SUMMARY
The pyruvate kinase isoforms PKM1 and PKM2 are alternatively spliced products of the PKM2 gene. PKM2, but not PKM1, alters glucose metabolism in cancer cells and contributes to tumorigenesis by mechanisms that are not explained by its known biochemical activity. We show that PKM2 gene transcription is activated by hypoxia-inducible factor 1 (HIF-1). PKM2 interacts directly with the HIF-1α subunit and promotes transactivation of HIF-1 target genes by enhancing HIF-1 binding and p300 recruitment to hypoxia response elements, whereas PKM1 fails to regulate HIF-1 activity. Interaction of PKM2 with prolyl hydroxylase 3 (PHD3) enhances PKM2 binding to HIF-1α and PKM2 coactivator function. Mass spectrometry and anti-hydroxyproline antibody assays demonstrate PKM2 hydroxylation on proline-403/408. PHD3 knockdown inhibits PKM2 coactivator function, reduces glucose uptake and lactate production, and increases O2 consumption in cancer cells. Thus, PKM2 participates in a positive feedback loop that promotes HIF-1 transactivation and reprograms glucose metabolism in cancer cells.
INTRODUCTION
The glycolytic pathway involves conversion of glucose to lactate and the generation of ATP. Pyruvate kinase (PK), which catalyzes the reaction of phosphoenolpyruvate (PEP) + ADP → pyruvate + ATP, is a key enzyme that determines glycolytic activity. PKM1 and PKM2 are alternatively spliced products of the primary RNA transcript that contain sequences encoded by exon 9 or exon 10, respectively, of the PKM2 gene (Noguchi et al., 1986). Heterogeneous nuclear ribonucleoproteins (hnRNP) I, A1, and A2 bind to RNA sequences encoded by exon 9 and inhibit PKM1 mRNA splicing (David et al., 2010). The oncoprotein c-Myc activates transcription of hnRNPI, hnRNPA1, and hnRNPA2, resulting in preferential PKM2 isoform expression (David et al., 2010).
Many cancer cells have increased glycolysis and lactate production and decreased O2 consumption compared to non-transformed cells, a phenomenon known as the Warburg effect (Gatenby and Gillies, 2004). PKM2 promotes the Warburg effect and tumorigenesis (Christofk et al., 2008; Hitosugi et al., 2009). Despite intensive studies, the mechanism by which PKM2 facilitates lactate production and blocks mitochondrial oxidative phosphorylation in cancer cells has remained a mystery.
Activation of hypoxia-inducible factor 1 (HIF-1), which commonly occurs in human cancers either as a result of hypoxia or genetic alterations (Harris, 2002; Semenza, 2010), leads to a switch from oxidative to glycolytic metabolism (Seagroves et al., 2001; Wheaton and Chandel, 2011). HIF-1 is a transcription factor that consists of an O2-regulated HIF-1α subunit and a constitutively expressed HIF-1β subunit (Wang et al., 1995). In well-oxygenated cells, HIF-1α is hydroxylated at proline (Pro) 402 and 564 (Kaelin and Ratcliffe, 2008). Three prolyl hydroxylases, PHD1-3, which require O2, Fe2+, 2-oxoglutarate, and ascorbate for their catalytic activity, have been shown to hydroxylate HIF-1α when overexpressed (Epstein et al., 2001). PHD2 is primarily responsible for regulating basal HIF-1α levels in cancer cells (Berra et al., 2003). Prolyl hydroxylated HIF-1α is bound by the von Hippel-Lindau (VHL) tumor suppressor protein, which recruits the Elongin C-Elongin B-Cullin 2-E3-ubiquitin-ligase complex, leading to proteasomal degradation of HIF-1α. Under hypoxic conditions, HIF-1α prolyl hydroxylation is inhibited, thereby stabilizing HIF-1α protein (Kaelin and Ratcliffe, 2008). In the nucleus, HIF-1α dimerizes with HIF-1β and binds to the consensus nucleotide sequence 5′-RCGTG-3′, which is present within the hypoxia response element (HRE) of target genes (Semenza et al., 1996). Hydroxylation of HIF-1α at asparagine-803, which is catalyzed by the asparaginyl hydroxylase FIH-1 in normoxic cells, blocks the binding of the transcriptional coactivator p300 to HIF-1α (Lando et al., 2002). Under hypoxic conditions, p300 catalyzes the acetylation of lysine residues on the N-terminal tail of core histones at HIF-1 target genes, leading to changes in chromatin structure that promote HIF-1-dependent gene transcription (Arany et al., 1996).
HIF-1 activates transcription of genes encoding proteins that are involved in key aspects of cancer biology, including angiogenesis, metabolism, cell survival, invasion, and metastasis (Harris, 2002; Melillo, 2007; Semenza, 2010). HIF-1 target genes include those encoding: the glucose transporter GLUT1, which increases glucose uptake; lactate dehydrogenase A (LDHA), which converts pyruvate to lactate; and pyruvate dehydrogenase kinase 1 (PDK1), which inactivates pyruvate dehydrogenase, thereby shunting pyruvate away from the mitochondria and inhibiting O2 consumption (Wheaton and Chandel, 2011).
In the present study, we demonstrate that PKM2 functions as a coactivator that stimulates HIF-1 transactivation of target genes encoding GLUT1, LDHA, and PDK1 in cancer cells. PHD3 binds to PKM2 and stimulates its function as a HIF-1 coactivator. The effect of PHD3 on PKM2 is dependent upon its hydroxylase activity and the presence of two Pro residues in PKM2. PHD3 knockdown reduces glucose uptake and lactate production and increases O2 consumption in VHL-null renal cancer cells. HIF-1 activates transcription of the genes encoding PKM2 and PHD3, which provides a feedforward mechanism that amplifies HIF-1-dependent metabolic reprogramming, thus providing a molecular basis for the observed effects of PKM2 on tumor metabolism.
RESULTS
PKM2 is a HIF-1 Target Gene
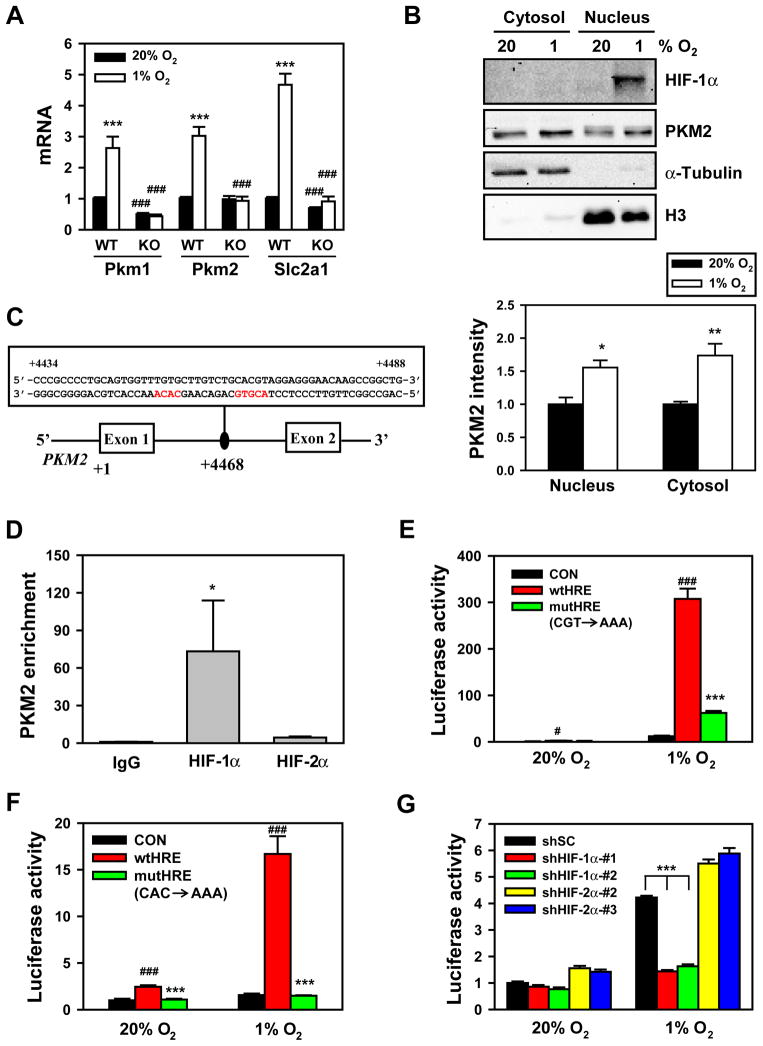
Previous studies demonstrated that hypoxia induces PKM mRNA expression (Semenza et al., 1994). To determine whether mRNA encoding PKM1 or PKM2 is regulated by HIF-1, wild-type (WT) mouse embryonic fibroblasts (MEFs) and HIF-1α-knockout (KO) MEFs were exposed to 20% or 1% O2 for 24 h. Quantitative real-time RT-PCR (qRT-PCR) assays with primers specific for mouse Pkm1 or Pkm2 mRNA demonstrated that both Pkm1 and Pkm2 mRNA levels were significantly increased in response to hypoxia in WT, but not in KO, MEFs (Figure 1A). Expression of the known direct HIF-1 target gene Slc2a1 encoding Glut1 was similarly induced by hypoxia in WT, but not in KO, MEFs (Figure 1A). PKM2 protein levels were increased by 1.6- and 1.7-fold in nuclear and cytosolic extracts, respectively, of hypoxic HeLa cells (Figure 1B). Taken together, these data indicate that Pkm1 and Pkm2 mRNA are both induced in response to hypoxia and regulated by HIF-1.
