL (1997) TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem 272: 27678–27685 [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Clarke DC, Betterton MB, Liu X (2006) Systems theory of Smad signalling. Syst Biol (Stevenage) 153: 412–424 [DOI] [PubMed] [Google Scholar]
- Clarke DC, Brown ML, Erickson RA, Shi Y, Liu X (2009) Transforming growth factor beta depletion is the primary determinant of Smad signaling kinetics. Mol Cell Biol 29: 2443–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DC, Liu X (2008) Decoding the quantitative nature of TGF-beta/Smad signaling. Trends Cell Biol 18: 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto MB, Li Y, Panus JF, Howe DJ, Xiong Y, Wang XF (1995) Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA 92: 5545–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL (2003) Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol 5: 410–421 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr (2008) Feedback regulation of opposing enzymes generates robust, all-or-none bistable responses. Curr Biol 18: R244–R245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE, Xiong W (2001) Bistability in cell signaling: how to make continuous processes discontinuous, and reversible processes irreversible. Chaos (Woodbury, NY) 11: 227–236 [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE Jr (1981) An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci USA 78: 6840–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE Jr (1982) Sensitivity amplification in biochemical systems. Q Rev Biophys 15: 555–591 [DOI] [PubMed] [Google Scholar]
- Green J (2002) Morphogen gradients, positional information, and Xenopus: interplay of theory and experiment. Dev Dyn 225: 392–408 [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Bourillot PY (2001) Morphogen gradient interpretation. Nature 413: 797–803 [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P (1997) TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390: 465–471 [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS (2002) Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell 10: 283–294 [DOI] [PubMed] [Google Scholar]
- Kahlem P, Newfeld SJ (2009) Informatics approaches to understanding TGFbeta pathway regulation. Development (Cambridge, England) 136: 3729–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell 6: 1365–1375 [DOI] [PubMed] [Google Scholar]
- Koshland DE Jr, Goldbeter A, Stock JB (1982) Amplification and adaptation in regulatory and sensory systems. Science (New York, NY) 217: 220–225 [DOI] [PubMed] [Google Scholar]
- Lander AD (2007) Morpheus unbound: reimagining the morphogen gradient. Cell 128: 245–256 [DOI] [PubMed] [Google Scholar]
- Legewie S, Bluthgen N, Herzel H (2005) Quantitative analysis of ultrasensitive responses. FEBS J 272: 4071–4079 [DOI] [PubMed] [Google Scholar]
- Li C, Donizelli M, Rodriguez N, Dharuri H, Endler L, Chelliah V, Li L, He E, Henry A, Stefan MI, Snoep JL, Hucka M, Le Novere N, Laibe C (2010) BioModels Database: an enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst Biol 4: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF (1997) Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci USA 94: 10669–10674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun Y, Weinberg RA, Lodish HF (2001) Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev 12: 1–8 [DOI] [PubMed] [Google Scholar]
- Luo K (2004) Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev 14: 65–70 [DOI] [PubMed] [Google Scholar]
- Marshall CJ (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80: 179–185 [DOI] [PubMed] [Google Scholar]
- Massague J (1998) TGF-beta signal transduction. Annu Rev Biochem 67: 753–791 [DOI] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P (1997) Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 389: 631–635 [DOI] [PubMed] [Google Scholar]
- Nicolas FJ, Hill CS (2003) Attenuation of the TGF-beta-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-beta-induced growth arrest. Oncogene 22: 3698–3711 [DOI] [PubMed] [Google Scholar]
- Reddy CC, Wells A, Lauffenburger DA (1996) Receptor-mediated effects on ligand availability influence relative mitogenic potencies of epidermal growth factor and transforming growth factor alpha. J Cell Physiol 166: 512–522 [DOI] [PubMed] [Google Scholar]
- Santos SD, Verveer PJ, Bastiaens PI (2007) Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol 9: 324–330 [DOI] [PubMed] [Google Scholar]
- Schmierer B, Tournier AL, Bates PA, Hill CS (2008) Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci USA 105: 6608–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113: 685–700 [DOI] [PubMed] [Google Scholar]
- Shimizu K, Gurdon JB (1999) A quantitative analysis of signal transduction from activin receptor to nucleus and its relevance to morphogen gradient interpretation. Proc Natl Acad Sci USA 96: 6791–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin CH (1997) Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J Biol Chem 272: 28107–28115 [DOI] [PubMed] [Google Scholar]
- Thron CD (1994) Theoretical dynamics of the cyclin B-MPF system: a possible role for p13suc1. Biosystems 32: 97–109 [DOI] [PubMed] [Google Scholar]
- Vilar JM, Jansen R, Sander C (2006) Signal processing in the TGF-beta superfamily ligand-receptor network. PLoS Comput Biol 2: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Itoh S, Goto T, Ohnishi E, Inamitsu M, Itoh F, Satoh K, Wiercinska E, Yang W, Shi L, Tanaka A, Nakano N, Mommaas AM, Shibuya H, Ten Dijke P, Kato M (2010) TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol Cell 37: 123–134 [DOI] [PubMed] [Google Scholar]
- Zi Z (2011) SBML-PET-MPI: a parallel parameter estimation tool for Systems Biology Markup Language based models. Bioinformatics (Oxford, England) 27: 1028–1029 [DOI] [PubMed] [Google Scholar]
- Zi Z, Klipp E (2006) SBML-PET: a Systems Biology Markup Language-based parameter estimation tool. Bioinformatics (Oxford, England) 22: 2704–2705 [DOI] [PubMed] [Google Scholar]
- Zi Z, Klipp E (2007a) Cellular signaling is potentially regulated by cell density in receptor trafficking networks. FEBS Lett 581: 4589–4595 [DOI] [PubMed] [Google Scholar]
- Zi Z, Klipp E (2007b) Constraint-based modeling and kinetic analysis of the Smad dependent TGF-beta signaling pathway. PloS one 2: e936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi Z, Zheng Y, Rundell AE, Klipp E (2008) SBML-SAT: a systems biology markup language (SBML) based sensitivity analysis tool. BMC Bioinformatics 9: 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information, Supplementary Tables S1-4, Supplementary Figures S1-9