Abstract
The outbreak of the novel swine-origin H1N1 influenza in the spring of 2009 took epidemiologists, immunologists, and vaccinologists by surprise and galvanized a massive worldwide effort to produce millions of vaccine doses to protect against this single virus strain. Of particular concern was the apparent lack of pre-existing antibody capable of eliciting cross-protective immunity against this novel virus, which fueled fears this strain would trigger a particularly far-reaching and lethal pandemic. Given that disease caused by the swine-origin virus was far less severe than expected, we hypothesized cellular immunity to cross-conserved T cell epitopes might have played a significant role in protecting against the pandemic H1N1 in the absence of cross-reactive humoral immunity. In a published study, we used an immunoinformatics approach to predict a number of CD4+ T cell epitopes are conserved between the 2008–2009 seasonal H1N1 vaccine strain and pandemic H1N1 (A/California/04/2009) hemagglutinin proteins. Here, we provide results from biological studies using PBMCs from human donors not exposed to the pandemic virus to demonstrate that pre-existing CD4+ T cells can elicit cross-reactive effector responses against the pandemic H1N1 virus. As well, we show our computational tools were 80–90% accurate in predicting CD4+ T cell epitopes and their HLA-DRB1-dependent response profiles in donors that were chosen at random for HLA haplotype. Combined, these results confirm the power of coupling immunoinformatics to define broadly reactive CD4+ T cell epitopes with highly sensitive in vitro biological assays to verify these in silico predictions as a means to understand human cellular immunity, including cross-protective responses, and to define CD4+ T cell epitopes for potential vaccination efforts against future influenza viruses and other pathogens.
Keywords: influenza, pandemic, H1N1, informatics, human, T cells, cross-reactive
INTRODUCTION
The recent pandemic swine-origin influenza A virus (S-OIV1; H1N1) presented a major worldwide public health threat during the 2009/2010 influenza season. Fears over this unique virus were fueled, in part, by its strong propensity to replicate and cause more severe disease in naïve laboratory animals than other seasonal H1N1 viruses [1]. Additionally, scientists predicted herd immunity would provide minimal protection against the S-OIV because its envelope hemagglutinin (HA) and neuraminidase proteins, which are the primary targets of influenza-specific antibody, were derived from a classical swine lineage that evolved pronounced antigenic variations following its split from the human seasonal H1N1 viruses during the 1918 pandemic [2–4]. This supposition was confirmed by a series of studies showing most individuals (those under 60 years of age) lack pre-existing cross-protective humoral immunity against the S-OIV and vaccination with the 2008/2009 seasonal trivalent influenza vaccine (TIV) rarely elicited neutralizing antibody responses against this unique H1N1 virus [3, 5].
Despite research studies suggesting the S-OIV is highly pathogenic in laboratory animals and shares few B cell epitopes with most seasonal H1N1 viruses [6], it triggered only mild symptoms in many patients and failed to evolve into a major global pandemic [7]. A potential explanation for this unexpected observation was that pre-existing (memory) influenza-specific T cells generated cross-reactive effector responses against the S-OIV capable of limiting disease severity and virus spread in individuals lacking pre-existing cross-protective antibody. A recent study in support of this theory showed ferrets immunized with the seasonal trivalent influenza vaccine (TIV) were protected from S-OIV-induced disease, but lacked sterilizing immunity against this novel virus [8]. This claim was further substantiated by an in silico evaluation and meta-analyses performed by our group [9] and Greenbaum et al. [6], respectively, demonstrating the pandemic and seasonal H1N1 viruses share highly conserved T cell epitopes.
While our computational tools provided strong evidence for the existence of shared T cell epitopes between the S-OIV and seasonal H1N1 strains, we sought a practical (biological) assessment of these predictions. This was accomplished in the current study by using a highly sensitive in vitro DC-based CD4+ T cell culture assay developed at sanofi pasteur – VaxDesign campus [10, 11] to examine the capacity of human influenza-specific T helper cells from donors not previously exposed to S-OIV to generate cross-reactive effector responses against these immunogens. Though our published in silico study provided a comprehensive evaluation of potentially cross-reactive H1N1 CD4+ and CD8+ T cell epitopes derived from both the influenza HA and NA proteins, we specifically focused the current evaluation on CD4+ T cells against the HA protein of the virus since we were interested in understanding whether vaccination with the seasonal TIV (comprised principally of the HA protein and poorly able to elicit CD8+ T cells) might be capable of generating cross-reactive CD4+ T cell responses against the S-OIV. As such, the target epitopes chosen for the biological evaluation included a series of synthetic HA peptides that are highly conserved between the pandemic H1N1 virus, A/California/07/2009, and the 2009/2010 seasonal H1N1 vaccine strain, A/Brisbane/59/2007, and were predicted by us to generate strong T cell responses by binding promiscuously to eight HLA-DRB1 alleles that cover 99% of the population [9, 12].
Using this approach, we readily generated effector T helper cells against ten HA epitopes that are highly conserved between S-OIV and other H1N1 viruses. Furthermore, we demonstrated the EpiMatrix T cell epitope algorithm was 80–90% accurate in predicting CD4+ T cell epitopes and their HLA-DRB1-dependent response profiles in donors that were chosen at random for HLA haplotype. As a whole, these results strongly support the hypothesis that pre-existing, cross-reactive CD4+ T cell immunity limited the spread and severity of disease resulting from the S-OIV pandemic. As well, this study highlights the utility of coupling robust in vitro immunoassays with computational techniques to better understand human immunity, including cross-protective responses, and to define CD4+ T cell epitopes for potential vaccination efforts against future influenza viruses and other pathogens.
MATERIALS AND METHODS
Human donors and PBMC isolations
PBMCs used in the assays were acquired from normal healthy donors who provided informed consent and were enrolled in a sanofi pasteur – VaxDesign Campus apheresis study program (protocol CRRI 0906009). Blood collections were performed at Florida’s Blood Centers (Orlando, FL) using standard techniques approved by their institutional review board. Within hours following their harvest from the donor, the enriched leukocytes were centrifuged over a ficoll-paque PLUS (GE Healthcare, Piscataway, NJ) density gradient [10, 11]. PBMCs at the interface were collected, washed, and cryopreserved in IMDM media (Lonza, Walkersville, MD) containing autologous serum and DMSO (Sigma-Aldrich, St. Louis, MO). All PBMCs used in this assay were collected from subjects in our donor program prior to the outbreak of the S-OIV in Florida in May of 2009. The ages of the donors (in years) follows: 1142 (25), 1010 (21), 940 (25), 923 (25), 720 (46), 548 (23), 208 (39), and 182 (40).
Peptides and reagents
Synthetic peptides matching sequences of the HA protein from S-OIV were generated by BioSynthesis, Inc. (Lewisville, TX) to a purity of at least 95%. (The sequences can be found in Table 1.) The tetanus toxoid P30 peptide (TT947–967; FNNFTVSFWLRVPKVSASHLE) was synthesized by Elim Biopharmaceuticals (Hayward, CA) to a purity of at least 95%. The influenza A/Brisbane/59/2007 (H1N1) hemagglutinin (Brisbane HA) protein and influenza A/California/07/2009 (H1N1) hemagglutinin (California HA) protein were purchased from Protein Sciences (Meriden, CT). The Fluvirin 2008–2009 TIV and influenza A H1N1 2009/2010 monovalent S-OIV vaccine were procured from Novartis International AG (Basel, Switzerland).
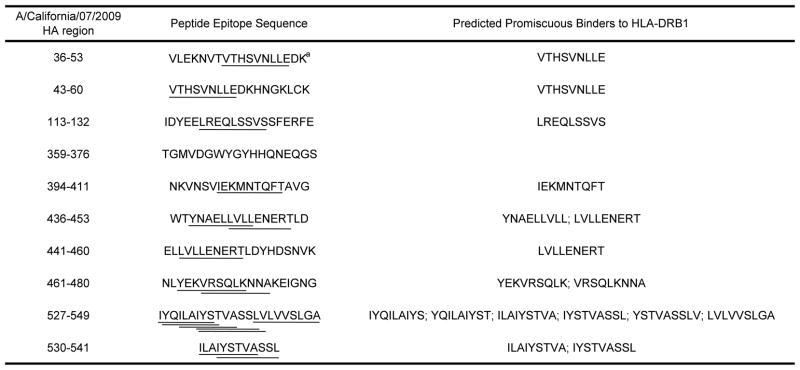
Table 1.