Figure 1. HIF-1 Regulates Hypoxia-Induced Expression of PKM1 and PKM2.
(A) qRT-PCR analysis of Pkm1, Pkm2 and Slc2a1 mRNA in wild-type (WT) and HIF-1α knockout (KO) mouse embryo fibroblasts (MEFs) exposed to 20% or 1% O2 for 24 h (mean ± SEM, n = 3–5). ***p<0.001 vs 20% O2. ###p<0.001 vs WT MEFs.
(B) Immunoblot assays of HIF-1α, PKM2, histone H3, and α-tubulin protein in nuclear and cytosolic lysates prepared from HeLa cells exposed to 20% or 1% O2 for 24 h. The intensity of PKM2 was quantified by densitometry and normalized to 20% O2 (mean ± SEM, n = 3–4). *p<0.05, **p<0.01 vs 20% O2.
(C) Nucleotide sequence of the hypoxia-response element (HIF-1 binding site 5′-ACGTG-3′ and 5′-CACA-3′ site shown in red) within intron 1 of the human PKM2 gene. The transcription initiation site is designated +1. Exons and intron are not drawn to scale.
(D) Chromatin was precipitated from DMOG-treated HeLa cells (4 h) with antibodies against HIF-1α, HIF-2α, or IgG, and analyzed by qPCR (mean ± SEM, n = 4–5). *p<0.05 vs IgG.
(E and F) HeLa cells were co-transfected with pGL2p control (CON), pGL2p-WT PKM2 HRE (wtHRE), or pGL2p-mutant PKM2 HRE (mutHRE) with CGT→AAA (E) or CAC→AAA (F), and pSV-Renilla, and exposed to 20% or 1% O2 for 24 h. The ratio of FLuc:RLuc activity was normalized to CON at 20% O2 (mean ± SEM, n = 4). #p<0.05, ###p<0.001 vs CON; ***p<0.001 vs wtHRE.
(G) HeLa cells were transfected with: vector encoding a short hairpin (sh) RNA targeting HIF-1α, HIF-2α, or a scrambled control (SC); pGL2p-wtHRE; and pSV-Renilla, and exposed to 20% or 1% O2 for 24 h. The ratio of FLuc:RLuc activity was normalized to shSC at 20% O2 (mean ± SEM, n = 4). ***p<0.001 vs shSC.
See also Figure S1.
Analysis of the human PKM2 gene sequence revealed a candidate HRE within the first intron containing (on the antisense strand) the HIF-1 binding site 5′-ACGTG-3′ followed by a 5′-CACA-3′ sequence (Figure 1C), which is found in many HREs (Fukuda et al., 2007). To determine whether HIF-1 binds at this site, chromatin (Ch) immunoprecipitation (IP) assays were performed in HeLa cells treated with the PHD inhibitor dimethyloxalylglycine (DMOG) to stabilize HIF-1α protein. ChIP with HIF-1α antibody enriched the putative HRE sequence >70-fold compared to IP with IgG (Figure 1D). In contrast, PKM2 occupancy by HIF-2α was not above background levels (Figure 1D). These data indicate that HIF-1α, but not HIF-2α, binds directly to the PKM2 gene.
To test whether this putative HRE in the PKM2 gene is functional, a 55-bp fragment encompassing the HRE (Figure 1C) was inserted downstream of firefly luciferase (FLuc) coding sequences controlled by a basal SV40 promoter in the reporter plasmid pGL2-promoter. HeLa cells were transfected with PKM2 HRE reporter and a control reporter, pSV-Renilla (in which Renilla luciferase [RLuc] expression is driven by the SV40 promoter alone), and exposed to 20% or 1% O2 for 24 h. The PKM2 HRE significantly increased Fluc activity in hypoxic HeLa cells (Figure 1E). Mutation of 5′-CGT-3′ to 5′-AAA-3′ at the core binding site, which eliminates HIF-1 binding (Semenza et al., 1996), significantly decreased hypoxia-induced FLuc activity (Figure 1E). Mutation of 5′-CAC-3′ to 5′-AAA-3′ also abolished PKM2 HRE-driven FLuc activity (Figure 1F). Transfection of either of two different HIF-1α short hairpin RNAs (shRNAs) or either of two different HIF-2α shRNAs knocked down HIF-1α or HIF-2α protein levels, respectively (Figure S1), but only HIF-1α shRNAs significantly reduced PKM2 HRE-dependent FLuc activity in hypoxic HeLa cells (Figure 1G). Taken together, these data indicate that HIF-1 binds to an HRE within the first intron of PKM2 and directly activates its transcription.
PKM2 Interacts with HIF-1α
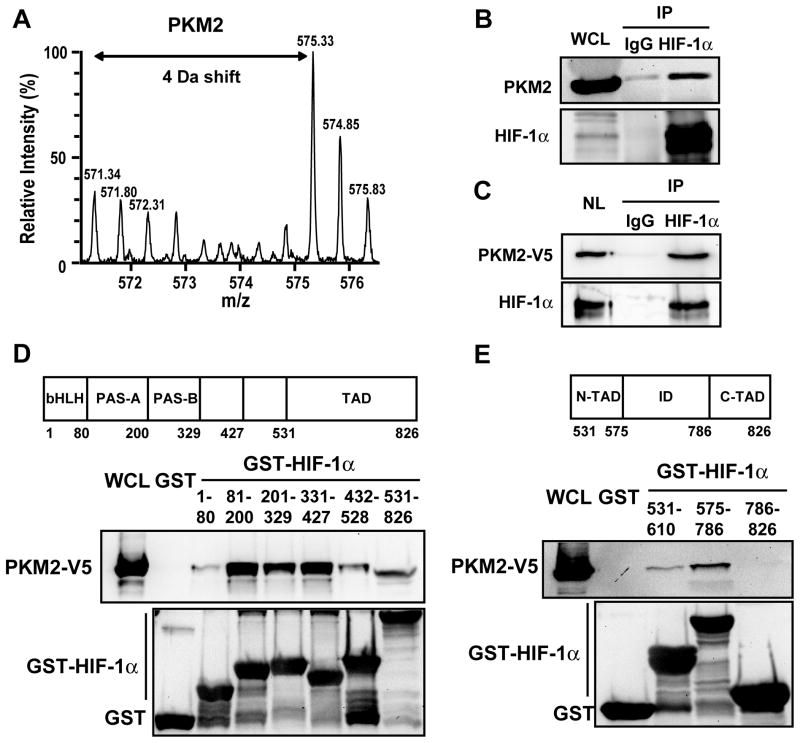
A large body of data indicates that HIF-1 and PKM2 contribute to the Warburg effect and tumor growth (Christofk et al., 2008; Gatenby and Gillies, 2004; Melillo, 2007; Semenza, 2010). The presence of PKM2 in the nucleus (Figure 1B) suggested the intriguing possibility that PKM2 may enhance glucose metabolism by regulating HIF-1. To test this hypothesis, we investigated whether HIF-1α interacts with PKM2. A stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomic screen (Luo et al., 2010) revealed that PKM2 preferentially bound to a GST-HIF-1α (531–826) fusion protein containing amino acid residues 531–826 of HIF-1α as compared to GST alone (Figure 2A), suggesting that PKM2 is a novel HIF-1α-interacting protein. To confirm this proteomic finding, co-IP assays were performed in HeLa cells exposed to 1% O2 for 24 h. Endogenous PKM2 was specifically precipitated by anti-HIF-1α antibody, but not by control IgG (Figure 2B). To determine whether PKM2 specifically interacts with HIF-1α in the nucleus, HeLa cells were transfected with an expression vector encoding V5 epitope-tagged PKM2, exposed to 1% O2 for 24 h to induce HIF-1α expression, and nuclear extracts were prepared. PKM2-V5 was efficiently co-immunoprecipitated with HIF-1α (Figure 2C). These data demonstrate that PKM2 physically interacts with HIF-1α in the nuclei of hypoxic human cancer cells.
Figure 2. PKM2 Interacts with HIF-1α.
(A) GST pull-down assays were performed with GST or GST-HIF-1α (531–826) and lysate prepared from HEK293 cells grown in media containing light isotopes (12C6-14N2-Lys and 12C6-14N4-Arg) or heavy isotopes (13C6-15N2-Lys and 13C6-15N4-Arg), respectively. The precipitated proteins were analyzed by Q-TOF mass spectrometry (Luo et al., 2010). The mass spectrum of a tryptic PKM2 peptide is shown. Monoisotopic peaks derived from PKM2 bound to GST-HIF-1α (531–826) are 4 Da greater in their mass/charge ratio (m/z) and have higher relative intensity compared to the corresponding peaks due to non-specific binding of PKM2 to GST.
