Abstract
The vomeronasal organ (VNO) or Jacobson's organ is responsible in terrestrial vertebrates for the sensory perception of pheromones, chemicals that elicit stereotyped behaviors among individuals of the same species. Pheromone-induced behaviors and a functional VNO have been described in a number of mammals, but the existence of this sensory system in human is still debated. Recently, two nonhomologous gene families, V1R and V2R, encoding pheromone receptors have been identified in rat. These receptors belong to the seven-transmembrane domain G-protein-coupled receptor superfamily. We sought to characterize V1R-like genes in the human genome. We have identified seven different human sequences by PCR and library screening with rodent sequences. These human sequences exhibit characteristic features of V1R receptors and show 52%–59% of amino acid sequence identity with the rat sequences. Using PCR on a monochromosomal somatic cell hybrid panel and/or FISH, we demonstrate that these V1R-like sequences are distributed on chromosomes 7, 16, 20, 13, 14, 15, 21, and 22 and possibly on additional chromosomes. One sequence hybridizes to pericentromeric locations on all the acrocentric chromosomes (13, 14, 15, 21, and 22). All of the seven V1R-like sequences analyzed show interrupted reading frames, indicating that they represent nonfunctional pseudogenes. The preponderence of pseudogenes among human V1R sequences and the striking anatomical differences between rodent and human VNO raise the possibility that humans may have lost the V1R/VNO-mediated sensory functions of rodents.[Sequence data from this article have been deposited with the DDBJ/EMBL/GenBank Data Libraries under accession nos. U73852–73853 and AF253312–253316.]
The olfactory sensory system allows animals to recognize odorants and respond with appropriate behavior. In mammals, the sense of smell is mediated by at least two anatomically distinct sensory organs: the main olfactory epithelium (MOE) and the vomeronasal organ (VNO) (Bargmann 1997). Ablation of the VNO in rodents interferes with the perception of pheromones, chemicals communicated between animals of the same species that convey information primarily regarding reproductive and social status (Wysocki and Lepri 1991). In mammals, the olfactory receptors (ORs) of the MOE are encoded by a large family of ∼500–1000 receptor genes belonging to the superfamily of seven-transmembrane-domain G-protein-coupled receptors. Two multigene families of G-protein receptors, named V1Rs and V2Rs, have been recently identified in VNO neurons of the rat (Dulac and Axel 1995; Herrada and Dulac 1997; Ryba and Tirindelli 1997) and mouse (Matsunami and Buck 1997). V1Rs are restricted to Gαi2-expressing neurons, and V2Rs are restricted to Gα0-expressing neurons in the VNO. On the basis of their structure and tissue distribution, these receptors are likely mediators of the response to pheromones.
V1R and V2R constitute novel families of seven-transmembrane-domain receptors with no sequence similarity with the ORs. The rat V1R family is estimated to contain 30–40 genes that share 50%–90% sequence identity but show no significant homology to other known proteins. So far, no pseudogene has been found among the seven V1R sequences examined from the rat genome (Dulac and Axel 1995). The V2Rs contain a large extracellular N-terminal domain and share similarity to extracellular calcium-sensing receptors and metabotropic glutamate receptors. The rat genome is estimated to contain ∼100 V2R-like genes, including a large number of pseudogenes (Herrada and Dulac 1997; Matsunami and Buck 1997).
It is not known if humans possess a functional VNO-mediated sensory system. Even the existence of a human VNO has been controversial. There is anatomical evidence for a foetal VNO in humans, but the VNO is typically described as an atrophied organ of vestigial function in the adult (Tirindelli et al. 1998). However, several groups have recently reported finding a structurally intact VNO (Garcia-Velasco and Mondragon 1991; Moran et al. 1991; Stensaas et al. 1991). Human pheromones have not been clearly identified. A recent study concluded that synchronization of menstrual cycles among women living together involves chemical signals, but it has not yet been demonstrated that this communication is mediated through the VNO (McClintock 1971; Stern and McClintock 1998).
We have taken a genomic-based approach to explore the possibility that humans possess functional V1R pheromone receptors. We have determined the sequence and chromosomal locations of a number of human V1R-like sequences. Our results shed some light on the evolution of this gene family and on this form of chemical communication in humans.