A/California/07/2009 HA peptide sequences included in this evaluation.
|
Lines are used to highlight core 9-mer regions within the peptides that have been predicted to bind strongly to one or more HLA-DRB1 alleles.
Generation of cytokine-derived dendritic cells
Dendritic cells (DCs) were prepared using our published methodology [10]. Briefly, monocytes were purified from total PBMCs by positive magnetic bead selection (Miltenyi Biotec, Cologne, Germany), cultured for 7 days in X-VIVO 15 (Lonza) serum-free media supplemented with GM-CSF (R&D Systems, Minneapolis, MN) and IL-4 (R & D Systems), and then harvested for assay usage. In all assay conditions described below where vaccines or purified proteins were used as antigens, the DCs were pulsed for at least 2 hrs with the immunogen and washed prior to being used in the assay. In cases where synthetic peptides were used to stimulate T cell responses, the epitopes were added directly to the assay wells (the DCs were not pre-pulsed).
IFNγELISPOT
CD4+ T cells were enriched from frozen PBMCs by negative magnetic bead selection (Miltenyi Biotec) and cocultured with autologous DCs at a 10:1 ratio in 96-Well Multiscreen HTS plates (Millipore, Billerica, MA) that had been pre-coated with anti-human IFNγ Ab (BD Pharmingen). As noted above, some DCs were pre-treated with a 1:500 dilution of the 2008/2009 seasonal TIV (which contained the Brisbane H1N1 virus) or S-OIV vaccine formulation or 1.8 μg/ml Brisbane HA or California HA proteins. Otherwise, the DCs were added to the T cells in the presence of 1.0 μg/ml pooled or individual S-OIV HA peptides or the negative control tetanus (TT947–967) epitope. After a 24-hr coculture, the plates were processed using the IFNγ ELISPOT kit (BD Pharmingen) and quantified by colorimetric evaluation using hardware and software analysis from AID EliSpot (Strassberg, Germany).
CD4+ T cell stimulation assay
CD4+ T cell stimulation assays were performed using protocols established at sanofi pasteur – VaxDesign Campus [10, 11]. Briefly, autologous CD4+ T cells were enriched from frozen PBMCs by negative magnetic bead selection (Miltenyi Biotec) and then co-cultured with autologous DCs that either had been untouched (control wells) or pre-pulsed for at least 2 hrs with 1.8 μg/ml Brisbane HA protein. After a 12-day incubation period, the lymphocytes were harvested and evaluated for effector activity in an intracellular cytokine staining assay (ICCS) where fresh autologous DCs served as APCs. For the readout/ICCS assay, the DCs were pre-pulsed with the Brisbane HA protein or cultured with the T cells directly in the presence of 1 μg/ml synthetic peptide. The T cells and target DCs were co-cultured for 7 h; 1 μg/ml brefeldin A (Sigma-Aldrich) was added to the wells during the final 5 h of culture to prevent protein egress from the Golgi apparatus. Following the incubation period, the cells were labeled with the Live/Dead Fixable Stain Kit (Invitrogen, Carlsbad, CA), treated with cytofix/cytoperm and perm/wash reagents from BD Biosciences (San Jose, CA), and then labeled with eBioscience (San Diego, CA) antibodies specific for human CD4 (SK3), IFNγ (B27), and CD154 (TRAP1). The samples were acquired on an LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Immunoinformatics analyses
EpiMatrix, a T-cell epitope mapping algorithm developed by EpiVax, screens protein sequences for 9–10 amino acid peptide segments predicted to bind to one or more MHC alleles [13, 14]. EpiMatrix uses the pocket profile method, which was first described by Sturniolo and Hammer [15], to make these predictions. For reasons of efficiency and simplicity, the predictions are limited to the eight most common HLA-DRB1 class II alleles [16]. Raw EpiMatrix values are normalized with respect to a score distribution derived from a large set of randomly generated peptide sequences. Any peptide/MHC allele scoring above 1.64 on the EpiMatrix “Z” scale (the top ~5% of the distribution of a random peptide set) has a significant chance of binding to the MHC molecule for which it was predicted; peptides scoring above 2.32 on the scale (the top ~1%) are extremely likely to bind. The scores of most well-known T cell epitopes fall within this range [17–19]. After screening for putative MHC class II epitopes, we used the ClustiMer algorithm to identify protein segments with high T cell epitope density. 93% of these epitope predictions have proven to be accurate in prospective biological evaluations [17–21].
individualized T cell epitope measure (iTEM)
The iTEM, or individualized T cell epitope measure, predicts how individuals will respond to specific T cell epitopes. Our original method for calculating iTEM scores was published and validated [22]; here, we used an improved algorithm, which produces even better predictions of subject-specific immune responses, to predict S-OIV HA epitope immunogenicity. Briefly, an iTEM score is generated by first evaluating a peptide for predicted binding to a subject’s HLA-DRB1 alleles using the EpiMatrix system (see above). Combining the EpiMatrix scores for each peptide-HLA-DRB1 allelic combination in the haplotype yields an iTEM score for a particular individual. This is achieved by ranking EpiMatrix scores for each allele by magnitude: the highest 9-mer score is weighted 100% and each subsequent positive score for the same allele is underweighted by a factor of 1/n, where n is the rank. This discount factor addresses the fact that a given HLA class II molecule’s ability to bind only one 9-mer peptide at a time, which results in lower-affinity peptides occupying a given peptide binding groove for comparatively less time than higher-affinity peptides.
In a series of retrospective studies completed during the development of the iTEM formula, a value of 2.06 gave the best balance between sensitivity and specificity [22]; as such, iTEM responses of 2.06 or higher in the current study were considered positive. The iTEM scores calculated for each of the peptide-HLA-DRB1 pairs were then compared to the in vitro T cell assay results generated using methods described above. (Specific CD4+ T cell responses 2-fold greater than the background (no antigen) condition were considered responsive in the in vitro stimulation assay.) Based on this evaluation, the peptide-HLA pairs were divided into one of four categories (true positives, true negatives, false positives, and false negatives). In this context, true-positive peptides were predicted and in vitro-validated as immunogenic, while true-negative peptides were predicted and biologically validated to be non-immunogenic. False-negative peptides were predicted to be non-immunogenic, yet produced a positive response; false-positive peptides were predicted to be immunogenic but produced no response in the in vitro T cell assay.
RESULTS
In an effort to better understand whether pre-existing cellular immunity might have generated protective responses against the S-OIV, we examined the capacity of human influenza-specific T helper cells to elicit cross-reactive effector responses against highly conserved epitopes from this novel virus. We specifically chose for this evaluation frozen PBMCs from our stock that had been collected from eight subjects prior to the emergence of the S-OIV in Florida in May of 2009 to try to eliminate the possibility they had been exposed to this virus. The T cell epitopes chosen for this study met two criteria: First, they contained one or more MHC class II-restricted core epitopes that are highly conserved between the S-OIV and Brisbane H1N1 viruses and were predicted to bind strongly to HLA-DRB1 molecules [9]. Second, they share sequence homology with CD4+ T cell epitopes identified in various seasonal influenza H1N1 viruses [6, 9]. This evaluation also included one additional published MHC class II peptide, HA359–376, that is cross-conserved between the pandemic and seasonal H1N1 viruses [6], but was not predicted by us to contain any core HLA-DRB1-restricted epitopes [9]. The sequences and corresponding promiscuous core 9-mer MHC class II-binding sequences are shown in Table 1.