(B) Co-IP was performed in HeLa cells exposed to 1% O2 for 24 h.
(C) Co-IP was performed using nuclear lysates (NL) prepared from HeLa cells transfected with PKM2-V5 vector and exposed to 1% O2 for 24 h.
(D and E) GST pull-down assays were performed with GST or GST fusion protein containing the indicated amino acid residues of HIF-1α (upper panels) and whole cell lysates (WCLs) from HeLa cells expressing PKM2-V5. bHLH, basic helix-loop-helix; PAS, Per-ARNT-Sim; TAD, transactivation domain; ID, inhibitory domain.
To localize sites of interaction with PKM2, we utilized a panel of GST-HIF-1α fusion proteins. In vitro GST pull-down assays revealed that PKM2-V5 strongly bound to GST-HIF-1α (81–200), (201–329), and (331–427), and also associated weakly with GST-HIF-1α (1–80), (432–528), and (531–826) (Figure 2D). The lack of interaction of PKM2-V5 with GST confirmed the specificity of binding (Figure 2D). These data indicate that PKM2 binds to multiple domains of HIF-1α in vitro. We further mapped PKM2 binding within the transactivation domain of HIF-1α and found that PKM2-V5 mainly bound to GST-HIF-1α (575–786) (Figure 2E), which encodes an inhibitory domain associated with regulation of the HIF-1α transactivation domain (Jiang et al., 1997).
PKM2 Regulates HIF-1 Transcriptional Activity
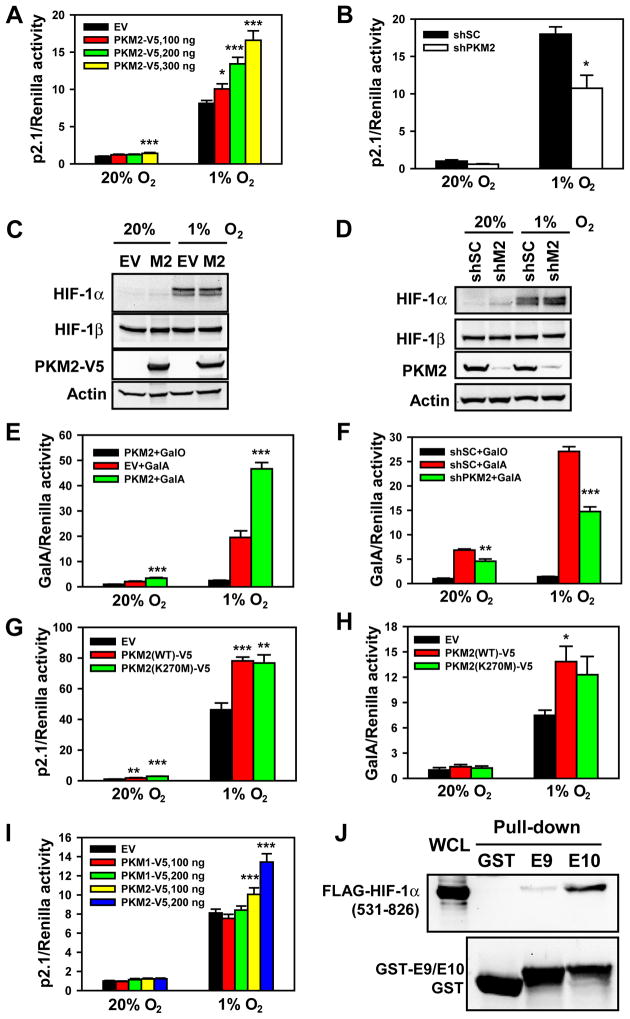
To test whether PKM2 regulates HIF-1 transcriptional activity, HeLa cells were cotransfected with: HIF-1 reporter plasmid p2.1, which contains an HRE from the human ENO1 gene upstream of SV40 promoter and FLuc coding sequences (Semenza et al., 1996); pSV-Renilla; and empty vector (EV) or PKM2-V5 expression vector. Transfected cells were exposed to 20% or 1% O2 for 24 h. PKM2-V5 significantly increased HIF-1 transcriptional activity in a concentration-dependent manner (Figure 3A). Conversely, knockdown of endogenous PKM2 expression by shRNA (shPKM2) significantly reduced HIF-1 transcriptional activity in hypoxic HeLa cells (Figure 3B). Similar results were observed in Hep3B hepatoblastoma cells (Figures S2A and S2B). These data indicate that PKM2 promotes HIF-1 transcriptional activity in human cancer cells.
Figure 3. PKM2 Promotes HIF-1 Transactivation.
(A, B, G and I) HeLa cells were transfected with p2.1, pSV-Renilla, and empty vector (EV) or the indicated expression vector, and exposed to 20% or 1% O2 for 24 h. The ratio of FLuc:RLuc activity was normalized to EV (A, G and I) or shSC (B) at 20% O2 (mean ± SEM, n = 4). *p<0.05, **p<0.01, ***p<0.001 vs EV or shSC.
(C) Immunoblot assays of HIF-1α, HIF-1β, PKM2-V5, and actin in HeLa cells transfected with EV or PKM2-V5 (M2) expression vector and exposed to 20% or 1% O2 for 4 h.
(D) HeLa cells were transduced with a retrovirus encoding shSC or two retroviruses encoding different shRNAs targeting PKM2 (shM2), exposed to 20% or 1% O2 for 4 h, and WCLs were subjected to immunoblot assays.
(E, F and H) HeLa cells were transfected with GalA or GalO, pG5E1bLuc, and pSV-Renilla, and the indicated expression vector, and exposed to 20% or 1% O2 for 24 h. The ratio of FLuc:RLuc activity was normalized to GalO (E and F) or EV (H) at 20% O2 (mean ± SEM, n = 4). *p<0.05, **p<0.01, ***p<0.001 vs EV or shSC.
(J) GST pull-down assays were performed with GST, GST-E9, or GST-E10 and WCLs from HeLa cells expressing FLAG-HIF-1α (531–826).
See also Figure S2.
To determine whether PKM2 influences HIF-1 transcriptional activity by increasing HIF-1α protein levels, HeLa cells were transfected with EV or PKM2-V5 expression vector and exposed to 20% or 1% O2 for 4 h. PKM2-V5 expression did not affect HIF-1α or HIF-1β protein levels (Figure 3C). PKM2 knockdown also failed to alter protein levels of HIF-1α or HIF-1β in HeLa cells (Figure 3D). These data rule out an effect on HIF-1α protein stability as the cause of PKM2-stimulated HIF-1 transcriptional activity.
To directly investigate the effect of PKM2 on HIF-1α transactivation domain function, we employed a reporter assay utilizing expression vector GalA, which encodes the GAL4 DNA-binding domain fused to the HIF-1α transactivation domain (531–826), and reporter plasmid pG5E1bLuc, which contains five GAL4-binding sites and a TATA box upstream of FLuc coding sequences (Jiang et al., 1997). HeLa cells were cotransfected with GalA or GalO (encoding GAL4 DNA-binding domain alone); pG5E1bLuc; pSV-Renilla; and EV or PKM2-V5 expression vector. PKM2-V5 significantly increased GalA transcriptional activity in HeLa cells exposed to 20% or 1% O2 for 24 h (Figure 3E). GalA protein levels were not affected by PKM2-V5 (data not shown). PKM2-V5 did not induce FLuc activity when cotransfected with GalO (Figure 3E), indicating that PKM2 specifically stimulates HIF-1α transactivation domain function. Knockdown of endogenous PKM2 significantly decreased GalA activity in HeLa cells (Figure 3F). PKM2 also regulated GalA activity in Hep3B cells (Figures S2C and S2D). Taken together, these data indicate that PKM2 directly stimulates HIF-1α transactivation domain function.
To determine whether PKM2 enzymatic activity is required to stimulate HIF-1α transactivation, we expressed catalytically inactive PKM2(K270M), which is deficient in phosphoryl transfer from PEP to ADP (Dombrauckas et al., 2005). p2.1 reporter assays demonstrated that PKM2(K270M) increased HIF-1 transcriptional activity in HeLa cells, similar to WT PKM2 (Figure 3G). GalA activity was also stimulated by catalytically inactive PKM2(K270M) (Figure 3H). We conclude that stimulation of HIF-1α transactivation by PKM2 is independent of its catalytic activity as a glycolytic enzyme.
We asked whether PKM2 can also regulate HIF-2 transcriptional activity. PKM2-V5 expression significantly enhanced p2.1 transcriptional activity in HeLa cells cotransfected with HIF-2α expression vector (Figure S2E). Co-IP assays using anti-HIF-2α antibody demonstrated that HIF-2α interacted with PKM2-V5 in hypoxic cells (Figure S2F). Thus, PKM2 also promotes HIF-2 transcriptional activity.