RESULTS
To isolate putative human pheromone receptor genes, we designed degenerate oligonucleotide primers (Ph1F and Ph2R) to recognize regions conserved among rat V1R sequences (Dulac and Axel 1995). As the VIR genes analyzed so far lack introns in their protein-coding regions, it is possible to generate a 373-bp product by PCR using genomic DNA as a template. When genomic DNA was used as a template with Ph1F/Ph2R, a smear of PCR products with bands at different sizes was obtained (not shown). This population was subcloned. Of 96 recombinant clones analyzed by PCR using M13 vector primers, 50 containing inserts of the expected size were sequenced with vector primers. Only two clones, PhH5 and PhH8, showed homology to V1R genes. PhH5 and PhH8 are 73% identical at the nucleotide level (NSI). They show 55.9% and 57.6% NSI with the most homologous known rat V1R gene, VN7 and VN4, respectively.
At the predicted protein level, both PhH5 and PhH8 appear to encode pseudogenes. Both sequences contain a frameshift because of “indels” (insertion/deletion events) causing stop codons at positions 50 (extracellular loop II) and 30 (transmembrane domain IV), respectively. The PhH8 sequence also shows a 3-nt deletion corresponding to amino acid 46. The two predicted proteins share 44% amino acid identity (ASI). These two pseudogenes are clearly members of the V1R family, as they show >30% ASI with rat V1R genes (Fig. 1A). Some portions of the PhH5 and PhH8 sequences, such as transmembrane domain V, are highly conserved and more similar to the rat sequences (55% ASI).
Figure 1.
Human V1R-like sequences obtained by PCR using consensus rat V1R primers. (A) The predicted sequences of human PhH5 and PhH8 proteins combined with rat V1R sequences (Dulac and Axel 1995). The sequence alignment was performed using the PileUp program from the GCG package. The aminoacids of the rat sequences are numbered according to their complete sequences, and the aminoacids of the partial human sequences are arbitrarily numbered from 1 to 110. The predicted positions of transmembrane domains IV and V are indicated by horizontal lines. Aminoacids common to at least six of the nine sequences and including at least one human sequence are highlighted in grey. Stop codons are represented by asterisks. Insertions or deletions leading to frameshifts are indicated as vertical bars. (B) Identity dendrogram of sequences presented above using the PileUp program. Families (amino acid sequence identity or ASI > 40%) are marked by circled numbers and subfamilies (ASI > 60%) by squared letters. The percentage of ASI was estimated by pairwise sequence comparisons using the GAP program (GCG package) and is indicated along the bottom.
Figure 1B presents a dendrogram in which the two human V1R-like sequences are compared with rat V1R sequences VN1 to VN7 (Dulac and Axel 1995). Several V1R-like families and subfamilies can be defined according to the nomenclature used for the olfactory receptors (a family being defined as a group of proteins having an ASI > 40% and a subfamily having an ASI > 60%; Buck and Axel 1991; Ben-Arie et al. 1994; Rouquier et al. 1998). The seven rat V1Rs form one family, which is divided into three subfamilies. The two human sequences assort into two subfamilies of a second family. To identify any additional V1R-like sequences among the 46 clones that were generated with the degenerate-primed PCR but not sequenced, PhH5 and PhH8 clones were used as a probe to hybridize at low stringency a Southern blot containing these clones. No other positive clone was found.
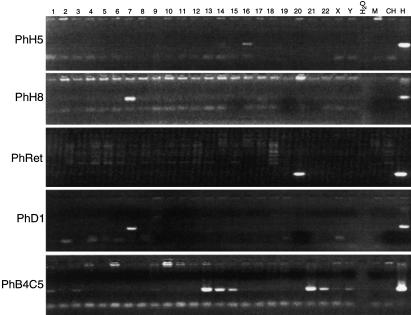
To determine the chromosomal localization of the PhH5 and PhH8 sequences in the human genome, we screened a human-rodent somatic cell hybrid panel by PCR using gene-specific primers. To discriminate human from rodent sequences, we designed the primer pairs for each human sequence in the regions that diverged most from the rodent sequences. As shown in Figure 2, PCR products of the expected size, equivalent in length to those produced with the human total genomic DNA control, assign PhH5 to human chromosome 16 and PhH8 to human chromosome 7. A very faint amplification product could be also observed for chromosome 14 using the PhH5 primer set, suggesting that a closely related sequence may exist on this chromosome.
Figure 2.
Chromosomal assignments of human V1R-like sequences by PCR amplification using a human–rodent somatic cell hybrid panel (NIGMS, Coriell Cell Repositories, Mapping Panel 2). The human chromosome present in each of the 24 somatic cell hybrids is shown above the lanes (chromosomes 1–22, X and Y). Lanes M, CH, and H contain mouse, Chinese hamster, and human DNA, respectively. Lane H2O is a negative control with no DNA. An example of the different chromosomal assignments is shown for sequences PhH5, PhH8, human retina cDNA clone PhRet, PhD1, and PhB4C5.