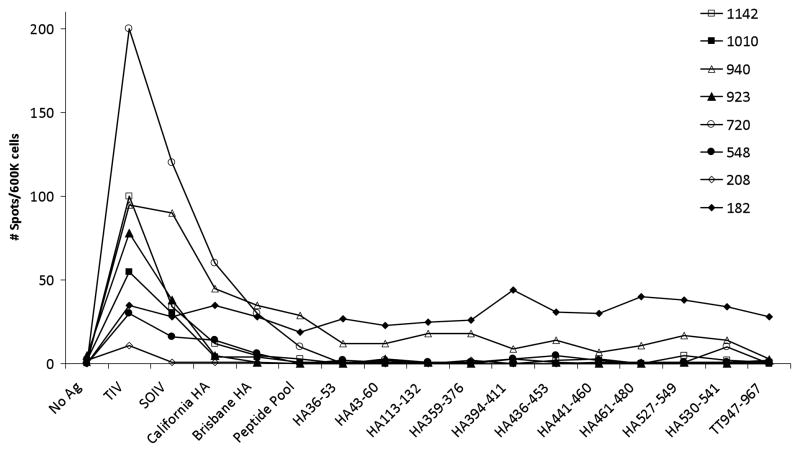
In an attempt to directly quantify the frequency of circulating influenza-specific CD4+ T cells capable of cross-reacting against S-OIV in eight donors, we stimulated purified lymphocytes with DCs for 24 hr in an IFNγ ELISPOT assay. (The DCs were either pre-pulsed with purified protein or peptides were added directly to the assay well.) By design, this short 1-day antigen encounter is intended to trigger cytokine production by antigen-specific T cells without inducing any cell divisions that would otherwise alter the precursor frequency determination. Using the seasonal 2008/2009 seasonal TIV (Brisbane H1N1) as a benchmark in this analysis, we were not surprised to find the frequency of the responding T cell population varied by as much as 10-fold (20–200 TIV-specific CD4+ T cells/600,000 total CD4+ T cells) since each donor’s unique genetic background and immune history with influenza would impact the magnitude of the responding population (Figure 1). The S-OIV vaccine also generated a detectable CD4+ T cell response in all but one donor, though in most cases this vaccine elicited a lower-magnitude response than the TIV. The purified Brisbane and S-OIV HA proteins elicited even weaker, but reasonably equivalent, CD4+ T cell populations in only a subset of the donors (Figure 1). (This might reflect a strong potential for T cell cross-reactivity between the Brisbane and pandemic H1N1 viruses.) Taken as a whole, this hierarchy of responses is perhaps not surprising since the three-component (H3N2, influenza B, and seasonal H1N1) seasonal TIV offered a broader complement of antigens for T cell recognition than the single-component S-OIV prophylactic and the purified proteins lack the inflammatory potential of the formulated vaccines. Despite our observation of positive and detectable CD4+ T cell responses in ELISPOT wells stimulated with vaccines and purified proteins, the T helper cell responses elicited by the single or pooled synthetic peptides were modest at best, with only two donors generating populations with measurable significance over the background (no antigen) control (Figure 1).
Figure 1.
Demonstration of influenza-specific CD4+ T cell cross-reactivity against the S-OIV. CD4+ T cells were isolated from frozen PBMCs and cultured with purified autologous DCs for 24 hrs. Where indicated, the DCs were left untouched or pre-pulsed overnight with vaccine or protein. Peptides, individually or together as a pool, were added directly to the DC/T cell coculture well. Thereafter, antigen-specific T cells were detected by IFNγ ELISPOT assay. Eight donors were included in this evaluation.
We believe the ELISPOT results described above provide compelling evidence that pre-existing influenza-specific CD4+ T cells can generate cross-reactive T helper cell responses against the novel S-OIV. However, we were unable to use this approach to examine the fine specificity of the circulating T helper cells on a single-epitope basis because of the low frequency of influenza-specific CD4+ T cells in PBMC samples evaluated directly ex vivo. To circumvent this issue, we performed a highly sensitive DC-based T cell coculture assay developed at sanofi pasteur – VaxDesign campus [10, 11] to amplify the influenza-specific population prior to evaluating the fine epitope specificities of the virus-specific lymphocytes. Briefly, purified CD4+ T cells from the same eight donors described in Figure 1 were stimulated for 12 days with autologous DCs that had been pre-pulsed with the Brisbane HA protein. (DCs not pulsed with the foreign protein served as a negative control.) Thereafter, the cultured T cells were harvested and evaluated for their potential to respond to the same HA protein or individual S-OIV HA peptide epitopes in a short-term (7-hr) ICCS assay. In this way, we could readily address whether the 2009/2010 seasonal TIV Brisbane H1N1 strain could amplify T helper cells against specific peptide epitopes derived from the S-OIV HA protein.
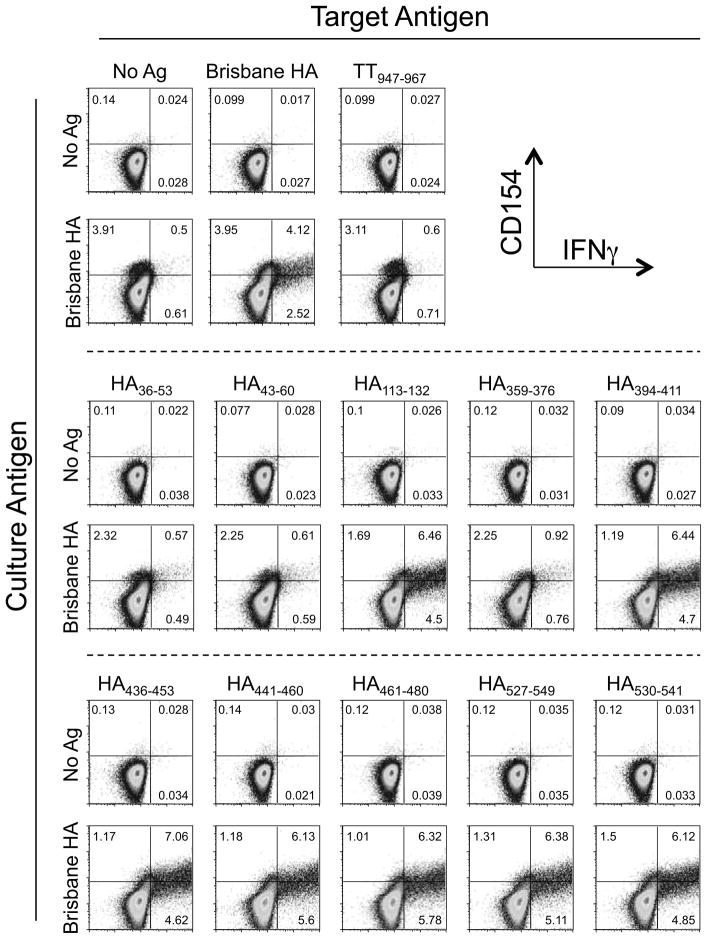
Focusing on donor 940 as a representative example of the datasets generated with this approach, we found the T helper cells from this subject were highly active against the Brisbane HA protein, eliciting a CD4+CD154+IFNγ+ T cell population against the matched target that was more than 8-fold over background in the no-antigen control target (Figure 2). (Of note, we use the dual expression of CD154 and IFNγ as a stringent readout of antigen specificity in the ICCS assay.) Likewise, this donor also responded vigorously against several of the pandemic H1N1 peptides, but not a universal T cell epitope from tetanus toxoid (TT947–967), which served as a negative control in these experiments (Figure 2). This latter result indicates the in vitro proliferative response was specific to the influenza protein and not global/non-specific in nature.
Figure 2.
Demonstration of robust cross-reactive influenza-specific CD4+ T cells effector responses against predicted S-OIV HA epitope sequences. DCs were pulsed with no antigen or Brisbane HA overnight and then co-cultured with purified autologous CD4+ T cells. After 12 days, the cultures were harvested and restimulated with fresh autologous DCs that had, in some cases, been pre-pulsed overnight with Brisbane HA. Some assay wells were pulsed directly with S-OIV HA peptides or the negative control TT peptide, TT947–967. After 7 hr, the lymphocytes were evaluated by flow cytometry using a conventional intracellular staining protocol. The dot plots show live/CD4-gated lymphocytes.