PKM1 is an alternative splice product of the PKM2 gene that does not mediate the Warburg effect (Christofk et al., 2008). We tested whether PKM1 also regulates HIF-1 transcriptional activity. PKM1-V5 protein levels were comparable to PKM2-V5 protein levels in HeLa cells (Figure S2G). However, PKM1-V5 failed to activate HIF-1-dependent p2.1 transcriptional activity (Figure 3I) or GalA-dependent transactivation (Figure S2H) in HeLa cells. In vitro binding assays revealed that FLAG-HIF-1α(531–826) strongly bound to a GST-E10 fusion protein, which contains the PKM2-specific amino acid residues encoded by PKM2 exon 10, but did not bind to GST and bound very weakly to GST-E9 fusion protein, which contains the residues encoded by PKM1-specific exon 9 (Figure 3J). Taken together, these data indicate that PKM2, but not PKM1, stimulates HIF-1 transcriptional activity, and that physical association between the HIF-1α transactivation domain and the alternatively spliced domain of PKM2 is critical for stimulation of HIF-1α transactivation.
PHD3 Potentiates the Effect of PKM2 on HIF-1α Transactivation
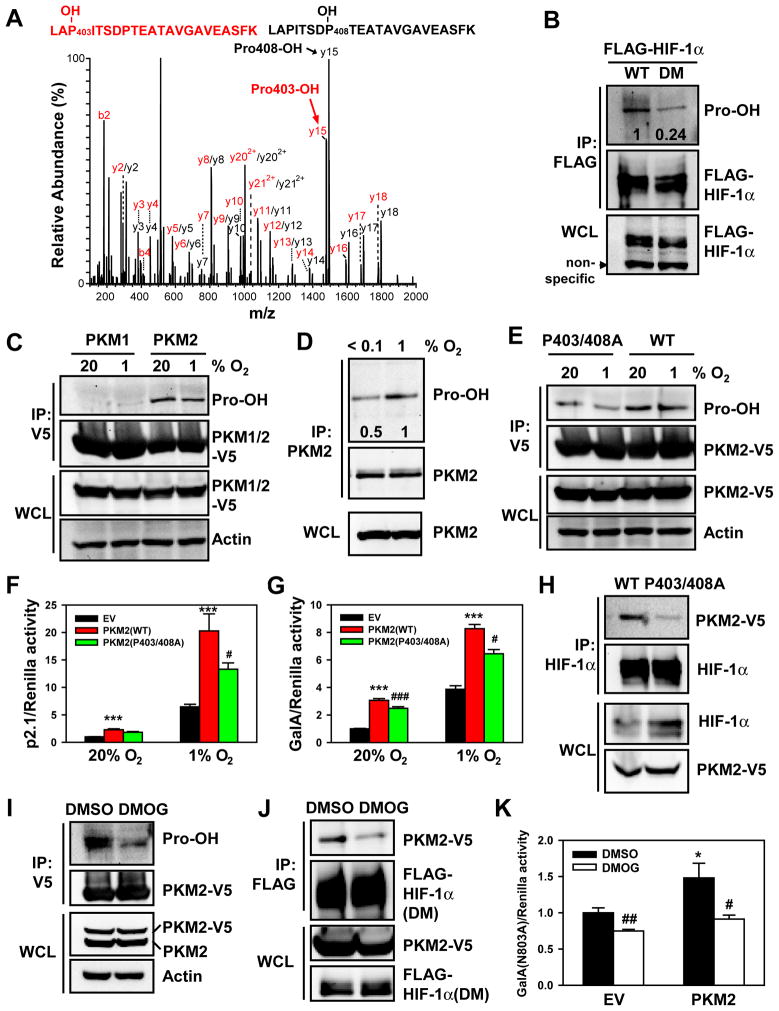
Analysis of the amino acid sequence of the alternatively spliced domain of PKM2 (Figure S3A) revealed a match with the prolyl hydroxylation motif LXXLAP403, which is present in all vertebrate HIF-1α and HIF-2α isoforms (Epstein et al., 2001). This prolyl hydroxylation motif is not present in PKM1 (Figure S3A). We hypothesized that hydroxylation of PKM2 at Pro-403 may regulate HIF-1α binding and stimulation of transactivation. To test this hypothesis, we first investigated prolyl hydroxylation of PKM2 by LTQ Orbitrap Velos mass spectrometry. GST-E10 fusion protein was purified from bacteria and incubated for 1 h with cell lysate that was supplemented with PHD cofactors (FeCl2, 2-oxoglutarate, and ascorbate). Mass spectrometry analysis revealed hydroxylation of Pro-403 as well as Pro-408 (Figure 4A). Prolyl hydroxylation of PKM2 was further verified using an antibody raised against hydroxyproline (Pro-OH). The anti-Pro-OH antibody preferentially bound to WT FLAG-HIF-1α as compared to double mutant (DM) FLAG-HIF-1α (P402/564A), in which both hydroxylatable Pro residues have been changed to alanine (Figure 4B). These results validated the selectivity of the anti-Pro-OH antibody. The residual antibody binding to FLAG-HIF-1α(DM) may represent low-affinity binding to non-hydroxylated Pro residues or spontaneous (non-catalytic) oxidation of other Pro residues in HIF-1α(DM). Analysis of immunoprecipitated PKM2-V5 protein revealed that PKM2-V5 was prolyl hydroxylated in HeLa cells cultured at 20% O2 (Figure 4C). Remarkably, prolyl hydroxylation of PKM2-V5 was also detected in HeLa cells cultured at 1% O2. In contrast, PKM1-V5 was not hydroxylated (Figure 4C), again demonstrating the selectivity of the antibody. Endogenous PKM2 was also hydroxylated in hypoxic HeLa cells (1% O2), but hydroxylation was reduced when cells was exposed to near-anoxic conditions (<0.1% O2) for 4 h (Figure 4D). Double mutation of Pro-403/408 to alanine significantly reduced Pro-OH antibody binding by 50% (Figures 4E and S3B). Together, these data indicate that PKM2 is hydroxylated at Pro-403/408 within the domain that is not present in PKM1.
Figure 4. Prolyl Hydroxylation of PKM2 Stimulates HIF-1 Transactivation.
(A) GST-E10 was incubated with WCLs to induce hydroxylation and tryptic peptides were analyzed by mass spectrometry. The fragmentation spectrum of 401LAPITSDPTEATAVGAVEASFK422 revealed the presence of peptides with hydroxylation of Pro-403 or Pro-408.
(B) HEK293 cells were transfected with WT FLAG-HIF-1α or double mutant (DM) FLAG-HIF-1α(P402A/P564A) and treated with the proteasome inhibitor MG132 (10 μM) for 4 h. IP was performed with anti-FLAG antibody, followed by immunoblot assay. The intensity of hydroxylated FLAG-HIF-1α was quantified by densitometry and normalized to WT.
(C) HeLa cells were transfected with vector encoding PKM2-V5 or PKM1-V5 and exposed to 20% or 1% O2 for 24 h. IP was performed with anti-V5 agarose, followed by immunoblot assay.
(D) HeLa cells were exposed to 1% or <0.1% O2 for 4 h. IP was performed with anti-PKM2 antibody, followed by immunoblot assay. The intensity of hydroxylated PKM2 was quantified by densitometry and normalized to 1% O2.
(E) HeLa cells were transfected with vector encoding WT PKM2-V5 or PKM2(P403/408A)-V5 and exposed to 20% or 1% O2 for 24 h. IP was performed with anti-V5 agarose, followed by immunoblot assay.
(F and G) HeLa cells were co-transfected with p2.1 (F) or GalA(P564A) and pG5E1bLuc (G); pSV-Renilla; and the indicated expression vector, and exposed to 20% or 1% O2 for 24 h. The ratio of FLuc:RLuc activity was normalized to EV at 20% O2 (mean ± SEM, n = 4). ***p<0.001 vs EV; #p<0.05, ###p<0.001 vs PKM2(WT).
(H) Co-IP was performed in HeLa cells transfected with vector encoding WT PKM2-V5 or PKM2(P403/408A)-V5 and exposed to 1% O2 for 4 h.
(I) HeLa cells were transfected with PKM2-V5 vector and treated with DMSO or DMOG (100 μM) for 4 h. IP was performed with anti-V5 agarose, followed by immunoblot assay.
(J) Co-IP was performed in HeLa cells co-transfected with FLAG-HIF-1α(DM) and PKM2-V5 vectors and treated with DMSO or DMOG for 4 h.
(K) HeLa cells were co-transfected with GalA(N803A) vector, pG5E1bLuc, pSV-Renilla, and EV or PKM2-V5 vector, and treated with DMSO or DMOG for 24 h. The ratio of FLuc:RLuc activity was normalized to EV + DMSO (mean ± SEM, n = 4). *p<0.05 vs EV; #p<0.05, ##p<0.01 vs DMSO.
See also Figure S3.
To investigate whether prolyl hydroxylation of PKM2 regulates HIF-1α transactivation, we compared the effect of WT PKM2-V5 and PKM2(P403/408A)-V5 in reporter assays. The p2.1 and GalA assays revealed that double mutation of Pro-403/408 significantly reduced PKM2-mediated HIF-1 transactivation in HeLa cells (Figures 4F and 4G), despite the fact that PKM2(P403/408A)-V5 was detected in the nucleus at levels similar to WT PKM2-V5 (Figure S3C). Co-IP assays revealed that double mutation of Pro-403/408 markedly reduced the binding of PKM2-V5 to endogenous HIF-1α in hypoxic cells (Figure 4H). Taken together, these data suggest that prolyl hydroxylation of PKM2 enhances HIF-1α:PKM2 interaction, thereby promoting HIF-1α transactivation.