A database search with PhH5 and PhH8 sequences identified two IMAGE human retina cDNA clones (IMAGp998A20830, AA012849–50-EMBL; IMAGp998P15837) that share ∼77% NSI with PhH8 and ∼53% NSI with VN6. We fully sequenced these cDNA clones and showed that they are identical. The 5′ part of the cDNA clone is missing, and the remaining sequence contains three in-frame stop codons and one frameshift. A primer pair was designed and used to assign the sequence to chromosome 20 using the hybrid panel (Fig. 2).
To identify additional V1R-like sequences in the human genome, the PhH5 and PhH8 clones were used together to screen a genomic library made in bacteriophage λ EMBL3. Twelve positive clones were found among 500,000 plaques probed (2.5 genome equivalents). These clones were confirmed to contain PhH5- or PhH8-like sequences by hybridization of a Southern blot of the EcoRI-digested phages with the mixed probe used for the library screening (not shown). The phages were analyzed by restriction-digest fingerprinting and were grouped on the basis of the similarity of their restriction pattern (not shown). Duplicate clones were eliminated, and analysis was pursued on seven clones: Ph2, PhA2, PhA4, PhB1, PhD1, PhB4, and PhC5. Southern blot analyses suggested that each phage contained only one V1R-like squence (not shown). The degenerate primers Ph1F and Ph2R were able to amplify products only from PhA2, PhB1, and PhD1. These PCR products were subcloned and sequenced. The three sequences are identical and are referred collectively to as PhD1. Restriction fragments of clones Ph2, PhA4, PhB4, and PhC5 hybridizing to the V1R-like sequences were subcloned and sequenced. Only partial V1R-like sequences were present in each fragment. Sequences obtained from clones PhB4 and PhC5 were identical and, henceforth, are referred to as PhB4C5. Based on their chromosomal localization by FISH and PCR (chromosome 7 or acrocentric chromosomes, see below), full-length sequences Ph2 and PhB4C5 were established. Like PhH5 and PhH8, the four additional V1R-like sequences obtained from these seven λ clones contain stop codons and/or frameshifts, indicating that they correspond to pseudogenes.
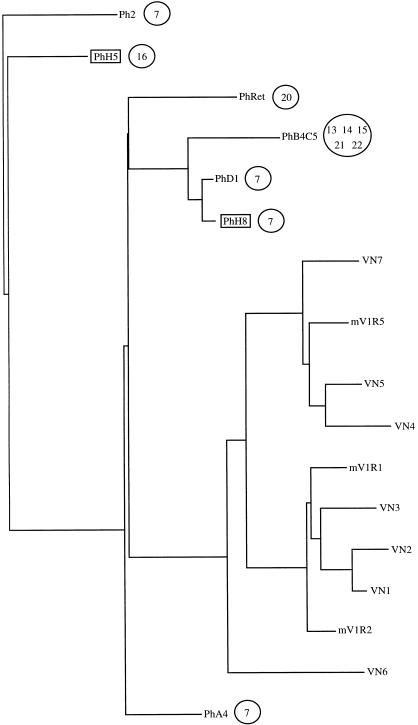
Although the proteins deduced from the six human V1R-like sequences are similar to V1Rs, the inferred amino acid sequence was uncertain in a few regions for some of the clones because of the many frame-shift-causing indels. We therefore present in Figure 3 the relationship among them in the form of a phylogenetic tree based on their nucleotide sequences. The tree includes the seven rat V1R sequences, three mouse V1R sequences (Rodriguez et al. 1999; Saito et al. 1998), and the human retinal cDNA clone. Human and rodent sequences form two distinct groups. The human sequences are very similar, with an average NSI of 76%. PhD1 and PhH8 share 96.9% NSI. Of the human sequences, PhA4 is the most similar to rodent sequences and shares 59.5% NSI with VN6.
Figure 3.
Phylogenetic tree placing the human V1R-like nucleotide sequences relative to rat V1R sequences VN1 to VN7 (Dulac and Axel 1995) and mouse mVR sequences (mV1R1 and mV1R2 [Saito et al. 1998] and mV1R5 [Rodriguez et al. 1999]). Sequences were aligned using CLUSTAL W version 1.7. The human sequences PhH5 and PhH8 that were used as probes to screen for the other human clones except PhRet are highlighted with boxes. Circled numbers indicate the chromosomal localization of the human sequences.