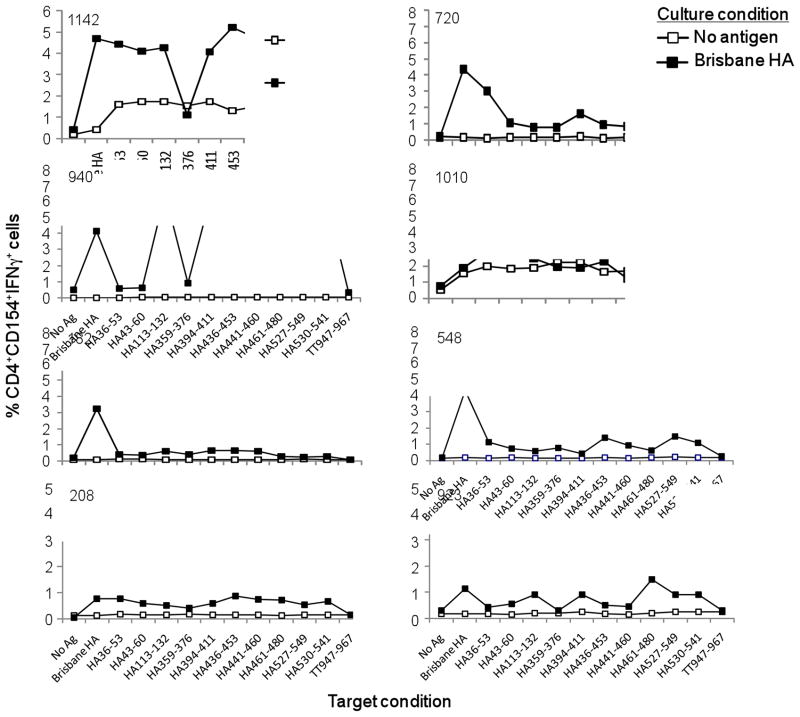
Extending this evaluation to include all eight donors, which are shown graphically in Figure 3, we found a considerable (up to 14-fold) increase in the frequency of Brisbane HA-specific CD4+ T cells in the antigen-stimulated cultures (closed symbols) in seven out of eight donors, which clearly illustrates the capacity of the lymphocytes to respond to specific antigen stimulation in the in vitro CD4+ T cell assay. As well, the fact that the S-OIV HA peptides could be used to elicit strong T helper cell responses from cultures stimulated with the Brisbane HA protein suggest these highly conserved epitopes are generated during the natural processing of the full-length HA for MHC class II presentation since the whole protein was used as an antigen source in the original 12-day stimulation assay. Similar to what we showed for donor 940 (Figure 2), the universal tetanus peptide generated no response over background in any subjects included in this evaluation (Figure 3). This point, taken together with our observation that not all influenza peptides elicited T cell activity in all donors, suggests the virus-specific CD4+ T cell responses are valid. It is notable that nine of the S-OIV HA peptide sequences included in this study were predicted to have a strong capacity to induce potent CD4+ T cell responses via their capacity for high-affinity and promiscuous interactions with multiple HLA-DRB1 alleles [17]. Therefore, we were not surprised to find most of the peptides elicited positive (2-fold over background/no-antigen control) responses in the majority of donors shown here (Figure 3). Given that these peptide sequences were chosen because of their strong homology between the Brisbane H1N1 and pandemic H1N1 viruses, these results suggest vaccination with the 2009/2010 seasonal TIV could have elicited potentially cross-protective CD4+ T cell responses against the S-OIV.
Figure 3.
Highly conserved S-OIV HA peptides elicit strong CD4+ T cell responses from donors not previously exposed to the pandemic H1N1 virus. CD4+ T cells and autologous DCs were cocultured and assessed by ICCS using the method described in Figure 2. The raw data, as generated in Figure 2, was plotted as line graphs for all eight donors.
To examine the fidelity of the computational tools used by Epivax to map T cell epitopes and MHC class II restriction profiles, we directly compared the human T cell data shown in Figure 3 with our published S-OIV HA peptide binding predictions [17]. To begin, each core 9-mer sequence was evaluated for its capacity to engage the HLA-DRB1 alleles expressed by the donors in this study; those sequences expected to have a high probability of inducing a response (binding potential ranked in the top 5th percentile) were marked with a large X in Table 2. We also assessed the HLA-DRB1 binding potential of each full-length peptide, with 5th percentile values indicated by a small X in Table 2. It is notable that peptide HA359–376, which is not predicted to have any promiscuous core 9-mer peptides (Table 1), nevertheless might have the potential to induce responses by engaging four predominant HLA-DRB1 molecules (Table 2).
Table 2.
Predicted binding of the S-OIV HA peptide sequences to particular HLA-DRB1 alleles.
| A/California/07/2009 HA region | Peptide Epitope Sequence | Predicted binding restriction to HLA-DRB1 alleles (top 5% score) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 101 | 301 | 401 | 701 | 801 | 1101 | 1301 | 1501 | ||
| 36–53 | VLEKNVTVTHSVNLLEDKa | x | X | X | x | X | X | x | |
| 43–60 | VTHSVNLLEDKHNGKLCK | X | X | X | X | ||||
| 113–132 | IDYEELREQLSSVSSFERFE | X | X | X | X | X | X | X | |
| 359–376 | TGMVDGWYGYHHQNEQGS | x | x | x | x | ||||
| 394–411 | NKVNSVIEKMNTQFTAVG | X | X | X | X | x | X | ||
| 436–453 | WTYNAELLVLLENERTLD | X | X | X | X | X | X | X | X |
| 441–460 | ELLVLLENERTLDYHDSNVK | X | x | X | X | X | X | X | |
| 461–480 | NLYEKVRSQLKNNAKEIGNG | X | X | X | X | X | X | X | X |
| 527–549 | IYQILAIYSTVASSLVLVVSLGA | X | X | X | X | X | X | X | X |
| 530–541 | ILAIYSTVASSL | X | X | X | X | X | |||
Whole-peptide binding predictions for HLA-DRB1 alleles are shown. Peptides are scored according to their potential to bind a particular allele. Scores in the top 5th percentile are marked with an X: a large X denotes binding of the peptide is enhanced by the core 9-mer binding peptides shown in Table 1; a small X indicates predicted binding without the contribution of a core 9-mer peptide.
Next, we directly correlated these in silico predictions with the in vitro biological data shown in Figure 3. When compiled together into Table 3, we found a strong correlation between the 5th percentile binding predictions (gray shading) and the true capacity of the peptides to induce specific T helper cell responses at least 2-fold over background (marked with an X). In fact, 95% (62 out of 65) of the total positive biological responses were predicted accurately at the 5th percentile ranking. As well, it is clear from this analysis that a 1% cutoff provides a very conservative prediction of influenza-specific CD4+ T cell epitopes since it markedly underestimated positive responses in the T cell assays. Increasing the number of predictions to include those in the 10th percentile slightly increased the number of hits (64 out of 65), but also introduced additional false-positives (Table 3). Of note, the fact that 8 of the 9 S-OIV HA peptides included in this evaluation were predicted to induce positive CD4+ T cell responses in the majority of the eight subjects included in this study reflects our interest in choosing peptides with highly promiscuous core 9-mer sequences that should be able to interact with most individuals in a population.
Table 3.