To determine whether prolyl hydroxylase activity stimulates HIF-1 transactivation, HeLa cells were transfected with PKM2-V5 vector and treated with the hydroxylase inhibitor DMOG or vehicle DMSO for 4 h. DMOG largely inhibited prolyl hydroxylation of PKM2-V5 (Figure 4I). PKM2-V5 binding to FLAG-HIF-1α(DM) was also reduced by DMOG treatment of HeLa cells (Figure 4J), thereby eliminating any potential effects of DMOG on prolyl hydroxylation of HIF-1α. To eliminate any potential effects of DMOG that were due to inhibition of asparaginyl hydroxylation by FIH-1, reporter assays were performed using GalA(N803A). DMOG significantly decreased basal GalA(N803A) activity as well as PKM2-enhanced GalA(N803A) activity in HeLa cells (Figure 4K). These results indicate that inhibition of prolyl hydroxylase activity impairs HIF-1α transactivation.
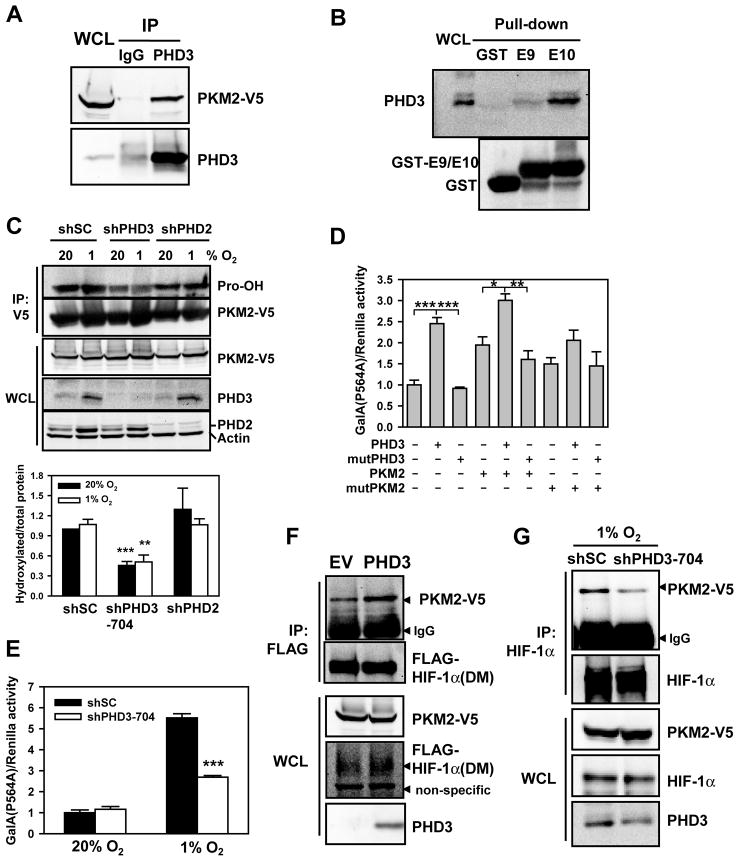
Next, we investigated which PHD isoform is involved in the hydroxylation of PKM2. PHD3 interacted with purified GST-PKM2 in vitro (Figure S4A) and co-immunoprecipitated with PKM2-V5 from transfected cells (Figure 5A). Moreover, PHD3 specifically bound to GST-E10, but not to GST or GST-E9 (Figure 5B), suggesting that PHD3 may mediate prolyl hydroxylation of PKM2. Immunoblot assays indicated that prolyl hydroxylation of PKM2-V5 was inhibited by 50% in HeLa cells transduced with retrovirus encoding PHD3 shRNA (shPHD3) (Figure 5C), which was similar to the effect of exposing cells to near-anoxic conditions (Figure 4D) or mutation of P403/408A (Figure 4E). Hypoxia dramatically increased PHD3 levels, which compensated for the decreased hydroxylase activity at 1% O2 (Epstein et al., 2001), resulting in similar levels of prolyl hydroxylation of PKM2 in cells incubated at 20% and 1% O2 for 24 h (Figure 5C). Overexpression of PHD3 dramatically reduced FLAG-HIF-1α levels in hypoxic HeLa cells (Figure S4B), confirming that PHD3 retains hydroxylase activity at 1% O2. PKM2-V5 protein levels were not altered by shPHD3 (Figure 5C).
Figure 5. PHD3 Stimulates the Coactivator Function of PKM2.
(A) Co-IP was performed in HeLa cells co-transfected with PKM2-V5 and PHD3 vectors.
(B) GST pull-down assays were performed with purified GST, GST-E9, or GST-E10 and WCLs from HeLa cells exposed to 1% O2 for 24 h.
(C) HeLa-shSC, HeLa-shPHD3-704, and HeLa-shPHD2 cells were transfected with PKM2-V5 vector and exposed to 20% or 1% O2 for 24 h. IP was performed with anti-V5 agarose, followed by immunoblot assay. Hydroxylated PKM2-V5 was quantified by densitometry and normalized to total immunoprecipitated PKM2-V5 (mean ± SEM, n = 3). **p<0.01, ***p<0.001 vs shSC.
(D) RCC4 cells were co-transfected with GalA(P564A) vector, pG5E1bLuc, pSV-Renilla, and the indicated expression vector for 24 h. The ratio of FLuc:RLuc activity was normalized to EV (mean ± SEM, n = 4). *p<0.05; **p<0.01; ***p<0.001. mutPHD3, PHD3(H135A/D137A); mutPKM2, PKM2(P403/408A).
(E) HeLa cells were co-transfected with GalA(P564A) vector, pG5E1bLuc, and pSV-Renilla, and the indicated expression vector and exposed to 20% or 1% O2 for 24 h. The ratio of FLuc:RLuc activity was normalized to shSC at 20% O2 (mean ± SEM, n = 4). ***p<0.001 vs shSC.
(F) Co-IP was performed in HeLa cells co-transfected with vector encoding double mutant (DM) FLAG-HIF-1α(P402A/P564A), EV or PHD3, and PKM2-V5.
(G) Co-IP was performed in HeLa cells co-transfected with vectors encoding PKM2-V5 and either shSC or shPHD3 and exposed to 1% O2 for 24 h.
See also Figure S4.
To examine whether PHD3 potentiates PKM2-mediated HIF-1α transactivation, we generated a mutant GalA(P564A) construct, so that only PKM2 would be a target for prolyl hydroxylation. GalA(P564A) assays demonstrated that PHD3 increased HIF-1α transactivation in RCC4 renal carcinoma cells, which constitutively express HIF-1α under non-hypoxic conditions due to VHL loss of function, whereas catalytically inactive PHD3(H135A/D137A) failed to increase HIF-1α transactivation (mutPHD3, Figure 5D). PHD3 overexpression in RCC4 cells enhanced HIF-1α transactivation mediated by WT PKM2 but not PKM2(P403/408A) (mutPKM2, Figure 5D). PHD3, but not PHD3(H135A/D137A), also potentiated PKM2-mediated HIF-1α transactivation in HeLa cells (Figure S4C). Thus, the ability of PHD3 to stimulate PKM2 coactivator function is dependent upon PHD3 catalytic activity and Pro residues (putative hydroxylation sites) in the PKM2-specific domain.
PHD3 knockdown significantly reduced HIF-1α transactivation in hypoxic HeLa cells (Figure 5E). PHD3 overexpression enhanced the interaction of PKM2-V5 with FLAG-HIF-1α(DM) in transfected HeLa cells (Figure 5F), but did not stimulate the interaction between PKM2(P403/408A)-V5 and FLAG-HIF-1α(DM) (Figure S4D). Conversely, PHD3 knockdown decreased PKM2-V5 interaction with endogenous HIF-1α in hypoxic HeLa cells (Figure 5G). The data presented in Figure 5 strongly suggest that PHD3 induces hydroxylation of PKM2 at Pro-403/408, thereby increasing the interaction of PKM2 with HIF-1α and potentiating PKM2-mediated HIF-1α transactivation.
We further investigated whether other PHDs regulate PKM2. FLAG-PHD1 failed to interact with PKM2-V5 (Figure S4E). Although PHD2 co-immunoprecipitated with PKM2 (Figure S4F), PHD2 knockdown failed to affect prolyl hydroxylation of PKM2 in HeLa cells transduced with lentivirus encoding an shRNA that effectively reduced PHD2 protein levels (Figure 5C). FLAG-PHD2 overexpression also had no effect on HIF-1α transactivation domain function (Figure S4G). Therefore, PHD3 is a specific regulator of PKM2-stimulated HIF-1 transcriptional activity.