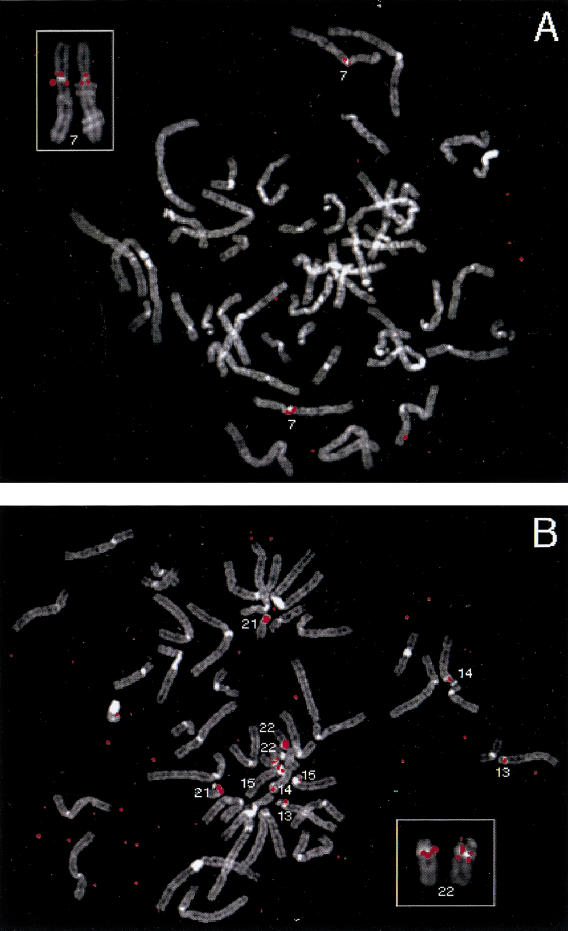
To map these V1R-like sequences on human chromosomes, fluorescence in situ hybridization (FISH) was performed with each phage clone (Fig. 4). Two major hybridization patterns were observed. Clones Ph2, PhA4, and PhD1 map near the centromere on chromosome 7 (Fig. 4A), with two pericentric locations labeled in some metaphases (Fig. 4A, inset). PhD1 and PhA2 also produced faint signals at low frequency on other chromosomes (not shown). Clones PhB4 and PhC5 hybridize very close to the centromere on the acrocentric chromosomes (chromosomes 13, 14, 15, 21, and 22; Fig. 4B). In some metaphases, two sets of signals could be resolved, suggesting the presence of more than one copy of these sequences in some pericentromeric regions (Fig. 4B, inset).
Figure 4.
Chromosomal locations of clones containing human V1R-like sequences determined by FISH. DAPI banding patterns are displayed using a gray scale; hybridization signals are displayed in red. (A) Clone Ph2 hybridizes to the pericentromeric region of chromosome 7 (main panel and inset, respectively). In the inset, hybridization signals are evident on both sides of the centromere of chromosome 7. The red spot on chromosome in the lower right is background and was not seen in other metaphases. (B) Clones PhB4 and PhC5 hybridize to the pericentromeric regions of chromosomes 13, 14, 15, 21, and 22 (main panel and inset, respectively). In the inset, PhC5 hybridization signals are evident on both sides of the centromere of chromosome 22.
The two FISH patterns are consistent with PCR analyses of the hybrid panel. Specific primer pairs derived from the coding part of Ph2, PhA4, and PhD1 amplified a single band from only the hybrid cell line containing chromosome 7 (Fig. 2 shows results for PhD1). Specific primers derived from PhB4/C5 amplified strong products from chromosomes 13, 14, 15, 21, and 22 and faint products from chromosomes 1, 3, 9, and Y (Fig. 2). Using PhB4C5-specific primers flanking a frameshift mutation, PCR products from either total genomic DNA (10 clones) or acrocentric chromosomes 13, 14, 15, 21, and 22 (two clones per chromosome) were subcloned and sequenced. These sequences are pseudogenes nearly identical to PhB4C5 and contain the same frameshift mutation, indicating that the gene-specific primers amplified similar sequences that were probably distributed in the genome by multiple rounds of duplication (not shown). The strong gene-specific PCR products demonstrate that the cross hybridization seen with FISH is at least in part caused by the presence of V1R-like sequences on each of the acrocentric chromosomes.