Comparison of predicted and actual S-OIV HA peptide immunogenicity.
| Donor ID | HLA-DRB1 alleles | A/California/7/2009 HA Peptide Sequence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HA36–53 | HA43–60 | HA113–132 | HA359–376 | HA394–411 | HA436–453 | HA441–460 | HA461–480 | HA527–549 | HA530–541 | |||
| 1st percentile | 1010 | 1301, 1302 | Xa | X | X | X | X | X | X | X | X | |
| 720 | 0101, 0701 | X | X | X | X | X | X | X | X | X | X | |
| 1142 | 0301, 0407 | X | X | X | X | X | X | X | X | X | X | |
| 182 | 0101, 0901 | X | X | X | X | X | ||||||
| 548 | 0701, 1302 | X | X | X | X | X | X | X | X | X | X | |
| 940 | 1201, 1501 | X | X | X | X | X | X | X | ||||
| 923 | 0101, 0103 | X | X | X | X | X | ||||||
| 208 | 0401, 0701 | X | X | X | X | X | X | X | X | X | X | |
| 5th percentile | Donor ID | HLA-DRB1 alleles | A/California/7/2009 HA Peptide Sequence | |||||||||
| HA36–53 | HA43–60 | HA113–132 | HA359–376 | HA394–411 | HA436–453 | HA441–460 | HA461–480 | HA527–549 | HA530–541 | |||
| 1010 | 1301, 1302 | X | X | X | X | X | X | X | X | X | ||
| 720 | 0101, 0701 | X | X | X | X | X | X | X | X | X | X | |
| 1142 | 0301, 0407 | X | X | X | X | X | X | X | X | X | X | |
| 182 | 0101, 0901 | X | X | X | X | X | ||||||
| 548 | 0701, 1302 | X | X | X | X | X | X | X | X | X | X | |
| 940 | 1201, 1501 | X | X | X | X | X | X | X | ||||
| 923 | 0101, 0103 | X | X | X | X | X | ||||||
| 208 | 0401, 0701 | X | X | X | X | X | X | X | X | X | X | |
| 10th percentile | Donor ID | HLA-DRB1 alleles | A/California/7/2009 HA Peptide Sequence | |||||||||
| HA36–53 | HA43–60 | HA113–132 | HA359–376 | HA394–411 | HA436–453 | HA441–460 | HA461–480 | HA527–549 | HA530–541 | |||
| 1010 | 1301, 1302 | X | X | X | X | X | X | X | X | X | ||
| 720 | 0101, 0701 | X | X | X | X | X | X | X | X | X | X | |
| 1142 | 0301, 0407 | X | X | X | X | X | X | X | X | X | X | |
| 182 | 0101, 0901 | X | X | X | X | X | ||||||
| 548 | 0701, 1302 | X | X | X | X | X | X | X | X | X | X | |
| 940 | 1201, 1501 | X | X | X | X | X | X | X | ||||
| 923 | 0101, 0103 | X | X | X | X | X | ||||||
| 208 | 0401, 0701 | X | X | X | X | X | X | X | X | X | X | |
Epitopes predicted to elicit a positive response in a particular donor in either the top first, fifth, or tenth percentile are indicated by gray shading. Those peptides that generated positive T helper cell responses at least 2-fold over background in the biological assays are indicated by an X.
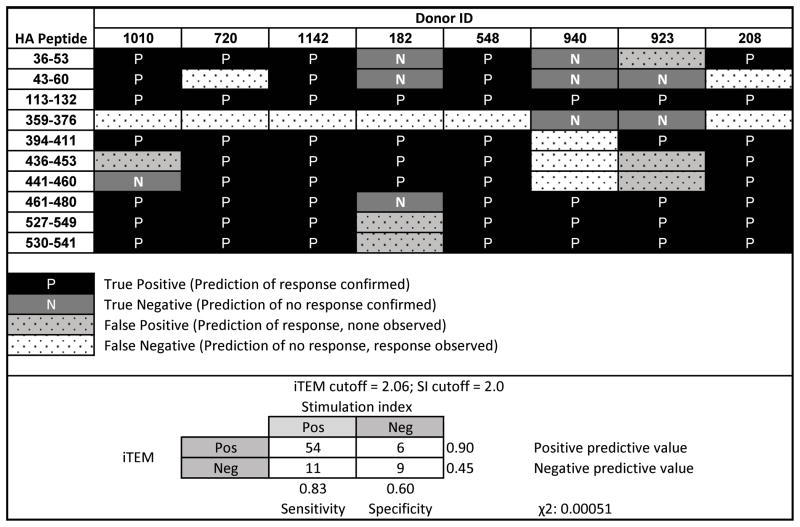
Broadening this in silico evaluation, we also used the iTEM algorithm (described in the Materials and Methods section) to calculate the probability that a given peptide sequence will elicit a CD4+ T cell immune response in a particular donor. This approach provides a more comprehensive forecast of T cell epitope immunogenicity since it takes into account the binding profile of all the predicted epitopes within a particular peptide sequence against the collective whole of an individual’s HLA-DRB1 haplotype. For this study, iTEM scores were calculated for each peptide-donor combination, and those yielding values greater than 2.06 (a predetermined threshold for this technique [22]) were evaluated for correlation with positive CD4+ T cell responses (2-fold over background) in the in vitro assays (Tables 4 and 5). Of the 80 peptide-HLA-DRB1 (8 donors × 10 peptides) combinations derived from this study, 65 of these yielded a significant CD4+ T cell response (SI =>2). Of these 65 combinations, 54 (83%) had iTEM scores > 2.06, which meant they were predicted to elicit significant T cell responses (Tables 4 and 5). In the 15 instances where the peptide-HLA combination did not generate a significant T cell response in the in vitro assay, 9 of these (60%) were also predicted to fail to generate responses in the iTEM analysis (score <2.06). One interesting outlier is HA359–376, a confirmed MHC class II peptide [6] that was not predicted to contain any core 9-mer binding motifs (Tables 1 and 2). While the in silico binding scores for this peptide fall below the threshold of significance, it did produce a modest response in vitro that was responsible for 6 of 11 false negative predictions. Upon further evaluation, we noted the strongest-scoring 9-mer within this sequence, YHHQNEQGS, contains a glutamic acid at position 6 that negatively impacts the Epimatrix scores since it is typically highly disfavored in this position. In this case, however, the adverse effect of the glutamic acid must be outweighed by the positive effects related to the surrounding amino acids. Taken together, the results of this study confirm an iTEM score greater than 2.06 provides a strong indicator of the capacity of a particular peptide to elicit a specific CD4+ T cell response. While it is possible for peptides with iTEM scores less than 2.06 to trigger responses, setting a high/conservative cutoff value reduces the chance the algorithm will be used to generate false-positive predictions. Indeed, the fact that 54 out of 60 (90%) predicted positive responses were confirmed by the biologic assessments provided in this analysis (Table 5) is perhaps the most important metric of success since one would not want to overestimate the capacity of the population to respond to a peptide vaccine.
Table 4.
Correlation between in silico prediction of donor responsiveness and in vitro biological assay results.
| Peptide | Donor ID
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1010a | 720 | 1142 | 182 | 548 | 940 | 923 | 208 | |||||||||
| SI | ITEM | SI | ITEM | SI | ITEM | SI | ITEM | SI | ITEM | SI | ITEM | SI | ITEM | SI | ITEM | |
| HA36–53 | 3.95 | 4.36 | 23.23 | 4.93 | 10.26 | 5.26 | 1.95 | 1.72 | 5.38 | 5.39 | 1.14 | 1.75 | 1.40 | 3.44 | 11.47 | 6.07 |
| HA43–60 | 5.01 | 4.36 | 8.15 | 0.00 | 9.53 | 5.15 | 1.75 | 0.00 | 3.67 | 2.18 | 1.22 | 0.00 | 1.83 | 0.00 | 8.82 | 1.66 |
| HA113–132 | 3.31 | 4.88 | 5.92 | 2.11 | 9.86 | 4.15 | 3.05 | 2.11 | 2.86 | 2.44 | 12.92 | 3.09 | 3.00 | 4.22 | 7.50 | 2.30 |
| HA359–376 | 2.64 | 0.00 | 6.00 | 1.70 | 2.56 | 1.68 | 2.00 | 0.00 | 3.76 | 1.70 | 1.84 | 0.00 | 1.00 | 0.00 | 6.32 | 1.70 |
| HA394–411 | 2.57 | 3.76 | 12.46 | 4.13 | 9.42 | 2.98 | 3.10 | 2.31 | 2.14 | 3.70 | 12.88 | 1.83 | 3.00 | 4.62 | 8.68 | 4.80 |
| HA436–453 | 1.65 | 3.64 | 7.00 | 7.44 | 12.09 | 4.26 | 3.10 | 3.35 | 6.71 | 5.91 | 14.00 | 1.84 | 1.67 | 6.69 | 12.79 | 6.27 |
| HA441–460 | 1.59 | 0.00 | 6.23 | 5.09 | 10.95 | 4.75 | 3.00 | 2.36 | 4.43 | 2.73 | 12.26 | 1.84 | 1.50 | 4.72 | 11.03 | 5.76 |
| HA461–480 | 4.81 | 8.89 | 4.85 | 5.09 | 10.65 | 5.90 | 1.40 | 2.04 | 2.76 | 7.49 | 12.64 | 3.06 | 4.93 | 4.08 | 10.74 | 5.17 |
| HA527–549 | 3.77 | 7.72 | 5.62 | 11.29 | 9.88 | 6.94 | 1.15 | 5.10 | 7.19 | 10.05 | 12.76 | 5.55 | 3.00 | 10.20 | 7.94 | 11.26 |
| HA530–541 | 5.92 | 6.01 | 7.46 | 6.77 | 2.16 | 2.22 | 1.35 | 3.19 | 5.33 | 6.59 | 12.24 | 4.11 | 3.00 | 6.38 | 9.85 | 5.80 |
For each donor/peptide combination, the actual T helper cell response stimulation indices (SI) and corresponding predicted immunogenicity values (iTEM) are shown.