PKM2 Enhances HIF-1 Binding to the HREs of Target Genes
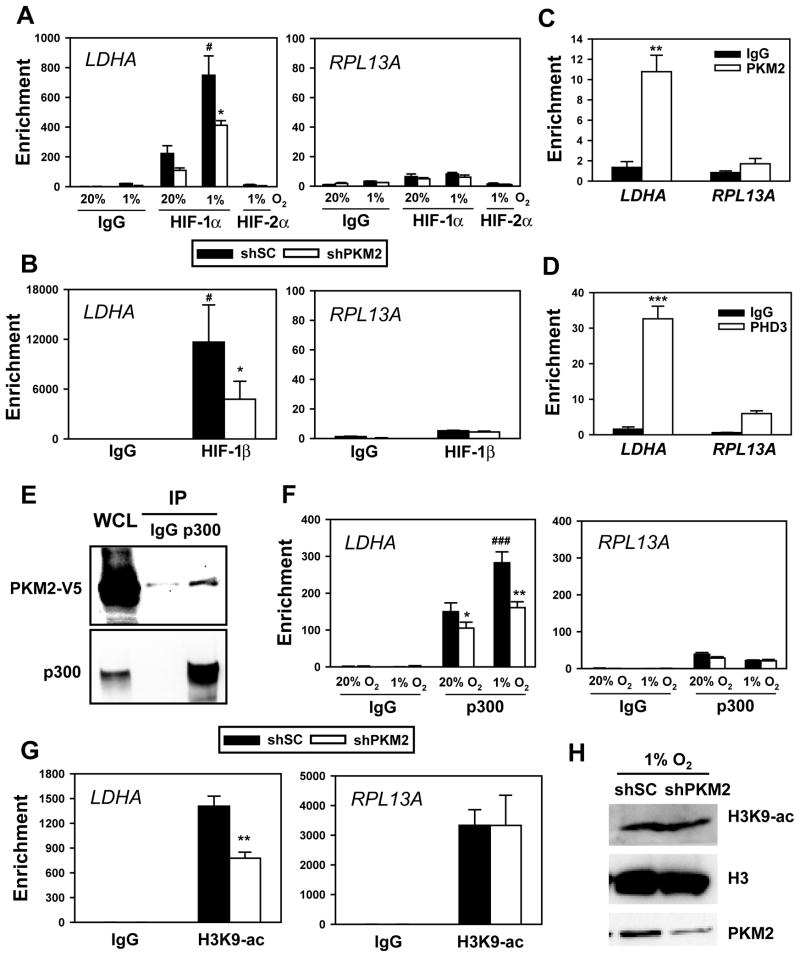
To determine whether PKM2 enhances HIF-1 DNA-binding activity, HeLa cells were transfected with vector encoding scrambled control shRNA (shSC) or shPKM2 and exposed to 20% or 1% O2 for 4 h. ChIP assays demonstrated that, relative to IgG background signals, HIF-1α occupancy was detected at the HRE of HIF-1 target genes, including LDHA and PDK1, but not at the non-HIF-1 target gene RPL13A, in non-hypoxic HeLa cells transfected with shSC vector (Figures 6A and S5A). Hypoxia significantly increased HIF-1α occupancy at the LDHA and PDK1 HREs. The lack of HIF-2α occupancy on HIF-1-selective target genes LDHA and PDK1 demonstrated the specificity of the assay (Figures 6A and S5A). Knockdown of endogenous PKM2 significantly reduced HIF-1α occupancy at the LDHA and PDK1 HREs in hypoxic HeLa cells, but not at the RPL13A gene (Figures 6A and S5A), indicating that PKM2 specifically enhances HIF-1α binding at the HRE of target genes. HIF-1β occupancy at the PDK1 and LDHA genes, but not at the RPL13A gene, was also significantly reduced by PKM2 knockdown in hypoxic HeLa cells (Figures 6B and S5B). PKM2 knockdown had no effect on HIF-1α or HIF-1β protein levels in HeLa cells (Figures 3D and S5D). Moreover, neither PKM2-V5 overexpression nor PKM2 knockdown affected HIF-1α/HIF-1β dimerization in hypoxic HeLa cells (Figures S5C and S5D). In contrast to LDHA and PDK1, VEGF gene transcription is regulated by both HIF-1 and HIF-2, and occupancy of the VEGF HRE by both HIF-1α and HIF-2α was significantly decreased by PKM2 knockdown (Figure S5E). Taken together, these data indicate that PKM2 enhances HIF binding to HREs within target genes, thereby promoting transactivation.
Figure 6. PKM2 Enhances HIF-1 Binding and p300 Recruitment to HREs.
(A) HeLa cells were transfected with shSC or shPKM2-#1 vector and exposed to 20% or 1% O2 for 4 h. ChIP assays were performed with IgG, anti-HIF-1α, or anti-HIF-2α antibody (mean ± SEM, n = 4). #p<0.05 vs shSC-20% O2; *p<0.05 vs shSC-1% O2.
(B) HeLa cells were transfected with shSC or shPKM2-#1 vector, and exposed to 1% O2 for 4 h. ChIP assays were performed with IgG or anti-HIF-1β antibody (mean ± SEM, n = 3). #p<0.05 vs IgG; *p<0.05 vs shSC.
(C and D) HeLa cells were exposed to 1% O2 for 24 h. ChIP assays were performed with IgG, anti-PKM2 (C), or anti-PHD3 (D) antibody (mean ± SEM, n = 3). **p<0.01, ***p<0.001 vs IgG.
(E) Co-IP was performed in HeLa cells transfected with PKM2-V5 vector for 24 h.
(F) HeLa cells expressing shSC or shPKM2-#1 were exposed to 20% or 1% O2 for 24 h. ChIP assays were performed with IgG or anti-p300 antibody (mean ± SEM, n = 3). *p<0.05, **p<0.01 vs shSC; ###p<0.001 vs shSC-20% O2.
(G) HeLa cells were transfected with shSC or shPKM2-#1 vector and exposed to 1% O2 for 24 h. ChIP assays were performed with IgG or anti-acetylated H3K9 (H3K9-ac) antibody (mean ± SEM, n = 3). **p<0.01 vs shSC.
(H) Immunoblot assays of total H3K9-ac, histone H3, and PKM2 in HeLa cells transfected with shSC or shPKM2-#1 vector and exposed to 1% O2 for 24 h.
See also Figure S5.
Next, we determined whether PKM2 is recruited to the HRE of HIF-1 target genes. ChIP assays demonstrated PKM2 occupancy at the HRE of HIF-1 target genes LDHA and PDK1, but not at the RPL13A gene in hypoxic HeLa cells (Figures 6C and S5F). PHD3 occupancy at the HRE of LDHA and PDK1 genes was also detected in hypoxic HeLa cells (Figures 6D and S5G). These data indicate that PKM2 and PHD3 both co-localize with HIF-1α at the HRE of HIF-1 target genes.
PKM2 Stimulates p300 Recruitment to the HRE of HIF-1 Target Genes
The histone acetyltransferase p300 is a coactivator that regulates HIF-1 transactivation (Arany et al., 1996). We investigated whether PKM2 contributes to p300 recruitment to the HRE of HIF-1 target genes. Co-IP assays demonstrated that PKM2-V5 physically interacted with p300 in HeLa cells (Figure 6E). ChIP assays revealed that p300 bound to the LDHA HRE, and hypoxia increased p300 occupancy in HeLa cells transfected with shSC vector (Figure 6F). However, PKM2 knockdown significantly reduced p300 occupancy at the LDHA HRE in non-hypoxic and hypoxic HeLa cells (Figure 6F). Levels of histone H3 acetylated at lysine-9 (H3K9-ac) at the LDHA HRE were significantly decreased in hypoxic HeLa cells transfected with shPKM2 vector (Figure 6G), whereas PKM2 knockdown did not affect H3K9-ac levels at the RPL13A gene (Figure 6G) or total histone H3 occupancy at the LDHA or RPL13A gene (Figures S5H and S5I). PKM2 knockdown also had no effect on total cellular H3K9-ac or H3 levels (Figure 6H). These data indicate that PKM2 specifically enhances p300 recruitment to the HRE of HIF-1 target genes and promotes H3K9 acetylation, which is an epigenetic modification that is associated with gene transcription.