DISCUSSION
The vomeronasal organ is a chemosensory organ present in most tetrapod species. It is important in intraspecific chemical (pheromone) communication. The cell bodies of vomeronasal sensory neurons reside in the VNO epithelium, and their axons project to the accessory olfactory bulb (AOB), a structure distinct from the main olfactory bulb. In humans, there is no discernible AOB (Keverne 1999), and the question remains open as to whether humans possess a functional VNO. There are two families of putative pheromone receptors expressed in rodents VNO, V1R and V2R, none of which has been characterized in detail in humans. In rodents, the V1R family is smaller than the V2R family, with ∼30–40 and 100 predicted members, respectively. So far, no pseudogene has been identified among seven rat and five mouse cDNA V1R sequences, whereas many V2R pseudogenes have been identified (Dulac and Axel 1995; Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997).
We sought to isolate putative human V1R homologs to approach the question of human VNO functionality. We identified seven human V1R-like sequences using a combined approach of degenerate PCR, low stringency screening of genomic libraries, and database searching. These sequences show a NSI ranging from ∼62% to ∼97% and collectively map to several human chromosomes: 7, 13, 14, 15, 16, 20, 21, and 22 and possibly additional chromosomes.
The human V1R-like family appears to be much smaller than the rodent family. We obtained only seven V1R-containing clones by screening a library representing 2.5-times coverage of the human genome. In addition, only single bands were revealed on low-stringency genomic Southern blots probed with Ph2 and PhC5 (not shown). It is possible that each band represents multiple copies of genes residing at different sites (e.g., PhC5 was shown by FISH and PCR to reside in at least five locations). Also, the gene number determined by Southern blot could be underestimated if other V1R sequences in the human genome have accumulated numerous mutations and consequently fail to cross-hybridize with our probes.
One group of human V1R-like sequences is located in the pericentromeric region of chromosome 7. These sequences (PhH8 or PhD1, PhA4, Ph2) show ∼72.5% NSI on average, suggesting that these multiple genes may have arisen through local duplications followed by gene divergence. PhD1 and PhH8, share 96.9% NSI and may represent alleles of the same gene, or alternatively, genes more recently duplicated as described for the olfactory receptor genes (Rouquier et al. 1998; Brand-Arpon et al. 1999).
A second set of sequences, represented by PhB4C5, resides near the centromeres of all acrocentric chromosomes (13, 14, 15, 21, and 22). BLAST searches against public databases reveal that PhB4C5 is 100% identical to a sequence contained in an 85-kb genomic sequence derived from the juxta-centromeric region of human chromosome 21 (GenBank accession no. AP000023) and to BAC B11L7C8 (GenBank accession no. AL078472), which overlaps this region (Ruault et al. 1999). This BAC has been FISH-mapped to the centromeric region of chromosomes 13, 14, 15, 21, and 22 (A. de Sario, pers. comm.). PhB4C5 is also 98% identical to a sequence from chromosome 22q11 (GenBank accession no. AC006548). Cross hybridization among these sequences was observed both by PCR and FISH. PCR also suggested that additional V1Rs reside on chromosomes 1, 3, 9, and Y. The pericentromeric/acrocentric distribution is particularly interesting in that it is reminiscent of the pattern seen with other families of genes and nonprocessed pseudogenes. For example, parts of the NF1 gene (17q11.2) are found in the pericentromeric regions of chromosomes 14, 15, 18, 21, and 22 and in 2q21, which contains a relic of an ancient centromere (Regnier et al. 1997). Many of the OR-like sequences also lie in pericentromeric regions (Rouquier et al. 1998). Notably, a DNA family called chAB4 has been found to be repeated in the pericentromeric regions of chromosomes 1, 3, 4, 9, 13, 14, 15, 21, 22, and Y (Wohr et al. 1996; Assum et al. 1998). This pattern corresponds, with the exception of chromosome 4, to the distribution that we observed for the human V1R-like sequence PhB4C5. The 85-kb genomic sequence from the juxtacentromeric region of chromosome 21 (GenBank accession no. AP000023) was recently analyzed (Ruault et al. 1999). They identified ORFs using GRAIL and annotated EST matches using GAIA and Genscan. Their analysis showed that this region is rich in retrotransposed pseudogenes and gene fragments transferred by chromosome duplications. Although they performed detailed analyses on this sequence, the V1R-like sequence was not detected; probably because it contains many mutations leading to short ORFs. We performed an analogous analysis (GRAIL and BLAST searches), but no putative pheromone receptor sequences other than PhB4C5 were found in AP000023. This genomic sequence contains several pseudogenes for which functional copies are located on other chromosomes. Thus, our results add V1R sequences to a pattern of pericentromeric duplications that has been observed for multiple processed or nonprocessed pseudogenes.