Table 5.
Analysis of the efficacy of in silico predictions of T helper cell responsiveness against the S-OIV.
|
DISCUSSION
In light of the fact that pre-existing antibodies offered only limited protection against the pandemic S-OIV of 2009, we and others postulated cross-reactive T lymphocytes elicited by the 2009/2010 seasonal TIV or prior exposure to other H1N1 viruses might have played an important role in limiting disease and the spread of this novel virus by generating direct antiviral effector functions or accelerating the induction of naïve virus-specific B cell responses against novel S-OIV antigens upon subsequent infection. Shortly after the emergence of this virus in 2009, we used in silico techniques to define T cell epitopes that are highly conserved between the S-OIV and the seasonal vaccine strain, Brisbane H1N1, and were predicted to bind promiscuously to the most prominent HLA-DRB1 alleles [17]. In the current study, by performing a biological evaluation of cross-reactive S-OIV-specific CD4+ T cells in the circulation of eight donors chosen at random for HLA-DR genotype, we demonstrated that the computational methods were highly accurate in defining CD4+ T cell epitopes that were broadly reactive, i.e., capable of eliciting responses from nearly all the donors included in this evaluation. In the process of completing this evaluation, we also confirmed that pre-existing CD4+ T cells can generate cross-reactive effector responses against the S-OIV virus, which bolsters the argument that cellular immunity might have engendered some protection against disease resulting from pandemic H1N1 infection.
Our demonstration of cross-reactive T helper cells against S-OIV is consistent with a series of studies that have evaluated potentially cross-protective influenza-specific T cell immunity against this novel virus. For example, two independent studies demonstrated CTLs and CD4+ T cells raised against the seasonal H1N1 viruses, A/Brisbane/59/2007 and A/New Caledonia/20/99, respectively, were capable of responding against whole protein antigens from the S-OIV [23, 24]. In addition, a study by Ge et al. [25] provided evidence of cross-reactive human T helper cell responses against defined epitopes from the HLA-DR4 molecule. Our study was complementary to these prior reports, but differed from them in two ways. First, we were particularly interested in addressing whether vaccination with the conventional seasonal TIV might elicit protective immunity against S-OIV, so we focused our evaluation on cross-reactive T helper cells specific for the primary vaccine antigen, HA. Second, we too were interested in assessing peptide-specific T cell responses, but did not limit our evaluation to any particular HLA-DR haplotype. In fact, as mentioned above, we targeted our evaluation of cross-reactive T helper cell epitopes against a panel of eight HLA-DR allele supertypes that cover 99% of the population [9].
While there is a long history of research examining cross-reactive T cells against influenza, most of these studies targeted the non-structural/more conserved proteins of the virus, such as NP, M1, and PB1, since they offer the greatest potential for eliciting long-lived cross-protective immunity against influenza. For example, recent studies have provided evidence for the existence of T cells reactive against several non-structural proteins from seasonal virus strains that can generate cross-protective responses against avian influenza (H5N1) [26, 27]. As well, individuals not previously exposed to H5N1 viruses were shown to exhibit cross-reactive T cell against both the structural and non-structural proteins from an avian (Hong Kong H5N1) influenza strain [28]. As mentioned above, we specifically focused our evaluation on the structural HA protein of the S-OIV because it is the primary component of the seasonal influenza vaccine and, thus, is likely the primary target of T cell immunity following immunization with a split virus vaccine. While HA is not a dominant target of cellular immunity during natural infection, recent studies with a DR1-transgenic mouse model and tetramer staining of human peripheral blood leukocytes suggest the presence of shared HA T cell epitopes between seasonal H1N1 viruses and the S-OIV [29–31].
Although CD8+ T cells are considered a key player in anti-influenza cellular immunity, we limited our investigation to CD4+ T cells because we were specifically interested in whether the 2009/2010 seasonal TIV, which is poor at generating CD8+ T cell immunity, had the capacity to elicit a T helper cell response against the S-OIV. Though we cannot make definitive statements from our data regarding the role of virus-specific CD4+ T cells in limiting S-OIV infection, a strong body of experimental evidence suggests influenza-specific T helper cells can limit influenza disease, particularly in the absence of an efficacious humoral response. A number of conclusions can be drawn from these published studies [8, 32–42]: (1) the rate of viral clearance upon secondary infection slows considerably, beyond that seen during the primary infection, in the absence of functional memory CD4+ T cells, (2) T cell help is required for the generation of high virus-specific IgG antibody titers, (3) vaccine efficacy is improved when cross-reactive helper T cell populations are present from prior infection and/or vaccination, (4) memory T helper cells specific for a previous influenza strain contribute to cross-strain antibody responses and confer direct protection against heterologous infection, and (5) effective vaccination can elicit protective cellular immune responses capable of secreting cytokines and cytolytic activity. For these reasons, we believe pre-existing CD4+ T cells elicited against the 2009/2010 seasonal TIV could have the capacity to limit S-OIV disease severity.
Our goal, as we embarked on this study, was to employ a short-term (24-hr) IFNγ ELISPOT assay to evaluate the frequency of circulating human T cells capable of responding against the novel S-OIV in donors with no prior exposure to this novel virus. Though we were quite successful using this highly sensitive assay to evaluate T cell responses when the autologous DC targets were pulsed with vaccine formulations or intact viral HA protein (each contain a multitude of potential T cell epitopes), we failed to detect specific T cell effectors in five of eight donors when single peptide antigens were added to the assay wells (Figure 1). While this result might be taken to suggest the peptides do not represent dominant HA epitopes, we do not think it is possible to broadly judge the relative strength of the peptides in this evaluation because the sum of the individual epitopes also exceeded the whole protein and pooled peptide responses by at least 10-fold for donors 182 and 208 (Figure 1). Furthermore, these results are consistent with our observations in other experimental systems that the sum of individual peptide-specific responses often does not match the total T cell response (A. DeGroot et al., unpublished results). Notwithstanding these observations, it is very likely the ten peptides included in this evaluation represent only a subset of the potentially cross-reactive peptides that are shared between Brisbane and California HA proteins. In fact, we chose for this evaluation only those peptides that contained sequences we had predicted were the most broadly cross-reactive (amongst multiple HLA class II alleles) and highly conserved between the Brisbane and California H1 viruses [9].