PKM2 and PHD3 Regulate HIF-1-dependent Metabolic Reprogramming
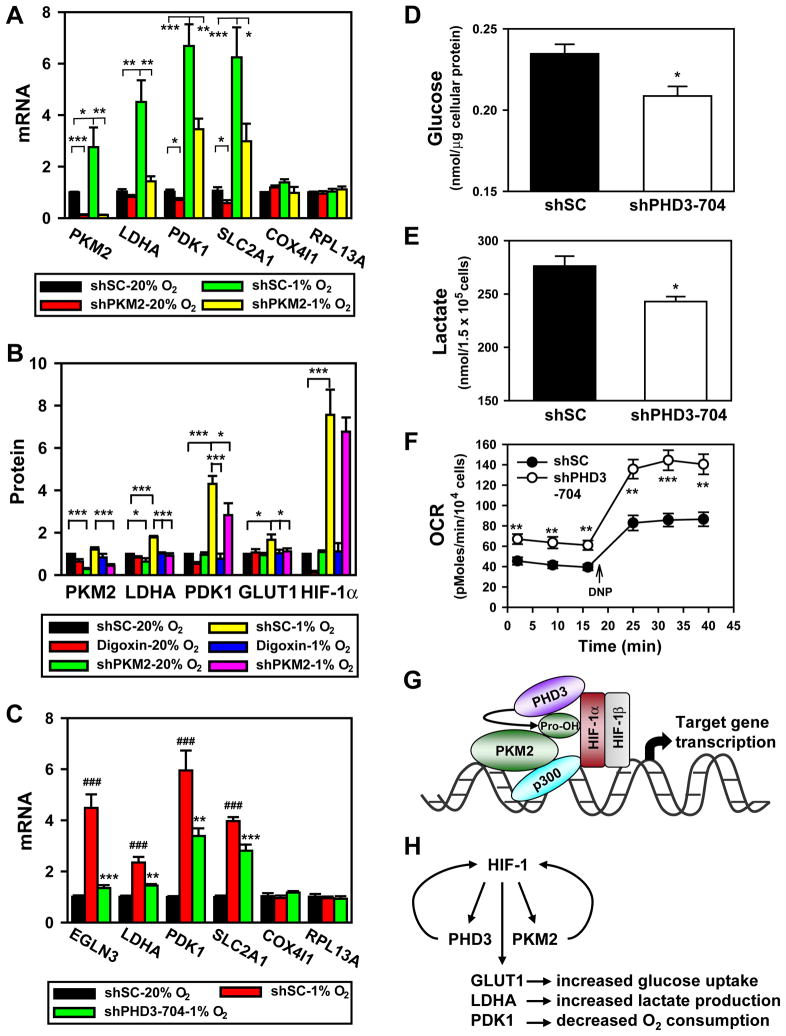
To determine whether PKM2 regulates HIF-1-dependent expression of genes encoding proteins in the glycolytic pathway, HeLa cells were transfected with shSC or shPKM2 vector, and exposed to 20% or 1% O2 for 24 h. qRT-PCR assays revealed that shPKM2 significantly decreased expression of the glycolytic genes LDHA, PDK1, SLC2A1, HK1 and the angiogenesis gene VEGF in HeLa cells, whereas expression of the non-HIF-1 target genes COX4I1, LDHB, and RPL13A was not affected (Figures 7A and S6A). LDHA, PDK1, and GLUT1 protein levels were also significantly reduced in HeLa cells transduced with shPKM2 retrovirus, which was comparable to the effect of treatment with digoxin (Figures 7B and S6B), a drug that inhibits HIF-1α synthesis (Zhang et al., 2008). Thus, PKM2 coactivates expression of HIF-1 target genes encoding glucose transporter and glycolytic enzymes.
Figure 7. PKM2 and PHD3 Promote HIF-1-dependent Glycolytic Metabolism.
(A) qRT-PCR analysis of indicated mRNAs in HeLa cells transfected with shSC or shPKM2-#1 vector and exposed to 20% or 1% O2 for 24 h (mean ± SEM, n = 4). *p<0.05, **p<0.01, ***p<0.001.
(B) HeLa cells were transduced with shSC or shPKM2-#1 and shPKM2-#2 retrovirus, and exposed to 20% or 1% O2 in the absence or presence of digoxin (100 nM) for 24 h. Levels of the indicated protein were determined by immunoblot assay, quantified by densitometry, and normalized to actin (mean ± SEM, n = 4). *p<0.05, ***p<0.001.
(C) qRT-PCR analysis of indicated mRNAs in HeLa cells transduced with shSC or shPHD3-704 retrovirus and exposed to 20% or 1% O2 for 24 h (mean ± SEM, n = 3–4). ###p<0.001 vs shSC-20% O2; **p<0.01, ***p<0.001 vs shSC-1% O2.
(D–F) RCC4 cells were transduced with shSC or shPHD3-704 retrovirus. Glucose was measured in lysates and normalized to total cellular protein amount (mean ± SEM, n = 3; D); lactate was measured in the culture media and normalized to cell number (mean ± SEM, n = 3; E); O2 consumption rate (OCR) was measured and normalized to cell number (mean ± SEM, n = 4–8; F). *p<0.05, **p<0.01, ***p<0.01 vs shSC.
(G) PKM2 is prolyl hydroxylated by PHD3 and interacts with HIF-1α and p300, thereby enhancing HRE occupancy by HIF-1 and p300, and increasing histone acetylation.
(H) Feed-forward mechanism for HIF-1 activity. HIF-1 activates transcription of genes encoding PHD3 and PKM2, which interact with HIF-1α to stimulate transactivation of target genes encoding GLUT1, LDHA, PDK1, and other metabolic enzymes that mediate the Warburg effect in cancer cells.
See also Figure S6.
To determine whether PHD3 activity stimulates expression of HIF-1 target genes, we performed qRT-PCR assays, which demonstrated that LDHA, PDK1, and SLC2A1 gene expression was significantly decreased in HeLa cells transduced with shPHD3 retrovirus, compared to cells transduced with shSC retrovirus (Figure 7C). However, expression of COX4I1 and RPL13A, which are not regulated by HIF-1, was not affected by PHD3 knockdown (Figure 7C). HIF-1α levels were not altered in PHD3 knockdown HeLa cells (Figure S6C), indicating that reduced HIF-1 transactivation accounts for the inhibition of HIF-1 target gene expression in PHD3 knockdown cells.
LDHA, PDK1, and GLUT1 mRNA and protein levels were also reduced by PHD3 knockdown in RCC4 cells (Figures S6D and S6E). Consistent with altered expression of glucose transporters and glycolytic enzymes, intracellular glucose and extracellular lactate levels were significantly decreased in RCC4 cells transduced with shPHD3 retrovirus, compared to cells transduced with shSC retrovirus (Figures 7D and 7E). PHD3 knockdown significantly increased the basal O2 consumption rate (OCR) and maximal OCR stimulated by 2,4-dinitrophenol (DNP, 20 μM) in RCC4 cells (Figure 7F). Similar results on glucose uptake, lactate production and OCR were also observed in RCC4 cells transduced with lentivirus encoding a different shRNA (Figures S6F–H), which completely knocked down PHD3 protein levels (Figure S6I). Thus, PHD3 knockdown, which inhibits PKM2 coactivator function (Figure 5D) and PKM2 binding to HIF-1α (Figure 5G), also reduces glucose uptake and lactate production, and increases O2 consumption in cancer cells.
DISCUSSION
In the present study, we demonstrate that PKM2 is a direct HIF-1 target gene and that expression of PKM1 and PKM2, the alternative isoenzymes of the PKM2 gene, is controlled by HIF-1. Remarkably, PKM2 interacts with HIF-1α within multiple domains. PKM2 binding to the transactivation domain of HIF-1α stimulates HIF-1 transcriptional activity. Although PKM2 also binds strongly to the PAS domain of HIF-1α, it does not affect the two known biological functions of the PAS domain, HIF-1α/HIF-1β dimerization and HIF-1α protein stability. ChIP data indicate that PKM2 stabilizes binding of HIF-1 to DNA and further studies are required to determine whether this property is dependent upon interaction of PKM2 with the PAS domain of HIF-1α.
PKM2 localizes in the nucleus, is recruited with HIF-1 to HREs, and enhances HIF-1 occupancy, p300 recruitment, and H3K9 acetylation, thereby promoting transactivation of genes encoding glucose transporters and glycolytic enzymes in cancer cells (Figure 7G). In contrast, PKM1 does not regulate HIF-1 transcriptional activity. Together, these findings delineate a molecular mechanism underlying the shift from oxidative to glycolytic metabolism that is associated with the expression of PKM2 in cancer cells. PKM2 also stimulates HIF-1/HIF-2-dependent VEGF gene expression in hypoxic HeLa cells, suggesting that through its function as a HIF-1 coactivator PKM2 may play a far broader role in promoting cancer progression than has been appreciated heretofore.
HIF-1α and HIF-2α were the first PHD3 substrates identified (Epstein et al., 2001), although PHD3 is less active in mediating HIF-1α hydroxylation than PHD2, which is the primary regulator of HIF-1α hydroxylation and proteasomal degradation in well-oxygenated cells (Berra et al., 2003). PHD3 was subsequently shown to promote degradation of the β2-adrenergic receptor (Xie et al., 2009) and the transcription factor ATF-4 (Köditz et al., 2007). Two Pro residues in the β2-adrenergic receptor were hydroxylated by PHD3 (Xie et al., 2009). Here we identified a putative PHD hydroxylation motif LRRLAP403 within the unique exon 10 domain of PKM2. Hydroxylation of Pro-403 and Pro-408 in PKM2 was demonstrated by mass spectrometry. The presence of acidic (Asp/Glu) residues near the Pro residue appears to be important for hydroxylation by PHD3 (Li et al., 2004; Xie et al., 2009) and several acidic residues are present on either side of Pro-403/408 (Figure S3A). Double mutation of Pro-403/408 reduced PKM2 hydroxylation to a degree that was similar to knockdown of PHD3 or exposure to near-anoxic conditions. The similarity in the degree of PKM2 hydroxylation (as measured by anti-Pro-OH antibody binding) in cells incubated at 20% vs 1% O2 for 24 h is due to the dramatic increase in PHD3 protein levels under hypoxic conditions (Figure 5C), which compensates for the reduction in hydroxylase activity. Quantitative mass spectrometric data comparing the prolyl hydroxylation of PKM2 in cells expressing or deficient in PHD3 cannot be obtained because the relevant PKM2 tryptic peptide is subject to multiple modifications in addition to prolyl hydroxylation (data not shown). Nevertheless, experimental evidence presented above indicates that PKM2 is hydroxylated by PHD3.