All seven of the human V1R-like sequences we have sequenced have incurred deleterious mutations (stop codons or frameshifts), indicating that they do not encode full-length V1R-like receptors. These results are consistent with other reports that the human V1R-like sequences that have been characterized so far correspond to pseudogenes, although these sequences are not available for comparison to our collection (Dulac and Axel 1995; Tirindelli et al. 1998). The question as to whether a functional copy of a human V1R exists in the human genome remains open. Given the preponderence of pseudogenes and the vestigial nature of the VNO in adult catarrhines and apes (Keverne 1999; Stoddart 1980), it is possible that no functional V1R genes will be found in the human genome. Indeed, sequencing of PCR products isolated from acrocentric chromosomes using PhB4C5-specific primers demonstrated that all these sequences are copies of the PhB4C5 pseudogene. In addition, sensory transduction in the VNO is thought to involve ion channels of the transient receptor family (TRP). One of these candidates, TRP2, has been cloned in rat, but the human ortholog is a pseudogene (Liman et al. 1999). Our results do not rule out the possibility that other types of pheromone receptors or other sensory pathways exist in human that would mimic the pheromone-sensing pathway. However, the anatomical and molecular evidence argues that it is unlikely that a chemosensory pathway mediated by V1Rs and VNO is functional in humans.
METHODS
PCR with Degenerate Primers
Degenerate PCR primers were designed to recognize regions conserved among the seven VR1 receptors known from rat (Dulac and Axel 1995). Ph1F is located at the predicted border of the third transmembrane domain and extracellular loop 4 (amino acid sequence LSPRSSC; degenerate sequence, 5′- CT(C/T) AG(T/C) (T/C)C(T/C) A(G/A)A A(G/A)(A/C) TCC TG). Ph2R is located between the fifth and the sixth transmembrane domains (SPEQRA; 5′- TGG CCC TTT G (C/T)T (G/C)TG G(G/A)G). Ph1F and Ph2R were used in PCR with human genomic DNA as template. The cycling conditions consisted of an initial denaturation for 5 min at 94°C, followed by 30 cycles of 94°C for 15 sec, 55°C for 30 sec, 72∼C for 45 sec, and a final extension step of 10 min at 72°C. PCR products were subcloned in PCR product cloning systems (the LigATor vector, R&D Systems or TA cloning kit, Invitrogen). Recombinant clones were analyzed by PCR using M13-vector primers. Clones were sequenced with −21M13 and M13rev dye primers using the PRISM Ready Reaction AmpliTaq FS kit (Perkin Elmer) on an ABI 373 automated sequencer (Applied Biosystems).
Screening of a Human Genomic Library
Two PCR products corresponding to putative human V1R sequences were labeled by PCR with [α32P]dCTP. Filters containing 500,000 phage plaques from a human genomic DNA library cloned in phage vector λEMBL3 (gift of Catherine N'Guyen, Centre d'Immunologie Marseille-Luminy) were hybridized in duplicate with the probe mixture in 6× SSS, 0.5× Denhardt, 0.1% SDS at 55°C for 18 h and washed under low-stringency conditions in 2× SSC, 0.1% SDS at 55°C. Filters were subjected to autoradiography at −80°C for 3 d. For the first screening, despite low stringency, the signals were very weak. These clones were subjected to a second round of screening and produced a strong signal indicating they were indeed homologous to the probes and not artifacts.
Phage Clone Analysis and Subcloning
Phage DNA was purified according to standard methods (Sambrook et al. 1989) or using a QIAGEN kit and analyzed by digestion with various restriction enzymes in buffers supplied by the manufacturer (Life Technologies). The restriction digests were analyzed by electrophoresis on 0.8% agarose gels. The gels were alkali blotted as described (Gemmill et al. 1987) onto nylon membranes (Hybond N+; Amersham) and used for hybridization as described above. Fragments containing putative V1R sequences were subcloned into pBluescript KS- (Stratagene). Sequencing was carried out by a combination of vector and custom-made primers using dye-labeled dideoxy terminator chemistry on an Applied Biosystems 373A automated DNA sequencer following the manufacturer's procedure. To complete the sequence of clones Ph2 and PhB4/C5, directional sequencing was performed directly on phage DNA using the following modified procedure: the volume of a standard cycle-sequencing terminator reaction was doubled from 20 to 40 μL, so that each reaction contained 16 μL of reaction mixture, 50 pmole of primer, and 10 μL of DNA corresponding to 1–2 μg. Cycling was performed following the manufacturer's instructions but with an added initial denaturation step at 96°C for 4 min. Multiple alignment was performed using the GCG package (University of Wisconsin) and the CLUSTAL W v.1.7 software package (Thompson et al. 1994).