Our inability to detect individual peptide-specific responses by ELISPOT is not surprising given other researchers have experienced similar difficulties using this technique to evaluate direct ex vivo T cell responses in humans [43, 44]. In fact, past studies aimed at evaluating individual peptide-specific T cell responses from humans have resorted to long-term in vitro stimulation periods (up to 10 day) to trigger the proliferation/accumulation of the specific lymphocytes to a number where they could be detected by ELISPOT assay [28]. In a similar approach, we employed a highly sensitive T cell stimulation/challenge assay developed in our laboratory to evaluate S-OIV peptide-specific CD4+ T cell responses. This technique has been successfully employed to evaluate primary and recall T cell responses against a variety of protein antigens and a formulated yellow fever vaccine [10, 11]. As well, we believe this was a critical component of the study presented here because it addresses one of the limitations of in silico epitope-mapping techniques, namely, that they predict epitopes based on the capacity of a peptide sequence to bind MHC, but do not evaluate the capacity of the MHC machinery of cells to yield that particular sequence for MHC presentation. To this point, the two-stage stimulation technique – where one round of DCs are pulsed with the HA protein (12-day coculture), and then the second round of DCs present the individual peptides (ICCS analysis) – provides direct evidence that the native protein was processed through the DCs machinery to give rise to the peptide epitopes of interest in this evaluation. It is notable that sum of the T cell responses elicited by the individual peptides often exceeded the response elicited by the native HA protein in the ICCS assay (Figures 2 and 3), though this observation is perhaps not surprising since pulsing DCs with the native HA protein may be inefficient compared to directly loading the APCs with high concentrations of individual peptides. Nevertheless, given the strength of the peptide-specific responses in the ICCS assay against either the whole protein or individual epitopes, we think it is safe to conclude the DCs did efficiently process the HA protein to generate the peptide epitopes included in this evaluation.
As demonstrated here, bioinformatics offers a powerful approach for predicting T cell epitopes, though the critical step for epitope-driven vaccine design remains the in vitro and in vivo validation of such predictions. Towards this goal, the algorithm used here has been successfully applied to the analysis of previously published epitopes and in the prospective selection of peptides from HIV, Mycobacterium tuberculosis, Tularemia, and vaccinia virus [21, 45–48]. In the current study, we sought to comprehensively assess the reactivity of influenza CD4+ T cell epitopes as a function of individuals’ specific HLA haplotypes, which can be predicted via iTEM calculations. Here, we utilized a refined methodology in which iTEM scores more closely correlate with in vitro responses to successfully model the wide spectrum of HLA haplotypes found in eight randomly chosen donors. In this case, we were able to predict immune responses using iTEM with high sensitivity and minimal false-positive predictions. The refined iTEM method may afford us an improved capacity to predict immune responses in the context of larger antigen sets; this would need to be investigated in future studies involving multiple protein antigens.
An epitope-driven approach towards vaccine design shows great promise with influenza and other infectious diseases and could overcome challenges facing both subunit and poly-epitopic vaccines. Indeed, merging in silico and in vitro strategies to define potential epitopes has led to the discovery of immunogenic tuberculosis specific T-cell epitopes which may have application in vaccines against this pathogen. So as to improve current influenza vaccine strategies, future pandemic epitope-based formulations could (1) expand the generation of cross-reactive T cell epitopes, (2) exploit sequence conservation within circulating influenza strains, and (3) expand HLA population coverage of cross-reactive epitopes. To improve the immunogenicity of this type of formulation, we feel it would be important to choose peptides that induce multifunctional T cell responses in human PBMCs. As well, we believe the successful implementation of this strategy would require a careful selection of vaccine delivery vehicle, route, and formulation strategy.
CONCLUSIONS
In summary, this study confirmed the capacity of circulating CD4+ T cells to generate cross-reactive effector responses against the S-OIV and validated our previous predictions of highly immunogenic HA-derived T cell epitopes that are shared between seasonal and pandemic H1N1 viruses. The implication of these results are clear, namely, that priming with the 2009/2010 seasonal TIV might have generated cross-conserved T helper cells capable of providing enhanced protection against subsequent S-OIV infection via direct anti-viral effects or accelerating the induction of naïve antibody responses against the novel virus. Going a step further, we think these observations lends support to the notion that vaccines which “arm the immune system” via cellular/T cell-dependent defenses against influenza virus might provide an alternative to current prophylactic strategies [49] since vaccines that stimulate effective antibody response must be developed on a seasonal basis in a costly and sometimes inefficient process. This hypothesis is supported by the study of Ellebedy et al. [8] demonstrating a correlation between the strength of T cell responses against cross-reactive epitopes and attenuation of influenza symptoms in H1N1-infected humans, ferrets, and mice. Finally, these observations lend support to the integration of in silico and sensitive in vitro testing methods for defining and assessing cross-reactive T cell response in preparation for the next influenza virus pandemic and other infectious diseases.
Acknowledgments
We are thankful to T. Kamala and J. Moser for critically reviewing the manuscript. As well, we appreciate the contribution of F. Terry in performing computational analyses. This work was funded by a DARPA/DSO (BAA09-310) project (#70023) entitled, “Immune Analysis of Brisbane and California H1N1 in Human Sera and the MIMIC® System.”
Footnotes
CONTRIBUTORS
BCS participated in the design of the biological assays, performed laboratory experiments, and co-authored the manuscript.
ADG conceived of the cross-reactive H1N1 project at EpiVax, directed the selection of epitopes, proposed the iTEM analysis, reviewed the data and literature, and co-wrote the manuscript.
MA performed iTEM analyses, assisted in modifying the original concept and methodology of iTEM, constructed the tables relevant to all bioinformatics analysis, and assisted in revising and editing the manuscript.
EM analyzed in silico data and contributed to writing and editing the manuscript.
LM analyzed in silico data and contributed to writing and editing the manuscript.
WDM supervised bioinformatic analyses and development of iTEM scores, analyzed data, and contributed to writing and editing the manuscript.
VW contributed to design of the study, interpretation of data, and editing of the manuscript.
WLW assisted with the design of assays and editing the manuscript.
DRD assisted in the design of the study, interpretation of data, and co-authored the manuscript.
Abbreviations: Swine-origin influenza A virus, S-OIV; seasonal trivalent vaccine, TIV; dendritic cells, DCs; individualized T cell epitope measure, iTEM; hemagglutinin, HA; stimulation index, SI.