Intratumoral hypoxia is commonly found in aggressive solid cancers, leading to HIF-1α accumulation (Harris, 2002; Semenza, 2003). However, HIF-1α is also highly expressed in well-oxygenated cancer cells with loss of function of certain tumor suppressors, most notably VHL (Melillo, 2007; Semenza, 2010). Although HIF-1α accumulates in VHL-null cells under aerobic conditions, FIH-1-mediated hydroxylation of the HIF-1α transactivation domain should block recruitment of p300 and inhibit transactivation. The interaction of PKM2 with HIF-1α and p300 may provide a mechanism to bypass negative regulation by FIH-1. We have shown that in VHL-null RCC4 cells, PKM2 promotes HIF-1 transactivation of target genes that mediate increased glucose uptake (GLUT1), increased lactate production (LDHA), and decreased oxidative metabolism (PDK1). This novel coactivator function of PKM2 provides a molecular mechanism by which it can act with HIF-1 to reprogram glucose metabolism in cancer cells, thus answering a major question regarding the role of PKM2 in cancer progression (Figure 7H). Our data on PKM2 and previous studies of the EGLN3 gene, which encodes PHD3 (Pescador et al., 2005), indicate that PKM2 and EGLN3 are both direct HIF-1 target genes, resulting in a positive feedback loop that amplifies HIF-1 activity and may accelerate metabolic reprogramming and other critical aspects of cancer progression that are mediated by HIF-1 (Figure 7H).
EXPERIMENTAL PROCEDURES
Detailed procedures are described in the Supplemental Experimental Procedures.
Cell Culture and Transfection
HeLa, MEFs, RCC4, HEK293, HEK293T, and Hep3B cells were cultured in DMEM with 10% heat-inactivated FBS, and 1 mM sodium pyruvate and 1% non-essential amino acid (MEFs only) at 37ºC in a 5% CO2/95% air incubator. Hypoxic cells (1% O2) were placed in a modular incubator chamber (Billups-Rothenberg). Anoxic cells (<0.1% O2) were placed in a controlled atmosphere chamber (Plas-Labs). Cells were transfected using Fugene-6 (Roche), Lipofectamine 2000 (Invitrogen), or PolyJet (SignaGen).
Virus Production
shSC, shNT, shPKM2, or shPHD3 retrovirus or lentivirus was generated by transfection of HEK293T cells with transducing vector, and packaging vectors pMD.G and pNGVL3-gag/pol (for retrovirus) or pCMVR8.91 (for lentivirus). After 48 h, virus particles in the medium were harvested, filtered, and transduced into HeLa or RCC4 cells.
In Vitro Binding Assays
GST fusion proteins were expressed in E. coli BL21-Gold (DE3) and purified. Mammalian cells were lysed in RIPA buffer. Co-IP, GST pull-down, and immunoblot assays were performed as described (Luo et al., 2010).
In Vitro Hydroxylation Assays
The immobilized GST-E10 protein was incubated at 30°C for 1 h with HEK293T cell lysates supplemented with 100 μM FeCl2, 5 mM ascorbate and 1 mM α-ketoglutarate, washed, fractionated by SDS-PAGE, and stained with colloidal blue staining kit (Invitrogen). GST-E10 protein was digested in-gel with trypsin and the tryptic peptides were analyzed by LTQ Orbitrap Velos mass spectrometry (Thermo Scientific).
Subcellular Fractionation
HeLa cells were lysed in hypotonic buffer by a Dounce homogenizer (40 strokes). Intact cells were removed by centrifugation at 50 g for 10 min. The nuclei were collected by centrifugation at 800 g for 10 min, washed, and lysed in isotonic buffer by sonication. The supernatant was taken as the cytosolic fraction.
Luciferase Reporter Assays
Cells were seeded onto 48-well plates, transfected with p2.1 reporter (Semenza et al., 1996), PKM2 HRE reporter, or GalA/GalO (Jiang et al., 1997) and reporter plasmid pG5E1bLuc; control reporter pSV-Renilla; together with PKM2, PKM1, PHD3, or PHD2 expression vector, and exposed to 20% or 1% O2 for 24 h. FLuc and RLuc activities were determined using the Dual-Luciferase Assay System (Promega).
qRT-PCR
Total RNA was isolated using Trizol (Invitrogen) and reverse-transcribed. qRT-PCR assays were performed as described (Fukuda et al., 2007). Primer sequences are listed in Table S1.
ChIP Assays
ChIP assays were performed using ChIP Assay Kit (Upstate). HeLa cells were crosslinked with 1% formaldehyde for 20 min at 37°C, and quenched in 0.125 M glycine. DNA was immunoprecipitated from the sonicated cell lysates and quantified using SYBR Green Real-time PCR analysis (Bio-Rad). Primer sequences are listed in Table S1. Fold enrichment was calculated based on Ct as 2−Δ(ΔCt), where ΔCt = CtIP − CtInput and Δ(ΔCt) = ΔCtantibody − ΔCtIgG.
Metabolism Assays
RCC4 cells were transduced with retrovirus encoding shSC or shPHD3-704, or with lentivirus encoding shNT or shPHD3-766. The intracellular glucose was measured in the cell lysates with glucose assay kit (BioVision). The extracellular lactate was measured in the medium with lactate assay kit (BioVision). The OCR was measured using a cartridge containing an optical fluorescent O2 sensor in a Seahorse Bioscience XF24 Extracellular Flux Analyzer.
Statistical Analysis
Data were expressed as mean ± SEM. Differences were examined by Student’s t-test between two groups or one-way analysis of variance within multiple groups.
Supplementary Material
Acknowledgments
We thank Karen Padgett (Novus Biologicals) for providing antibodies against HIF-1β, HIF-2α, PKM2, PHD2, PHD3, GLUT1, PDK1, LDHA, histone H3, p300 and rabbit IgG; Vickram Srinivas (Thomas Jefferson University) for FLAG-PHD1 vector; Frank S. Lee (University of Pennsylvania) for FLAG-PHD2 vector; Ted Dawson (The Johns Hopkins University) for shNT vector; and Kevin Bittman (Seahorse Bioscience, Inc) for assistance with OCR assays. This work was supported by contract N01-HV28180. G.L.S. is the C. Michael Armstrong Professor at The Johns Hopkins University School of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, Bunn HF, Livingston DM. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1α: modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Köditz J, Nesper J, Wottawa M, Stiehl DP, Camenisch G, Franke C, Myllyharju J, Wenger RH, Katschinski DM. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood. 2007;110:3610–3617. doi: 10.1182/blood-2007-06-094441. [DOI] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- Li D, Hirsila M, Koivunen P, Brenner MC, Xu L, Yang C, Kivirikko KI, Myllyharju J. Many amino acid substitutions in a hypoxia-inducible transcription factor (HIF)-1α-like peptide cause only minor changes in its hydroxylation by the HIF prolyl 4-hydroxylases: substitution of 3,4-dehydroproline or azetidine-2-carboxylic acid for the proline leads to a high rate of uncoupled 2-oxoglutarate decarboxylation. J Biol Chem. 2004;279:55051–55059. doi: 10.1074/jbc.M410287200. [DOI] [PubMed] [Google Scholar]
- Luo W, Zhong J, Chang R, Hu H, Pandey A, Semenza GL. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J Biol Chem. 2010;285:3651–3663. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo G. Targeting hypoxia cell signaling for cancer therapy. Cancer Metastasis Rev. 2007;26:341–352. doi: 10.1007/s10555-007-9059-x. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- Pescador N, Cuevas Y, Naranjo S, Alcaide M, Villar D, Landazuri MO, Del Peso L. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem J. 2005;390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seagroves TN, Ryan HE, Lu H, Wouters BG, Knapp M, Thibault P, Laderoute K, Johnson RS. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol. 2001;21:3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton WW, Chandel NS. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol. 2011;300:C385–393. doi: 10.1152/ajpcell.00485.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Xiao K, Whalen EJ, Forrester MT, Freeman RS, Fong G, Gygi SP, Lefkowitz RJ, Stamler JS. Oxygen-regulated β2-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal. 2009;2:ra33. doi: 10.1126/scisignal.2000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qian DZ, Tan YS, Lee K, Gao P, Ren YR, Rey S, Hammers H, Chang D, Pili R, et al. Digoxin and other cardiac glycosides inhibit HIF-1α synthesis and block tumor growth. Proc Natl Acad Sci U S A. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.