Chromosomal Assignments
Fluorescence In Situ Hybridization (FISH)
Metaphase cell preparations were prepared from human PHA–stimulated peripheral blood of a normal male donor. DNA from phage was biotinylated by nick-translation and hybridized in the presence of human Cot1 DNA to metaphase cells fixed on slides. Methods for preparation of the slides and probe, hybridization, washing, detection with FITC, fluorescent banding, and analysis are described elsewhere (Trask 1999). Chromosomes were banded (a QFH-like pattern) with DAPI staining. The FITC and DAPI fluorescence images were collected in registration and displayed in false colors using a Princeton cooled CCD camera, ChromaTechnology spectral filters, and image-analysis software (IP Lab Spectrum). The DAPI fluorescence image was in some cases inverted to display a Giemsa-like banding pattern.
Typing of a Monochromosomal Hybrid Panel
To map the location of human sequences using a monochromosomal rodent × human hybrid panel, gene-specific primer pairs were chosen in the coding parts that appeared to diverge the most from their rat counterparts (primer set A9F2/A9R2 for PhH5: 5′-ATC CAC AAA TTA CAT TTT GCA TG; 5′-AGA CAT ACT TGG ATA ATG TCA C; primer set B4F2/B4R for PhH8: 5′-AGT GTG ACC CAG ATA ATT CTG; 5′-TCT GGA TAA CGA CAG AGT AAC; primer set VN-A4F1/VNA4R1 for PhA4: 5′-GAA TGT CAG TAA ACA CTG CTC; 5′- CAT AAC AGG ATT GAG GAT GTT; primer set VN-2F/VN-2R for Ph2: 5′-TCA TTA TCC AGA GAT GTG TCC; 5′-GAG AGA CTC AAA TCC ATC TGG; primer set BC-5′F/BC-5′R for PhB4C5: 5′-TAT TTG CAC AAG GTG ATG AGG; 5′-GTA GAT TAT CAT GTC ACT ACTG; primer set VN-D1F/VN-D1R for PhD1: 5′- CAA CAG TGA CAT GAT AAT CTA C; 5′- ACA AGA GAA TCA CCA TGA AGG; primer set RF2/RR2 for the human retina cDNA clone R1: 5′-CAT CTT CAC ACT CCT TCA GG;5′-AGC TTT CCA ACC AGA AGT TGC). PhB4C5-specific sequences from the acrocentric chromosomes (13, 14, 15, 21, and 22, using the monochromosomal mapping panel) as well as from total human genomic DNA were established using PhB4C5-specific primers BC-5′F and BC-2R (LLSSVYM; 5′-ATG TAT ACA CTT GAG AGC AGC-3′) chosen ∼160 bp 3′ to BC-5′R in order to flank a frameshift mutation. PCR reactions of 25 μL were conducted following conventional procedures using 40 ng of DNA from each cell line of the NIGMS monochromosomal hybrid panel (Coriell Cell Repositories, Mapping Panel 2), with an annealing temperature of 55°C. PCR products were analyzed on ethidium bromide–stained 1% agarose gels.
Acknowledgments
This research was supported by a grant from the Fondation pour la Recherche Médicale to S.R. and D.G., and NIH grants GM57070 and DC04209 to B.T.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rouquier@igh.cnrs.fr; FAX 33-499-619-901.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.146700
REFERENCES
- Assum G, Pasantes J, Gläser B, Schempp W, Wöhr G. Concerted evolution of members of the multisequence family chAB4 located on various nonhomologous chromosomes. Mamm Genome. 1998;9:58–63. doi: 10.1007/s003359900680. [DOI] [PubMed] [Google Scholar]
- Bargmann C. Olfactory receptors, vomeronasal receptors, and the organization of olfactory information. Cell. 1997;90:585–587. doi: 10.1016/s0092-8674(00)80518-8. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter DH, Carrozzo R, Patel K, Sheer D, Lehrach H, et al. Olfactory receptor gene cluster on human chromosome 17: Possible duplication of an ancestral receptor repertoire. Hum Mol Genet. 1994;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- Brand-Arpon V, Rouquier S, Massa H, de Jong P, Ferraz C, Ioannou P, Demaille J, Trask B, Giorgi D. A genomic region encompassing a cluster of olfactory receptor genes and a myosin light chain kinase (MYLK) gene is duplicated on human chromosome regions 3q13–21 and 3p13. Genomics. 1999;56:98–110. doi: 10.1006/geno.1998.5690. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Garcia-Velasco J, Mondragon M. The incidence of the vomeronasal organ in 1000 human subjects and its possible clinical significance. J Steroid Biochem Mol Biol. 1991;39:561–564. doi: 10.1016/0960-0760(91)90253-2. [DOI] [PubMed] [Google Scholar]