References
- 1.Memoli MJ, et al. An early ‘classical’ swine H1N1 influenza virus shows similar pathogenicity to the 1918 pandemic virus in ferrets and mice. Virology. 2009;393(2):338–45. doi: 10.1016/j.virol.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garten RJ, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock K, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361(20):1945–52. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 4.Itoh Y, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460(7258):1021–5. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep. 2009;58(19):521–4. [PubMed] [Google Scholar]
- 6.Greenbaum JA, et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci U S A. 2009;106(48):20365–70. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain SKL, Bramley AM, Schmitz AM, Benoit SR, Louie J, Sugerman DE, Druckenmiller JK, Ritger KA, Chugh R, Jasuja S, Deutscher M, Chen S, Walker JD, Duchin JS, Lett S, Soliva S, Wells EV, Swerdlow D, Uyeki TM, Fiore AE, Olsen SJ, Fry AM, Bridges CB, Finelli L. The 2009 Pandemic Influenza A (H1N1) Virus Hospitalizations Investigation Team. N Engl J Med. 2009 [Google Scholar]
- 8.Ellebedy AH, et al. Impact of prior seasonal influenza vaccination and infection on pandemic A(H1N1) influenza virus replication in ferrets. Vaccine. 2010 doi: 10.1016/j.vaccine.2010.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Groot AS, et al. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008–2009 conventional influenza vaccine. Vaccine. 2009;27(42):5740–7. doi: 10.1016/j.vaccine.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 10.Moser JM, et al. Optimization of a dendritic cell-based assay for the in vitro priming of naive human CD4+ Tcells. J Immunol Methods. 2010;353(1–2):8–19. doi: 10.1016/j.jim.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Schanen BC, Drake DR., 3rd A novel approach for the generation of human dendritic cells from blood monocytes in the absence of exogenous factors. J Immunol Methods. 2008;335(1–2):53–64. doi: 10.1016/j.jim.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 12.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50(3–4):201–12. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 13.De Groot AS, et al. An interactive Web site providing major histocompatibility ligand predictions: application to HIV research. AIDS Res Hum Retroviruses. 1997;13(7):529–31. doi: 10.1089/aid.1997.13.529. [DOI] [PubMed] [Google Scholar]
- 14.Schafer JR, et al. Prediction of well-conserved HIV-1 ligands using a matrix-based algorithm, EpiMatrix. Vaccine. 1998;16(19):1880–4. doi: 10.1016/s0264-410x(98)00173-x. [DOI] [PubMed] [Google Scholar]
- 15.Sturniolo T, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17(6):555–61. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 16.De Groot AS, Knopp PM, Martin W. De-immunization of therapeutic proteins by T-cell epitope modification. Dev Biol (Basel) 2005;122:171–94. [PubMed] [Google Scholar]
- 17.De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin Immunol. 2009;131(2):189–201. doi: 10.1016/j.clim.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 18.De Groot AS, et al. Developing an epitope-driven tuberculosis (TB) vaccine. Vaccine. 2005;23(17–18):2121–31. doi: 10.1016/j.vaccine.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 19.Moise L, et al. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009;27(46):6471–9. doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Groot AS, et al. Identification of immunogenic HLA-B7 “Achilles’ heel” epitopes within highly conserved regions of HIV. Vaccine. 2008;26(24):3059–71. doi: 10.1016/j.vaccine.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMurry JA, et al. Diversity of Francisella tularensis Schu4 antigens recognized by T lymphocytes after natural infections in humans: identification of candidate epitopes for inclusion in a rationally designed tularemia vaccine. Vaccine. 2007;25(16):3179–91. doi: 10.1016/j.vaccine.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 22.Cohen T, et al. A method for individualizing the prediction of immunogenicity of protein vaccines and biologic therapeutics: individualized T cell epitope measure (iTEM) J Biomed Biotechnol. 2010 doi: 10.1155/2010/961752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards KA, et al. Cutting Edge: CD4 T Cells Generated from Encounter with Seasonal Influenza Viruses and Vaccines Have Broad Protein Specificity and Can Directly Recognize Naturally Generated Epitopes Derived from the Live Pandemic H1N1 Virus. J Immunol. 2010 doi: 10.4049/jimmunol.1001395. [DOI] [PubMed] [Google Scholar]
- 24.Tu W, et al. Cytotoxic T lymphocytes established by seasonal human influenza cross-react against 2009 pandemic H1N1 influenza virus. J Virol. 2010;84(13):6527–35. doi: 10.1128/JVI.00519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge X, et al. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J Virol. 2010;84(7):3312–9. doi: 10.1128/JVI.02226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cusick MF, Wang S, Eckels DD. In vitro responses to avian influenza H5 by human CD4 T cells. J Immunol. 2009;183(10):6432–41. doi: 10.4049/jimmunol.0901617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gioia C, et al. Cross-subtype immunity against avian influenza in persons recently vaccinated for influenza. Emerg Infect Dis. 2008;14(1):121–8. doi: 10.3201/eid1401.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jameson J, et al. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J Immunol. 1999;162(12):7578–83. [PubMed] [Google Scholar]
- 29.Richards KA, et al. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol. 2007;81(14):7608–19. doi: 10.1128/JVI.02834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards KA, Chaves FA, Sant AJ. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol. 2009;83(13):6566–77. doi: 10.1128/JVI.00302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roti M, et al. Healthy human subjects have CD4+ Tcells directed against H5N1 influenza virus. J Immunol. 2008;180(3):1758–68. doi: 10.4049/jimmunol.180.3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider C, Van Regenmortel MH. Immunogenicity of free synthetic peptides corresponding to T helper epitopes of the influenza HA 1 subunit. Induction of virus cross reacting CD4+ T lymphocytes in mice. Arch Virol. 1992;125(1–4):103–19. doi: 10.1007/BF01309631. [DOI] [PubMed] [Google Scholar]
- 33.Alexander J, et al. Universal influenza DNA vaccine encoding conserved CD4+ Tcell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine. 2010;28(3):664–72. doi: 10.1016/j.vaccine.2009.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belz GT, et al. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J Virol. 2002;76(23):12388–93. doi: 10.1128/JVI.76.23.12388-12393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brooks JW, et al. Requirement for CD40 ligand, CD4(+) T cells, and B cells in an infectious mononucleosis-like syndrome. J Virol. 1999;73(11):9650–4. doi: 10.1128/jvi.73.11.9650-9654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardin RD, et al. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J Exp Med. 1996;184(3):863–71. doi: 10.1084/jem.184.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellebedy AH, et al. Contemporary seasonal A (H1N1) influenza virus infection primes for a more robust response to split inactivated pandemic A(H1N1) vaccination in ferrets. Clin Vaccine Immunol. 2010 doi: 10.1128/CVI.00247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen IB, et al. The principle of delivery of T cell epitopes to antigen-presenting cells applied to peptides from influenza virus, ovalbumin, and hen egg lysozyme: implications for peptide vaccination. Proc Natl Acad Sci U S A. 2001;98(18):10296–301. doi: 10.1073/pnas.181336898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall D, et al. TH cells primed during influenza virus infection provide help for qualitatively distinct antibody responses to subsequent immunization. J Immunol. 1999;163(9):4673–82. [PubMed] [Google Scholar]
- 40.Boon AC, et al. Recognition of homo- and heterosubtypic variants of influenza A viruses by human CD8+ T lymphocytes. J Immunol. 2004;172(4):2453–60. doi: 10.4049/jimmunol.172.4.2453. [DOI] [PubMed] [Google Scholar]
- 41.Kreijtz JH, et al. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine. 2007;25(4):612–20. doi: 10.1016/j.vaccine.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 42.Ulmer JB, et al. Protective CD4+ and CD8+ Tcells against influenza virus induced by vaccination with nucleoprotein DNA. J Virol. 1998;72(7):5648–53. doi: 10.1128/jvi.72.7.5648-5653.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goy K, et al. Heterosubtypic T-cell responses against avian influenza H5 haemagglutinin are frequently detected in individuals vaccinated against or previously infected with human subtypes of influenza. Influenza Other Respi Viruses. 2008;2(4):115–25. doi: 10.1111/j.1750-2659.2008.00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zinckgraf JW, et al. Identification of HLA class II H5N1 hemagglutinin epitopes following subvirion influenza A (H5N1) vaccination. Vaccine. 2009;27(39):5393–401. doi: 10.1016/j.vaccine.2009.06.081. [DOI] [PubMed] [Google Scholar]
- 45.Otero M, et al. Efficacy of novel plasmid DNA encoding vaccinia antigens in improving current smallpox vaccination strategy. Vaccine. 2006;24(21):4461–70. doi: 10.1016/j.vaccine.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Meister GE, et al. Two novel T cell epitope prediction algorithms based on MHC-binding motifs; comparison of predicted and published epitopes from Mycobacterium tuberculosis and HIV protein sequences. Vaccine. 1995;13(6):581–91. doi: 10.1016/0264-410x(94)00014-e. [DOI] [PubMed] [Google Scholar]
- 47.Bond KB, et al. An HLA-directed molecular and bioinformatics approach identifies new HLA-A11 HIV-1 subtype E cytotoxic T lymphocyte epitopes in HIV-1-infected Thais. AIDS Res Hum Retroviruses. 2001;17(8):703–17. doi: 10.1089/088922201750236988. [DOI] [PubMed] [Google Scholar]
- 48.Dong Y, et al. HLA-A2-restricted CD8+-cytotoxic-T-cell responses to novel epitopes in Mycobacterium tuberculosis superoxide dismutase, alanine dehydrogenase, and glutamine synthetase. Infect Immun. 2004;72(4):2412–5. doi: 10.1128/IAI.72.4.2412-2415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McMurry JA, Johansson BE, De Groot AS. A call to cellular & humoral arms: enlisting cognate T cell help to develop broad-spectrum vaccines against influenza A. Hum Vaccin. 2008;4(2):148–57. doi: 10.4161/hv.4.2.5169. [DOI] [PubMed] [Google Scholar]