- Gemmill RM, Coyle-Morris JF, McPeek FD, Ware-Uribe LF, Hecht F. Construction of long-range restriction maps in human DNA using pulsed field gel electrophoresis. Gene Anal Technol. 1987;4:119–131. doi: 10.1016/0735-0651(87)90010-0. [DOI] [PubMed] [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Keverne EB. The Vomeronasal organ. Science. 1999;286:716–720. doi: 10.1126/science.286.5440.716. [DOI] [PubMed] [Google Scholar]
- Liman ER, Corey DP, Dulac C. TRP2: A candidate transduction channel for mammalian pheromone sensory signaling. Proc Natl Acad Sci. 1999;96:5791–5796. doi: 10.1073/pnas.96.10.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Buck L. A putative family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- McClintock MK. Menstrual synchrony and suppression. Nature. 1971;229:244–245. doi: 10.1038/229244a0. [DOI] [PubMed] [Google Scholar]
- Moran DT, Jafek BW, Rowley JC., III The vomeronasal (Jacobson's) organ in man: Ultrastructure and frequency of occurrence. J Steroid Biochem Mol Biol. 1991;39:545–552. doi: 10.1016/0960-0760(91)90251-y. [DOI] [PubMed] [Google Scholar]
- Regnier V, Meddeb M, Lecointre G, Richard F, Duverger A, Nguyen VC, Dutrillaux B, Bernheim A, Danglot G. Emergence and scattering of multiple neurofibromatosis (NF1)-related sequences during hominoid evolution suggest a process of pericentromeric interchromosomal transposition. Hum Mol Genet. 1997;6:9–16. doi: 10.1093/hmg/6.1.9. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Feinstein P, Mombaerts P. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell. 1999;97:199–208. doi: 10.1016/s0092-8674(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Rouquier S, Taviaux S, Trask B, Brand-Arpon V, van den Engh G, Demaille J, Giorgi D. Distribution of olfactory receptor genes in the human genome. Nature Genet. 1998;18:243–250. doi: 10.1038/ng0398-243. [DOI] [PubMed] [Google Scholar]
- Ruault M, Trichet V, Gimenez S, Boyle S, Gardiner K, Rolland M, Roizes G, De Sario A. Juxta-centromeric region of human chromosome 21 is enriched for pseudogenes and gene fragments. Gene. 1999;239:55–64. doi: 10.1016/s0378-1119(99)00381-9. [DOI] [PubMed] [Google Scholar]
- Ryba N, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Saito H, Mimmack ML, Keverne EB, Kishimoto J, Emson PC. Isolation of mouse vomeronasal receptor genes and their co-localization with specific G-protein messenger RNAs. Brain Res Mol Brain Res. 1998;60:215–227. doi: 10.1016/s0169-328x(98)00183-1. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Stensaas LJ, Lavker RM, Monti-Bloch L, Grossier BI, Berliner DL. Ultrastucture of the human vomeronasal organ. J Steroid Biochem Mol Biol. 1991;39:553–560. doi: 10.1016/0960-0760(91)90252-z. [DOI] [PubMed] [Google Scholar]
- Stern K, McClintock M. Regulation of ovulation by human pheromones. Nature. 1998;392:177–179. doi: 10.1038/32408. [DOI] [PubMed] [Google Scholar]
- Stoddart D. The ecology of vertebrate olfaction. New York: Chapman and Hall; 1980. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirindelli R, Mucignat-Caretta C, Ryba N. Molecular aspects of pheromonal communication via the vomeronasal organ of mammals. Trends Neurosci. 1998;21:482–486. doi: 10.1016/s0166-2236(98)01274-0. [DOI] [PubMed] [Google Scholar]
- Trask BJ. In: Fluorescence in situ hybridization. In: Genome Analysis, a Laboratory Manual. Birren B, Green E, Hieter P, Myers R, editors. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1999. [Google Scholar]
- Wohr G, Fink T, Assum G. A palindromic structure in the pericentromeric region of various human chromosomes. Genome Res. 1996;6:267–279. doi: 10.1101/gr.6.4.267. [DOI] [PubMed] [Google Scholar]
- Wysocki CJ, Lepri JJ. Consequence of removing the vomeronasal organ. J Steroid Biochem Mol Biol. 1991;39:661–669. doi: 10.1016/0960-0760(91)90265-7. [DOI] [PubMed] [Google Scholar]